By Caleb O’Brien | MU Bond Life Sciences Center
Seeing the whole picture can mean a lot when it comes to figuring out HIV.
Researchers at the University of Missouri Bond Life Sciences Center are gaining a clearer idea of what a key protein in HIV looks like, which will help explain the flexible protein’s vital role in the virus life cycle.
The protein the researchers imaged is a building block that forms the virus’ capsid, a protective shell surrounding the virus’ genes. The journal Science published their findings online June 4.

“The capsid acts as an invisibility cloak that hides the virus’ genetic information, the genome, while it is being copied in a hostile environment for the virus,” said Stefan Sarafianos, a virologist at Bond LSC and lead author of the study. “Fine-tuned capsid stability is critical for successful infection: too stable a capsid shell and the cargo is never delivered properly; not stable enough and the contents are detected by our immune defenses, triggering an antiviral response. Capsid stability is a key to the puzzle, and to solve it you have to understand its structure.”
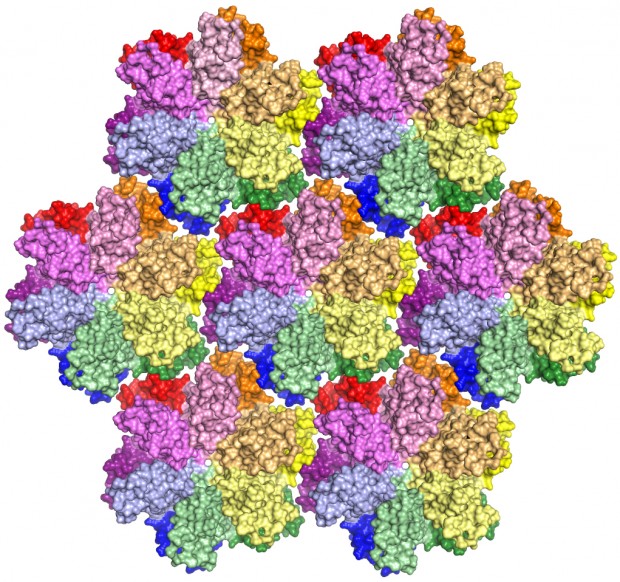
This is the most complete model yet of an HIV-1 capsid protein. In a virus, the protein combines in groups of five or six — called pentamers and hexamers, respectively — that assemble into a mosaic that forms the capsid shell. Roughly 1,500 copies of the protein, grouped into about 250 hexamers and 12 pentamers, comprise the capsid.
HIV, or human immunodeficiency virus, is the retrovirus that leads to AIDS — acquired immunodeficiency syndrome. Roughly 1.2 million people live with HIV in the United States, according to the Centers for Disease Control and Prevention. Globally, about 35 million people were living with HIV in 2013.
A lucky break
Over the years, scientists have employed various techniques and tricks to figure out the structure of the capsid protein. But until now, the clearest image had been made of a mutated version of the protein. It was a compromise: the mutation made the protein stable enough that the scientists could get a good snapshot, but they couldn’t see the detailed interactions between hexamers.
Sarafianos’ lab figured out how to get the full picture: a detailed image of the unmodified proteins that filled all the gaps between hexamers.
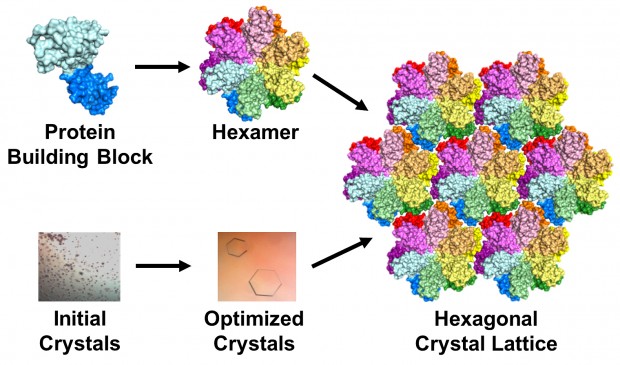
The team used a technique called X-ray crystallography to unravel the protein’s secrets. Basically, they took many copies of the protein and coaxed them into forming a patterned, crystalline lattice. Next they shot high-powered X-ray beams at the crystal. By interpreting how the X-rays scattered when they ricocheted off the proteins, the researchers made a 3-D map of the protein.

“But it doesn’t make sense until we make an atomic model of the protein to fit in that map,” said Karen Kirby, a research scientist at Bond LSC. “The map is just a grid that you can’t really interpret unless you put a model into it to see ‘Ok, it looks like this part is here, and that part is there, and this is how the protein is put together.’”
The researchers altered, tested and honed their 3-D model until it exactly matched the map produced by the X-ray diffraction pattern. This can be difficult and painstaking, but the researchers’ greatest challenge was creating the protein crystals in the first place: Scientists had been trying to crystallize the unmodified version of the HIV protein for decades without success.
To make a crystal, proteins are suspended in a liquid then slowly precipitated out, just like a “grow your own crystals” kit. But there are a lot of variables that control the process, from salts and additives in the liquid to the amount of protein in the mixture.
“It’s a very delicate balance to grow crystals,” Kirby said. “Many people call it more of an art than a science. It’s frustrating because you can never predict which solution will grow crystals. There are a large number of variables.”
Initially, most arrangements the researchers tried resulted in useless brown junk, Kirby said, caused by the proteins forming solids too quickly. Anna Gres, an MU chemistry grad student who led the project, used a crystallization robot to screen roughly 2,500 conditions.
That was the easy part, Gres said: “The real challenge begins afterwards, as one needs to manually optimize the initial crystallization conditions to find the one that will produce protein crystals of desired quality. This process can take years. In our case, I think we were lucky: It took approximately 500 manual screenings and about 6 month.” But the hard work paid off when she was finally able to produce lovely, hexagonal crystals. Surprisingly, the crystals formed in groups of six proteins, which matched their formation in the viral capsid.
The transition from tiny, useless particulate to invaluable crystals was tremendously exciting, Kirby said. But even to Kirby and Sarafianos, why their attempts succeeded when many others failed remains a little mysterious.
“I still don’t know what are the fine details that made the difference,” Sarafianos said.
“That’s the million dollar question,” said Kirby. “We really don’t have a good answer for that.”
Although solving the enigmatic crystal structure of the native full-length capsid protein was really rewarding, Gres said, she will continue to tinker with her technique: “I am still trying to optimize crystallization conditions, hoping to improve the quality of the crystals and diffraction.”
Water, water everywhere
Once the researchers got a good look at the interactions between hexamers, they were surprised by what they found.
Based on the genetic sequence of the protein, scientists speculated that they would be hydrophobic, or repel water. Instead, they found that “ordered” water molecules at specific sites played a crucial structural role by bridging interactions at the interface between hexamers.
“We thought, ‘How could these lowly waters really be of consequence?’” Sarafianos said. “But if you think about it, there’s 256 of these hexamers in the whole capsid and all kinds of interfaces among them: There’s thousands of water molecules that stabilize the whole structure. We hypothesize that this is an essential part of the stability of the whole capsid molecule.”
To test that hypothesis, they took the crystals, dehydrated them and checked to see if their shape changed. Although the protein lattices may look like sturdy crystals, they’re more like jello, Sarafianos said.
“The protein molecules are precariously touching each other and forming a lattice that is very, very sensitive. It’s held together in this case by water molecules in addition to other interactions.”
The change in shape suggested that water molecules are important in that they allow the capsid to assume different shapes. Moreover, Sarafianos said, the capsid’s malleability and plasticity could be critical to the life cycle of the virus and allow it to act as a multi-functional molecular Swiss army knife.
Onward with research
A clearer image of the capsid protein, could help Sarafianos’ lab gain a better understanding of how the body combats the virus and to discover new ways to disrupt the viral capsid.
“Now we have a system to study effects of capsid-targeting compounds with novel mechanism of action,” Gres said.
Working with a medicinal chemist, Sarafianos’ lab will undertake an iterative process of making compounds, solving their structures, testing them against HIV and then refining the molecules, with the ultimate aim of producing new and effective antiviral drugs.
Sarafianos is an associate professor of molecular microbiology and immunology and Chancellor’s Chair of Excellence in molecular virology with MU’s School of Medicine and an associate professor of biochemistry in the MU College of Agriculture, Food and Natural Resources.
The study, “X-Ray Crystal Structures of Native HIV-1 Capsid Protein Reveal Conformational Variability,” was recently published in the journal Science. Research funding has been provided by the National Institutes of Health (Grants AI112417, AI120860, GM103368, AI076119, AI099284, and AI100890 (SGS), and GM066087 (OP)). The content is solely the responsibility of the authors and does not necessarily represent the official views of the funding agencies.


Регистрация в binance
February 21, 2024 @ 2:25 pm
Thank you for your sharing. I am worried that I lack creative ideas. It is your article that makes me full of hope. Thank you. But, I have a question, can you help me? https://www.binance.com/ru-UA/join?ref=53551167
最佳Binance推荐代码
February 23, 2024 @ 8:56 pm
Your point of view caught my eye and was very interesting. Thanks. I have a question for you. https://www.binance.com/zh-CN/join?ref=P9L9FQKY
binance Inscreva-se
March 7, 2024 @ 3:12 am
Can you be more specific about the content of your article? After reading it, I still have some doubts. Hope you can help me.
бнанс акаунт
March 11, 2024 @ 3:28 am
Can you be more specific about the content of your article? After reading it, I still have some doubts. Hope you can help me.
Prihlásit se a získat 100 USDT
March 15, 2024 @ 5:31 am
Your article helped me a lot, is there any more related content? Thanks!
Kode Referal Binance
March 23, 2024 @ 12:29 pm
I don’t think the title of your article matches the content lol. Just kidding, mainly because I had some doubts after reading the article.
Registrasi Binance
March 24, 2024 @ 11:22 am
Thank you for your sharing. I am worried that I lack creative ideas. It is your article that makes me full of hope. Thank you. But, I have a question, can you help me?
registrace na binance
March 30, 2024 @ 4:09 am
I don’t think the title of your article matches the content lol. Just kidding, mainly because I had some doubts after reading the article.
https://mostbetcasino.blogspot.com/2021/10/explore-basics-of-gambling.html
April 3, 2024 @ 6:27 pm
When I originally commented I clicked the “Notify me when new comments are added” checkbox and
now each time a comment is added I get four e-mails witrh the same comment.
Is there any way you can remove me from that service?Bless you!
my page: https://mostbetcasino.blogspot.com/2021/10/explore-basics-of-gambling.html
Anonymous
April 14, 2024 @ 8:54 am
sex ロボット Oksexdollで同意できる合理的なセックスドールを購入するシリコンドールを請求する必要がある理由拡張されたセックスドールの信じられないほどの利点Irontechdollの新しい配置と新しい進歩
Anonymous
April 21, 2024 @ 9:36 pm
I don’t think the title of your article matches the content lol. Just kidding, mainly because I had some doubts after reading the article.
シリコンドール
April 29, 2024 @ 8:22 am
YOKIDOLLリアルシリコンドールは高度的な自然、人に信じられる見た目を備えます。シリコンラブドールはもっと真実に感じられて、リアルな人間を驚くほど似たような質感があります。つまり、シリコンはセックスドールの完璧な材質です,リアルで生き生きとした、人に満足できるセックスドールを探す方はシリコンドールの注文を検討する必要があります。
explainer video company india
May 9, 2024 @ 3:24 am
I am in fact pleased to glance at this website posts which contains plenty of
useful data, thanks for providing these kinds of information.
Here is my blog post :: explainer video company india
explainer videos
May 14, 2024 @ 1:47 pm
Superb blog! Do you have any suggestions for aspiring writers?
I’m hoping to start my own site soon but I’m a little lost on everything.
Would you propose starting with a free platform like
Wordpress or go for a paid option? There are so many choices out there that I’m completely overwhelmed ..
Any ideas? Kudos!
Also visit my page … explainer videos
ラブドール
May 15, 2024 @ 3:45 am
I’m curious to find out what blog platform
you’re using? I’m experiencing some small security
issues with my latest website and I would like to find something more risk-free.
Do you have any solutions?ラブドール エロ
Roosevelt
May 16, 2024 @ 2:01 pm
I really like your blog.. very nice colors & theme.
Did you design this website yourself or did you hire
someone to do it for you? Plz answer back as I’m
looking to design my own blog and would like to know where u got this from.
thanks a lot
My site: Roosevelt
geinoutime.com
May 23, 2024 @ 12:05 pm
geinoutime.com
그러나 때때로 규칙을 어기는 사람들이 항상 있을 것입니다.
seo services india
May 24, 2024 @ 1:43 am
Hello there! This is my first visit to your blog! We
are a collection of volunteers and starting a new project in a community in the
same niche. Your blog provided us valuable information to work on. You
have done a outstanding job!
Take a look at my website – seo services india
binance bonus za napotitev
May 25, 2024 @ 12:40 am
I don’t think the title of your article matches the content lol. Just kidding, mainly because I had some doubts after reading the article.
geinoutime.com
May 25, 2024 @ 5:08 am
geinoutime.com
“예, 삼천 명의 제자와 무수한 제자와 손자가 있다고 합니다.”
Tumerpag
May 25, 2024 @ 1:42 pm
Hello.
This post was created with 2ssdsd3222aa.com
geinoutime.com
May 26, 2024 @ 9:28 pm
geinoutime.com
네트워크는 원래 힘에 의해 지원되고 가족은 쇠퇴하고 있기 때문에 누가 당신을 걱정합니다.모든 해안선은 수천 마일에서 수백 마일에 이르기까지 길기 때문입니다.
Leon
May 27, 2024 @ 4:44 am
I like what you guys tend to be up too. Such clever work and coverage!
Keep up the great works guys I’ve you guys to my blogroll.
Website monthly maintenance packages
Feel free to surf to my homepage … Leon
k8 カジノ
May 28, 2024 @ 12:20 am
アナザーゴッドハーデス-奪われたZEUSver
現実的で実用的な情報が多く、読む価値がありました。
k8 カジノ
May 29, 2024 @ 9:16 am
k8 カジノ 誕生日ボーナス
非常に有意義な内容でした。また読みたいと思います。
geinoutime.com
May 30, 2024 @ 6:07 am
geinoutime.com
Liu Jian은 말을 마친 후 쉭쉭 거리며 홀을 뛰쳐 나갔다.
k8 カジノ
May 30, 2024 @ 8:23 am
アイムジャグラーEX (2発1)(V2.2)
非常にインスピレーションを受ける記事でした。素晴らしい!
Создать личный аккаунт
May 31, 2024 @ 12:24 am
Can you be more specific about the content of your article? After reading it, I still have some doubts. Hope you can help me.
k8 カジノ
May 31, 2024 @ 6:35 am
吉宗
この記事から多くを学びました。非常に役立つ情報です。
k8 カジノ
June 2, 2024 @ 10:08 pm
北斗の拳 修羅の国篇(V2.2)
このブログは常に私に多くのことを教えてくれます。大好きです!
geinoutime.com
June 7, 2024 @ 10:57 pm
geinoutime.com
그러나 일단 이것이 축적되면 많은 구리 부족이 발생할 것입니다.
seo services
June 9, 2024 @ 8:12 am
Remarkable! Its genuinely awesome article, I have got
much clear idea concerning from this paragraph.
Feel free to visit my blog: seo services
슬롯
June 11, 2024 @ 9:35 am
geinoutime.com
Fang Jifan은 “아직 시간이 있으니 열심히해야합니다. “라고 손가락을 세었습니다.
슬롯
June 14, 2024 @ 6:46 am
geinoutime.com
그는 Fang Jifan이 안목이 있음을 인정했지만 말은 때때로 넘어졌습니다.
Top Explainer Video Agency in India - Explainervideo.in
June 18, 2024 @ 7:17 am
Remarkable! Its in fact awesome post, I have got much clear idea
on the topic of from this post.
Explainer Video Company from India explainervideo.in https://www.dealerbaba.com/suppliers/business-services/explainer-video-company-india.Html
bonus za registraci na binance
June 19, 2024 @ 4:14 pm
Your point of view caught my eye and was very interesting. Thanks. I have a question for you.
슬롯
June 22, 2024 @ 8:23 am
와일드 바운티 쇼다운
그는 이번 수해를 직접 경험했기 때문에 자연스럽게 잘 알고 있다.
Cheap Explainer Video Company
June 23, 2024 @ 9:34 am
Hmm is anyone else having problems with the pictures on this blog loading?
I’m trying to figure out if its a problem on my end or if it’s
the blog. Any feed-back would be greatly appreciated.
my web page – Cheap Explainer Video Company
파라오카지노
June 25, 2024 @ 3:17 am
I’ve done a lot of online casino information, we’ve helped a lot with the casino, and we’ve never scammed anyone. All I can say is your post is beautiful, thanks for sharing.
https://main7.top/thenine/
medication
June 27, 2024 @ 6:44 am
[url=http://xlyrica.online/]lyrica cheap price[/url]
슬롯
June 27, 2024 @ 7:36 am
sm 슬롯
이상하게도 Zhang Heling과 다른 사람들은 감염되지 않았습니다.
Utwórz konto na Binance
June 28, 2024 @ 2:41 am
Thanks for sharing. I read many of your blog posts, cool, your blog is very good.
DonaldPoild
June 29, 2024 @ 5:19 am
mexican mail order pharmacies
http://cmqpharma.com/# mexican pharmaceuticals online
mexican border pharmacies shipping to usa
Stephenced
June 29, 2024 @ 7:40 am
buying prescription drugs in mexico online: mexican pharmacy – mexican mail order pharmacies
슬롯
June 29, 2024 @ 7:35 pm
에그 카지노
Xiao Jing과 Mou Bin은 머리를 묻고 여전히 분노를 표출하지 못했습니다.
DonaldPoild
June 29, 2024 @ 7:43 pm
pharmacies in mexico that ship to usa
https://cmqpharma.online/# purple pharmacy mexico price list
mexican pharmaceuticals online
Donaldiodix
July 2, 2024 @ 7:45 am
Профессиональные seo https://seo-optimizaciya-kazan.ru услуги для максимизации онлайн-видимости вашего бизнеса. Наши эксперты проведут глубокий анализ сайта, оптимизируют контент и структуру, улучшат технические аспекты и разработают индивидуальные стратегии продвижения.
슬롯
July 2, 2024 @ 8:44 am
마블 슬롯
Liu Jian과 다른 사람들은 고개를 저을 수밖에 없었습니다. “노신은 전하의 말에 동의하지 않습니다.”
Stephantheow
July 4, 2024 @ 5:51 am
Top sports news https://idman-azerbaycan.com.az photos and blogs from experts and famous athletes, as well as statistics and information about matches of leading championships.
HectorVed
July 4, 2024 @ 5:58 am
Latest news and details about the NBA in Azerbaijan https://nba.com.az. Hot events, player transfers and the most interesting events. Explore the world of the NBA with us.
JamesAnype
July 4, 2024 @ 6:05 am
The latest top football news https://futbol.com.az today. Interviews with football players, online broadcasts and match results, analytics and football forecasts, photos and videos.
EddieAffit
July 4, 2024 @ 6:06 am
Discover the fascinating world of online games with GameHub Azerbaijan https://online-game.com.az. Get the latest news, reviews and tips for your favorite games. Join our gaming community today!
DesmondBus
July 4, 2024 @ 9:02 am
Каталог рейтингов хостингов https://pro-hosting.tech на любой вкус и под любые, даже самые сложные, задачи.
JordanInfek
July 4, 2024 @ 9:06 am
https://santekhnik-moskva.blogspot.com — вызов сантехника на дом в Москве и Московской области в удобное для вас время.
HarryCraph
July 4, 2024 @ 9:36 am
Play PUBG Mobile https://pubg-mobile.com.az an exciting world of high-quality mobile battle royale. Unique maps, strategies and intense combat await you in this exciting mobile version of the popular game.
Oscarcrife
July 4, 2024 @ 9:37 am
The Dota 2 website https://dota2.com.az Azerbaijan provides the most detailed information about the latest game updates, tournaments and upcoming events. We have all the winning tactics, secrets and important guides.
LeroyFUs
July 4, 2024 @ 12:12 pm
Check out the latest news, guides and in-depth reviews of the available options for playing Minecraft Az https://minecraft.com.az. Find the latest information about Minecraft Download, Pocket Edition and Bedrock Edition.
BruceHon
July 4, 2024 @ 12:13 pm
Latest news about games for Android https://android-games.com.az, reviews and daily updates. Read now and get the latest information on the most exciting games
Raphaelmoown
July 4, 2024 @ 12:14 pm
The most popular sports site https://sports.com.az of Azerbaijan, where the latest sports news, forecasts and analysis are collected.
BarryusemI
July 4, 2024 @ 12:24 pm
Latest news and analytics of the Premier League https://premier-league.com.az. Detailed descriptions of matches, team statistics and the most interesting football events. EPL Azerbaijan is the best place for football fans.
Charlesrek
July 4, 2024 @ 12:26 pm
Хотите сделать в квартире ремонт? Тогда советуем вам посетить сайт https://stroyka-gid.ru, где вы найдете всю необходимую информацию по строительству и ремонту.
Victorren
July 4, 2024 @ 4:58 pm
https://loveflover.ru — сайт посвященный комнатным растениям. Предлагает подробные статьи о выборе, выращивании и уходе за различными видами комнатных растений. Здесь можно найти полезные советы по созданию зелёного уголка в доме, руководства по декору и решению распространённых проблем, а также информацию о подходящих горшках и удобрениях. Платформа помогает создавать уютную атмосферу и гармонию в интерьере с помощью растений.
MatthewRot
July 4, 2024 @ 5:04 pm
1xbet https://1xbet.best-casino-ar.com with withdrawal without commission. Register online in a few clicks. A large selection of slot machines in mobile applications and convenient transfers in just a few minutes.
GeorgeSciet
July 4, 2024 @ 5:34 pm
Pin-up Casino https://pin-up.admsov.ru/ is an online casino licensed and regulated by the government of Curacao . Founded in 2016, it is home to some of the industry’s leading providers, including NetEnt, Microgaming, Play’n GO and others. This means that you will be spoiled for choice when it comes to choosing a game.
RobertNeods
July 4, 2024 @ 5:36 pm
Pin Up official https://pin-up.adb-auto.ru website. Login to your personal account and register through the Pin Up mirror. Slot machines for real money at Pinup online casino.
Colinbes
July 4, 2024 @ 6:52 pm
Pin Up Casino https://pin-up.noko39.ru Registration and Login to the Official Pin Up Website. thousands of slot machines, online tables and other branded entertainment from Pin Up casino. Come play and get big bonuses from the Pinup brand today
MichaelKic
July 4, 2024 @ 6:53 pm
Pin Up online casino https://pin-up.webrabota77.ru/ is the official website of a popular gambling establishment for players from the CIS countries. The site features thousands of slot machines, online tables and other branded entertainment from Pin Up casino.
JeffreyRerry
July 4, 2024 @ 8:26 pm
Реальные анкеты проституток https://prostitutki-213.ru Москвы с проверенными фото – от элитных путан до дешевых шлюх. Каталог всех индивидуалок на каждой станции метро с реальными фотографиями без ретуши и с отзывами реальных клиентов.
MatthewRot
July 4, 2024 @ 8:29 pm
Смотрите онлайн сериал Отчаянные домохозяйки https://domohozyayki-serial.ru в хорошем качестве HD 720 бесплатно, рейтинг сериала: 8.058, режиссер сериала: Дэвид Гроссман, Ларри Шоу, Дэвид Уоррен.
슬롯
July 4, 2024 @ 8:39 pm
프라그마틱 슬롯 추천
그런데 지금은 하늘도 크고 땅도 크고 생각이 다릅니다.
JeffreyMed
July 4, 2024 @ 8:51 pm
Buy TikTok followers https://tiktok-followers-buy.com to get popular and viral with your content. All packages are real and cheap — instant delivery within minutes. HQ followers for your TikTok. 100% real users. The lowest price for TikTok followers on the market
VictorBloth
July 4, 2024 @ 10:01 pm
Pin Up Casino https://pin-up.sibelshield.ru official online casino website for players from the CIS countries. Login and registration to the Pin Up casino website is open to new users with bonuses and promotional free spins.
JamesEveva
July 4, 2024 @ 10:49 pm
Изготовление памятников и надгробий https://uralmegalit.ru по низким ценам. Собственное производство. Высокое качество, широкий ассортимент, скидки, установка.
GeorgeDob
July 4, 2024 @ 11:03 pm
Pin Up Casino https://pin-up.ergojournal.ru приглашает игроков зарегистрироваться на официальном сайте и начать играть на деньги в лучшие игровые автоматы, а на зеркалах онлайн казино Пин Ап можно найти аналогичную витрину слотов
Terryrounc
July 4, 2024 @ 11:07 pm
Pin-up casino https://pin-up.jes-design.ru популярное онлайн-казино и ставки на спорт. Официальный сайт казино для доступа к играм и другим функциям казино для игры на деньги.
AaronJed
July 5, 2024 @ 3:47 am
Pin Up https://pin-up.fotoevolution.ru казино, которое радует гемблеров в России на протяжении нескольких лет. Узнайте, что оно подготовило посетителям. Описание, бонусы, отзывы о легендарном проекте. Регистрация и вход.
JesseOxync
July 5, 2024 @ 4:55 am
Открой мир карточных игр в Pin-Up https://pin-up.porsamedlab.ru казино Блэкджек, Баккара, Хило и другие карточные развлечения. Регистрируйтесь и играйте онлайн!
RobertCrync
July 5, 2024 @ 5:00 am
Официальный сайт Pin Up казино https://pin-up.nasledie-smolensk.ru предлагает широкий выбор игр и щедрые бонусы для игроков. Уникальные бонусные предложения, онлайн регистрация.
Williamcolla
July 5, 2024 @ 5:02 am
Pinup казино https://pin-up.vcabinet.kz это не просто сайт, а целый мир азартных развлечений, где каждый может найти что-то свое. От традиционных игровых автоматов до прогнозов на самые популярные спортивные события.
MichaelBow
July 5, 2024 @ 5:18 am
Latest Diablo news https://diablo.com.az game descriptions and guides. Diablo.az is the largest Diablo portal in the Azerbaijani language.
Marvinnup
July 5, 2024 @ 7:00 am
Latest World of Warcraft (WOW) tournament news https://wow.com.az, strategies and game analysis. The most detailed gaming portal in Azerbaijani language
Willieler
July 5, 2024 @ 9:03 am
Azerbaijan NFL https://nfl.com.az News, analysis and topics about the latest experience, victories and records. A portal where the most beautiful NFL games in the world are generally studied.
RobertCrync
July 5, 2024 @ 9:09 am
Discover exciting virtual football in Fortnite https://fortnite.com.az. Your central hub for the latest news, expert strategies and interesting e-sports reports. Collecting points with us!
Jamesskany
July 5, 2024 @ 9:10 am
The latest analysis, tournament reviews and the most interesting features of the Spider-Man game https://spider-man.com.az series in Azerbaijani.
WilliamJadia
July 5, 2024 @ 9:47 am
Read the latest Counter-Strike 2 news https://counter-strike.net.az, watch the most successful tournaments and become the best in the world of the game on the CS2 Azerbaijan website.
Bryonwew
July 5, 2024 @ 12:47 pm
Mesut Ozil https://mesut-ozil.com.az latest news, statistics, photos and much more. Get the latest news and information about one of the best football players Mesut Ozil.
Rogercling
July 5, 2024 @ 12:54 pm
Explore the extraordinary journey of Kilian Mbappe https://kilian-mbappe.com.az, from his humble beginnings to global stardom. Delve into his early years, meteoric rise through the ranks, and impact on and off the football field.
ArthurGef
July 5, 2024 @ 12:55 pm
Latest news, statistics, photos and much more about Pele https://pele.com.az. Get the latest news and information about football legend Pele.
DavidInfat
July 5, 2024 @ 1:34 pm
Latest boxing news https://boks.com.az, Resul Abbasov’s achievements, Tyson Fury’s fights and much more. All in Ambassador Boxing.
Robertodus
July 5, 2024 @ 1:47 pm
Sergio Ramos Garcia https://sergio-ramos.com.az Spanish footballer, defender. Former Spanish national team player. He played for 16 seasons as a central defender for Real Madrid, where he captained for six seasons.
Jeffreywhoff
July 5, 2024 @ 4:22 pm
Gianluigi Buffon https://buffon.com.az Italian football player, goalkeeper. Considered one of the best goalkeepers of all time. He holds the record for the number of games in the Italian Championship, as well as the number of minutes in this tournament without conceding a goal.
Jamesdoork
July 5, 2024 @ 4:31 pm
Paulo Bruno Ezequiel Dybala https://dybala.com.az Argentine footballer, striker for the Italian club Roma and the Argentina national team. World champion 2022.
WilliamHam
July 5, 2024 @ 4:35 pm
Paul Labille Pogba https://pogba.com.az French footballer, central midfielder of the Italian club Juventus. Currently suspended for doping and unable to play. World champion 2018.
Williamimate
July 5, 2024 @ 5:52 pm
Канал для того, чтобы знания и опыт, могли помочь любому человеку сделать ремонт https://tvin270584.livejournal.com в своем жилище, любой сложности!
KennethHeImi
July 5, 2024 @ 5:53 pm
Kevin De Bruyne https://kevin-de-bruyne.liverpool-fr.com Belgian footballer, born 28 June 1991 years in Ghent. He has had a brilliant club career and also plays for the Belgium national team. De Bruyne is known for his spectacular goals and brilliant assists.
Georgevaf
July 5, 2024 @ 7:42 pm
Mohamed Salah Hamed Mehrez Ghali https://mohamed-salah.liverpool-fr.com Footballeur egyptien, attaquant du club anglais de Liverpool et l’equipe nationale egyptienne. Considere comme l’un des meilleurs joueurs du monde.
Billygah
July 5, 2024 @ 7:45 pm
Paul Labille Pogba https://paul-pogba.psg-fr.com Footballeur francais, milieu de terrain central du club italien de la Juventus. Champion du monde 2018. Actuellement suspendu pour dopage et incapable de jouer.
WilliamHam
July 5, 2024 @ 7:46 pm
The young talent who conquered Paris Saint-Germain: how Xavi Simons became https://xavi-simons.psg-fr.com leader of a superclub in record time.
Davidkaw
July 6, 2024 @ 4:06 am
Kylian Mbappe https://kylian-mbappe.psg-fr.com Footballeur, attaquant francais. Il joue pour le PSG et l’equipe de France. Ne le 20 decembre 1998 a Paris. Mbappe est francais de nationalite. La taille de l’athlete est de 178 cm.
KennethSmend
July 6, 2024 @ 4:17 am
Kevin De Bruyne https://liverpool.kevin-de-bruyne-fr.com Belgian footballer, born 28 June 1991 years in Ghent. He has had a brilliant club career and also plays for the Belgium national team. De Bruyne is known for his spectacular goals and brilliant assists.
Billygah
July 6, 2024 @ 4:22 am
Paul Pogba https://psg.paul-pogba-fr.com is a world-famous football player who plays as a central midfielder. The player’s career had its share of ups and downs, but he was always distinguished by his perseverance and desire to win.
Michaelhag
July 6, 2024 @ 4:40 am
Kylian Mbappe https://psg.kylian-mbappe-fr.com Footballeur, attaquant francais. L’attaquant de l’equipe de France Kylian Mbappe a longtemps refuse de signer un nouveau contrat avec le PSG, l’accord etant en vigueur jusqu’a l’ete 2022.
LucioEpils
July 6, 2024 @ 5:26 am
Изготовление, сборка и ремонт мебели https://shkafy-na-zakaz.blogspot.com для Вас, от эконом до премиум класса.
KennethSmend
July 6, 2024 @ 7:39 am
Thibaut Nicolas Marc Courtois https://thibaut-courtois.real-madrid-ar.com Footballeur belge, gardien de but du Club espagnol “Real Madrid”. Lors de la saison 2010/11, il a ete reconnu comme le meilleur gardien de la Pro League belge, ainsi que comme joueur de l’annee pour Genk. Trois fois vainqueur du Trophee Ricardo Zamora, decerne chaque annee au meilleur gardien espagnol
RobertFelia
July 6, 2024 @ 7:43 am
Forward Rodrigo https://rodrygo.real-madrid-ar.com is now rightfully considered a rising star of Real Madrid. The talented Santos graduate is compared to Neymar and Cristiano Ronaldo, but the young talent does not consider himself a star.
SamuelAxiob
July 6, 2024 @ 8:10 am
Saud Abdullah Abdulhamid https://saud-abdulhamid.real-madrid-ar.com Saudi footballer, defender of the Al -Hilal” and the Saudi Arabian national team. Asian champion in the age category up to 19 years. Abdulhamid is a graduate of the Al-Ittihad club. On December 14, 2018, he made his debut in the Saudi Pro League in a match against Al Bateen
DonaldOnelm
July 6, 2024 @ 8:22 am
Khvicha Kvaratskhelia https://khvicha-kvaratskhelia.real-madrid-ar.com midfielder of the Georgian national football team and the Italian club “Napoli”. Became champion of Italy and best player in Serie A in the 2022/23 season. Kvaratskhelia is a graduate of Dynamo Tbilisi and played for the Rustavi team.
Martinaperb
July 6, 2024 @ 10:52 am
Vinicius Junior https://vinisius-junior.com.az player news, fresh current and latest events for today about the player of the 2024 season
RodolfoBob
July 6, 2024 @ 10:55 am
Latest news and information about Marcelo https://marcelo.com.az on this site! Find Marcelo’s biography, career, playing stats and more. Find out the latest information about football master Marcelo with us!
AnthonySault
July 6, 2024 @ 11:19 am
Khabib Abdulmanapovich Nurmagomedov https://khabib-nurmagomedov.com.az Russian mixed martial arts fighter who performed under the auspices of the UFC. Former UFC lightweight champion.
Kelvinlat
July 6, 2024 @ 11:38 am
Welcome to our official site! Get to know the history, players and latest news of Inter Miami Football Club https://inter-miami.com.az. Discover with us the successes and great performances of America’s newest and most exciting soccer club.
ArturomeT
July 6, 2024 @ 11:43 am
Conor Anthony McGregor https://conor-mcgregor.com.az Irish mixed martial arts fighter who also performed in professional boxing. He performs under the auspices of the UFC in the lightweight weight category. Former UFC lightweight and featherweight champion.
Anthonylep
July 6, 2024 @ 1:52 pm
Оперативный вывод из запоя https://www.liveinternet.ru/users/laralim/post505923855/ на дому. Срочный выезд частного опытного нарколога круглосуточно. При необходимости больного госпитализируют в стационар.
Craigfog
July 6, 2024 @ 2:10 pm
Видеопродакшн студия https://humanvideo.ru полного цикла. Современное оборудование продакшн-компании позволяет снимать видеоролики, фильмы и клипы высокого качества. Создание эффективных видеороликов для рекламы, мероприятий, видеоролики для бизнеса.
JustinGrink
July 6, 2024 @ 3:42 pm
Заказать вывоз мусора https://musorovozzz.ru в Москве и Московской области, недорого и в любое время суток в мешках или контейнерами 8 м?, 20 м?, 27 м?, 38 м?, собственный автопарк. Заключаем договора на вывоз мусора.
ArturomeT
July 6, 2024 @ 3:50 pm
Реальные анкеты снять проститутку Москвы с проверенными фото – от элитных путан до дешевых шлюх. Каталог всех индивидуалок на каждой станции метро с реальными фотографиями без ретуши и с отзывами реальных клиентов.
AnthonyRit
July 6, 2024 @ 3:53 pm
Совсем недавно открылся новый интернет портал BlackSprut (Блекспрут) https://bs2cite.cc в даркнете, который предлагает купить нелегальные товары и заказать запрещенные услуги. Самая крупнейшая площадка СНГ. Любимые шопы и отзывчивая поддержка.
RaymondJorgo
July 6, 2024 @ 5:00 pm
Welcome to the site dedicated to Michael Jordan https://michael-jordan.com.az, a basketball legend and symbol of world sports culture. Here you will find highlights, career, family and news about one of the greatest athletes of all time.
Jerryspoig
July 6, 2024 @ 5:05 pm
Diego Armando Maradona https://diego-maradona.com.az Argentine footballer who played as an attacking midfielder and striker. He played for the clubs Argentinos Juniors, Boca Juniors, Barcelona, ??Napoli, and Sevilla.
DennisJow
July 6, 2024 @ 7:11 pm
Gucci купить http://thebestluxurystores.ru по низкой цене в интернет-магазине брендовой одежды. Одежда и обувь бренда Gucci c доставкой.
Enriquerhids
July 6, 2024 @ 7:15 pm
Muhammad Ali https://muhammad-ali.com.az American professional boxer who competed in the heavy weight category; one of the most famous boxers in the history of world boxing.
ChrisBew
July 6, 2024 @ 7:29 pm
Монтаж систем отопления https://fectum.pro, водоснабжения, вентиляции, канализации, очистки воды, пылеудаления, снеготаяния, гелиосистем в Краснодаре под ключ.
Bretttow
July 7, 2024 @ 2:05 am
Lev Ivanovich Yashin https://lev-yashin.com.az Soviet football player, goalkeeper. Olympic champion in 1956 and European champion in 1960, five-time champion of the USSR, three-time winner of the USSR Cup.
Phillipcrark
July 7, 2024 @ 2:19 am
Usain St. Leo Bolt https://usain-bolt.com.az Jamaican track and field athlete, specialized in short-distance running, eight-time Olympic champion and 11-time world champion (a record in the history of this competition among men).
Edgarmic
July 7, 2024 @ 2:20 am
Al-Nasr https://al-nasr.com.az your source of news and information about Al-Nasr Football Club . Find out the latest results, transfer news, player and manager interviews, fixtures and much more.
Donaldgon
July 7, 2024 @ 2:27 am
You have a source of the latest and most interesting sports news from Kazakhstan: “Kazakhstan sports news https://sports-kazahstan.kz: Games and records” ! Follow us to receive updates and interesting news every minute!
RobertClils
July 7, 2024 @ 2:29 am
Game World https://kz-games.kz offers the latest online gaming news, game reviews, gameplay and ideas, gaming tactics and tips . Start playing our most popular and amazing games and get ready to become the leader in the online gaming world!
Charlesrof
July 7, 2024 @ 5:40 am
Latest news and information about the NBA https://basketball-kz.kz in Kazakhstan. Hot stories, player transfers and highlights. Watch the NBA world with us.
Rodneyrhymn
July 7, 2024 @ 5:41 am
Top sports news https://sport-kz-news.kz, photos and blogs from experts and famous athletes, as well as statistics and information about matches of leading championships.
HarrysaK
July 7, 2024 @ 5:43 am
The latest top football news https://football-kz.kz today. Interviews with football players, online broadcasts and match results, analytics and football forecasts, photos and videos.
Leslietalia
July 7, 2024 @ 11:20 am
Latest news about games for Android https://android-games.kz, reviews and daily updates. Read now and get the latest information about the most exciting games
Thomasscold
July 7, 2024 @ 11:20 am
Check out Minecraft kz https://minecraft-kz.kz for the latest news, guides, and in-depth reviews of the game options available. Find the latest information on Minecraft Download, Pocket Edition and Bedrock Edition.
Jefferypaf
July 7, 2024 @ 11:22 am
Latest news from World of Warcraft https://wow-kz.kz (WOW) tournaments, strategy and game analysis. The most detailed gaming portal in the language.
BrettCub
July 7, 2024 @ 11:37 am
Latest news and analysis of the Premier League https://premier-league.kz. Full descriptions of matches, team statistics and the most interesting football events. Premier Kazakhstan is the best place for football fans.
ThomasMeete
July 7, 2024 @ 2:35 pm
Доставка груза и грузоперевозки https://tamozhennyy-deklarant.blogspot.com по России через транспортную компанию автотранспортом доступна и для частных лиц. Перевозчик отправит или доставит ваш груз: выгодные тарифы индивидуальный подход из рук в руки 1 машиной.
Michaelstawl
July 7, 2024 @ 2:40 pm
Зеркала интерьерные https://zerkala-mag.ru в интернет-магазине «Зеркала с подсветкой» Самые низкие цены на зеркала!
JamesEnlat
July 7, 2024 @ 2:41 pm
Предлагаем купить гаражное оборудование https://profcomplex.pro, автохимию, технику и уборочный инвентарь для клининговых компаний. Доставка по Москве и другим городам России.
Kennethagoge
July 7, 2024 @ 3:17 pm
Купить зеркала https://zerkala-m.ru по низким ценам. Более 1980 моделей, купить недорого в интернет-магазине в Москве с доставкой по России. Удобный каталог, низкие цены, качественные фото.
Lewismaync
July 7, 2024 @ 5:57 pm
Spider-Man https://spiderman.kz the latest news, articles, reviews, dates, spoilers and other latest information. All materials on the topic “Spider-Man”
HerbertHex
July 7, 2024 @ 5:58 pm
The latest top football news https://football.sport-news-eg.com today. Interviews with football players, online broadcasts and match results, analytics and football forecasts, photos and videos.
StaceyEuror
July 7, 2024 @ 5:59 pm
Latest Counter-Strike 2 news https://counter-strike-kz.kz, watch the most successful tournaments and be the best in the gaming world.
AlbertoFah
July 7, 2024 @ 6:19 pm
Discover the dynamic world of Arab sports https://sports-ar.com through the lens of Arab sports news. Your premier source for breaking news, exclusive interviews, in-depth analysis and live coverage of everything happening in sports.
JasonVof
July 7, 2024 @ 9:09 pm
NHL news https://nhl-ar.com (National Hockey League) – the latest and most up-to-date NHL news for today.
PatrickGok
July 7, 2024 @ 9:09 pm
Интернет магазин электроники https://techno-line.store и цифровой техники по доступным ценам. Доставка мобильной электроники по Москве и Московской области.
KermitJeorn
July 7, 2024 @ 9:19 pm
UFC news https://ufc-ar.com, schedule of fights and tournaments 2024, ratings of UFC fighters, interviews, photos and videos. Live broadcasts and broadcasts of tournaments, statistics, forums and fan blogs.
Ronaldmup
July 7, 2024 @ 9:23 pm
The most important sports news https://bein-sport-egypt.com, photos and blogs from experts and famous athletes, as well as statistics and information about matches of leading leagues.
RichardceD
July 7, 2024 @ 9:28 pm
News and events of the American Basketball League https://basketball-eg.com in Egypt. Hot events, player transfers and the most interesting events. Explore the world of the NBA with us.
Williamlal
July 8, 2024 @ 12:54 am
Discover the wonderful world of online games https://game-news-ar.com. Get the latest news, reviews and tips for your favorite games.
Felixfah
July 8, 2024 @ 12:59 am
Minecraft news https://minecraft-ar.com, guides and in-depth reviews of the gaming features available in Minecraft Ar. Get the latest information on downloading Minecraft, Pocket Edition and Bedrock Edition.
Brandongah
July 8, 2024 @ 1:02 am
News, tournaments, guides and strategies about the latest GTA games https://gta-ar.com. Stay tuned for the best GTA gaming experience
RobertGuits
July 8, 2024 @ 1:05 am
Latest news https://android-games-ar.com about Android games, reviews and daily updates. The latest information about the most exciting games.
Ronnieexawn
July 8, 2024 @ 8:47 am
Открытие для себя Ерлинг Хааланда https://manchestercity.erling-haaland-cz.com, a talented player of «Manchester City». Learn more about his skills, achievements and career growth.
ScottHurne
July 8, 2024 @ 9:01 am
The path of 21-year-old Jude Bellingham https://realmadrid.jude-bellingham-cz.com from young talent to one of the most promising players in the world, reaching new heights with Dortmund and England.
Charlessnick
July 8, 2024 @ 9:07 am
The site is dedicated to football https://fooball-egypt.com, football history and news. Latest news and fresh reviews of the world of football
Aaronopemy
July 8, 2024 @ 9:13 am
Harry Kane’s journey https://bavaria.harry-kane-cz.com from Tottenham’s leading striker to Bayern’s leader and Champions League champion – this is the story of a triumphant ascent to the football Olympus.
VictorChemy
July 8, 2024 @ 9:13 am
French prodigy Kylian Mbappe https://realmadrid.kylian-mbappe-cz.com is taking football by storm, joining his main target, ” Real.” New titles and records are expected.
RobertZinge
July 8, 2024 @ 12:42 pm
Изготовим для Вас изделия из металла https://smith-moskva.blogspot.com, по вашим чертежам или по нашим эскизам.
Jamesraf
July 8, 2024 @ 12:46 pm
Промышленные насосы https://superomsk.ru/news/137099/pogrujne_nasos/ Wilo предлагают широкий ассортимент решений для различных отраслей промышленности, включая водоснабжение, отопление, вентиляцию, кондиционирование и многие другие. Благодаря своей высокой производительности и эффективности, насосы Wilo помогают снизить расходы на энергию и обслуживание, что делает их идеальным выбором для вашего бизнеса.
Thomasbailm
July 8, 2024 @ 1:14 pm
https://rolaks.com отделочные материалы для фасада – интернет-магазин
Patricknig
July 8, 2024 @ 1:38 pm
The fascinating story of the rise of Brazilian prodigy Vinicius Junior https://realmadrid.vinicius-junior-cz.com to the heights of glory as part of the legendary Madrid “Real”
WilliamBrick
July 8, 2024 @ 2:09 pm
Mohamed Salah https://liverpool.mohamed-salah-cz.com, who grew up in a small town in Egypt, conquered Europe and became Liverpool star and one of the best players in the world.
HermanJEW
July 8, 2024 @ 4:20 pm
The inspiring story of how talented Kevin De Bruyne https://manchestercity.kevin-de-bruyne-cz.com became the best player of Manchester City and the Belgium national team. From humble origins to the leader of a top club.
BrianNug
July 8, 2024 @ 4:37 pm
Bernardo Mota Veiga de Carvalho e Silva https://manchestercity.bernardo-silva-cz.com Portuguese footballer, club midfielder Manchester City and the Portuguese national team.
Orvillepycle
July 8, 2024 @ 4:41 pm
Lionel Messi https://intermiami.lionel-messi-cz.com, one of the best football players of all time, moves to Inter Miami” and changes the face of North American football.
JamesraIte
July 8, 2024 @ 6:06 pm
Antoine Griezmann https://atlticomadrid-dhb.antoine-griezmann-cz.com Atletico Madrid star whose talent and decisive goals helped the club reach the top of La Liga and the UEFA Champions League.
WilliamBraro
July 8, 2024 @ 8:05 pm
Son Heung-min’s https://tottenhamhotspur.son-heung-min-cz.com success story at Tottenham Hotspur and his influence on the South Korean football, youth inspiration and changing the perception of Asian players.
Bernardnus
July 8, 2024 @ 8:10 pm
The story of Robert Lewandowski https://barcelona.robert-lewandowski-cz.com, his impressive journey from Poland to Barcelona, ??where he became not only a leader on the field, but also a source of inspiration for young players.
Raymonddrilm
July 8, 2024 @ 8:16 pm
The impact of the arrival of Cristiano Ronaldo https://annasr.cristiano-ronaldo-cz.com at Al-Nasr. From sporting triumphs to cultural changes in Saudi football.
CurtisFeria
July 8, 2024 @ 8:18 pm
We explore the path of Luka Modric https://realmadrid.luka-modric-cz.com to Real Madrid, from a difficult adaptation to legendary Champions League triumphs and personal awards.
IrvinDag
July 9, 2024 @ 12:12 am
mexico drug stores pharmacies: mexican mail order pharmacies – mexican pharmacy
IrvinDag
July 9, 2024 @ 1:30 am
п»їbest mexican online pharmacies: online mexican pharmacy – mexico drug stores pharmacies
Anthonyelila
July 9, 2024 @ 3:52 am
Find out how Pedri https://barcelona.pedri-cz.com becomes a key figure for Barcelona – his development, influence and ambitions determine the club’s future success in world football.
Robertduasy
July 9, 2024 @ 3:53 am
How Karim Benzema https://alIttihad.karim-benzema-cz.com changed the game of Al-Ittihad and Saudi football: new tactics, championship success, increased viewership and commercial success.
Bradleygaify
July 9, 2024 @ 3:53 am
A study of the influence of Rodrigo https://realmadrid.rodrygo-cz.com on the success and marketing strategy of Real Madrid: analysis of technical skills, popularity in Media and commercial success.
Jasperemile
July 9, 2024 @ 4:01 am
Find out about Alisson https://liverpool.alisson-becker-cz.com‘s influence on Liverpool’s success, from his defense to personal achievements that made him one of the best goalkeepers in the world.
WayneRek
July 9, 2024 @ 4:07 am
Find out how Pedro Gavi https://barcelona.gavi-cz.com helped Barcelona achieve success thanks to his unique qualities, technique and leadership, becoming a key player in the team.
TravisBalge
July 9, 2024 @ 11:18 am
r7 casino войти https://mabiclub.ru
DennisUtera
July 9, 2024 @ 11:35 am
buy instagram followers buy instagram likes
Georgemix
July 9, 2024 @ 11:47 am
Thibaut Courtois https://realmadrid.thibaut-courtois-cz.com the indispensable goalkeeper of “Real”, whose reliability, leadership and outstanding The game made him a key figure in the club.
JamesLig
July 9, 2024 @ 12:24 pm
Find out how Virgil van Dijk https://liverpool.virgil-van-dijk-cz.com became an integral part of style игры «Liverpool», ensuring the stability and success of the team.
RonaldStype
July 9, 2024 @ 12:51 pm
Find out how Bruno Guimaraes https://newcastleunited.bruno-guimaraes-cz.com became a catalyst for the success of Newcastle United thanks to his technical abilities and leadership on the field and beyond.
Joshuaglymn
July 9, 2024 @ 3:40 pm
Study of the playing style of Toni Kroos https://real-madrid.toni-kroos-cz.com at Real Madrid: his accurate passing, tactical flexibility and influence on the team’s success.
Curtisjen
July 9, 2024 @ 3:45 pm
Romelu Lukaku https://chelsea.romelu-lukaku-cz.com, one of the best strikers in Europe, returns to Chelsea to continue climbing to the top of the football Olympus.
Walterpes
July 9, 2024 @ 3:49 pm
The young Uruguayan Darwin Nunez https://liverpool.darwin-nunez-cz.com broke into the elite of world football, and he became a key Liverpool player.
Jimmyfah
July 9, 2024 @ 3:51 pm
Star Brazilian striker Gabriel Jesus https://arsenal.gabriel-jesus-cz.com put in a superb performance to lead Arsenal to new heights after moving from Manchester City.
Jaimeron
July 9, 2024 @ 4:06 pm
A fascinating story about how David Alaba https://realmadrid.david-alaba-cz.com after starting his career at the Austrian academy Vienna became a key player and leader of the legendary Real Madrid.
pill
July 9, 2024 @ 4:39 pm
[url=https://zithromaxl.online/]cost for azithromycin tablets 250mg[/url]
Kennethgen
July 9, 2024 @ 11:49 pm
The story of how the incredibly talented footballer Riyad Mahrez https://alahli.riyad-mahrez-cz.com reached new heights in career, moving to Al Ahly and leading the team to victory.
Keithtruct
July 9, 2024 @ 11:55 pm
The fascinating story of Antonio Rudiger’s transfer https://real-madrid.antonio-rudiger-cz.com to Real Madrid and his rapid rise as a key player at one of the best clubs in the world.
Zacherydix
July 9, 2024 @ 11:56 pm
The fascinating story of Marcus Rashford’s ascent https://manchester-united.marcus-rashford-cz.com to glory in the Red Devils: from a young talent to one of the key players of the team.
RobertUnsub
July 10, 2024 @ 3:53 am
Fascinating event related to this Keanu Reeves helped him in the role of the iconic John Wick characters https://john-wick.keanu-reeves.cz, among which there is another talent who has combat smarts with inappropriate charisma.
Calvinfes
July 10, 2024 @ 3:55 am
Try to make a fascinating actor Johnny Depp https://secret-window.johnny-depp.cz, who will become the slave of his strong hero Moudriho Creeps in the thriller “Secret Window”.
RichardRer
July 10, 2024 @ 4:00 am
Jackie Chan https://peakhour.jackie-chan.cz from a poor boy from Hong Kong to a world famous Hollywood stuntman. The incredible success story of Jackie Chan.
DavidAgrig
July 10, 2024 @ 4:09 am
Follow Liam Neeson’s career https://hostage.liam-neeson.cz as he fulfills his potential as Brian Mills in the film “Taken” and becomes one of the leading stars of Hollywood action films.
RobertDweld
July 10, 2024 @ 4:12 am
Emily Olivia Laura Blunt https://oppenheimer.emily-blunt.cz British and American actress. Winner of the Golden Globe (2007) and Screen Actors Guild (2019) awards.
GarryRip
July 10, 2024 @ 8:13 am
The inspiring story of Zendaya’s rise https://spider-man.zendaya-maree.cz, from her early roles to her blockbuster debut in Marvel Cinematic Universe.
Derricknum
July 10, 2024 @ 8:15 am
The inspiring story of the ascent of the young actress Anya Taylor https://queensmove.anya-taylor-joy.cz to fame after her breakthrough performance in the TV series “The Queen’s Move”. Conquering new peaks.
EdwardListe
July 10, 2024 @ 8:16 am
An indomitable spirit, incredible skills and five championships – how Kobe Bryant https://losangeles-lakers.kobe-bryant.cz became an icon of the Los Angeles Lakers and the entire NBA world.
DariusLef
July 10, 2024 @ 8:17 am
Carlos Vemola https://oktagon-mma.karlos-vemola.cz Czech professional mixed martial artist, former bodybuilder, wrestler and member Sokol.
RaymondFlaps
July 10, 2024 @ 8:29 am
Witness the thrilling story of Jiri Prochazka’s https://ufc.jiri-prochazka-ufc.cz rapid rise to the top of the UFC’s light heavyweight division, marked by his dynamic fighting style and relentless determination.
HenryTosse
July 10, 2024 @ 11:54 am
An article about the triumphant 2023 Ferrari https://ferrari.charles-leclerc.cz and their star driver Charles Leclerc, who became the Formula world champion 1.
Stacybreag
July 10, 2024 @ 11:56 am
Jon Jones https://ufc.jon-jones.cz a dominant fighter with unrivaled skill, technique and physique who has conquered the light heavyweight division.
DouglasgAf
July 10, 2024 @ 12:08 pm
The legendary Spanish racer Fernando Alonso https://formula-1.fernando-alonso.cz returns to Formula 1 after several years.
EarnestZer
July 10, 2024 @ 12:14 pm
Young Briton Lando Norris https://mclaren.lando-norris.cz is at the heart of McLaren’s Formula 1 renaissance, regularly achieving podium finishes and winning.
JamesMeexy
July 10, 2024 @ 1:10 pm
Activision and Call of Duty https://activision.call-of-duty.cz leading video game publisher and iconic shooter with a long history market dominance.
Josephcow
July 10, 2024 @ 3:48 pm
properties in montenegro https://montenegro-real-estate-prices.com
Williammethy
July 10, 2024 @ 3:48 pm
the most popular sports website https://sports-forecasts.com in the Arab world with the latest sports news, predictions and analysis in real time.
AaronAdopy
July 10, 2024 @ 3:57 pm
Latest news and analysis of the English Premier League https://epl-ar.com. Detailed descriptions of matches, team statistics and the most interesting football events.
Robertmon
July 10, 2024 @ 3:58 pm
Free movies https://www.moviesjoy.cc and TV streaming online, watch movies online in HD 1080p.
JamesCivok
July 10, 2024 @ 4:03 pm
Latest Diablo news https://diablo-ar.com, detailed game descriptions and guides. Diablo.az – The largest Diablo information portal in Arabic.
DavidTup
July 10, 2024 @ 7:01 pm
Latest World of Warcraft tournament news https://ar-wow.com (WOW), strategies and game analysis. The most detailed gaming portal in Arabic.
RosarioMycle
July 10, 2024 @ 7:09 pm
The latest analysis, reviews of https://spider-man-ar.com tournaments and the most interesting things from the “Spider-Man” series of games in Azerbaijani language. It’s all here!
EddieAbave
July 10, 2024 @ 7:12 pm
NFL https://nfl-ar.com News, analysis and topics about the latest practices, victories and records. A portal that explores the most beautiful games in the NFL world in general.
Jamesrib
July 10, 2024 @ 7:15 pm
Discover exciting virtual football https://fortnite-ar.com in Fortnite. Your central hub for the latest news, expert strategy and exciting eSports reporting.
Robertmut
July 10, 2024 @ 7:19 pm
Latest Counter-Strike 2 news https://counter-strike-ar.com, watch the most successful tournaments and be the best in the gaming world on CS2 ar.
HenryElory
July 10, 2024 @ 9:55 pm
purple pharmacy mexico price list: cmq pharma – medicine in mexico pharmacies
RaymondPutty
July 11, 2024 @ 1:04 am
Latest boxing news, achievements of Raisol Abbasov https://boxing-ar.com, Tyson Fury fights and much more. It’s all about the boxing ambassador.
KennethVes
July 11, 2024 @ 1:10 am
Latest news from the world of boxing https://boks-uz.com, achievements of Resul Abbasov, Tyson Fury’s fights and much more. Everything Boxing Ambassador has.
ScottKex
July 11, 2024 @ 1:16 am
Discover the wonderful world of online games https://onlayn-oyinlar.com with GameHub. Get the latest news, reviews and tips for your favorite games. Join our gaming community today!
JoshuaNup
July 11, 2024 @ 1:17 am
Sports news https://gta-uzbek.com the most respected sports site in Uzbekistan, which contains the latest sports news, forecasts and analysis.
Jamesseaft
July 11, 2024 @ 1:17 am
Latest GTA game news https://gta-uzbek.com, tournaments, guides and strategies. Stay tuned for the best GTA gaming experience
JamesRerce
July 11, 2024 @ 4:54 am
Explore the extraordinary journey of Kylian Mbappe https://mbappe-real-madrid.com, from his humble beginnings to global stardom.
Phillipbeend
July 11, 2024 @ 4:58 am
Latest news about Pele https://mesut-ozil-uz.com, statistics, photos and much more. Get the latest news and information about football legend Pele.
Stevensquib
July 11, 2024 @ 5:04 am
Get the latest https://mesut-ozil-uz.com Mesut Ozil news, stats, photos and more.
StevenAming
July 11, 2024 @ 5:07 am
Serxio Ramos Garsiya https://serxio-ramos.com ispaniyalik futbolchi, himoyachi. Ispaniya terma jamoasining sobiq futbolchisi. 16 mavsum davomida u “Real Madrid”da markaziy himoyachi sifatida o’ynadi.
Robertvop
July 11, 2024 @ 5:36 am
Ronaldo de Asis Moreira https://ronaldinyo.com braziliyalik futbolchi, yarim himoyachi va hujumchi sifatida o’ynagan. Jahon chempioni (2002). “Oltin to’p” sovrindori (2005).
Orlandomaype
July 11, 2024 @ 8:33 am
Официальный сайт онлайн-казино Vavada https://vavada-kz-game.kz это новый адрес лучших слотов и джекпотов. Ознакомьтесь с бонусами и играйте на реальные деньги из Казахстана.
AnthonyLoula
July 11, 2024 @ 8:39 am
Marcus Lilian Thuram-Julien https://internationale.marcus-thuram-fr.com French footballer, forward for the Internazionale club and French national team.
JosephNergo
July 11, 2024 @ 8:49 am
Legendary striker Cristiano Ronaldo https://an-nasr.cristiano-ronaldo-fr.com signed a contract with the Saudi club ” An-Nasr”, opening a new chapter in his illustrious career in the Middle East.
Edwardunlow
July 11, 2024 @ 8:51 am
Manchester City and Erling Haaland https://manchester-city.erling-haaland-fr.com explosive synergy in action. How a club and a footballer light up stadiums with their dynamic play.
Warrenfup
July 11, 2024 @ 8:57 am
Lionel Messi https://inter-miami.lionel-messi-fr.com legendary Argentine footballer, announced his transfer to the American club Inter Miami.
Davidcounk
July 11, 2024 @ 12:39 pm
The official website where you can find everything about the career of Gianluigi Buffon https://gianluigi-buffon.com. Discover the story of this legendary goalkeeper who left his mark on football history and relive his achievements and unforgettable memories with us.
RockySluse
July 11, 2024 @ 12:50 pm
Website dedicated to football player Paul Pogba https://pogba-uz.com. Latest news from the world of football.
JamesFaG
July 11, 2024 @ 12:54 pm
Welcome to our official website! Go deeper into Paulo Dybala’s https://paulo-dybala.com football career. Discover Dybala’s unforgettable moments, amazing talents and fascinating journey in the world of football on this site.
PeterGab
July 11, 2024 @ 2:54 pm
Latest news on the Vinicius Junior fan site https://vinisius-junior.com. Vinicius Junior has been playing since 2018 for Real Madrid (Real Madrid). He plays in the Left Winger position.
Josephreisk
July 11, 2024 @ 2:54 pm
Coffeeroom https://coffeeroom.by – магазин кофе, чая, кофетехники, посуды, химии и аксессуаров в Минске для дома и офиса.
Everettblept
July 11, 2024 @ 4:08 pm
Прокат и аренда автомобилей https://autorent.by в Минске 2019-2022. Сутки от 35 руб.
Scottbougs
July 11, 2024 @ 4:20 pm
Find the latest information on Khabib Nurmagomedov https://khabib-nurmagomedov.uz news and fights. Check out articles and videos detailing Khabib UFC career, interviews, wins, and biography.
TimothyPak
July 11, 2024 @ 4:27 pm
Latest news and information about Marcelo https://marselo-uz.com on this site! Find Marcelo’s biography, career, game stats and more.
RobertTup
July 12, 2024 @ 12:04 am
Discover how Riyad Mahrez https://al-ahli.riyad-mahrez.com transformed Al-Ahli, becoming a key player and catalyst in reaching new heights in world football.
AngelRap
July 12, 2024 @ 12:11 am
Find the latest information on Conor McGregor https://conor-mcgregor.uz news, fights, and interviews. Check out detailed articles and news about McGregor’s UFC career, wins, training, and personal life.
WilliamLax
July 12, 2024 @ 12:15 am
Explore the dynamic world of sports https://noticias-esportivas-br.org through the lens of a sports reporter. Your source for breaking news, exclusive interviews, in-depth analysis and live coverage of all sports.
Davidincet
July 12, 2024 @ 12:21 am
Get to know the history, players and latest news of the Inter Miami football club https://inter-miami.uz. Join us to learn about the successes and great performances of America’s newest and most exciting soccer club.
Jamesasype
July 12, 2024 @ 12:23 am
A site dedicated to Michael Jordan https://michael-jordan.uz, a basketball legend and symbol of world sports culture. Here you will find highlights, career, family and news about one of the greatest athletes of all time.
Jorgegelry
July 12, 2024 @ 3:41 am
The latest top football news https://futebol-ao-vivo.net today. Interviews with football players, online broadcasts and match results, analytics and football forecasts
WilliamBic
July 12, 2024 @ 3:50 am
Site with the latest news, statistics, photos of Pele https://edson-arantes-do-nascimento.com and much more. Get the latest news and information about football legend Pele.
Jimmygaw
July 12, 2024 @ 3:50 am
Welcome to our official website, where you will find everything about the career of Gianluigi Buffon https://gianluigi-buffon.org. Discover the story of this legendary goalkeeper who made football history.
Charlesmup
July 12, 2024 @ 3:51 am
If you are a fan of UFC https://ufc-hoje.com the most famous organization in the world, come visit us. The most important news and highlights from the UFC world await you on our website.
Leonardjes
July 12, 2024 @ 5:48 am
The best site dedicated to the football player Paul Pogba https://pogba.org. Latest news from the world of football.
Gabrielmeape
July 12, 2024 @ 6:46 am
Vinicius Junior https://vinicius-junior.org all the latest current and latest news for today about the player of the 2024 season
AllenUsato
July 12, 2024 @ 6:59 am
Analysis of Arsenal’s impressive revival https://arsenal.bukayo-saka.biz under the leadership of Mikel Arteta and the key role of young star Bukayo Saki in the club’s return to the top.
JesseNoism
July 12, 2024 @ 7:03 am
Gavi’s success story https://barcelona.gavi-fr.com at Barcelona: from his debut at 16 to a key role in club and national team of Spain, his talent inspires the world of football.
Gregginduh
July 12, 2024 @ 7:05 am
Pedri’s story https://barcelona.pedri-fr.com from his youth in the Canary Islands to becoming a world-class star in Barcelona, ??with international success and recognition.
Henrytromo
July 12, 2024 @ 12:00 pm
Discover the journey of Charles Leclerc https://ferrari.charles-leclerc-fr.com, from young Monegasque driver to Ferrari Formula 1 leader, from his early years to his main achievements within the team.
WilliamBrape
July 12, 2024 @ 12:08 pm
Discover Pierre Gasly’s https://alpine.pierre-gasly.com journey through the world of Formula 1, from his beginnings with Toro Rosso to his extraordinary achievements with Alpine.
JeffreyElard
July 12, 2024 @ 12:24 pm
Discover the story of Rudy Gobert https://minnesota-timberwolves.rudy-gobert.biz, the French basketball player whose defensive play and leadership transformed the Minnesota Timberwolves into a powerhouse NBA team.
JamesNup
July 12, 2024 @ 12:30 pm
From childhood teams to championship victories, the path to success with the Los Angeles Lakers https://los-angeles-lakers.lebron-james-fr.com requires not only talent, but also undeniable dedication and work.
JamesGuh
July 12, 2024 @ 12:37 pm
Leroy Sane’s https://bavaria.leroy-sane-ft.com success story at FC Bayern Munich: from adaptation to influence on the club’s results. Inspiration for hard work and professionalism in football.
Rodgerner
July 12, 2024 @ 4:47 pm
The story of the Moroccan footballer https://al-hilal.yassine-bounou.com, who became a star at Al-Hilal, traces his journey from the streets of Casablanca to international football stardom and his personal development.
DonaldSar
July 12, 2024 @ 4:50 pm
Victor Wembanyama’s travel postcard https://san-antonio-spurs.victor-wembanyama.biz from his career in France to his impact in the NBA with the San Antonio Spurs.
StanleyMef
July 12, 2024 @ 4:55 pm
Neymar https://al-hilal.neymar-fr.com at Al-Hilal: his professionalism and talent inspire young people players, taking the club to new heights in Asian football.
AnthonyCoith
July 12, 2024 @ 5:00 pm
The history of Michael Jordan’s Chicago Bulls https://chicago-bulls.michael-jordan-fr.com extends from his rookie in 1984 to a six-time NBA championship.
ZacharymaK
July 12, 2024 @ 5:05 pm
Golden State Warriors success story https://golden-state-warriors.stephen-curry-fr.com Stephen Curry: From becoming a leader to creating a basketball dynasty that redefined the game.
ThomasVog
July 12, 2024 @ 8:49 pm
Del Mar Energy is an international industrial holding company engaged in the extraction of oil, gas, and coal
BrianFup
July 12, 2024 @ 8:55 pm
The success story of the French footballer https://juventus.thierry-henry.biz at Juventus: from his career at the club to leadership on the field , becoming a legend and a source of inspiration for youth.
BruceSeish
July 12, 2024 @ 9:02 pm
The story of the great Kobe Bryant https://los-angeles-lakers.kobe-bryant-fr.com with ” Los Angeles Lakers: his path to the championship, his legendary achievements.
WilliamRousy
July 12, 2024 @ 9:03 pm
Novak Djokovic’s https://tennis.novak-djokovic-fr.biz journey from childhood to the top of world tennis: early years, first victories, dominance and influence on the sport.
ClaudeBal
July 12, 2024 @ 9:11 pm
Find out the story of Jon Jones https://ufc.jon-jones-fr.biz in the UFC: his triumphs, records and controversies, which made him one of the greatest fighters in the MMA world.
HaroldQuogs
July 13, 2024 @ 8:50 am
Jannik Sinner https://tennis.jannik-sinner-fr.biz an Italian tennis player, went from starting his career to entering the top 10 of the ATP, demonstrating unique abilities and ambitions in world tennis.
RandyAxome
July 13, 2024 @ 8:57 am
Carlos Alcaraz https://tennis.carlos-alcaraz-fr.biz from a talented junior to the ATP top 10. His rise is the result of hard work, support and impressive victories at major world tournaments.
Dennystalm
July 13, 2024 @ 8:59 am
The fascinating story of Daniil Medvedev’s https://tennis.daniil-medvedev-fr.biz rise to world number one. Find out how a Russian tennis player quickly broke into the elite and conquered the tennis Olympus.
CarlosThalk
July 13, 2024 @ 9:02 am
The fascinating story of Alexander Zverev’s https://tennis.alexander-zverev-fr.biz rapid rise from a junior star to one of the leaders of modern tennis.
Thomasnaw
July 13, 2024 @ 9:07 am
Discover Casper Ruud’s https://tennis.casper-ruud-fr.com journey from his Challenger debut to the top 10 of the world tennis rankings. A unique success.
Tariorrnn
July 13, 2024 @ 4:17 pm
[u][b] Здравствуйте![/b][/u]
Где заказать диплом специалиста?
Приобрести диплом о высшем образовании.
http://onlineboxing.net/jforum/user/profile/271811.page
[u][b] Рады оказаться полезными![u][b]
Diplomi_vakr
July 13, 2024 @ 4:33 pm
купить дипломы в россии [url=https://ast-diplomas.com/]купить дипломы в россии[/url] .
BrianPeext
July 13, 2024 @ 5:54 pm
[u][b] Добрый день![/b][/u]
Заказать диплом о высшем образовании.
Мы предлагаем купить диплом высокого качества, который не отличить от оригинала без использования дорогостоящего оборудования и опытного специалиста.
[b]Где приобрести диплом специалиста?[/b]
http://ks4yumuo.listbb.ru/viewtopic.php?f=18&t=631
[b]Успешной учебы![/b]
Lunrxbf
July 13, 2024 @ 6:35 pm
[u][b] Добрый день![/b][/u]
Мы изготавливаем дипломы любых профессий по приятным ценам.
Мы готовы предложить документы техникумов, которые находятся на территории всей России. Можно заказать диплом за любой год, в том числе документы СССР. Документы выпускаются на бумаге самого высокого качества. Это позволяет делать настоящие дипломы, не отличимые от оригиналов. Они будут заверены всеми требуемыми печатями и штампами.
Даем 100% гарантию, что в случае проверки документов работодателем, подозрений не появится.
[b]Где купить диплом специалиста?[/b]
[url=http://belydom.ru/forum/user/37523/]kadastr.bestbb.ru/viewtopic.php?id=680#p1467[/url]
Charlescal
July 13, 2024 @ 9:57 pm
The story of Fernando Alonso https://formula-1.fernando-alonso-fr.com in Formula 1: a unique path to success through talent, tenacity and strategic decisions, inspiring and exciting.
JustinBonry
July 13, 2024 @ 9:57 pm
The fascinating story of how Lewis Hamilton https://mercedes.lewis-hamilton-fr.biz became a seven-time Formula 1 world champion after signing with Mercedes.
Timothydip
July 13, 2024 @ 9:58 pm
The legendary boxing world champion Mike Tyson https://ufc.mike-tyson-fr.biz made an unexpected transition to the UFC in 2024, where he rose to the top, becoming the oldest heavyweight champion.
Frankcig
July 13, 2024 @ 10:20 pm
The fascinating story of the creation and rapid growth of Facebook https://facebook.mark-zuckerberg-fr.biz under the leadership of Mark Zuckerberg, who became one of the most influential technology entrepreneurs of our time.
russkoeporno365.pro
July 14, 2024 @ 12:59 am
This site definitely has all of the information I needed about this subject
My website: analporno.club
Mariondar
July 14, 2024 @ 1:14 am
Kim Kardashian’s https://the-kardashians.kim-kardashian-fr.com incredible success story, from sex scandal to pop culture icon and billion-dollar fortune.
Richardspags
July 14, 2024 @ 1:24 am
Max Verstappen and Red Bull Racing’s https://red-bull-racing.max-verstappen-fr.com path to success in Formula 1. A story of talent, determination and team support leading to a championship title.
RobertNoinc
July 14, 2024 @ 1:25 am
The astonishing story of Emmanuel Macron’s https://president-of-france.emmanuel-macron-fr.com political rise from bank director to the highest office in France.
Thomaspreby
July 14, 2024 @ 1:26 am
The story of Joe Biden’s https://president-of-the-usa.joe-biden-fr.com triumphant journey, overcoming many obstacles on his path to the White House and becoming the 46th President of the United States.
ErwinDus
July 14, 2024 @ 1:36 am
Une ascension fulgurante au pouvoir Donald Trump https://usa.donald-trump-fr.com et son empire commercial
Tariormby
July 14, 2024 @ 2:18 am
[u][b] Привет![/b][/u]
Где купить диплом по актуальной специальности?
Приобрести диплом о высшем образовании.
https://www.ecmo.ru/users/danilowwitek
[u][b] Окажем помощь![u][b]
Diplomi_wkkr
July 14, 2024 @ 2:45 am
купить диплом бакалавра в челябинске [url=https://ast-diplomas.com/]ast-diplomas.com[/url] .
BrianPeext
July 14, 2024 @ 4:04 am
[u][b] Добрый день![/b][/u]
Приобрести диплом университета.
Наша компания предлагает заказать диплом высокого качества, который невозможно отличить от оригинального документа без использования дорогостоящего оборудования и опытного специалиста.
[b]Где купить диплом специалиста?[/b]
http://ks4yumuo.listbb.ru/viewtopic.php?f=18&t=631
[b]Успехов в учебе![/b]
Lunrrfl
July 14, 2024 @ 4:33 am
[u][b] Здравствуйте![/b][/u]
Мы предлагаем дипломы любой профессии по выгодным ценам.
Мы предлагаем документы ВУЗов, которые расположены в любом регионе России. Вы можете купить качественный диплом от любого заведения, за любой год, включая документы старого образца СССР. Документы делаются на “правильной” бумаге самого высокого качества. Это дает возможность делать государственные дипломы, которые невозможно отличить от оригиналов. Они заверяются всеми требуемыми печатями и подписями.
Даем гарантию, что в случае проверки документа работодателями, подозрений не появится.
[b]Где приобрести диплом специалиста?[/b]
[url=http://prof-komplekt.com/club/user/4861/blog/10343/]wow-tour.ru/kupit-diplom-vash-klyuch-k-perspektivnoy-karere-i-professionalnomu-rostu[/url]
RobertDiuby
July 14, 2024 @ 4:38 am
Parisian PSG https://paris.psg-fr.com is one of the most successful and ambitious football clubs in Europe. Find out how he became a global football superstar.
Matthewsmash
July 14, 2024 @ 4:44 am
Travel to the pinnacle of French football https://stadede-bordeaux.bordeaux-fr.org at the Stade de Bordeaux, where the passion of the game meets the grandeur of architecture.
Larryfrods
July 14, 2024 @ 4:51 am
The fascinating story of Gigi Hadid’s rise to Victoria’s Secret Angel https://victorias-secret.gigi-hadid-fr.com status and her journey to the top of the modeling industry.
JimmyMaync
July 14, 2024 @ 4:53 am
Olympique de Marseille https://liga1.marseilles-fr.com after several years in the shadows, once again becomes champion of France. How did they do it and what prospects open up for the club
Craigriz
July 14, 2024 @ 6:33 am
The fascinating story of the creation and meteoric rise of Amazon https://amazon.jeff-bezos-fr.com from its humble beginnings as an online bookstore to its dominant force in the world of e-commerce.
Uazryej
July 14, 2024 @ 7:56 am
[u][b] Добрый день![/b][/u]
Приобрести диплом о высшем образовании
[b]Наши специалисты предлагают[/b] быстро и выгодно заказать диплом, который выполнен на бланке ГОЗНАКа и заверен печатями, штампами, подписями. Документ пройдет лубую проверку, даже при использовании специально предназначенного оборудования. Решайте свои задачи максимально быстро с нашими дипломами.
[b]Где приобрести диплом по актуальной специальности?[/b]
http://testruslit.ru/forum/posting.php?mode=post&f=2
[u][b] Всегда вам поможем![u][b].
Diplomi_uvsr
July 14, 2024 @ 8:42 am
дипломы купить родителям [url=https://asxdiplomik.com/]asxdiplomik.com[/url] .
Andreerymn
July 14, 2024 @ 9:02 am
The inspiring story of Travis Scott’s https://yeezus.travis-scott-fr.com rise from emerging artist to one of modern hip-hop’s brightest stars through his collaboration with Kanye West.
Diplomi_yoSa
July 14, 2024 @ 9:03 am
купить диплом машиниста [url=https://diplomyx.com/]diplomyx.com[/url] .
Danielnof
July 14, 2024 @ 9:04 am
A fascinating story about how Elon Musk https://spacex.elon-musk-fr.com and his company SpaceX revolutionized space exploration, opening new horizons for humanity.
Williamskymn
July 14, 2024 @ 9:11 am
An exploration of Nicole Kidman’s https://watch.nicole-kidman-fr.com career, her notable roles, and her continued quest for excellence as an actress.
Marvincen
July 14, 2024 @ 9:15 am
How Taylor Swift https://midnights.taylor-swift-fr.com reinvented her sound and image on the intimate and reflective album “Midnights,” revealing new dimensions of her talent.
Diplomi_upmi
July 14, 2024 @ 11:03 am
купить диплом в одинцово [url=https://ast-diploms.com/]ast-diploms.com[/url] .
Diplomi_qpKt
July 14, 2024 @ 11:10 am
диплом техникума купить [url=https://diplomasx.com/]диплом техникума купить[/url] .
Thomastuh
July 14, 2024 @ 2:41 pm
regent porto montenegro https://www.weather-webcam-in-montenegro.com
EddieCew
July 14, 2024 @ 3:01 pm
Единственная в России студия кастомных париков https://wigdealers.ru, где мастера индивидуально подбирают структуру волос и основу по форме головы, после чего стригут, окрашивают, делают укладку и доводят до идеала ваш будущий аксессуар.
BrianImire
July 14, 2024 @ 3:06 pm
The iconic Anfield https://enfield.liverpool-fr.com stadium and the passionate Liverpool fans are an integral part of English football culture.
BobbyNum
July 14, 2024 @ 3:12 pm
An exploration of the history of Turin’s https://turin.juventus-fr.org iconic football club – Juventus – its rivalries, success and influence on Italian football.
Robertpeago
July 14, 2024 @ 3:22 pm
The new Premier League https://premier-league.chelsea-fr.com season has gotten off to an intriguing start, with a new-look Chelsea looking to return to the Champions League, but serious challenges lie ahead.
Trefjqy
July 14, 2024 @ 3:55 pm
[b]Здравствуйте[/b]!
Задался вопросом: можно ли на самом деле купить диплом государственного образца в Москве? Был приятно удивлен — это реально и легально!
Сначала искал информацию в интернете на тему: купить диплом медицинского училища, купить диплом тренера, купить диплом врача, купить диплом в миассе , купить диплом во владимире и получил базовые знания. В итоге остановился на материале: http://forum.jeep-club.by/index.php?/blog/43/entry-3012-%D0%BA%D1%83%D0%BF%D0%B8%D1%82%D1%8C-%D0%B4%D0%B8%D0%BF%D0%BB%D0%BE%D0%BC-%D0%B2-%D0%B2%D0%BE%D0%BB%D0%B3%D0%BE%D0%B3%D1%80%D0%B0%D0%B4%D0%B5/
Успешной учебы!
Uazrixp
July 14, 2024 @ 5:06 pm
[u][b] Здравствуйте![/b][/u]
Купить диплом любого университета
[b]Наши специалисты предлагают[/b] максимально быстро приобрести диплом, который выполнен на оригинальной бумаге и заверен печатями, водяными знаками, подписями должностных лиц. Документ способен пройти любые проверки, даже с применением специального оборудования. Решите свои задачи быстро и просто с нашей компанией.
[b]Где купить диплом по необходимой специальности?[/b]
http://ekonomimvmeste.ukrbb.net/viewtopic.php?f=8&t=57863
[u][b] Всегда вам поможем![u][b].
Diplomi_wqsr
July 14, 2024 @ 5:54 pm
купить диплом о среднем специальном цена [url=https://asxdiplomik.com/]купить диплом о среднем специальном цена[/url] .
Diplomi_zdSa
July 14, 2024 @ 6:31 pm
купить диплом о полном [url=https://diplomyx.com/]diplomyx.com[/url] .
Diplomi_sfmi
July 14, 2024 @ 9:22 pm
купить диплом повара цена [url=https://ast-diploms.com/]ast-diploms.com[/url] .
Diplomi_zuKt
July 14, 2024 @ 9:48 pm
купить диплом института в спб [url=https://diplomasx.com/]купить диплом института в спб[/url] .
MelvinAssug
July 14, 2024 @ 10:55 pm
Inter Miami FC https://mls.inter-miami-fr.com has become a major player in MLS thanks to its star roster, economic growth and international influence.
Matthewdus
July 14, 2024 @ 10:56 pm
Tyson Fury https://wbc.tyson-fury-fr.com is the undefeated WBC world champion and reigns supreme in boxing’s heavyweight division.
DerrickSucky
July 14, 2024 @ 10:57 pm
Explore the career and significance of Monica Bellucci https://malena.monica-bellucci-fr.com in Malena (2000), which explores complex themes of beauty and human strength in wartime.
Brucesleem
July 14, 2024 @ 10:59 pm
Discover Rafael Nadal’s https://mls.inter-miami-fr.com impressive rise to the top of world tennis, from his debut to his career Grand Slam victory.
Anthonynug
July 14, 2024 @ 11:05 pm
The story of Kanye West https://the-college-dropout.kanye-west-fr.com, starting with his debut album “The College Dropout,” which changed hip-hop and became his cultural legacy.
GabrielLof
July 15, 2024 @ 2:01 am
Преимущества аренды склада https://efrprograms.ru/preimushhestva-arendy-sklada-kak-arenda-skladskih-pomeshhenij-mozhet-uluchshit-vash-biznes/, как аренда складских помещений может улучшить ваш бизнес
Trefhjz
July 15, 2024 @ 2:03 am
[b]Привет, друзья[/b]!
Всегда считал, что покупка диплома о высшем образовании — это миф. Но, оказалось, что все возможно! Сначала искал информацию по теме: купить диплом в сыктывкаре, купить диплом в воронеже, купить диплом в красноярске, купить диплом в москве, купить диплом логиста, а затем перешел на дипломы вузов. Подробнее можно узнать здесь: http://forummsk.getbb.ru/viewtopic.php?f=11&t=2163
Оказалось, что все возможно и официально, с упрощенными программами обучения. Теперь у меня диплом московского вуза нового образца, и я рекомендую вам воспользоваться этим шансом!
Удачи!
Thomasswori
July 15, 2024 @ 2:10 am
Rivaldo, or Rivaldo https://barcelona.rivaldo-br.com, is one of the greatest football players to ever play for Barcelona.
Anthonyspike
July 15, 2024 @ 2:12 am
The fascinating story of the phenomenal rise and meteoric fall of Diego Maradona https://napoli.diegomaradona.biz, who became a cult figure at Napoli in the 1980s.
JasonEpiff
July 15, 2024 @ 2:14 am
A fascinating story about Brazilian veteran Thiago Silva’s https://chelsea.thiago-silva.net difficult path to the top of European football as part of Chelsea London.
Diplomi_ixkr
July 15, 2024 @ 2:18 am
купить диплом ссср в ростове на дону [url=https://ast-diplomas.com/]ast-diplomas.com[/url] .
Rodneyjoina
July 15, 2024 @ 2:19 am
Explore the remarkable journey of Vinicius Junior https://real-madrid.vinicius-junior.net, the Brazilian prodigy who conquered the world’s biggest stage with his dazzling skills and unparalleled ambition at Real Madrid.
RobertFrelp
July 15, 2024 @ 9:04 am
Cristiano Ronaldo https://al-nassr.cristianoronaldo-br.net one of the greatest football players of all time, begins a new chapter in his career by joining An Nasr Club.
Stevenwoody
July 15, 2024 @ 9:07 am
The story of Luka Modric’s rise https://real-madrid.lukamodric-br.com from young talent to one of the greatest midfielders of his generation and a key player for the Royals.
LouisGag
July 15, 2024 @ 9:09 am
The fascinating story of Marcus Rashford’s rise https://manchester-united.marcusrashford-br.com from academy youth to the main striker and captain of Manchester United. Read about his meteoric rise and colorful career.
RobertMak
July 15, 2024 @ 9:14 am
From academy product to captain and leader of Real Madrid https://real-madrid.ikercasillas-br.com Casillas became one of the greatest players in the history of Real Madrid.
Donaldpanda
July 15, 2024 @ 9:29 am
Follow Bernardo Silva’s impressive career https://manchester-city.bernardosilva.net from his debut at Monaco to to his status as a key player and leader of Manchester City.
Diplomi_urkr
July 15, 2024 @ 12:40 pm
диплом о высшем медицинском образовании купить [url=https://ast-diplomas.com/]диплом о высшем медицинском образовании купить[/url] .
Williamtem
July 15, 2024 @ 1:26 pm
вход Rio Bet Casino Rio Bet Casino
TimothyArili
July 15, 2024 @ 1:28 pm
рио бет казино вход Rio Bet Casino
JosephRor
July 15, 2024 @ 1:30 pm
регистрация драгон мани казино бонус драгон мани казино
MauriceEncaf
July 15, 2024 @ 1:35 pm
реристрация Dragon Money Casino официальный сайт драгон мани казино
Ronniereole
July 15, 2024 @ 2:00 pm
Хотите научиться готовить самые изысканные и сложные торты? В этом https://v1.skladchik.org/tags/tort/ разделе вы найдете множество подробных пошаговых рецептов самых трендовых и известных тортов с возможностью получить их за сущие копейки благодаря складчине. Готовьте с удовольствием и открывайте для себя новые рецепты вместе с Skladchik.org
Chuckscova
July 15, 2024 @ 4:18 pm
Лучшие пансионаты для пожилых людей https://ernst-neizvestniy.ru в Самаре – недорогие дома для престарелых в Самарской области
Williambelty
July 15, 2024 @ 4:19 pm
Пансионаты для пожилых людей https://moyomesto.ru в Самаре по доступным ценам. Специальные условия по уходу, индивидуальные программы.
Charlesmus
July 15, 2024 @ 4:21 pm
The fascinating story of Sergio Ramos’ https://seville.sergioramos.net rise from Sevilla graduate to one of Real Madrid and Spain’s greatest defenders.
Jesusbof
July 15, 2024 @ 4:27 pm
Ousmane Dembele’s https://paris-saint-germain.ousmanedembele-br.com rise from promising talent to key player for French football giants Paris Saint-Germain. An exciting success story.
Diplomi_ivOa
July 15, 2024 @ 5:43 pm
купить диплом фармацевта [url=www.diploms-x.com/]www.diploms-x.com/[/url] .
Danrhvn
July 15, 2024 @ 6:53 pm
[u][b] Привет![/b][/u]
[b]Где купить диплом по нужной специальности?[/b]
[b]Мы изготавливаем дипломы[/b] психологов, юристов, экономистов и прочих профессий по доступным тарифам.
[url=http://www.camry-club.ru/member.php/61231-sonnick84?tab=activitystream&type=user&page=2]www.camry-club.ru/member.php/61231-sonnick84?tab=activitystream&type=user&page=2[/url]
Williamasymn
July 15, 2024 @ 11:42 pm
The incredible success story of 20-year-old Florian Wirtz https://bayer-04.florianwirtz-br.com, who quickly joined the Bayer team and became one of the best young talents in the world.
Donaldpoify
July 15, 2024 @ 11:44 pm
The compelling story of Alisson Becker’s https://bayer-04.florianwirtz-br.com meteoric rise from young talent to key figure in Liverpool’s triumphant era under Jurgen Klopp.
CarlosHulty
July 15, 2024 @ 11:50 pm
The incredible story https://napoli.khvichakvaratskhelia-br.com of a young Georgian talent’s transformation into an Italian Serie A star. Khvicha Kvaraeshvili is a rising phenomenon in European football.
Charlesket
July 15, 2024 @ 11:52 pm
O meio-campista Rafael Veiga leva https://palmeiras.raphaelveiga-br.com o Palmeiras ao sucesso – o campeonato brasileiro e a vitoria na Copa Libertadores aos 24 anos.
Justinchodo
July 15, 2024 @ 11:57 pm
Midfielder Rafael Veiga leads https://manchester-city.philfoden-br.com Palmeiras to success – the championship Brazilian and victory in the Copa Libertadores at the age of 24.
Trefgbx
July 16, 2024 @ 1:11 am
[b]Привет всем[/b])
Студенческая жизнь прекрасна, пока не приходит время писать диплом, как это случилось со мной. Не стоит отчаиваться, ведь существуют компании, которые помогают с написанием и защитой диплома на высокие оценки!
Сначала я искал информацию по теме: купить диплом в зеленодольске, купить диплом в новокузнецке, купить диплом в воронеже, купить диплом в майкопе, купить диплом в мичуринске, а потом наткнулся на http://rtpsouth.com/UserProfile/tabid/69/userId/105773/Default.aspx, где все мои учебные проблемы были решены!
Удачи!
Sazrsrp
July 16, 2024 @ 1:22 am
[u][b] Привет![/b][/u]
[b]Всё, что нужно знать о покупке аттестата о среднем образовании [/b]
[url=http://rst.adk.audio/company/personal/user/1577/forum/message/1953/1985/#message1985/]rst.adk.audio/company/personal/user/1577/forum/message/1953/1985/#message1985[/url]
[u][b] Окажем помощь![u][b].
Williamguern
July 16, 2024 @ 3:27 am
In-depth articles about the most famous football players https://zenit-saint-petersburg.wendel-br.com, clubs and events. Learn everything about tactics, rules of the game and football history.
Robertlix
July 16, 2024 @ 3:27 am
The rise of 20-year-old midfielder Jamal Musiala https://bavaria.jamalmusiala-br.com to the status of a winger in the Bayern Munich team. A story of incredible talent.
BrandonLaf
July 16, 2024 @ 3:34 am
A historia da jornada triunfante de Anitta https://veneno.anitta-br.com de aspirante a cantora a uma das interpretes mais influentes da musica moderna, incluindo sua participacao na serie de TV “Veneno”.
Williamrow
July 16, 2024 @ 3:40 am
Fabrizio Moretti https://the-strokes.fabriziomoretti-br.com the influential drummer of The Strokes, and his unique sound revolutionized the music scene, remaining icons of modern rock.
LarryAdeby
July 16, 2024 @ 3:43 am
Selena Gomez https://calm-down.selenagomez-br.net the story from child star to global musical influence, summarized in hit “Calm Down”, with Rema.
Diplomi_llOa
July 16, 2024 @ 4:05 am
купить диплом об образовании в спб [url=https://diploms-x.com]https://diploms-x.com[/url] .
Sazrdgf
July 16, 2024 @ 8:52 am
[u][b] Привет![/b][/u]
[b]Как приобрести аттестат о среднем образовании в Москве и других городах [/b]
[url=http://kvitka.ukrbb.net/viewtopic.php?f=58&t=23468/]kvitka.ukrbb.net/viewtopic.php?f=58&t=23468[/url]
[u][b] Рады оказать помощь![u][b].
Trefxlw
July 16, 2024 @ 11:15 am
[b]Здравствуйте[/b]!
Задался вопросом: можно ли на самом деле купить диплом государственного образца в Москве? Был приятно удивлен — это реально и легально!
Сначала искал информацию в интернете на тему: купить дипломы о высшем образовании цена, купить диплом в шадринске, куплю диплом, купить диплом в буденновске, купить диплом воспитателя и получил базовые знания. В итоге остановился на материале: https://notebooks.ru/forum/user/57149/
Успешной учебы!
Diplomi_soSi
July 16, 2024 @ 3:40 pm
купить диплом в назрани [url=https://asxdiplomik.com/kupit-diplom-moskva/]asxdiplomik.com/kupit-diplom-moskva[/url] .
Diplomi_tzSi
July 16, 2024 @ 6:03 pm
даркнет купить диплом [url=https://asxdiplomik.com/kupit-diplom-moskva/]asxdiplomik.com/kupit-diplom-moskva[/url] .
ErnestBar
July 16, 2024 @ 6:31 pm
Kobe Bryant https://los-angeles-lakers.kobebryant-br.net one of the greatest basketball players of all the times, left an indelible mark on the history of sport.
Gilbertpoump
July 16, 2024 @ 6:33 pm
Achraf Hakimi https://paris-saint-germain.ashrafhakimi.net is a young Moroccan footballer who quickly reached the football elite European in recent years.
Darrellrop
July 16, 2024 @ 6:41 pm
Bieber’s https://baby.justinbieber-br.com path to global fame began with his breakthrough success Baby, which became his signature song and one of the most popular music videos of all time.
Michaeltenia
July 16, 2024 @ 6:51 pm
Ayrton Senna https://mclaren.ayrtonsenna-br.com is one of the greatest drivers in the history of Formula 1.
EddieDit
July 16, 2024 @ 6:58 pm
In the world of professional tennis, the name of Gustavo Kuerten https://roland-garros.gustavokuerten.com is closely linked to one of the most prestigious Grand Slam tournaments – Roland Garros.
Fazrhlu
July 16, 2024 @ 9:50 pm
[u][b] Здравствуйте![/b][/u]
Мы изготавливаем дипломы психологов, юристов, экономистов и других профессий по разумным тарифам. Цена может зависеть от определенной специальности, года выпуска и университета.
Где приобрести диплом специалиста?
[b]Заказать диплом любого университета[/b]
[url=http://landik-diploms-srednee.ru/kupit-diplom-v-novosibirske ]landik-diploms-srednee.ru/kupit-diplom-v-novosibirske [/url]
[b]Успешной учебы![/b]
Dnrtjew
July 16, 2024 @ 10:49 pm
[u][b] Привет, друзья![/b][/u]
[b]Купить документ[/b] о получении высшего образования можно в нашей компании.
[url=http://ast-diplom24.ru/kupit-diplom-rostov-na-donu/]ast-diplom24.ru/kupit-diplom-rostov-na-donu[/url]
[b]Успешной учебы![/b]
Iariormiy
July 16, 2024 @ 11:40 pm
Полезные советы по безопасной покупке диплома о высшем образовании
[url=http://newsbig.timeforchangecounselling.com/rekomendacii-po-pokupki-dokumentov-v-seti/]newsbig.timeforchangecounselling.com/rekomendacii-po-pokupki-dokumentov-v-seti[/url]
Cazrhqq
July 16, 2024 @ 11:47 pm
[u][b] Здравствуйте![/b][/u]
Мы предлагаем документы техникумов, которые расположены на территории всей РФ. Можно купить качественный диплом от любого заведения, за любой год, указав актуальную специальность и хорошие оценки за все дисциплины. Дипломы и аттестаты выпускаются на бумаге самого высокого качества. Это позволяет делать настоящие дипломы, которые не отличить от оригинала. Документы будут заверены необходимыми печатями и штампами.
[b]Заказать диплом ВУЗа.[/b]
[url=http://antspride.com/read-blog/824/]antspride.com/read-blog/824[/url]
[b]Хорошей учебы![/b]
Fazrvgq
July 16, 2024 @ 11:58 pm
[u][b] Добрый день![/b][/u]
Мы можем предложить дипломы любых профессий по приятным тарифам. Стоимость будет зависеть от конкретной специальности, года получения и образовательного учреждения.
Где заказать диплом по актуальной специальности?
[b]Купить диплом о высшем образовании[/b]
[url=http://arusak-diploms-srednee.ru/diplom-sredne-spetcialnoe-obrazovanie ]arusak-diploms-srednee.ru/diplom-sredne-spetcialnoe-obrazovanie [/url]
[b]Успехов в учебе![/b]
Sazrpbb
July 17, 2024 @ 12:45 am
[u][b] Привет, друзья![/b][/u]
[b]Полезные советы по покупке диплома о высшем образовании без риска [/b]
[url=http://dvasadovoda.ru/forum/user/8843/]dvasadovoda.ru/forum/user/8843[/url]
[u][b] Всегда вам поможем![u][b].
Dnrtpou
July 17, 2024 @ 1:13 am
[u][b] Здравствуйте![/b][/u]
[b]Заказать документ[/b] о получении высшего образования вы можете у нас.
[url=http://diploms-x.com/kupit-diplom-vracha/]diploms-x.com/kupit-diplom-vracha[/url]
[b]Удачи![/b]
Lutherorila
July 17, 2024 @ 1:48 am
Anderson Silva https://killer-bees-muay-thai-college.andersonsilva.net was born in 1975 in Curitiba Brazil. From a young age he showed an interest in martial arts, starting to train in karate at the age of 5.
Davidvat
July 17, 2024 @ 2:06 am
Daniel Alves https://paris-saint-germain.danielalves.net is a name that symbolizes the greatness of the world of football.
DwainSar
July 17, 2024 @ 2:06 am
Rodrygo Silva de Goes https://real-madrid.rodrygo-br.com, known simply as Rodrygo, emerged as one of the the brightest young talents in world football.
Herbertwremy
July 17, 2024 @ 2:07 am
Bruno Miguel Borges Fernandes https://manchester-united.brunofernandes-br.com was born on September 8, 1994 in Maia, Portugal.
Jameszeple
July 17, 2024 @ 2:12 am
Nuno Mendes https://paris-saint-germain.nuno-mendes.com, a talented Portuguese left-back, He quickly became one of the key figures in the Paris Saint-Germain (PSG) team.
binance Препоръчителен код
July 17, 2024 @ 3:04 am
Thanks for sharing. I read many of your blog posts, cool, your blog is very good.
Sazregi
July 17, 2024 @ 3:13 am
[u][b] Добрый день![/b][/u]
[b]Официальная покупка диплома вуза с сокращенной программой обучения в Москве [/b]
[url=http://www.polosedan-club.com/search/search/]www.polosedan-club.com/search/search[/url]
[u][b] Рады оказать помощь![u][b].
Qazrmkw
July 17, 2024 @ 6:20 am
[u][b] Привет![/b][/u]
Где приобрести диплом специалиста?
Наша компания предлагает быстро и выгодно приобрести диплом, который выполняется на оригинальном бланке и заверен мокрыми печатями, штампами, подписями. Данный документ пройдет лубую проверку, даже с использованием специальных приборов. Достигайте свои цели максимально быстро с нашим сервисом.
[b]Купить диплом любого университета.[/b]
[url=http://www.euusedgoodstrading.com/read-blog/2792/]www.euusedgoodstrading.com/read-blog/2792[/url]
Travistem
July 17, 2024 @ 6:56 am
Earvin “Magic” Johnson https://los-angeles-lakers.magicjohnson.biz is one of the most legendary basketball players in history. NBA history.
HowardVit
July 17, 2024 @ 7:13 am
Vinicius Junior https://real-madrid.vinicius-junior-ar.com the Brazilian prodigy whose full name is Vinicius Jose Baixao de Oliveira Junior, has managed to win the hearts of millions of fans around the world in a short period of time.
Stacyfen
July 17, 2024 @ 7:21 am
Victor Osimhen https://napoli.victorosimhen-ar.com born on December 29, 1998 in Lagos, Nigeria, has grown from an initially humble player to one of the brightest strikers in modern football.
Leslieruide
July 17, 2024 @ 7:21 am
Toni Kroos https://real-madrid.tonikroos-ar.com the German midfielder known for his accurate passes and calmness on the field, has achieved remarkable success at one of the most prestigious football clubs in the world.
Jasonloura
July 17, 2024 @ 7:26 am
Robert Lewandowski https://barcelona.robertlewandowski-ar.com is one of the most prominent footballers of our time, and his move to Barcelona has become one of the most talked about topics in world football.
Iariorrvp
July 17, 2024 @ 9:53 am
Как получить диплом о среднем образовании в Москве и других городах
[url=http://ukrevents.ru/ofitsialnyie-diplomyi-garantiya-podlinnosti-i-kachestva//]ukrevents.ru/ofitsialnyie-diplomyi-garantiya-podlinnosti-i-kachestva/[/url]
Lariormbq
July 17, 2024 @ 11:23 am
Купить диплом старого образца, можно ли это сделать по быстрой схеме?
[url=http://ast-diplom.com/kupit-diplom-ekaterinbur/]ast-diplom.com/kupit-diplom-ekaterinbur[/url]
Diplomi_ubmi
July 17, 2024 @ 1:25 pm
купить диплом королев [url=https://ast-diploms.com/]ast-diploms.com[/url] .
Dnrtrsi
July 17, 2024 @ 1:32 pm
[u][b] Добрый день![/b][/u]
[b]Купить документ[/b] о получении высшего образования можно в нашей компании в Москве.
[url=http://ast-diplom.com/kupit-diplom-nizhnij-novgorod/]ast-diplom.com/kupit-diplom-nizhnij-novgorod[/url]
[b]Успехов в учебе![/b]
Qazrjjl
July 17, 2024 @ 2:15 pm
[u][b] Здравствуйте![/b][/u]
Где купить диплом по необходимой специальности?
Мы предлагаем выгодно купить диплом, который выполнен на оригинальной бумаге и заверен мокрыми печатями, водяными знаками, подписями должностных лиц. Данный диплом пройдет любые проверки, даже с применением профессионального оборудования. Решите свои задачи максимально быстро с нашим сервисом.
[b]Приобрести диплом любого ВУЗа.[/b]
[url=http://followgrown.com/read-blog/2700/]followgrown.com/read-blog/2700[/url]
Diplomi_hmSi
July 17, 2024 @ 2:32 pm
[u][b] Добрый день![/b][/u]
[b]Купить документ[/b] ВУЗа можно в нашем сервисе.
[url=http://asxdiplomik.com/kupit-diplom-nizhnij-novgorod/]asxdiplomik.com/kupit-diplom-nizhnij-novgorod[/url]
Dnrtmfb
July 17, 2024 @ 3:59 pm
[u][b] Привет, друзья![/b][/u]
[b]Заказать документ[/b] ВУЗа можно у нас в столице.
[url=http://ast-diplomas.com/kupit-diplom-krasnoyarsk/]ast-diplomas.com/kupit-diplom-krasnoyarsk[/url]
[b]Хорошей учебы![/b]
Diplomi_blSi
July 17, 2024 @ 4:59 pm
[u][b] Привет![/b][/u]
[b]Купить документ[/b] ВУЗа можно у нас.
[url=http://asxdiplomik24.ru/kupit-diplom-rostov-na-donu/]asxdiplomik24.ru/kupit-diplom-rostov-na-donu[/url]
Cazrvuz
July 17, 2024 @ 5:16 pm
[u][b] Привет![/b][/u]
[b]Заказать диплом любого университета.[/b]
Мы предлагаем документы ВУЗов, расположенных в любом регионе РФ. Можно приобрести диплом от любого учебного заведения, за любой год, указав подходящую специальность и хорошие оценки за все дисциплины. Дипломы и аттестаты выпускаются на бумаге высшего качества. Это позволяет делать настоящие дипломы, которые не отличить от оригиналов. Они заверяются необходимыми печатями и подписями.
[url=http://firstplat.com/read-blog/12286/]firstplat.com/read-blog/12286[/url]
Pazrxpf
July 17, 2024 @ 5:36 pm
[u][b] Добрый день![/b][/u]
Мы изготавливаем дипломы любой профессии по приятным ценам. Цена может зависеть от определенной специальности, года получения и образовательного учреждения.
[url=http://arusak-diploms-srednee.ru/kupit-diplom-medicinskogo-uchilishha ]arusak-diploms-srednee.ru/kupit-diplom-medicinskogo-uchilishha [/url]
[b]Успешной учебы![/b]
Sazrutg
July 17, 2024 @ 5:45 pm
[u][b] Привет![/b][/u]
[b]Официальная покупка диплома вуза с сокращенной программой обучения в Москве [/b]
[url=http://pyha.ru/forum/topic/13526.1#msg216473/]pyha.ru/forum/topic/13526.1#msg216473[/url]
[u][b] Поможем вам всегда![u][b].
Manrbxj
July 17, 2024 @ 6:29 pm
[u][b] Привет, друзья![/b][/u]
Где купить диплом по нужной специальности?
Мы предлагаем документы институтов, расположенных в любом регионе Российской Федерации. Можно заказать диплом за любой год, включая сюда документы старого образца. Дипломы делаются на бумаге высшего качества. Это позволяет делать настоящие дипломы, которые не отличить от оригинала. Они будут заверены всеми требуемыми печатями и подписями.
[b]Мы можем предложить дипломы[/b] любой профессии по приятным ценам.
[url=http://diploms-x.com/otzyvy]diploms-x.com/otzyvy[/url]
[u][b] Будем рады вам помочь![u][b]
Wazrxqt
July 17, 2024 @ 6:42 pm
[u][b] Привет![/b][/u]
[b]Мы можем предложить документы техникумов,[/b] расположенных на территории всей России. Вы имеете возможность заказать диплом за любой год, указав необходимую специальность и хорошие оценки за все дисциплины. Документы делаются на “правильной” бумаге высшего качества. Это позволяет делать настоящие дипломы, не отличимые от оригиналов. Документы будут заверены всеми необходимыми печатями и подписями.
[url=http://www.suicide-forum.com/member.php?12290-sonnick84&tab=visitor_messaging&page=1]www.suicide-forum.com/member.php?12290-sonnick84&tab=visitor_messaging&page=1[/url]
Cazrroj
July 17, 2024 @ 7:28 pm
[u][b] Привет, друзья![/b][/u]
[b]Заказать диплом любого университета.[/b]
Мы готовы предложить документы ВУЗов, расположенных на территории всей РФ. Можно заказать качественный диплом за любой год, указав необходимую специальность и хорошие оценки за все дисциплины. Дипломы и аттестаты печатаются на бумаге самого высокого качества. Это дает возможность делать настоящие дипломы, не отличимые от оригинала. Документы будут заверены необходимыми печатями и штампами.
[url=http://chatclub.mn.co/posts/61838312/]chatclub.mn.co/posts/61838312[/url]
Xazrfmo
July 17, 2024 @ 7:45 pm
[u][b] Здравствуйте![/b][/u]
Купить документ ВУЗа вы имеете возможность в нашем сервисе. Мы оказываем услуги по продаже документов об окончании любых ВУЗов РФ. Вы получите диплом по любой специальности, включая документы старого образца.
[url=http://thelspr.listbb.ru/viewtopic.php?f=13&t=361/]thelspr.listbb.ru/viewtopic.php?f=13&t=361[/url]
[url=http://mireait.listbb.ru/viewtopic.php?f=2&t=478/]mireait.listbb.ru/viewtopic.php?f=2&t=478[/url]
[url=http://sciencenewhop.maxbb.ru/viewtopic.php?f=44&t=764/]sciencenewhop.maxbb.ru/viewtopic.php?f=44&t=764[/url]
[url=http://umorforme.ru/nadezhnyie-diplomyi-ot-professionalov/]umorforme.ru/nadezhnyie-diplomyi-ot-professionalov[/url]
[url=http://ukrevents.ru/diplomyi-po-vsem-vostrebovannyim-spetsialnostyam//]ukrevents.ru/diplomyi-po-vsem-vostrebovannyim-spetsialnostyam/[/url]
Pazrbpx
July 17, 2024 @ 7:49 pm
[u][b] Добрый день![/b][/u]
Мы можем предложить дипломы любой профессии по разумным ценам. Цена может зависеть от той или иной специальности, года получения и образовательного учреждения.
[url=http://landik-diploms-srednee.ru/svidetelstvo-o-rozhdenii ]landik-diploms-srednee.ru/svidetelstvo-o-rozhdenii [/url]
[b]Успешной учебы![/b]
Sazrrka
July 17, 2024 @ 8:07 pm
[u][b] Добрый день![/b][/u]
[b]Быстрое обучение и получение диплома магистра – возможно ли это? [/b]
[url=http://g95334gq.beget.tech/2024/07/11/vash-diplom-bez-lishnih-hlopot-i-usiliy.html/]g95334gq.beget.tech/2024/07/11/vash-diplom-bez-lishnih-hlopot-i-usiliy.html[/url]
[u][b] Всегда вам поможем![u][b].
Lazrqvb
July 17, 2024 @ 8:12 pm
[u][b] Добрый день![/b][/u]
Где заказать диплом по актуальной специальности?
[b]Заказать диплом о высшем образовании.[/b]
[url=http://gamesfortop.ru/diplom-lyuboy-professii-kachestvenno-i-nadezhno/]gamesfortop.ru/diplom-lyuboy-professii-kachestvenno-i-nadezhno[/url]
Lazrxum
July 17, 2024 @ 8:17 pm
[u][b] Привет![/b][/u]
[b]Мы можем предложить дипломы[/b] любых профессий по доступным ценам.
[url=http://scientistsufo.ru/vyisshee-obrazovanie-dlya-vas-legalno-i-dostupno/]scientistsufo.ru/vyisshee-obrazovanie-dlya-vas-legalno-i-dostupno[/url]
Diplomi_nzSn
July 17, 2024 @ 8:18 pm
купить диплом тренера [url=https://asxdiplomik24.ru/]asxdiplomik24.ru[/url] .
Williamraics
July 17, 2024 @ 8:20 pm
[u][b] Привет![/b][/u]
[b]Наша компания предлагает[/b] быстро и выгодно купить диплом, который выполняется на оригинальной бумаге и заверен мокрыми печатями, штампами, подписями должностных лиц. Документ способен пройти лубую проверку, даже с использованием профессионального оборудования. Решите свои задачи быстро и просто с нашим сервисом.
[url=http://ekspoteh.ru/index.php?name=account&op=info&uname=yjocowif/]ekspoteh.ru/index.php?name=account&op=info&uname=yjocowif[/url]
[b]Удачи![/b]
Uazrfqb
July 17, 2024 @ 8:23 pm
[u][b] Здравствуйте![/b][/u]
[b]Купить диплом о высшем образовании.[/b]
[url=http://network-1062788.mn.co/posts/61748215/]network-1062788.mn.co/posts/61748215[/url]
Diplomi_wcOa
July 17, 2024 @ 8:41 pm
купить аттестат 9 классов [url=http://diploms-x.com]http://diploms-x.com[/url] .
Uazrhfr
July 17, 2024 @ 8:54 pm
[u][b] Здравствуйте![/b][/u]
Где заказать диплом по необходимой специальности?
[url=http://sklad-slabov.ru/forum/user/8263//]sklad-slabov.ru/forum/user/8263/[/url]
[b]Хорошей учебы![/b]
Lariorbmc
July 17, 2024 @ 9:35 pm
Как приобрести аттестат о среднем образовании в Москве и других городах
[url=http://ast-diploms24.ru/kupit-diplom-magistra/]ast-diploms24.ru/kupit-diplom-magistra[/url]
Danrstw
July 17, 2024 @ 9:40 pm
[u][b] Привет![/b][/u]
[b]Где заказать диплом по необходимой специальности?[/b]
[b]Мы готовы предложить дипломы[/b] любой профессии по доступным тарифам.
[url=http://www.pentictonsoccerclub.com/may-classic]www.pentictonsoccerclub.com/may-classic[/url]
Xazrqii
July 17, 2024 @ 10:18 pm
[u][b] Привет![/b][/u]
Приобрести документ института вы можете у нас в Москве. Мы оказываем услуги по изготовлению и продаже документов об окончании любых университетов России. Вы сможете получить диплом по любым специальностям, включая документы СССР.
[url=http://iqtorg.ru/forum/messages/forum2/topic632/message706/?result=new#message706/]iqtorg.ru/forum/messages/forum2/topic632/message706/?result=new#message706[/url]
[url=http://m9-com.ru/forums/viewtopic.php?f=3&t=23895/]m9-com.ru/forums/viewtopic.php?f=3&t=23895[/url]
[url=http://sensemi.getbb.ru/posting.php?mode=post&f=6&sid=14af900ac8145d2b22ec146f02c6270e/]sensemi.getbb.ru/posting.php?mode=post&f=6&sid=14af900ac8145d2b22ec146f02c6270e[/url]
[url=http://liquidationrama.com/read-blog/1030/]liquidationrama.com/read-blog/1030[/url]
[url=http://tripsitter.ixbb.ru/viewtopic.php?id=111#p111/]tripsitter.ixbb.ru/viewtopic.php?id=111#p111[/url]
Trefeig
July 17, 2024 @ 10:32 pm
[u][b] Привет, друзья![/b][/u]
[b]Вопросы и ответы: можно ли быстро купить диплом старого образца? [/b]
[url=http://webmoney.forumkz.ru/viewtopic.php?id=2764#p7865/]webmoney.forumkz.ru/viewtopic.php?id=2764#p7865[/url]
[u][b] Окажем помощь![u][b].
Yrefkni
July 17, 2024 @ 10:48 pm
[u][b] Добрый день![/b][/u]
[b]Купить диплом ВУЗа.[/b]
[url=http://allukrnews.ru/oformite-diplom-vsego-za-neskolko-shagov//]allukrnews.ru/oformite-diplom-vsego-za-neskolko-shagov/[/url]
[b]Успешной учебы![/b]
Diplomi_pwmi
July 17, 2024 @ 11:54 pm
купить диплом работу [url=https://ast-diploms.com/]ast-diploms.com[/url] .
Diplomi_dtKt
July 17, 2024 @ 11:57 pm
купить диплом в комсомольске-на-амуре [url=https://diplomasx.com/]diplomasx.com[/url] .
Dnrtwif
July 18, 2024 @ 3:19 am
[u][b] Добрый день![/b][/u]
[b]Купить документ[/b] о получении высшего образования можно у нас в Москве.
[url=http://ast-diplomy24.ru/kupit-diplom-sankt-peterburg/]ast-diplomy24.ru/kupit-diplom-sankt-peterburg[/url]
[b]Успешной учебы![/b]
PablovatTe
July 18, 2024 @ 3:27 am
Pedro Gonzalez Lopez https://barcelona.pedri-ar.com known as Pedri, was born on November 25, 2002 in the small town of Tegeste, located on Tenerife, one of the Canary Islands.
Razrelr
July 18, 2024 @ 3:41 am
[u][b] Здравствуйте![/b][/u]
[b]Мы предлагаем купить диплом[/b] в высоком качестве, который невозможно отличить от оригинального документа без участия специалистов высокой квалификации со специальным оборудованием.
[url=http://rytmika-gdansk.pl/profile.php?lookup=30035/]rytmika-gdansk.pl/profile.php?lookup=30035[/url]
[b]Успешной учебы![/b]
Vazrkdp
July 18, 2024 @ 3:44 am
[u][b] Добрый день![/b][/u]
Приобрести документ о получении высшего образования можно в нашем сервисе. Мы оказываем услуги по продаже документов об окончании любых ВУЗов РФ.
[url=http://forum.drustvogil-galad.si/index.php/topic,12726.new.html#new/]forum.drustvogil-galad.si/index.php/topic,12726.new.html#new[/url]
[url=http://mouur6.ru/forum/profile.php?action=show&member=3292/]mouur6.ru/forum/profile.php?action=show&member=3292[/url]
[url=http://biker-customshop.ru/review.php/]biker-customshop.ru/review.php[/url]
[url=http://kioskindustry.ru/index.php?subaction=userinfo&user=uhomyryj/]kioskindustry.ru/index.php?subaction=userinfo&user=uhomyryj[/url]
[url=http://berzarina28a.ru/index.php?/topic/25770-%D0%BA%D0%B0%D0%BA-%D1%81%D0%BE%D0%B7%D0%B4%D0%B0%D1%82%D1%8C-%D0%BD%D0%BE%D0%B2%D1%83%D1%8E-%D1%82%D0%B5%D0%BC%D1%83-berzarina28aru//]berzarina28a.ru/index.php?/topic/25770-%D0%BA%D0%B0%D0%BA-%D1%81%D0%BE%D0%B7%D0%B4%D0%B0%D1%82%D1%8C-%D0%BD%D0%BE%D0%B2%D1%83%D1%8E-%D1%82%D0%B5%D0%BC%D1%83-berzarina28aru/[/url]
MarceloZes
July 18, 2024 @ 3:44 am
The story of Leo Messi https://inter-miami.lionelmessi.ae‘s transfer to Inter Miami began long before the official announcement. Rumors about Messi’s possible departure from Barcelona appeared in 2020
Robertneeli
July 18, 2024 @ 3:49 am
Andreson Souza Conceicao https://al-nassr.talisca-ar.com known as Talisca, is one of the brightest stars of modern football.
GeorgeWaw
July 18, 2024 @ 3:59 am
Yacine Bounou https://al-hilal.yassine-bounou-ar.com known simply as Bono, is one of the most prominent Moroccan footballers of our time.
Manrtiz
July 18, 2024 @ 4:09 am
[u][b] Добрый день![/b][/u]
Где купить диплом по актуальной специальности?
Мы готовы предложить документы институтов, расположенных в любом регионе Российской Федерации. Вы можете приобрести качественно напечатанный диплом от любого высшего учебного заведения, за любой год, указав актуальную специальность и оценки за все дисциплины. Дипломы печатаются на бумаге самого высокого качества. Это дает возможность делать настоящие дипломы, которые не отличить от оригиналов. Они будут заверены всеми требуемыми печатями и подписями.
[b]Мы изготавливаем дипломы[/b] любой профессии по невысоким ценам.
[url=http://diplomasx24.ru/kupit-diplom-nizhnij-novgorod]diplomasx24.ru/kupit-diplom-nizhnij-novgorod[/url]
[u][b] Поможем вам всегда![u][b]
Wazrufo
July 18, 2024 @ 4:33 am
[u][b] Привет, друзья![/b][/u]
[b]Мы готовы предложить документы техникумов,[/b] расположенных на территории всей Российской Федерации. Вы сможете приобрести качественно сделанный диплом от любого заведения, за любой год, в том числе документы старого образца. Дипломы и аттестаты выпускаются на бумаге высшего качества. Это дает возможности делать настоящие дипломы, которые не отличить от оригинала. Они будут заверены необходимыми печатями и подписями.
[url=http://scientistsufo.ru/page/14]scientistsufo.ru/page/14[/url]
Dnrtvkx
July 18, 2024 @ 5:55 am
[u][b] Привет![/b][/u]
[b]Приобрести документ[/b] о получении высшего образования вы можете у нас.
[url=http://ast-diploms.com/kupit-diplom-sankt-peterburg/]ast-diploms.com/kupit-diplom-sankt-peterburg[/url]
[b]Удачи![/b]
Diplomi_vnSn
July 18, 2024 @ 6:43 am
дистанционное образование купить диплом [url=https://asxdiplomik24.ru/]asxdiplomik24.ru[/url] .
Williamraics
July 18, 2024 @ 6:43 am
[u][b] Привет![/b][/u]
[b]Наша компания предлагает[/b] быстро купить диплом, который выполнен на оригинальной бумаге и заверен печатями, штампами, подписями должностных лиц. Данный диплом способен пройти лубую проверку, даже при использовании специального оборудования. Достигайте свои цели быстро с нашими дипломами.
[url=http://club.7ya.ru/dilopluyy//]club.7ya.ru/dilopluyy/[/url]
[b]Успешной учебы![/b]
Diplomi_mgOa
July 18, 2024 @ 6:52 am
диплом какого вуза купить [url=http://www.diploms-x.com]диплом какого вуза купить[/url] .
Uazrozm
July 18, 2024 @ 7:00 am
[u][b] Здравствуйте![/b][/u]
[b]Приобрести диплом о высшем образовании.[/b]
[url=http://kinogonews.ru/nastoyashhie-diplomyi-bez-lishnih-voprosov/]kinogonews.ru/nastoyashhie-diplomyi-bez-lishnih-voprosov[/url]
https://gamebanana.com/members/3441923
July 18, 2024 @ 7:35 am
If some one wants expert view concerning blogging afterward i suggest
him/her to pay a visit this website, Keep up
the fastidious job. https://gamebanana.com/members/3441923
Horseracingnation.Com
July 18, 2024 @ 7:35 am
Interesting blog! Is your theme custom made or did you download
iit from somewhere? A theme like yours with a feww simple tweeks would really make my blog stand
out. Please let me know where you got your design. Cheers https://www.horseracingnation.com/user/aviator-game
https://www.universe.com/users/conner-hill-3KDP02
July 18, 2024 @ 7:38 am
I was wondering iif you ever thought of changing the page layout of your website?
Its very well written; I love what youve got to say.
But mayybe you coiuld a littyle mmore in the way of content so people could
connect witfh it better. Youve got an awful lot of text
for only having onee or two images. Maytbe you could space it out
better? https://www.universe.com/users/conner-hill-3KDP02
https://Www.bestjobs.Eu/
July 18, 2024 @ 7:39 am
Wonderful, what a blog it is! This webpage providees useful facts to us, keep
it up. https://www.bestjobs.eu/en/companies/aviator-2
https://www.tumblr.com/abbyholloway/746744730450362368/top-10-mistakes-beginners-make-in-casinos-how-to
July 18, 2024 @ 7:41 am
WOW just what I was searching for. Caame here bby searching
for casino https://www.tumblr.com/abbyholloway/746744730450362368/top-10-mistakes-beginners-make-in-casinos-how-to
https://linktr.ee/aviatorsgames
July 18, 2024 @ 7:44 am
Keep thijs going please, great job! https://linktr.ee/aviatorsgames
https://robertsspaceindustries.com/citizens/aviatorgameapp
July 18, 2024 @ 7:46 am
When I initially commented I clicked the “Notify me when new comments are added” checkbox and now each time a comment is
added I get several emails with thee same comment. Is
there any way you can remove people frm that service?
Cheers! https://robertsspaceindustries.com/citizens/aviatorgameapp
https://www.producthunt.com/@aviator_game
July 18, 2024 @ 7:46 am
Hey I know this iis off topic but I wwas wondering if you knew of any widgets I could aadd to my blog that automatically tweet my newest twitter updates.
I’ve been looking for a plug-in like this for quite some time
and was hoping maybe you would have some experience with something like
this. Please let me know if you ruun into anything. I truly
njoy reading your blog and I look forward to your new updates. https://www.producthunt.com/@aviator_game
https://blog.aajjo.com/post/sky-high-excitement-player-reviews-of-the-aviator-game-experience
July 18, 2024 @ 7:47 am
I wanted to thank you foor this wonderful read!! I absolutely enjoyed every
bitt of it. I have you saved as a favorite to look at neew stuff you post… https://blog.aajjo.com/post/sky-high-excitement-player-reviews-of-the-aviator-game-experience
https://linktr.ee/aviatorgame12
July 18, 2024 @ 7:51 am
Its not my first ttime to pay a quick visit this web
page, i am visitng thyis web page dailly aand get good
information from here alll the time. https://linktr.ee/aviatorgame12
Danrkhr
July 18, 2024 @ 7:54 am
[u][b] Привет, друзья![/b][/u]
[b]Где купить диплом по актуальной специальности?[/b]
[b]Мы изготавливаем дипломы[/b] любой профессии по приятным ценам.
[url=http://ediblehomegardensresort.com/index.php?do=/public/blog/view/id_119635/title_-2024/]ediblehomegardensresort.com/index.php?do=/public/blog/view/id_119635/title_-2024/[/url]
Trefkqy
July 18, 2024 @ 9:06 am
[u][b] Привет![/b][/u]
[b]Легальные способы покупки диплома о среднем полном образовании [/b]
[url=http://autoschool-progress.kz/index.php?subaction=userinfo&user=usefizaq/]autoschool-progress.kz/index.php?subaction=userinfo&user=usefizaq[/url]
[u][b] Поможем вам всегда![u][b].
Yrefoah
July 18, 2024 @ 9:32 am
[u][b] Привет, друзья![/b][/u]
[b]Приобрести диплом о высшем образовании.[/b]
[url=http://korrespondentweek.ru/kupite-diplom-dlya-uspeshnoy-kareryi//]korrespondentweek.ru/kupite-diplom-dlya-uspeshnoy-kareryi/[/url]
[b]Успешной учебы![/b]
Jamesbisse
July 18, 2024 @ 11:15 am
Harry Kane https://bayern.harry-kane-ar.com one of the most prominent English footballers of his generation, completed his move to German football club Bayern Munich in 2023.
Ronaldevene
July 18, 2024 @ 11:21 am
Brazilian footballer Neymar https://al-hilal.neymar-ar.com known for his unique playing style and outstanding achievements in world football, has made a surprise move to Al Hilal Football Club.
Sazrcxe
July 18, 2024 @ 11:22 am
[u][b] Привет![/b][/u]
[b]Как оказалось, купить диплом кандидата наук не так уж и сложно [/b]
[url=http://www.soad.msk.ru/forum/viewforum.php?f=16/]www.soad.msk.ru/forum/viewforum.php?f=16[/url]
[u][b] Окажем помощь![u][b].
Francisspesy
July 18, 2024 @ 11:29 am
Erling Haaland https://manchester-city.erling-haaland-ar.com born on July 21, 2000 in Leeds, England, began his football journey at an early age.
StephennaL
July 18, 2024 @ 11:39 am
Luka Modric https://real-madrid.lukamodric-ar.com can certainly be called one of the outstanding midfielders in modern football.
Richardfully
July 18, 2024 @ 12:08 pm
Football in Saudi Arabia https://al-hilal.ali-al-bulaihi-ar.com has long been one of the main sports, attracting millions of fans. In recent years, one of the brightest stars in Saudi football has been Ali Al-Bulaihi, defender of Al-Hilal Football Club.
Xazreiu
July 18, 2024 @ 12:42 pm
[u][b] Привет, друзья![/b][/u]
Приобрести документ ВУЗа вы имеете возможность в нашей компании. Мы оказываем услуги по продаже документов об окончании любых университетов РФ. Вы сможете получить диплом по любым специальностям, любого года выпуска, включая документы старого образца.
[url=http://forum.supermunchkin.org/viewtopic.php?f=1&t=4182/]forum.supermunchkin.org/viewtopic.php?f=1&t=4182[/url]
[url=http://newru.getbb.ru/viewtopic.php?f=29&t=1356/]newru.getbb.ru/viewtopic.php?f=29&t=1356[/url]
[url=http://mybaltika.info/ru/blogs/587/10399//]mybaltika.info/ru/blogs/587/10399/[/url]
[url=http://www.cyberpinoy.net/read-blog/35444/]www.cyberpinoy.net/read-blog/35444[/url]
[url=http://pytalovo.4admins.ru/posting.php?mode=post&f=15&sid=51d90f1a6f082c1795c9e0874429815f/]pytalovo.4admins.ru/posting.php?mode=post&f=15&sid=51d90f1a6f082c1795c9e0874429815f[/url]
Cazrxbp
July 18, 2024 @ 1:37 pm
[u][b] Здравствуйте![/b][/u]
[b]Мы предлагаем документы техникумов[/b], расположенных на территории всей Российской Федерации. Вы имеете возможность купить качественно напечатанный диплом от любого заведения, за любой год, указав подходящую специальность и хорошие оценки за все дисциплины. Документы выпускаются на “правильной” бумаге самого высшего качества. Это позволяет делать настоящие дипломы, которые не отличить от оригиналов. Они заверяются всеми требуемыми печатями и подписями.
[url=http://socialnetwork.cloudyzx.com/read-blog/12109/]socialnetwork.cloudyzx.com/read-blog/12109[/url]
Sazrgxf
July 18, 2024 @ 1:54 pm
[u][b] Привет, друзья![/b][/u]
[b]Аттестат 11 класса купить официально с упрощенным обучением в Москве [/b]
[url=http://school97.ru/vesti20/index.php?PAGE_NAME=profile_view&UID=210365/]school97.ru/vesti20/index.php?PAGE_NAME=profile_view&UID=210365[/url]
[u][b] Будем рады вам помочь![u][b].
Vazrkdx
July 18, 2024 @ 1:57 pm
[u][b] Привет![/b][/u]
Купить документ о получении высшего образования вы имеете возможность у нас. Мы предлагаем документы об окончании любых университетов Российской Федерации.
[url=http://www.diywiki.org/index.php?title=premiummdiplomms/]www.diywiki.org/index.php?title=premiummdiplomms[/url]
[url=http://arbitrajniki.ru/forums/topic/kupit-diplom-cena-w27f//]arbitrajniki.ru/forums/topic/kupit-diplom-cena-w27f/[/url]
[url=http://xn—-7sbabja7ekbaahnetbi5o6b.xn--p1ai/profile.php?lookup=33184/]xn—-7sbabja7ekbaahnetbi5o6b.xn--p1ai/profile.php?lookup=33184[/url]
[url=http://stavklad.ru/viewtopic.php?f=20&t=5126/]stavklad.ru/viewtopic.php?f=20&t=5126[/url]
[url=http://dianov.bget.ru/forum/thread57506.html#1281078/]dianov.bget.ru/forum/thread57506.html#1281078[/url]
Razrodh
July 18, 2024 @ 1:58 pm
[u][b] Здравствуйте![/b][/u]
[b]Мы предлагаем заказать диплом[/b] высочайшего качества, неотличимый от оригинала без использования специального оборудования и квалифицированного специалиста.
[url=http://amvnews.ru/forum/posting.php?mode=newtopic&f=44/]amvnews.ru/forum/posting.php?mode=newtopic&f=44[/url]
[b]Хорошей учебы![/b]
Jariornnn
July 18, 2024 @ 2:02 pm
[u][b] Привет, друзья![/b][/u]
Купить диплом ВУЗа
[b]Наша компания предлагает[/b] выгодно заказать диплом, который выполняется на оригинальном бланке и заверен печатями, водяными знаками, подписями. Диплом пройдет лубую проверку, даже при помощи профессиональных приборов. Достигайте своих целей быстро и просто с нашей компанией.
[b]Где заказать диплом специалиста?[/b]
[url=http://odessaforum.getbb.ru/viewtopic.php?f=5&t=20402/]odessaforum.getbb.ru/viewtopic.php?f=5&t=20402[/url]
[url=http://mosday.getbb.ru/viewtopic.php?f=12&t=1059/]mosday.getbb.ru/viewtopic.php?f=12&t=1059[/url]
[url=http://vaeaem.profiforum.ru/t3822-topic#7107/]vaeaem.profiforum.ru/t3822-topic#7107[/url]
[url=http://meldog.3nx.ru/viewtopic.php?p=23387#23387/]meldog.3nx.ru/viewtopic.php?p=23387#23387[/url]
[url=http://promintern.listbb.ru/viewtopic.php?f=16&t=474/]promintern.listbb.ru/viewtopic.php?f=16&t=474[/url]
Scottpoili
July 18, 2024 @ 3:05 pm
visit my website https://currencyconvert.net
Samuelanido
July 18, 2024 @ 3:14 pm
Сайт https://ps-likers.ru предлагает уроки по фотошоп для начинающих. На страницах сайта можно найти пошаговые руководства по анимации, созданию графики для сайтов, дизайну, работе с текстом и фотографиями, а также различные эффекты.
Xazrpsv
July 18, 2024 @ 3:20 pm
[u][b] Привет, друзья![/b][/u]
Приобрести документ о получении высшего образования вы сможете в нашей компании в Москве. Мы предлагаем документы об окончании любых университетов РФ. Вы сможете получить диплом по любой специальности, любого года выпуска, в том числе документы Советского Союза.
[url=http://peaceofficial.5nx.ru/viewtopic.php?f=111&t=812/]peaceofficial.5nx.ru/viewtopic.php?f=111&t=812[/url]
[url=http://dog-ola.ru/viewtopic.php?f=28&t=7451/]dog-ola.ru/viewtopic.php?f=28&t=7451[/url]
[url=http://cardinalparkmld.listbb.ru/viewtopic.php?f=3&t=472/]cardinalparkmld.listbb.ru/viewtopic.php?f=3&t=472[/url]
[url=http://chromerp.listbb.ru/ucp.php?mode=login/]chromerp.listbb.ru/ucp.php?mode=login[/url]
[url=http://mangorpp.getbb.ru/posting.php?mode=post&f=3&sid=db4846fe30f9834c2eea7ce682dd9949/]mangorpp.getbb.ru/posting.php?mode=post&f=3&sid=db4846fe30f9834c2eea7ce682dd9949[/url]
Williamguevy
July 18, 2024 @ 3:41 pm
N’Golo Kante https://al-ittihad.ngolokante-ar.com the French midfielder whose career has embodied perseverance, hard work and skill, has continued his path to success at Al-Ittihad Football Club, based in Saudi Arabia.
Diplomi_kzSi
July 18, 2024 @ 5:15 pm
[u][b] Добрый день![/b][/u]
[b]Купить документ[/b] о получении высшего образования вы сможете в нашем сервисе.
[url=http://diplomyx.com/kupit-diplom-krasnoyarsk/]diplomyx.com/kupit-diplom-krasnoyarsk[/url]
Charlesgrofs
July 18, 2024 @ 5:21 pm
Kobe Bryant https://los-angeles-lakers.kobebryant-ar.com also known as the “Black Mamba”, is one of the most iconic and iconic figures in NBA history.
Xazrjwv
July 18, 2024 @ 7:21 pm
[u][b] Здравствуйте![/b][/u]
Предлагаем документы техникумов, расположенных на территории всей Российской Федерации. Можно заказать диплом от любого заведения, за любой год, указав подходящую специальность и хорошие оценки за все дисциплины. Дипломы и аттестаты печатаются на “правильной” бумаге высшего качества. Это дает возможность делать государственные дипломы, не отличимые от оригиналов. Документы будут заверены необходимыми печатями и штампами.
[b]Мы изготавливаем дипломы[/b] любой профессии по доступным тарифам. Стоимость зависит от выбранной специальности, года получения и образовательного учреждения. Всегда стараемся поддерживать для клиентов адекватную ценовую политику. [b]Важно[/b], чтобы дипломы были доступны для подавляющей массы граждан.
[url=http://arusak-diploms-srednee.ru/kupit-diplom-kandidata-nauk ]arusak-diploms-srednee.ru/kupit-diplom-kandidata-nauk [/url]
[b]Успехов в учебе![/b]
Diplomi_egSi
July 18, 2024 @ 7:40 pm
[u][b] Добрый день![/b][/u]
[b]Купить документ[/b] университета вы имеете возможность в нашем сервисе.
[url=http://asxdiplomik.com/kupit-diplom-magistra/]asxdiplomik.com/kupit-diplom-magistra[/url]
Dnrtvpy
July 18, 2024 @ 7:58 pm
[u][b] Добрый день![/b][/u]
[b]Купить документ[/b] ВУЗа вы можете в нашей компании в Москве.
[url=http://ast-diplomas.com/kupit-diplom-omsk/]ast-diplomas.com/kupit-diplom-omsk[/url]
[b]Хорошей учебы![/b]
Xazrwlp
July 18, 2024 @ 9:28 pm
[u][b] Здравствуйте![/b][/u]
Предлагаем документы ВУЗов, расположенных в любом регионе России. Можно заказать качественно напечатанный диплом за любой год, включая сюда документы старого образца. Документы делаются на “правильной” бумаге самого высшего качества. Это дает возможность делать государственные дипломы, которые невозможно отличить от оригинала. Документы заверяются всеми обязательными печатями и штампами.
[b]Мы предлагаем дипломы[/b] любой профессии по приятным тарифам. Стоимость зависит от определенной специальности, года выпуска и образовательного учреждения. Всегда стараемся поддерживать для заказчиков адекватную ценовую политику. [b]Важно[/b], чтобы документы были доступными для большинства граждан.
[url=http://landik-diploms-srednee.ru/kupit-diplom-v-voronezhe ]landik-diploms-srednee.ru/kupit-diplom-v-voronezhe [/url]
[b]Удачи![/b]
Dnrtjyv
July 18, 2024 @ 10:15 pm
[u][b] Добрый день![/b][/u]
[b]Приобрести документ[/b] о получении высшего образования можно в нашей компании в Москве.
[url=http://diploms-x24.ru/kupit-diplom-vracha/]diploms-x24.ru/kupit-diplom-vracha[/url]
[b]Успешной учебы![/b]
CharlesOxync
July 19, 2024 @ 1:27 am
Karim Benzema https://al-ittihad.karimbenzema.ae is a name worthy of admiration and respect in the world of football.
RogererOwl
July 19, 2024 @ 1:32 am
Cristiano Ronaldo https://al-nassr.cristiano-ronaldo.ae is one of the greatest names in football history, with his achievements inspiring millions of fans around the world.
WalterGew
July 19, 2024 @ 1:39 am
In 2018, the basketball world witnessed one of the most remarkable transformations in NBA history. LeBron James https://los-angeles-lakers.lebronjames-ar.com one of the greatest players of our time, decided to leave his hometown Cleveland Cavaliers and join the Los Angeles Lakers.
Sazrqpl
July 19, 2024 @ 1:42 am
[u][b] Добрый день![/b][/u]
[b]Вопросы и ответы: можно ли быстро купить диплом старого образца? [/b]
[url=http://mf.getbb.ru/viewtopic.php?f=28&t=642/]mf.getbb.ru/viewtopic.php?f=28&t=642[/url]
[u][b] Окажем помощь![u][b].
Qazrtld
July 19, 2024 @ 3:15 am
[u][b] Привет, друзья![/b][/u]
[b]Как избежать рисков при покупке диплома колледжа или ВУЗа в России.[/b]
[b]Купить диплом университета.[/b]
[url=http://bayplore.com/read-blog/156/]bayplore.com/read-blog/156[/url]
https://dev2.emathisi.gr/blog/index.php?entryid=376
July 19, 2024 @ 3:24 am
Hello just wanted to give you a quick hads up. The
words in your article seem to be running off the screen in Safari.
I’mnot sure if this is a formatting issue or something to do with internet browser compatibility but I thought I’d post
to let you know. The design and style look great though!
Hope you get the problem solved soon. Cheers https://dev2.emathisi.gr/blog/index.php?entryid=376
https://enfogentraining.com/blog/index.php?entryid=67300
July 19, 2024 @ 3:26 am
Hey just wnted to give you a uick heads up. The words in your content
seem to be running off the screen in Ie. I’m not sure if
this is a format issue or something to do with web
browsesr compatibility but I figured I’d post
to let you know. The design look great though! Hope you get thhe problem fixed soon. Thanks https://enfogentraining.com/blog/index.php?entryid=67300
https://www.backloggd.com/u/brianwalsh/
July 19, 2024 @ 3:48 am
It’s going to be finish of mine day, except before end I am reading
this enormous paragraph to improve my know-how. https://www.backloggd.com/u/brianwalsh/
https://forum.pgbu.ir/profile.php?id=149
July 19, 2024 @ 3:54 am
Hello! Do you know iif they make anyy plugins to assist ith Search Engine Optimization? I’m
tryiing to get my bloog to rank for some targrted keywords
but I’m not seeing very good results. If you know of aany please
share. Kudos! https://forum.pgbu.ir/profile.php?id=149
https://eythar.org/blog/index.php?entryid=484905
July 19, 2024 @ 4:01 am
I love looking through a post that can make people think.
Also, thanks ffor allowing for me to comment! https://eythar.org/blog/index.php?entryid=484905
Sazrxuj
July 19, 2024 @ 4:09 am
[u][b] Привет![/b][/u]
[b]Можно ли купить аттестат о среднем образовании, основные моменты и вопросы [/b]
[url=http://ractis.ru/forums/index.php?autocom=gallery&req=si&img=2536/]ractis.ru/forums/index.php?autocom=gallery&req=si&img=2536[/url]
[u][b] Рады оказать помощь![u][b].
https://e-academy.navttc.gov.pk/blog/index.php?entryid=94448
July 19, 2024 @ 4:11 am
Thanks , I have recently been looking for info about thks subject for
a while and yours is the best I’ve came upon so far. However, what about the conclusion? Are you sure about the
source? https://e-academy.navttc.gov.pk/blog/index.php?entryid=94448
http://another-ro.com/forum/viewtopic.php?id=407672
July 19, 2024 @ 4:22 am
I’m really impressed with your writing skills and also with the
layoit on your weblog. Is thiis a paid theme oor did you
modify it yourself? Anyway keep up the excellent
quality writing, it is rare to see a great blog like this one today. http://another-ro.com/forum/viewtopic.php?id=407672
Terri
July 19, 2024 @ 4:33 am
Thankk you a lot for sharing this with all of us yyou actually recognize
what youu aree speaking approximately! Bookmarked.
Please additionally seek advice from my web site =).
We may have a hyperlink chage arrangement among us https://stemacumen.net/blog/index.php?entryid=2777
ChrisReifs
July 19, 2024 @ 4:38 am
Luis Diaz https://liverpool.luis-diaz-ar.com is a young Colombian striker who has enjoyed rapid growth since joining the ” Liverpool” in January 2022.
https://ashwinihydropneumatics.com/how-to-write-a-good-introduction-about-yourself-2/
July 19, 2024 @ 4:44 am
It’s remarkabl foor me to have a website, which is helpful
for my knowledge. thanks admin https://ashwinihydropneumatics.com/how-to-write-a-good-introduction-about-yourself-2/
https://promed-sd.com/blog/index.php?entryid=168343
July 19, 2024 @ 4:55 am
I just couldn’t go away your site prior to suggesting
tbat I actually loved the standard info a person profide on your guests?
Is goling to be again continuously inn order to check up on nnew posts https://promed-sd.com/blog/index.php?entryid=168343
Wesleyskync
July 19, 2024 @ 5:20 am
Kevin De Bruyne https://manchester-city.kevin-de-bruyne-ar.com is a name every football fan knows today.
Georgecaw
July 19, 2024 @ 5:48 am
Muhammad Al Owais https://al-hilal.mohammed-alowais-ar.com is one of the most prominent names in modern Saudi football. His path to success in Al Hilal team became an example for many young athletes.
RobertPus
July 19, 2024 @ 9:17 am
Maria Sharapova https://tennis.maria-sharapova-ar.com was born on April 19, 1987 in Nyagan, Russia. When Masha was 7 years old, her family moved to Florida, where she started playing tennis.
JohnnieGlall
July 19, 2024 @ 9:22 am
Roberto Firmino https://al-ahli.roberto-firmino-ar.com one of the most talented and famous Brazilian footballers of our time, has paved his way to success in different leagues and teams.
PhillipBed
July 19, 2024 @ 9:45 am
Angel Di Maria https://benfica.angel-di-maria-ar.com is a name that will forever remain in the memories of Benfica fans.
WestonVurce
July 19, 2024 @ 9:46 am
Khvicha Kvaratskhelia https://napoli.khvicha-kvaratskhelia-ar.com is a name that in recent years has become a symbol of Georgian football talent and ambition.
Andremem
July 19, 2024 @ 10:19 am
Football in Saudi Arabia https://al-hilal.saud-abdulhamid-ar.com is gaining more and more popularity and recognition on the international stage, and Saud Abdul Hamid, the young and talented defender of Al Hilal, is a shining example of this success.
Lariorjlx
July 19, 2024 @ 1:07 pm
Диплом пту купить официально с упрощенным обучением в Москве
[url=http://ast-diplomas24.ru/kupit-diplom-moskva/]ast-diplomas24.ru/kupit-diplom-moskva[/url]
Petericest
July 19, 2024 @ 3:14 pm
Казахский национальный технический университет https://satbayev.university им. К.Сатпаева
ShaunMof
July 19, 2024 @ 3:33 pm
Upcoming fantasy MOBA https://bladesofthevoid.com evolved by Web3. Gacha perks, AI and crafting in one swirling solution!
Lazrkdz
July 19, 2024 @ 3:52 pm
[u][b] Здравствуйте![/b][/u]
[b]Мы изготавливаем дипломы[/b] любой профессии по выгодным ценам.
[url=http://shockmusik.ru/legkiy-i-byistryiy-sposob-poluchit-diplom/]shockmusik.ru/legkiy-i-byistryiy-sposob-poluchit-diplom[/url]
KevinDrimi
July 19, 2024 @ 3:57 pm
Продажа новых автомобилей Hongqi
https://hongqi-krasnoyarsk.ru/models в Красноярске у официального дилера Хончи. Весь модельный ряд, все комплектации, выгодные цены, кредит, лизинг, трейд-ин
Daviduriva
July 19, 2024 @ 4:15 pm
Kylie Jenner https://kylie-cosmetics.kylie-jenner-ar.com is an American model, media personality, and businesswoman, born on August 10, 1997 in Los Angeles, California.
Lazrclm
July 19, 2024 @ 4:35 pm
[u][b] Привет, друзья![/b][/u]
Где приобрести диплом специалиста?
[b]Приобрести диплом университета.[/b]
[url=http://dev.spooo.ru/post/article/217862/]dev.spooo.ru/post/article/217862[/url]
Xazrcnl
July 19, 2024 @ 5:19 pm
[u][b] Здравствуйте![/b][/u]
Заказать документ ВУЗа можно у нас в Москве. Мы оказываем услуги по продаже документов об окончании любых ВУЗов Российской Федерации. Вы получите диплом по любой специальности, любого года выпуска, в том числе документы старого образца.
[url=http://dinsta-gram.com/read-blog/4913/]dinsta-gram.com/read-blog/4913[/url]
[url=http://ya.2bb.ru/viewtopic.php?id=19613#p241140/]ya.2bb.ru/viewtopic.php?id=19613#p241140[/url]
[url=http://vintfint.com/blogs/22839/Диплом-в-кратчайшие-сроки-качественно-и-безопасно/]vintfint.com/blogs/22839/Диплом-в-кратчайшие-сроки-качественно-и-безопасно[/url]
[url=http://86hm.ru/forum/flame/?topic_id=25104/]86hm.ru/forum/flame/?topic_id=25104[/url]
[url=http://forexrassia.ru/diplomyi-lyuboy-slozhnosti-byistro-i-bez-problem/]forexrassia.ru/diplomyi-lyuboy-slozhnosti-byistro-i-bez-problem[/url]
Xazrfhx
July 19, 2024 @ 5:19 pm
[u][b] Добрый день![/b][/u]
Ьожем предложить документы ВУЗов, расположенных в любом регионе Российской Федерации. Можно купить качественный диплом от любого заведения, за любой год, указав актуальную специальность и оценки за все дисциплины. Документы делаются на “правильной” бумаге высшего качества. Это позволяет делать настоящие дипломы, которые не отличить от оригинала. Документы заверяются необходимыми печатями и подписями.
[b]Мы изготавливаем дипломы[/b] психологов, юристов, экономистов и любых других профессий по невысоким ценам. Цена зависит от той или иной специальности, года получения и ВУЗа. Стараемся поддерживать для покупателей адекватную политику тарифов. [b]Важно[/b], чтобы документы были доступными для большинства граждан.
[url=http://landik-diploms-srednee.ru/diplom-bakalavra В ]landik-diploms-srednee.ru/diplom-bakalavra В [/url]
[b]Успехов в учебе![/b]
Diplomi_tnSi
July 19, 2024 @ 5:21 pm
[u][b] Здравствуйте![/b][/u]
[b]Купить документ[/b] о получении высшего образования можно у нас.
[url=http://diploms-x24.ru/kupit-diplom-sankt-peterburg/]diploms-x24.ru/kupit-diplom-sankt-peterburg[/url]
Dnrtcdq
July 19, 2024 @ 5:37 pm
[u][b] Добрый день![/b][/u]
[b]Заказать документ[/b] о получении высшего образования можно в нашем сервисе.
[url=http://ast-diplomy.com/kupit-diplom-magistra/]ast-diplomy.com/kupit-diplom-magistra[/url]
[b]Успешной учебы![/b]
Diplomi_dkKt
July 19, 2024 @ 6:10 pm
купить диплом специалиста в москве [url=https://diplomasx.com/]купить диплом специалиста в москве[/url] .
Diplomi_eomi
July 19, 2024 @ 6:11 pm
купить диплом настоящий [url=https://ast-diploms.com/]ast-diploms.com[/url] .
Cazrbrg
July 19, 2024 @ 6:27 pm
[u][b] Привет, друзья![/b][/u]
[b]Мы можем предложить документы ВУЗов[/b], расположенных на территории всей РФ. Можно купить диплом от любого учебного заведения, за любой год, указав актуальную специальность и оценки за все дисциплины. Дипломы и аттестаты делаются на “правильной” бумаге самого высокого качества. Это позволяет делать государственные дипломы, которые не отличить от оригиналов. Они заверяются всеми требуемыми печатями и подписями.
[url=http://b98385gb.beget.tech/2024/07/09/otlichnye-ceny-i-prekrasnoe-kachestvo-diplomy-onlayn.html/]b98385gb.beget.tech/2024/07/09/otlichnye-ceny-i-prekrasnoe-kachestvo-diplomy-onlayn.html[/url]
Lazrvrr
July 19, 2024 @ 6:51 pm
[u][b] Привет, друзья![/b][/u]
Где заказать диплом специалиста?
[b]Заказать диплом о высшем образовании.[/b]
[url=http://sonnick84.rusedu.net/post/10787/113369/]sonnick84.rusedu.net/post/10787/113369[/url]
Xazrhdy
July 19, 2024 @ 7:38 pm
[u][b] Привет, друзья![/b][/u]
Ьожем предложить документы ВУЗов, расположенных в любом регионе России. Вы имеете возможность приобрести качественно сделанный диплом от любого учебного заведения, за любой год, указав актуальную специальность и хорошие оценки за все дисциплины. Документы делаются на бумаге высшего качества. Это дает возможность делать настоящие дипломы, не отличимые от оригинала. Они заверяются всеми требуемыми печатями и штампами.
[b]Мы изготавливаем дипломы[/b] любой профессии по приятным ценам. Стоимость может зависеть от конкретной специальности, года выпуска и образовательного учреждения. Всегда стараемся поддерживать для покупателей адекватную ценовую политику. [b]Важно[/b], чтобы дипломы были доступны для большинства граждан.
[url=http://arusak-diploms-srednee.ru/kupit-diplom-v-krasnodare В ]arusak-diploms-srednee.ru/kupit-diplom-v-krasnodare В [/url]
[b]Успехов в учебе![/b]
Diplomi_snSi
July 19, 2024 @ 7:42 pm
[u][b] Здравствуйте![/b][/u]
[b]Заказать документ[/b] о получении высшего образования можно в нашем сервисе.
[url=http://ast-diplomas24.ru/kupit-diplom-nizhnij-novgorod/]ast-diplomas24.ru/kupit-diplom-nizhnij-novgorod[/url]
Xazrkjj
July 19, 2024 @ 7:52 pm
[u][b] Добрый день![/b][/u]
Приобрести документ о получении высшего образования вы сможете в нашей компании в Москве. Мы оказываем услуги по производству и продаже документов об окончании любых ВУЗов России. Вы получите диплом по любым специальностям, включая документы старого образца.
[url=http://thelspr.listbb.ru/viewtopic.php?f=13&t=361/]thelspr.listbb.ru/viewtopic.php?f=13&t=361[/url]
[url=http://openmotonews.ru/oformlenie-diplomov-s-garantiey-kachestva/]openmotonews.ru/oformlenie-diplomov-s-garantiey-kachestva[/url]
[url=http://mediamemorial.ru/club/user/125807/forum/message/3831/13313/#message13313/]mediamemorial.ru/club/user/125807/forum/message/3831/13313/#message13313[/url]
[url=http://rashin.4adm.ru/viewtopic.php?f=27&t=2029/]rashin.4adm.ru/viewtopic.php?f=27&t=2029[/url]
[url=http://vatrusha.maxbb.ru/viewtopic.php?f=2&t=701/]vatrusha.maxbb.ru/viewtopic.php?f=2&t=701[/url]
Dnrtmft
July 19, 2024 @ 7:59 pm
[u][b] Привет, друзья![/b][/u]
[b]Заказать документ[/b] о получении высшего образования можно в нашей компании в Москве.
[url=http://diploms-x.com/kupit-diplom-moskva/]diploms-x.com/kupit-diplom-moskva[/url]
[b]Хорошей учебы![/b]
Cazrksa
July 19, 2024 @ 8:48 pm
[u][b] Добрый день![/b][/u]
[b]Мы можем предложить документы ВУЗов[/b], которые расположены в любом регионе России. Можно купить качественно сделанный диплом за любой год, включая сюда документы старого образца СССР. Дипломы и аттестаты печатаются на “правильной” бумаге самого высшего качества. Это дает возможность делать настоящие дипломы, не отличимые от оригинала. Они будут заверены всеми необходимыми печатями и подписями.
[url=http://kopoluzhona.image-perth.org/bystro-priobretaem-diplom-v-lucsem-magazine-russian-diplom/]kopoluzhona.image-perth.org/bystro-priobretaem-diplom-v-lucsem-magazine-russian-diplom[/url]
Sazrflh
July 19, 2024 @ 9:15 pm
[u][b] Привет![/b][/u]
[b]Полезная информация как купить диплом о высшем образовании без рисков [/b]
[url=http://obshenie.flybb.ru/viewtopic.php?f=2&t=688/]obshenie.flybb.ru/viewtopic.php?f=2&t=688[/url]
[u][b] Окажем помощь![u][b].
Davidgrigh
July 19, 2024 @ 9:19 pm
medication from mexico pharmacy: mexico pharmacies prescription drugs – mexico drug stores pharmacies
Charlesskimi
July 19, 2024 @ 9:42 pm
mexican online pharmacies prescription drugs [url=https://foruspharma.com/#]best online pharmacies in mexico[/url] best online pharmacies in mexico
Michaeleduch
July 19, 2024 @ 9:53 pm
world pharmacy india: mail order pharmacy india – top online pharmacy india
Diplomi_hrSn
July 19, 2024 @ 10:43 pm
диплом медсестры с занесением в реестр купить [url=https://asxdiplomik24.ru/]asxdiplomik24.ru[/url] .
Edwardbus
July 19, 2024 @ 11:44 pm
https://foruspharma.com/# mexican online pharmacies prescription drugs
Larioruov
July 19, 2024 @ 11:44 pm
Легальная покупка диплома ПТУ с сокращенной программой обучения
[url=http://ast-diplom24.ru/kupit-diplom-sankt-peterburg/]ast-diplom24.ru/kupit-diplom-sankt-peterburg[/url]
Diplomi_ttOa
July 20, 2024 @ 12:20 am
купить аттестат образование [url=www.diploms-x.com]купить аттестат образование[/url] .
Edwardbus
July 20, 2024 @ 12:24 am
http://foruspharma.com/# mexico drug stores pharmacies
Charlesskimi
July 20, 2024 @ 12:48 am
best india pharmacy [url=https://indiapharmast.com/#]Online medicine order[/url] india online pharmacy
Michaeleduch
July 20, 2024 @ 1:16 am
buying from canadian pharmacies: canada drug pharmacy – canadian pharmacy india
Mazrhmq
July 20, 2024 @ 3:43 am
[u][b] Привет![/b][/u]
[b]Приобретение школьного аттестата с официальным упрощенным обучением в Москве[/b]
[url=http://allsportime.ru/oformite-diplom-onlayn-za-neskolko-minut/]allsportime.ru/oformite-diplom-onlayn-za-neskolko-minut[/url]
Richardcab
July 20, 2024 @ 4:20 am
Bella Hadid https://img-models.bella-hadid-ar.com is an American model who has emerged in recent years as one of the most influential figures in the world of fashion.
Dannyjed
July 20, 2024 @ 4:29 am
Brazilian footballer Ricardo Escarson https://orlando-city.kaka-ar.com dos Santos Leite, better known as Kaka, is one of the most famous and successful players in football history.
JasonNem
July 20, 2024 @ 4:31 am
Sadio Mane https://al-nassr.sadio-mane-ar.com the Senegalese footballer best known for his performances at clubs such as Southampton and Liverpool, has become a prominent figure in Al Nassr.
Diplomi_ynmi
July 20, 2024 @ 5:13 am
купить диплом в химках [url=https://ast-diploms.com/]ast-diploms.com[/url] .
Diplomi_nsKt
July 20, 2024 @ 5:15 am
купить диплом колледжа в омске [url=https://diplomasx.com/]diplomasx.com[/url] .
Trefyfh
July 20, 2024 @ 5:34 am
[u][b] Привет, друзья![/b][/u]
[b]Как купить аттестат 11 класса с официальным упрощенным обучением в Москве [/b]
[url=http://obmenka.forum2x2.ru/t17060-topic#38627/]obmenka.forum2x2.ru/t17060-topic#38627[/url]
[u][b] Рады помочь![u][b].
Lazrqpm
July 20, 2024 @ 6:28 am
[u][b] Добрый день![/b][/u]
Где купить диплом специалиста?
[b]Заказать диплом о высшем образовании.[/b]
[url=http://chrstms.ru/club/user/11503/blog/11039//]chrstms.ru/club/user/11503/blog/11039/[/url]
Michaeleduch
July 20, 2024 @ 6:33 am
canadian pharmacy victoza: ed drugs online from canada – rate canadian pharmacies
Dnrtjzr
July 20, 2024 @ 7:35 am
[u][b] Привет, друзья![/b][/u]
[b]Приобрести документ[/b] ВУЗа можно у нас в столице.
[url=http://diplomasx.com/kupit-diplom-ekaterinbur/]diplomasx.com/kupit-diplom-ekaterinbur[/url]
[b]Успехов в учебе![/b]
Louisfut
July 20, 2024 @ 8:42 am
Edson Arantes https://santos.pele-ar.com do Nascimento, known as Pele, was born on October 23, 1940 in Tres Coracoes, Minas Gerais, Brazil.
Tyroneutick
July 20, 2024 @ 8:57 am
Monica Bellucci https://dracula.monica-bellucci-ar.com one of the most famous Italian actresses of our time, has a distinguished artistic career spanning many decades. Her talent, charisma, and stunning beauty made her an icon of world cinema.
Yrefuyl
July 20, 2024 @ 9:17 am
[u][b] Привет, друзья![/b][/u]
[b]Приобрести диплом о высшем образовании.[/b]
[url=http://obozrevatelevents.ru/kupit-diplom-s-polnoy-podderzhkoy//]obozrevatelevents.ru/kupit-diplom-s-polnoy-podderzhkoy/[/url]
[b]Удачи![/b]
Xazrvbf
July 20, 2024 @ 9:29 am
Привет, друзья!
Заказать диплом о высшем образовании
[url=http://landik-diploms-srednee.ru/diplom-s-reestromkupit-kupit В ]landik-diploms-srednee.ru/diplom-s-reestromkupit-kupit В [/url]
Успехов в учебе!
Cazrczx
July 20, 2024 @ 9:37 am
[u][b] Здравствуйте![/b][/u]
[b]Мы предлагаем документы ВУЗов[/b], которые расположены на территории всей РФ. Можно приобрести диплом от любого заведения, за любой год, указав необходимую специальность и хорошие оценки за все дисциплины. Документы выпускаются на бумаге самого высшего качества. Это дает возможности делать государственные дипломы, не отличимые от оригинала. Они будут заверены необходимыми печатями и штампами.
[url=http://midiario.com.mx/read-blog/61777/]midiario.com.mx/read-blog/61777[/url]
Diplomi_wwSn
July 20, 2024 @ 9:38 am
купить защиту диплома [url=https://asxdiplomik24.ru/]asxdiplomik24.ru[/url] .
Diplomi_bzOa
July 20, 2024 @ 9:45 am
я хочу купить диплом высшем образовании [url=diploms-x.com]diploms-x.com[/url] .
Qazrrfn
July 20, 2024 @ 9:52 am
[u][b] Привет![/b][/u]
[b]Купить диплом о среднем полном образовании, в чем подвох и как избежать обмана?.[/b]
[b]Приобрести диплом ВУЗа.[/b]
[url=http://infinirealm.com/read-blog/322/]infinirealm.com/read-blog/322[/url]
Dnrtpzn
July 20, 2024 @ 10:03 am
[u][b] Привет, друзья![/b][/u]
[b]Приобрести документ[/b] института можно у нас.
[url=http://diploms-x24.ru/kupit-diplom-o-srednem-obrazovanii/]diploms-x24.ru/kupit-diplom-o-srednem-obrazovanii[/url]
[b]Успехов в учебе![/b]
Michaeleduch
July 20, 2024 @ 10:26 am
mexican rx online: mexican mail order pharmacies – п»їbest mexican online pharmacies
Xazrnkc
July 20, 2024 @ 11:06 am
[u][b] Привет, друзья![/b][/u]
Купить документ ВУЗа можно в нашей компании. Мы оказываем услуги по производству и продаже документов об окончании любых университетов России. Вы получите диплом по любой специальности, включая документы старого образца.
[url=http://cardinalparkmld.listbb.ru/viewtopic.php?f=3&t=472/]cardinalparkmld.listbb.ru/viewtopic.php?f=3&t=472[/url]
[url=http://1stroitelny.kz/faq/show/id/669248c6a489f940a43a1afc.html/]1stroitelny.kz/faq/show/id/669248c6a489f940a43a1afc.html[/url]
[url=http://ashinova.ru/category-5/t-1737.html#1737/]ashinova.ru/category-5/t-1737.html#1737[/url]
[url=http://angelladydety.getbb.ru/posting.php?mode=post&f=46&sid=8ab65b996ddaf75d3f14a18397d4f386/]angelladydety.getbb.ru/posting.php?mode=post&f=46&sid=8ab65b996ddaf75d3f14a18397d4f386[/url]
[url=http://roving.club/read-blog/1537/]roving.club/read-blog/1537[/url]
Iariormrt
July 20, 2024 @ 11:34 am
[u][b] Добрый день![/b][/u]
Приобрести документ о получении высшего образования можно в нашей компании в Москве. Мы оказываем услуги по продаже документов об окончании любых университетов России. Вы получите необходимый диплом по любым специальностям, любого года выпуска, включая документы Советского Союза. Даем гарантию, что в случае проверки документов работодателями, подозрений не возникнет.
[url=http://cittaviva.net/read-blog/56/]cittaviva.net/read-blog/56[/url]
[url=http://bn.tobase.ru/viewtopic.php?f=30&t=3343/]bn.tobase.ru/viewtopic.php?f=30&t=3343[/url]
[url=http://demo.advised360.com/read-blog/161564/]demo.advised360.com/read-blog/161564[/url]
[url=http://www.yagath.com/blogs/678064/Рекомендации-по-приобретению-документов-в-сети/]www.yagath.com/blogs/678064/Рекомендации-по-приобретению-документов-в-сети[/url]
[url=http://www.webofiice.ro/blogs/1895/Как-подобрать-надежный-магазин-с-документами-Обзор-специалиста/]www.webofiice.ro/blogs/1895/Как-подобрать-надежный-магазин-с-документами-Обзор-специалиста[/url]
Xazrwka
July 20, 2024 @ 11:51 am
Привет!
Купить диплом университета
[url=http://arusak-diploms-srednee.ru/kupit-diplom-v-cheliabinske В ]arusak-diploms-srednee.ru/kupit-diplom-v-cheliabinske В [/url]
Хорошей учебы!
Cazrren
July 20, 2024 @ 12:01 pm
[u][b] Добрый день![/b][/u]
[b]Мы предлагаем документы ВУЗов[/b], которые находятся на территории всей России. Вы сможете приобрести диплом от любого учебного заведения, за любой год, включая документы старого образца. Документы делаются на “правильной” бумаге высшего качества. Это позволяет делать настоящие дипломы, не отличимые от оригиналов. Документы заверяются всеми требуемыми печатями и подписями.
[url=http://ozoms.com/read-blog/514/]ozoms.com/read-blog/514[/url]
Trefnqn
July 20, 2024 @ 12:57 pm
[u][b] Добрый день![/b][/u]
[b]Официальная покупка диплома вуза с сокращенной программой в Москве [/b]
[url=http://qna.habr.com/user/dilopluyy/]qna.habr.com/user/dilopluyy[/url]
[u][b] Рады оказать помощь![u][b].
Edwardbus
July 20, 2024 @ 1:11 pm
https://canadapharmast.online/# canadianpharmacymeds com
Xazruar
July 20, 2024 @ 1:36 pm
[u][b] Здравствуйте![/b][/u]
Заказать документ ВУЗа можно в нашем сервисе. Мы предлагаем документы об окончании любых университетов России. Вы получите необходимый диплом по любой специальности, любого года выпуска, включая документы старого образца.
[url=http://www.paladiny.ru/forummess.dwar.php?TopicID=27649/]www.paladiny.ru/forummess.dwar.php?TopicID=27649[/url]
[url=http://mediamemorial.ru/club/user/125807/forum/message/3847/13329/#message13329/]mediamemorial.ru/club/user/125807/forum/message/3847/13329/#message13329[/url]
[url=http://cv53297-livestreet-1.tw1.ru/2024/07/13/bystroe-oformlenie-diploma-nadezhno-i-bezopasno.html/]cv53297-livestreet-1.tw1.ru/2024/07/13/bystroe-oformlenie-diploma-nadezhno-i-bezopasno.html[/url]
[url=http://4division.ru/viewtopic.php?f=13&t=8629/]4division.ru/viewtopic.php?f=13&t=8629[/url]
[url=http://forum-nine.mirbb.com/t11771-topic#31188/]forum-nine.mirbb.com/t11771-topic#31188[/url]
Clifford
July 20, 2024 @ 2:07 pm
Hello, i think that i saaw you visited my weblog thus i came to “return the
favor”.I’m trying to find things to enhance my web site!I suppose
its ok to use some of your ideas!! https://telegra.ph/Icelandic-Gambling-07-18
Razrbvf
July 20, 2024 @ 2:20 pm
[u][b] Здравствуйте![/b][/u]
[b]Наш сервис предлагает заказать диплом[/b] отличного качества, который не отличить от оригинала без использования дорогостоящего оборудования и квалифицированного специалиста.
[url=http://mail.unnewsusa.com/bbs/board.php?bo_table=free&wr_id=2286574/]mail.unnewsusa.com/bbs/board.php?bo_table=free&wr_id=2286574[/url]
[b]Успехов в учебе![/b]
Vazrsxm
July 20, 2024 @ 2:23 pm
[u][b] Привет, друзья![/b][/u]
Приобрести документ о получении высшего образования можно у нас. Мы оказываем услуги по производству и продаже документов об окончании любых университетов РФ.
[url=http://cookownfood.blogspot.com/2012/11/cold-bean-thread-noodles-with-spinach.html/]cookownfood.blogspot.com/2012/11/cold-bean-thread-noodles-with-spinach.html[/url]
[url=http://msk-service.spb.ru/index.php?subaction=userinfo&user=ynalif/]msk-service.spb.ru/index.php?subaction=userinfo&user=ynalif[/url]
[url=http://gmaii.ru/threads/2183//]gmaii.ru/threads/2183/[/url]
[url=http://sc-grandmaster.ru/forum/?PAGE_NAME=profile_view&UID=19204/]sc-grandmaster.ru/forum/?PAGE_NAME=profile_view&UID=19204[/url]
[url=http://”cf58051.tmweb.ru/index.php?action=post;topic=2257.0;last_msg=132488″/]”cf58051.tmweb.ru/index.php?action=post;topic=2257.0;last_msg=132488″[/url]
Diplomi_xySi
July 20, 2024 @ 2:25 pm
[u][b] Привет![/b][/u]
[b]Купить документ[/b] о получении высшего образования можно в нашей компании.
[url=http://ast-diplom.com/kupit-diplom-o-srednem-obrazovanii/]ast-diplom.com/kupit-diplom-o-srednem-obrazovanii[/url]
BrianErods
July 20, 2024 @ 2:34 pm
Brazilian footballer Malcom https://al-hilal.malcom-ar.com (full name Malcom Felipe Silva de Oliveira) achieved great success in Al Hilal, one of the leading football teams in Saudi Arabia and the entire Middle East.
Charlesskimi
July 20, 2024 @ 2:45 pm
canadian drug pharmacy [url=https://canadapharmast.com/#]best canadian pharmacy online[/url] canadian pharmacy mall
AndrewLow
July 20, 2024 @ 3:17 pm
Jackie Chan https://karate-kid.jackiechan-ar.com was born in 1954 in Hong Kong under the name Chan Kong San.
Manrcal
July 20, 2024 @ 3:21 pm
[u][b] Привет![/b][/u]
Где купить диплом специалиста?
Мы готовы предложить документы институтов, которые находятся на территории всей России. Можно приобрести качественно сделанный диплом от любого высшего учебного заведения, за любой год, указав актуальную специальность и оценки за все дисциплины. Дипломы делаются на бумаге высшего качества. Это позволяет делать государственные дипломы, не отличимые от оригинала. Они будут заверены всеми необходимыми печатями и штампами.
[b]Мы изготавливаем дипломы[/b] психологов, юристов, экономистов и других профессий по приятным тарифам.
[url=http://asxdiplomik24.ru/kupit-diplom-krasnoyarsk]asxdiplomik24.ru/kupit-diplom-krasnoyarsk[/url]
[u][b] Рады оказаться полезными![u][b]
PhillipSab
July 20, 2024 @ 3:26 pm
Jude Bellingham https://real-madrid.jude-bellingham-ar.com a young and talented English footballer, has enjoyed great success with Real Madrid since his arrival.
Williamraics
July 20, 2024 @ 4:00 pm
[u][b] Добрый день![/b][/u]
[b]Наша компания предлагает[/b] быстро приобрести диплом, который выполнен на оригинальном бланке и заверен печатями, водяными знаками, подписями должностных лиц. Диплом пройдет любые проверки, даже с использованием специальных приборов. Достигайте цели максимально быстро с нашей компанией.
[url=http://www.rcauto.pl/modules.php?name=Your_Account&op=userinfo&username=ypygo/]www.rcauto.pl/modules.php?name=Your_Account&op=userinfo&username=ypygo[/url]
[b]Успешной учебы![/b]
Michaeleduch
July 20, 2024 @ 4:03 pm
mexican border pharmacies shipping to usa: mexican border pharmacies shipping to usa – buying from online mexican pharmacy
Davidgrigh
July 20, 2024 @ 4:21 pm
mexican rx online: mexican border pharmacies shipping to usa – buying prescription drugs in mexico
Diplomi_xjSi
July 20, 2024 @ 4:53 pm
[u][b] Привет![/b][/u]
[b]Заказать документ[/b] института можно у нас.
[url=http://diplomasx.com/kupit-diplom-magistra/]diplomasx.com/kupit-diplom-magistra[/url]
Uazrtrn
July 20, 2024 @ 5:08 pm
[u][b] Привет, друзья![/b][/u]
[b]Заказать диплом любого ВУЗа.[/b]
[url=http://www.libkids51.ru/forum/viewtopic.php?f=16&t=5021/]www.libkids51.ru/forum/viewtopic.php?f=16&t=5021[/url]
Yreflji
July 20, 2024 @ 5:37 pm
[u][b] Привет, друзья![/b][/u]
[b]Заказать диплом любого ВУЗа.[/b]
[url=http://www.mylot.su/blogs/2282/]www.mylot.su/blogs/2282[/url]
[b]Хорошей учебы![/b]
Michaeleduch
July 20, 2024 @ 7:37 pm
best online pharmacy india: п»їlegitimate online pharmacies india – pharmacy website india
Tazrsdl
July 20, 2024 @ 8:36 pm
[u][b] Привет![/b][/u]
Заказать диплом университета
Мы можем предложить документы ВУЗов, которые расположены на территории всей РФ. Можно купить качественный диплом от любого учебного заведения, за любой год, включая сюда документы старого образца СССР. Документы печатаются на бумаге самого высокого качества. Это дает возможности делать настоящие дипломы, не отличимые от оригиналов. Они будут заверены всеми обязательными печатями и штампами.
[url=http://www.marqueze.net/miembros/billyoung/profile/classic/]www.marqueze.net/miembros/billyoung/profile/classic/[/url]
Рады оказаться полезными!.
Dnrttqf
July 20, 2024 @ 9:52 pm
[u][b] Привет, друзья![/b][/u]
[b]Заказать документ[/b] о получении высшего образования можно в нашей компании.
[url=http://diploms-x.com/kupit-diplom-moskva/]diploms-x.com/kupit-diplom-moskva[/url]
[b]Успешной учебы![/b]
Iariormnc
July 20, 2024 @ 9:53 pm
[u][b] Добрый день![/b][/u]
Приобрести документ о получении высшего образования вы имеете возможность в нашей компании в Москве. Мы оказываем услуги по изготовлению и продаже документов об окончании любых ВУЗов РФ. Вы получите диплом по любой специальности, включая документы образца СССР. Даем 100% гарантию, что в случае проверки документов работодателями, подозрений не появится.
[url=http://financetimenews.ru/legalnaya-pokupka-diplomov-dostupno-i-nadezhno//]financetimenews.ru/legalnaya-pokupka-diplomov-dostupno-i-nadezhno/[/url]
[url=http://www.hristianka.ru/forum/r/prev_loaded/1//]www.hristianka.ru/forum/r/prev_loaded/1/[/url]
[url=http://nc750.ru/member.php?u=2812/]nc750.ru/member.php?u=2812[/url]
[url=http://1776reloaded.us/blogs/62/Вы-хотите-узнать-как-приобрести-документы-в-сети-Ждем-Вас/]1776reloaded.us/blogs/62/Вы-хотите-узнать-как-приобрести-документы-в-сети-Ждем-Вас[/url]
[url=http://nfrb.pl/index.php?/gallery/image/55-ассортимент-и-методы-доставки-в-интернет-магазине-покупаем-диплом//]nfrb.pl/index.php?/gallery/image/55-ассортимент-и-методы-доставки-в-интернет-магазине-покупаем-диплом/[/url]
Xazrgko
July 20, 2024 @ 11:46 pm
Привет!
Приобрести диплом о высшем образовании
[url=http://arusak-diploms-srednee.ru В ]arusak-diploms-srednee.ru В [/url]
Успехов в учебе!
Dnrtwto
July 21, 2024 @ 12:14 am
[u][b] Здравствуйте![/b][/u]
[b]Купить документ[/b] о получении высшего образования можно у нас.
[url=http://ast-diploms.com/kupit-diplom-omsk/]ast-diploms.com/kupit-diplom-omsk[/url]
[b]Удачи![/b]
Sazrwpc
July 21, 2024 @ 12:17 am
[u][b] Добрый день![/b][/u]
[b]Пошаговая инструкция по официальной покупке диплома о высшем образовании [/b]
[url=http://mmix.ukrbb.net/viewtopic.php?f=23&t=22553/]mmix.ukrbb.net/viewtopic.php?f=23&t=22553[/url]
[u][b] Поможем вам всегда![u][b].
Edwardbus
July 21, 2024 @ 12:18 am
https://foruspharma.com/# mexican border pharmacies shipping to usa
Charlesskimi
July 21, 2024 @ 12:32 am
india pharmacy [url=https://indiapharmast.com/#]best india pharmacy[/url] best online pharmacy india
Cazrkah
July 21, 2024 @ 12:36 am
[u][b] Привет, друзья![/b][/u]
[b]Мы готовы предложить документы ВУЗов[/b], которые расположены в любом регионе России. Вы можете купить качественно напечатанный диплом от любого заведения, за любой год, включая документы старого образца СССР. Документы печатаются на бумаге высшего качества. Это позволяет делать государственные дипломы, которые невозможно отличить от оригиналов. Они заверяются необходимыми печатями и штампами.
[url=http://igpsclub.ru/social/read-blog/5482/]igpsclub.ru/social/read-blog/5482[/url]
Razrskl
July 21, 2024 @ 12:41 am
[u][b] Здравствуйте![/b][/u]
[b]Мы предлагаем заказать диплом[/b] в высочайшем качестве, который не отличить от оригинала без использования дорогостоящего оборудования и опытного специалиста.
[url=http://www.miranetwork.it/index.php?option=com_k2&view=itemlist&task=user&id=446460/]www.miranetwork.it/index.php?option=com_k2&view=itemlist&task=user&id=446460[/url]
[b]Хорошей учебы![/b]
Michaeleduch
July 21, 2024 @ 12:49 am
mexico drug stores pharmacies: п»їbest mexican online pharmacies – purple pharmacy mexico price list
Vazrfxn
July 21, 2024 @ 12:51 am
[u][b] Здравствуйте![/b][/u]
Купить документ о получении высшего образования можно в нашем сервисе. Мы оказываем услуги по продаже документов об окончании любых университетов РФ.
[url=http://rubukkit.org/threads/diplomandoci.192093//]rubukkit.org/threads/diplomandoci.192093/[/url]
[url=http://matripedia.de/doku.php?id=diploman/]matripedia.de/doku.php?id=diploman[/url]
[url=http://led119.ru/forum/user/107366//]led119.ru/forum/user/107366/[/url]
[url=http://romb4x4.ru/forum/threads/kupit-diplom-cena-i810p.3122//]romb4x4.ru/forum/threads/kupit-diplom-cena-i810p.3122/[/url]
[url=http://bumblebeeco.ru/index.php?subaction=userinfo&user=enevadiv/]bumblebeeco.ru/index.php?subaction=userinfo&user=enevadiv[/url]
Manrnfv
July 21, 2024 @ 1:39 am
[u][b] Привет, друзья![/b][/u]
Где купить диплом по необходимой специальности?
Мы готовы предложить документы ВУЗов, расположенных в любом регионе РФ. Можно заказать диплом от любого заведения, за любой год, указав необходимую специальность и хорошие оценки за все дисциплины. Дипломы выпускаются на “правильной” бумаге самого высшего качества. Это позволяет делать настоящие дипломы, не отличимые от оригинала. Документы будут заверены необходимыми печатями и штампами.
[b]Мы готовы предложить дипломы[/b] любой профессии по приятным ценам.
[url=http://ast-diplom24.ru/kupit-diplom-sankt-peterburg]ast-diplom24.ru/kupit-diplom-sankt-peterburg[/url]
[u][b] Поможем вам всегда![u][b]
Xazrxuh
July 21, 2024 @ 2:12 am
Добрый день!
Приобрести диплом любого ВУЗа
[url=http://arusak-diploms-srednee.ru/kupit-diplom-sssr В ]arusak-diploms-srednee.ru/kupit-diplom-sssr В [/url]
Хорошей учебы!
Williamraics
July 21, 2024 @ 2:29 am
[u][b] Привет, друзья![/b][/u]
[b]Наши специалисты предлагают[/b] быстро и выгодно приобрести диплом, который выполнен на бланке ГОЗНАКа и заверен печатями, водяными знаками, подписями. Наш диплом способен пройти любые проверки, даже при использовании профессиональных приборов. Решайте свои задачи быстро и просто с нашими дипломами.
[url=http://teplo-hata.com.ua/users/uqonuxuk/]teplo-hata.com.ua/users/uqonuxuk[/url]
[b]Хорошей учебы![/b]
Uazrpoe
July 21, 2024 @ 3:51 am
[u][b] Привет![/b][/u]
[b]Приобрести диплом любого ВУЗа.[/b]
[url=http://www.anytalkworld.com/read-blog/6488/]www.anytalkworld.com/read-blog/6488[/url]
MarikaJeX0502
July 21, 2024 @ 3:51 am
XEvil 6.0 automatically solve most kind of captchas,
Including such type of captchas: ReCaptcha v.2, ReCaptcha v.3, Hotmail (Microsoft), Google captcha, Solve Media, BitcoinFaucet, Steam, Amazon, Twitter, Microsoft, Twitch, Outlook, +12000
+ hCaptcha, ArkoseLabs FunCaptcha, ReCaptcha Enterprize supported in new XEvil 6.0!
1.) Fast, easy, precisionly
XEvil is the fastest captcha killer in the world. Its has no solving limits, no threads number limits
you can solve even 1.000.000.000 captchas per day and it will cost 0 (ZERO) USD! Just buy license for 59 USD and all!
2.) Several APIs support
XEvil supports more than 6 different, worldwide known API: 2captcha.com, anti-captcha (antigate), RuCaptcha, DeathByCaptcha, etc.
just send your captcha via HTTP request, as you can send into any of that service – and XEvil will solve your captcha!
So, XEvil is compatible with hundreds of applications for SEO/SMM/password recovery/parsing/posting/clicking/cryptocurrency/etc.
3.) Useful support and manuals
After purchase, you got access to a private tech.support forum, Wiki, Skype/Telegram online support
Developers will train XEvil to your type of captcha for FREE and very fast – just send them examples
4.) How to get free trial use of XEvil full version?
– Try to search in Google “Home of XEvil”
– you will find IPs with opened port 80 of XEvil users (click on any IP to ensure)
– try to send your captcha via 2captcha API ino one of that IPs
– if you got BAD KEY error, just tru another IP
– enjoy! 🙂
– (its not work for hCaptcha!)
WARNING: Free XEvil DEMO does NOT support ReCaptcha, hCaptcha and most other types of captcha!
http://xrumersale.site/
Michaeleduch
July 21, 2024 @ 4:45 am
п»їbest mexican online pharmacies: reputable mexican pharmacies online – medicine in mexico pharmacies
Uazrfet
July 21, 2024 @ 4:53 am
[u][b] Привет, друзья![/b][/u]
Где заказать диплом специалиста?
[url=http://battle-station.com/posting.php?mode=post&f=86/]battle-station.com/posting.php?mode=post&f=86[/url]
[b]Удачи![/b]
Lazrszi
July 21, 2024 @ 6:33 am
[u][b] Привет![/b][/u]
Где заказать диплом специалиста?
[b]Заказать диплом любого университета.[/b]
[url=http://logan.in.ua/people/user/7276/blog/7929//]logan.in.ua/people/user/7276/blog/7929/[/url]
MarikaJeX6532
July 21, 2024 @ 6:43 am
XEvil 6.0 automatically solve most kind of captchas,
Including such type of captchas: ReCaptcha-2, ReCaptcha v.3, Hotmail, Google captcha, SolveMedia, BitcoinFaucet, Steam, Amazon, Twitter, Microsoft, Twitch, Outlook, +12k
+ hCaptcha, ArkoseLabs FunCaptcha, ReCaptcha Enterprize supported in new XEvil 6.0!
1.) Fast, easy, precisionly
XEvil is the fastest captcha killer in the world. Its has no solving limits, no threads number limits
you can solve even 1.000.000.000 captchas per day and it will cost 0 (ZERO) USD! Just buy license for 59 USD and all!
2.) Several APIs support
XEvil supports more than 6 different, worldwide known API: 2captcha.com, anti-captcha (antigate), RuCaptcha, death-by-captcha, etc.
just send your captcha via HTTP request, as you can send into any of that service – and XEvil will solve your captcha!
So, XEvil is compatible with hundreds of applications for SEO/SMM/password recovery/parsing/posting/clicking/cryptocurrency/etc.
3.) Useful support and manuals
After purchase, you got access to a private tech.support forum, Wiki, Skype/Telegram online support
Developers will train XEvil to your type of captcha for FREE and very fast – just send them examples
4.) How to get free trial use of XEvil full version?
– Try to search in Google “Home of XEvil”
– you will find IPs with opened port 80 of XEvil users (click on any IP to ensure)
– try to send your captcha via 2captcha API ino one of that IPs
– if you got BAD KEY error, just tru another IP
– enjoy! 🙂
– (its not work for hCaptcha!)
WARNING: Free XEvil DEMO does NOT support ReCaptcha, hCaptcha and most other types of captcha!
http://XEvil.Net/
Xazrhdx
July 21, 2024 @ 7:14 am
[u][b] Добрый день![/b][/u]
Заказать документ о получении высшего образования вы можете у нас. Мы предлагаем документы об окончании любых университетов РФ. Вы сможете получить необходимый диплом по любым специальностям, любого года выпуска, в том числе документы старого образца.
[url=http://ratingforex.ru/forum-forex/viewtopic.php?f=18&t=45599&sid=b6fd21b907fc818d35ed6ff7e3b894f0/]ratingforex.ru/forum-forex/viewtopic.php?f=18&t=45599&sid=b6fd21b907fc818d35ed6ff7e3b894f0[/url]
[url=http://kids-news.ru/zakazhite-diplom-pryamo-seychas-i-nachnite-novuyu-kareru/]kids-news.ru/zakazhite-diplom-pryamo-seychas-i-nachnite-novuyu-kareru[/url]
[url=http://wiki.intradition.ru/index.php/Дипломы_от_лучших_учебных_заведений/]wiki.intradition.ru/index.php/Дипломы_от_лучших_учебных_заведений[/url]
[url=http://cocapal.com/read-blog/975/]cocapal.com/read-blog/975[/url]
[url=http://vnuci.listbb.ru/viewtopic.php?f=2&t=544/]vnuci.listbb.ru/viewtopic.php?f=2&t=544[/url]
MarikaJeX7658
July 21, 2024 @ 9:46 am
XEvil 5.0 automatically solve most kind of captchas,
Including such type of captchas: ReCaptcha-2, ReCaptcha v.3, Hotmail, Google, Solve Media, BitcoinFaucet, Steam, Amazon, Twitter, Microsoft, Twitch, Outlook, +12000
+ hCaptcha, ArkoseLabs FunCaptcha, ReCaptcha Enterprize supported in new XEvil 6.0!
1.) Fast, easy, precisionly
XEvil is the fastest captcha killer in the world. Its has no solving limits, no threads number limits
you can solve even 1.000.000.000 captchas per day and it will cost 0 (ZERO) USD! Just buy license for 59 USD and all!
2.) Several APIs support
XEvil supports more than 6 different, worldwide known API: 2Captcha, anti-captchas.com (antigate), RuCaptcha, death-by-captcha, etc.
just send your captcha via HTTP request, as you can send into any of that service – and XEvil will solve your captcha!
So, XEvil is compatible with hundreds of applications for SEO/SMM/password recovery/parsing/posting/clicking/cryptocurrency/etc.
3.) Useful support and manuals
After purchase, you got access to a private tech.support forum, Wiki, Skype/Telegram online support
Developers will train XEvil to your type of captcha for FREE and very fast – just send them examples
4.) How to get free trial use of XEvil full version?
– Try to search in Google “Home of XEvil”
– you will find IPs with opened port 80 of XEvil users (click on any IP to ensure)
– try to send your captcha via 2captcha API ino one of that IPs
– if you got BAD KEY error, just tru another IP
– enjoy! 🙂
– (its not work for hCaptcha!)
WARNING: Free XEvil DEMO does NOT support ReCaptcha, hCaptcha and most other types of captcha!
http://xrumersale.site/
Xazrwts
July 21, 2024 @ 10:44 am
[u][b] Добрый день![/b][/u]
Купить документ о получении высшего образования вы можете у нас в Москве. Мы оказываем услуги по изготовлению и продаже документов об окончании любых университетов Российской Федерации. Вы получите необходимый диплом по любой специальности, любого года выпуска, в том числе документы СССР.
[url=http://ya.forum.cool/viewtopic.php?id=5849#p17424/]ya.forum.cool/viewtopic.php?id=5849#p17424[/url]
[url=http://p91648f6.beget.tech/2024/07/13/oformlenie-diploma-pod-klyuch-bezopasno-i-legalno.html/]p91648f6.beget.tech/2024/07/13/oformlenie-diploma-pod-klyuch-bezopasno-i-legalno.html[/url]
[url=http://mf.getbb.ru/viewtopic.php?f=28&t=646/]mf.getbb.ru/viewtopic.php?f=28&t=646[/url]
[url=http://abitu.net/events/topic/create/subject/event_4172/]abitu.net/events/topic/create/subject/event_4172[/url]
[url=http://opaseke.com/users/1243/]opaseke.com/users/1243[/url]
ThomasVer
July 21, 2024 @ 10:58 am
https://paxloviddelivery.pro/# Paxlovid over the counter
MyronMalry
July 21, 2024 @ 11:17 am
https://ciprodelivery.pro/# ciprofloxacin
amoxicillin 200 mg tablet [url=http://amoxildelivery.pro/#]purchase amoxicillin 500 mg[/url] amoxicillin 500mg capsule buy online
Diplomi_aySi
July 21, 2024 @ 12:20 pm
[u][b] Добрый день![/b][/u]
[b]Заказать документ[/b] университета можно в нашей компании в Москве.
[url=http://ast-diplomas24.ru/kupit-diplom-omsk/]ast-diplomas24.ru/kupit-diplom-omsk[/url]
MarikaJeX8253
July 21, 2024 @ 12:51 pm
XEvil 6.0 automatically solve most kind of captchas,
Including such type of captchas: ReCaptcha v.2, ReCaptcha-3, Hotmail, Google captcha, SolveMedia, BitcoinFaucet, Steam, Amazon, Twitter, Microsoft, Twitch, Outlook, +12k
+ hCaptcha, ArkoseLabs FunCaptcha, ReCaptcha Enterprize supported in new XEvil 6.0!
1.) Fast, easy, precisionly
XEvil is the fastest captcha killer in the world. Its has no solving limits, no threads number limits
you can solve even 1.000.000.000 captchas per day and it will cost 0 (ZERO) USD! Just buy license for 59 USD and all!
2.) Several APIs support
XEvil supports more than 6 different, worldwide known API: 2Captcha, anti-captchas.com (antigate), RuCaptcha, DeathByCaptcha, etc.
just send your captcha via HTTP request, as you can send into any of that service – and XEvil will solve your captcha!
So, XEvil is compatible with hundreds of applications for SEO/SMM/password recovery/parsing/posting/clicking/cryptocurrency/etc.
3.) Useful support and manuals
After purchase, you got access to a private tech.support forum, Wiki, Skype/Telegram online support
Developers will train XEvil to your type of captcha for FREE and very fast – just send them examples
4.) How to get free trial use of XEvil full version?
– Try to search in Google “Home of XEvil”
– you will find IPs with opened port 80 of XEvil users (click on any IP to ensure)
– try to send your captcha via 2captcha API ino one of that IPs
– if you got BAD KEY error, just tru another IP
– enjoy! 🙂
– (its not work for hCaptcha!)
WARNING: Free XEvil DEMO does NOT support ReCaptcha, hCaptcha and most other types of captcha!
http://xrumersale.site/
Dnrtyhp
July 21, 2024 @ 1:05 pm
[u][b] Привет, друзья![/b][/u]
[b]Купить документ[/b] о получении высшего образования можно в нашей компании.
[url=http://diploms-x24.ru/kupit-diplom-sankt-peterburg/]diploms-x24.ru/kupit-diplom-sankt-peterburg[/url]
[b]Успешной учебы![/b]
Jamesbug
July 21, 2024 @ 2:20 pm
doxycycline buy online usa: buy doxycycline 100mg cheap – doxycycline 300 mg cost
Diplomi_fvSi
July 21, 2024 @ 2:53 pm
[u][b] Добрый день![/b][/u]
[b]Приобрести документ[/b] института вы сможете в нашей компании.
[url=http://ast-diplom24.ru/kupit-diplom-ekaterinbur/]ast-diplom24.ru/kupit-diplom-ekaterinbur[/url]
Xazriio
July 21, 2024 @ 3:06 pm
Здравствуйте!
Полезные советы по покупке диплома о высшем образовании без риска
[url=http://arusak-diploms-srednee.ru/kupit-diplom-magistra В ]arusak-diploms-srednee.ru/kupit-diplom-magistra В [/url]
Jamesbug
July 21, 2024 @ 3:25 pm
paxlovid for sale: paxlovid covid – paxlovid price
Dnrthpu
July 21, 2024 @ 3:32 pm
[u][b] Добрый день![/b][/u]
[b]Приобрести документ[/b] университета можно у нас.
[url=http://diplomasx24.ru/kupit-diplom-vracha/]diplomasx24.ru/kupit-diplom-vracha[/url]
[b]Удачи![/b]
MarikaJeX4144
July 21, 2024 @ 3:46 pm
XEvil 5.0 automatically solve most kind of captchas,
Including such type of captchas: ReCaptcha-2, ReCaptcha v.3, Hotmail (Microsoft), Google captcha, SolveMedia, BitcoinFaucet, Steam, Amazon, Twitter, Microsoft, Twitch, Outlook, +12k
+ hCaptcha, ArkoseLabs FunCaptcha, ReCaptcha Enterprize supported in new XEvil 6.0!
1.) Fast, easy, precisionly
XEvil is the fastest captcha killer in the world. Its has no solving limits, no threads number limits
you can solve even 1.000.000.000 captchas per day and it will cost 0 (ZERO) USD! Just buy license for 59 USD and all!
2.) Several APIs support
XEvil supports more than 6 different, worldwide known API: 2Captcha, anti-captchas.com (antigate), rucaptcha.com, death-by-captcha, etc.
just send your captcha via HTTP request, as you can send into any of that service – and XEvil will solve your captcha!
So, XEvil is compatible with hundreds of applications for SEO/SMM/password recovery/parsing/posting/clicking/cryptocurrency/etc.
3.) Useful support and manuals
After purchase, you got access to a private tech.support forum, Wiki, Skype/Telegram online support
Developers will train XEvil to your type of captcha for FREE and very fast – just send them examples
4.) How to get free trial use of XEvil full version?
– Try to search in Google “Home of XEvil”
– you will find IPs with opened port 80 of XEvil users (click on any IP to ensure)
– try to send your captcha via 2captcha API ino one of that IPs
– if you got BAD KEY error, just tru another IP
– enjoy! 🙂
– (its not work for hCaptcha!)
WARNING: Free XEvil DEMO does NOT support ReCaptcha, hCaptcha and most other types of captcha!
http://XEvil.Net/
Sazrfzs
July 21, 2024 @ 3:56 pm
[u][b] Здравствуйте![/b][/u]
[b]Парадокс, но купить диплом кандидата наук оказалось не так и сложно [/b]
[url=http://solo-it.ru/forum/?PAGE_NAME=profile_view&UID=3787/]solo-it.ru/forum/?PAGE_NAME=profile_view&UID=3787[/url]
[u][b] Всегда вам поможем![u][b].
Lariorryy
July 21, 2024 @ 4:07 pm
Приобретение школьного аттестата с официальным упрощенным обучением в Москве
[url=http://diplomyx.com/kupit-diplom-krasnoyarsk/]diplomyx.com/kupit-diplom-krasnoyarsk[/url]
ThomasVer
July 21, 2024 @ 4:19 pm
https://ciprodelivery.pro/# ciprofloxacin
Trefvbw
July 21, 2024 @ 4:46 pm
[u][b] Привет, друзья![/b][/u]
[b]Сколько стоит получить диплом высшего и среднего образования легально? [/b]
[url=http://vk.com/club16132629?w=wall-16132629_3527/]vk.com/club16132629?w=wall-16132629_3527[/url]
[u][b] Всегда вам поможем![u][b].
Xazrtcb
July 21, 2024 @ 5:32 pm
Привет, друзья!
Легальная покупка диплома ПТУ с сокращенной программой обучения
[url=http://arusak-diploms-srednee.ru ]arusak-diploms-srednee.ru [/url]
MyronMalry
July 21, 2024 @ 5:53 pm
https://doxycyclinedelivery.pro/# doxycycline buy online usa
п»їpaxlovid [url=http://paxloviddelivery.pro/#]paxlovid for sale[/url] paxlovid covid
MarikaJeX8269
July 21, 2024 @ 6:26 pm
XEvil 6.0 automatically solve most kind of captchas,
Including such type of captchas: ReCaptcha v.2, ReCaptcha v.3, Hotmail, Google captcha, Solve Media, BitcoinFaucet, Steam, Amazon, Twitter, Microsoft, Twitch, Outlook, +12000
+ hCaptcha, ArkoseLabs FunCaptcha, ReCaptcha Enterprize supported in new XEvil 6.0!
1.) Fast, easy, precisionly
XEvil is the fastest captcha killer in the world. Its has no solving limits, no threads number limits
you can solve even 1.000.000.000 captchas per day and it will cost 0 (ZERO) USD! Just buy license for 59 USD and all!
2.) Several APIs support
XEvil supports more than 6 different, worldwide known API: 2captcha.com, anti-captcha (antigate), rucaptcha.com, death-by-captcha, etc.
just send your captcha via HTTP request, as you can send into any of that service – and XEvil will solve your captcha!
So, XEvil is compatible with hundreds of applications for SEO/SMM/password recovery/parsing/posting/clicking/cryptocurrency/etc.
3.) Useful support and manuals
After purchase, you got access to a private tech.support forum, Wiki, Skype/Telegram online support
Developers will train XEvil to your type of captcha for FREE and very fast – just send them examples
4.) How to get free trial use of XEvil full version?
– Try to search in Google “Home of XEvil”
– you will find IPs with opened port 80 of XEvil users (click on any IP to ensure)
– try to send your captcha via 2captcha API ino one of that IPs
– if you got BAD KEY error, just tru another IP
– enjoy! 🙂
– (its not work for hCaptcha!)
WARNING: Free XEvil DEMO does NOT support ReCaptcha, hCaptcha and most other types of captcha!
http://XEvil.Net/
Sazrdbo
July 21, 2024 @ 6:28 pm
[u][b] Привет, друзья![/b][/u]
[b]Приобретение школьного аттестата с официальным упрощенным обучением в Москве [/b]
[url=http://zxz.listbb.ru/viewtopic.php?f=7&t=1524/]zxz.listbb.ru/viewtopic.php?f=7&t=1524[/url]
Рады оказаться полезными!
Cazrjvd
July 21, 2024 @ 6:47 pm
[u][b] Привет![/b][/u]
[b]Мы можем предложить документы техникумов[/b], которые расположены на территории всей России. Можно купить диплом за любой год, включая сюда документы СССР. Дипломы и аттестаты печатаются на бумаге самого высокого качества. Это позволяет делать настоящие дипломы, не отличимые от оригиналов. Они будут заверены необходимыми печатями и штампами.
[url=http://talktai.com/read-blog/1721_podrobnoe-opisanie-zakaza-diploma-v-populyarnom-magazine.html/]talktai.com/read-blog/1721_podrobnoe-opisanie-zakaza-diploma-v-populyarnom-magazine.html[/url]
ThomasVer
July 21, 2024 @ 7:21 pm
https://ciprodelivery.pro/# cipro for sale
Cazrooq
July 21, 2024 @ 9:06 pm
[u][b] Привет![/b][/u]
[b]Мы предлагаем документы ВУЗов[/b], расположенных в любом регионе Российской Федерации. Можно купить диплом от любого заведения, за любой год, в том числе документы старого образца СССР. Дипломы и аттестаты делаются на бумаге самого высокого качества. Это дает возможности делать настоящие дипломы, которые невозможно отличить от оригиналов. Документы будут заверены всеми необходимыми печатями и подписями.
[url=http://lucylink.com/read-blog/56/]lucylink.com/read-blog/56[/url]
MyronMalry
July 21, 2024 @ 9:07 pm
https://clomiddelivery.pro/# can i get clomid price
paxlovid covid [url=http://paxloviddelivery.pro/#]paxlovid buy[/url] paxlovid for sale
MarikaJeX9154
July 21, 2024 @ 9:22 pm
XEvil 6.0 automatically solve most kind of captchas,
Including such type of captchas: ReCaptcha v.2, ReCaptcha v.3, Hotmail (Microsoft), Google captcha, Solve Media, BitcoinFaucet, Steam, Amazon, Twitter, Microsoft, Twitch, Outlook, +12000
+ hCaptcha, ArkoseLabs FunCaptcha, ReCaptcha Enterprize supported in new XEvil 6.0!
1.) Fast, easy, precisionly
XEvil is the fastest captcha killer in the world. Its has no solving limits, no threads number limits
you can solve even 1.000.000.000 captchas per day and it will cost 0 (ZERO) USD! Just buy license for 59 USD and all!
2.) Several APIs support
XEvil supports more than 6 different, worldwide known API: 2Captcha, anti-captcha (antigate), rucaptcha.com, death-by-captcha, etc.
just send your captcha via HTTP request, as you can send into any of that service – and XEvil will solve your captcha!
So, XEvil is compatible with hundreds of applications for SEO/SMM/password recovery/parsing/posting/clicking/cryptocurrency/etc.
3.) Useful support and manuals
After purchase, you got access to a private tech.support forum, Wiki, Skype/Telegram online support
Developers will train XEvil to your type of captcha for FREE and very fast – just send them examples
4.) How to get free trial use of XEvil full version?
– Try to search in Google “Home of XEvil”
– you will find IPs with opened port 80 of XEvil users (click on any IP to ensure)
– try to send your captcha via 2captcha API ino one of that IPs
– if you got BAD KEY error, just tru another IP
– enjoy! 🙂
– (its not work for hCaptcha!)
WARNING: Free XEvil DEMO does NOT support ReCaptcha, hCaptcha and most other types of captcha!
http://xrumersale.site/
JamesNuach
July 21, 2024 @ 9:25 pm
Travis Scott https://astroworld.travis-scott-ar.com is one of the brightest stars in the modern hip-hop industry.
Donaldcaw
July 21, 2024 @ 9:33 pm
Zinedine Zidane https://real-madrid.zinedine-zidane-ar.com the legendary French footballer, entered the annals of football history as a player and coach.
Williamnip
July 21, 2024 @ 9:44 pm
The history of one of France’s https://france.paris-saint-germain-ar.com most famous football clubs, Paris Saint-Germain, began in 1970, when capitalist businessmen Henri Delaunay and Jean-Auguste Delbave founded the club in the Paris Saint-Germain-en-Laye area.
DonaldGot
July 21, 2024 @ 9:46 pm
Chelsea https://england.chelsea-ar.com is one of the most successful English football clubs of our time.
CalvinSweno
July 21, 2024 @ 9:47 pm
Juventus Football Club https://italy.juventus-ar.com is one of the most successful and decorated clubs in the history of Italian and world football.
Diplomi_gjmi
July 21, 2024 @ 10:55 pm
купить диплом курсовая работа [url=https://ast-diploms.com/]ast-diploms.com[/url] .
Diplomi_dlKt
July 21, 2024 @ 11:22 pm
куплю аттестат камень [url=https://diplomasx.com/]куплю аттестат камень[/url] .
Diplomi_ezOa
July 21, 2024 @ 11:28 pm
среднее специальное образование купить [url=http://diploms-x.com]среднее специальное образование купить[/url] .
Trefrqp
July 22, 2024 @ 12:12 am
[u][b] Здравствуйте![/b][/u]
[b]Можно ли купить аттестат о среднем образовании? Основные рекомендации [/b]
[url=http://smetdlysmet.ru/forum/viewtopic.php?f=8&t=41807&p=98711#p98711/]smetdlysmet.ru/forum/viewtopic.php?f=8&t=41807&p=98711#p98711[/url]
[u][b] Всегда вам поможем![u][b].
MarikaJeX3690
July 22, 2024 @ 12:17 am
XEvil 6.0 automatically solve most kind of captchas,
Including such type of captchas: ReCaptcha v.2, ReCaptcha v.3, Hotmail (Microsoft), Google, SolveMedia, BitcoinFaucet, Steam, Amazon, Twitter, Microsoft, Twitch, Outlook, +12k
+ hCaptcha, ArkoseLabs FunCaptcha, ReCaptcha Enterprize supported in new XEvil 6.0!
1.) Fast, easy, precisionly
XEvil is the fastest captcha killer in the world. Its has no solving limits, no threads number limits
you can solve even 1.000.000.000 captchas per day and it will cost 0 (ZERO) USD! Just buy license for 59 USD and all!
2.) Several APIs support
XEvil supports more than 6 different, worldwide known API: 2captcha.com, anti-captcha (antigate), rucaptcha.com, DeathByCaptcha, etc.
just send your captcha via HTTP request, as you can send into any of that service – and XEvil will solve your captcha!
So, XEvil is compatible with hundreds of applications for SEO/SMM/password recovery/parsing/posting/clicking/cryptocurrency/etc.
3.) Useful support and manuals
After purchase, you got access to a private tech.support forum, Wiki, Skype/Telegram online support
Developers will train XEvil to your type of captcha for FREE and very fast – just send them examples
4.) How to get free trial use of XEvil full version?
– Try to search in Google “Home of XEvil”
– you will find IPs with opened port 80 of XEvil users (click on any IP to ensure)
– try to send your captcha via 2captcha API ino one of that IPs
– if you got BAD KEY error, just tru another IP
– enjoy! 🙂
– (its not work for hCaptcha!)
WARNING: Free XEvil DEMO does NOT support ReCaptcha, hCaptcha and most other types of captcha!
http://xrumersale.site/
ThomasVer
July 22, 2024 @ 12:27 am
http://amoxildelivery.pro/# amoxicillin buy canada
Diplomi_gaSi
July 22, 2024 @ 1:30 am
[u][b] Добрый день![/b][/u]
[b]Заказать документ[/b] о получении высшего образования можно у нас в Москве.
[url=http://ast-diplomas24.ru/kupit-diplom-sankt-peterburg/]ast-diplomas24.ru/kupit-diplom-sankt-peterburg[/url]
Xazrkux
July 22, 2024 @ 1:41 am
[u][b] Привет, друзья![/b][/u]
Заказать документ ВУЗа можно у нас в Москве. Мы предлагаем документы об окончании любых ВУЗов Российской Федерации. Вы сможете получить необходимый диплом по любым специальностям, любого года выпуска, в том числе документы Советского Союза.
[url=http://jagplay.ekafe.ru/viewtopic.php?f=22&t=4165/]jagplay.ekafe.ru/viewtopic.php?f=22&t=4165[/url]
[url=http://a63.flybb.ru/viewtopic.php?f=16&t=3215/]a63.flybb.ru/viewtopic.php?f=16&t=3215[/url]
[url=http://brawlstarsacc.listbb.ru/viewtopic.php?f=3&t=486/]brawlstarsacc.listbb.ru/viewtopic.php?f=3&t=486[/url]
[url=http://aurorahcs.com/forum/posting.php?mode=post&f=8/]aurorahcs.com/forum/posting.php?mode=post&f=8[/url]
[url=http://odincovoschool16.flybb.ru/viewtopic.php?f=2&t=839/]odincovoschool16.flybb.ru/viewtopic.php?f=2&t=839[/url]
Yrefown
July 22, 2024 @ 1:53 am
[u][b] Привет, друзья![/b][/u]
[b]Приобрести диплом любого ВУЗа.[/b]
[url=http://wiki.gta-zona.ru/index.php/??????_??????:_????????_?_???_??????/]wiki.gta-zona.ru/index.php/??????_??????:_????????_?_???_??????[/url]
[b]Удачи![/b]
vcs
July 22, 2024 @ 2:13 am
Hello, just wanted to mention, I loved this blog post.
It was practical. Keep on posting!
Diplomi_nkSn
July 22, 2024 @ 2:28 am
купить диплом института в самаре [url=https://asxdiplomik24.ru/]asxdiplomik24.ru[/url] .
Larioragi
July 22, 2024 @ 2:58 am
Как оказалось, купить диплом кандидата наук не так уж и сложно
[url=http://diplomyx.com/kupit-diplom-omsk/]diplomyx.com/kupit-diplom-omsk[/url]
MyronMalry
July 22, 2024 @ 3:04 am
http://clomiddelivery.pro/# where can i buy clomid without rx
buy ciprofloxacin over the counter [url=http://ciprodelivery.pro/#]buy generic ciprofloxacin[/url] buy ciprofloxacin over the counter
MarikaJeX4347
July 22, 2024 @ 3:05 am
XEvil 6.0 automatically solve most kind of captchas,
Including such type of captchas: ReCaptcha-2, ReCaptcha v.3, Hotmail (Microsoft), Google captcha, SolveMedia, BitcoinFaucet, Steam, Amazon, Twitter, Microsoft, Twitch, Outlook, +12k
+ hCaptcha, ArkoseLabs FunCaptcha, ReCaptcha Enterprize supported in new XEvil 6.0!
1.) Fast, easy, precisionly
XEvil is the fastest captcha killer in the world. Its has no solving limits, no threads number limits
you can solve even 1.000.000.000 captchas per day and it will cost 0 (ZERO) USD! Just buy license for 59 USD and all!
2.) Several APIs support
XEvil supports more than 6 different, worldwide known API: 2Captcha, anti-captchas.com (antigate), rucaptcha.com, DeathByCaptcha, etc.
just send your captcha via HTTP request, as you can send into any of that service – and XEvil will solve your captcha!
So, XEvil is compatible with hundreds of applications for SEO/SMM/password recovery/parsing/posting/clicking/cryptocurrency/etc.
3.) Useful support and manuals
After purchase, you got access to a private tech.support forum, Wiki, Skype/Telegram online support
Developers will train XEvil to your type of captcha for FREE and very fast – just send them examples
4.) How to get free trial use of XEvil full version?
– Try to search in Google “Home of XEvil”
– you will find IPs with opened port 80 of XEvil users (click on any IP to ensure)
– try to send your captcha via 2captcha API ino one of that IPs
– if you got BAD KEY error, just tru another IP
– enjoy! 🙂
– (its not work for hCaptcha!)
WARNING: Free XEvil DEMO does NOT support ReCaptcha, hCaptcha and most other types of captcha!
http://xrumersale.site/
Jamesbug
July 22, 2024 @ 3:09 am
amoxicillin 1000 mg capsule: amoxicillin 875 125 mg tab – 875 mg amoxicillin cost
Dnrtjkt
July 22, 2024 @ 3:38 am
[u][b] Здравствуйте![/b][/u]
[b]Приобрести документ[/b] о получении высшего образования можно в нашей компании.
[url=http://ast-diplom24.ru/kupit-diplom-sankt-peterburg/]ast-diplom24.ru/kupit-diplom-sankt-peterburg[/url]
[b]Удачи![/b]
ThomasVer
July 22, 2024 @ 3:39 am
http://amoxildelivery.pro/# amoxicillin 500 mg without a prescription
tablet
July 22, 2024 @ 3:43 am
[url=http://modafinile.com/]buy provigil rx[/url]
Jamesbug
July 22, 2024 @ 4:09 am
buy cheap doxycycline uk: cost of generic doxycycline – doxycycline tablet 100 mg
Diplomi_xsSi
July 22, 2024 @ 4:09 am
[u][b] Привет![/b][/u]
[b]Приобрести документ[/b] института вы сможете в нашей компании в столице.
[url=http://ast-diploms24.ru/kupit-diplom-moskva/]ast-diploms24.ru/kupit-diplom-moskva[/url]
Raymonddus
July 22, 2024 @ 5:22 am
Автомобили Hongqi https://hongqi-krasnoyarsk.ru в наличии – официальный дилер Hongqi Красноярск
MatthewCib
July 22, 2024 @ 5:26 am
Liverpool https://england.liverpool-ar.com holds a special place in the history of football in England.
RobertElinc
July 22, 2024 @ 5:37 am
Priyanka Chopra https://baywatch.priyankachopra-ar.com is an Indian actress, singer, film producer and model who has achieved global success.
KennethIcows
July 22, 2024 @ 5:40 am
When Taylor Swift https://shake-it-off.taylor-swift-ar.com released “Shake It Off” in 2014, she had no idea how much the song would impact her life and music career.
MarikaJeX3704
July 22, 2024 @ 5:41 am
XEvil 5.0 automatically solve most kind of captchas,
Including such type of captchas: ReCaptcha v.2, ReCaptcha-3, Hotmail (Microsoft), Google captcha, SolveMedia, BitcoinFaucet, Steam, Amazon, Twitter, Microsoft, Twitch, Outlook, +12k
+ hCaptcha, ArkoseLabs FunCaptcha, ReCaptcha Enterprize supported in new XEvil 6.0!
1.) Fast, easy, precisionly
XEvil is the fastest captcha killer in the world. Its has no solving limits, no threads number limits
you can solve even 1.000.000.000 captchas per day and it will cost 0 (ZERO) USD! Just buy license for 59 USD and all!
2.) Several APIs support
XEvil supports more than 6 different, worldwide known API: 2captcha.com, anti-captchas.com (antigate), rucaptcha.com, death-by-captcha, etc.
just send your captcha via HTTP request, as you can send into any of that service – and XEvil will solve your captcha!
So, XEvil is compatible with hundreds of applications for SEO/SMM/password recovery/parsing/posting/clicking/cryptocurrency/etc.
3.) Useful support and manuals
After purchase, you got access to a private tech.support forum, Wiki, Skype/Telegram online support
Developers will train XEvil to your type of captcha for FREE and very fast – just send them examples
4.) How to get free trial use of XEvil full version?
– Try to search in Google “Home of XEvil”
– you will find IPs with opened port 80 of XEvil users (click on any IP to ensure)
– try to send your captcha via 2captcha API ino one of that IPs
– if you got BAD KEY error, just tru another IP
– enjoy! 🙂
– (its not work for hCaptcha!)
WARNING: Free XEvil DEMO does NOT support ReCaptcha, hCaptcha and most other types of captcha!
http://XEvil.Net/
Xazreby
July 22, 2024 @ 5:55 am
Привет, друзья!
Где и как купить диплом о высшем образовании без лишних рисков
[url=http://arusak-diploms-srednee.ru/kupit-diplom-v-krasnoiarske]arusak-diploms-srednee.ru/kupit-diplom-v-krasnoiarske[/url]
Dnrtmpd
July 22, 2024 @ 6:11 am
[u][b] Привет![/b][/u]
[b]Купить документ[/b] университета можно в нашей компании.
[url=http://diplomasx24.ru/otzyvy/]diplomasx24.ru/otzyvy[/url]
[b]Успехов в учебе![/b]
MyronMalry
July 22, 2024 @ 6:26 am
https://doxycyclinedelivery.pro/# doxycycline 100mg acne
paxlovid buy [url=https://paxloviddelivery.pro/#]paxlovid covid[/url] buy paxlovid online
Sazrdom
July 22, 2024 @ 7:07 am
[u][b] Привет, друзья![/b][/u]
[b]Рекомендации по безопасной покупке диплома о высшем образовании [/b]
[url=http://toolsrepair.ru/forum/viewtopic.php?f=5&t=2933455/]toolsrepair.ru/forum/viewtopic.php?f=5&t=2933455[/url]
Рады оказаться полезными!
RobertAcike
July 22, 2024 @ 7:54 am
Jennifer Lopez https://lets-get-loud.jenniferlopez-ar.com was born in 1969 in the Bronx, New York, to parents who were Puerto Rican immigrants.
Jariormwh
July 22, 2024 @ 8:06 am
[u][b] Привет![/b][/u]
Купить диплом ВУЗа
[b]Наши специалисты предлагают[/b] выгодно приобрести диплом, который выполняется на оригинальной бумаге и заверен мокрыми печатями, водяными знаками, подписями должностных лиц. Наш документ способен пройти любые проверки, даже при использовании специально предназначенного оборудования. Достигайте цели быстро с нашими дипломами.
[b]Где приобрести диплом по необходимой специальности?[/b]
[url=http://86hm.ru/forum/flame/?topic_id=25002/]86hm.ru/forum/flame/?topic_id=25002[/url]
[url=http://www.pets-ural.ru/content/forum/viewtopic.php?p=63537#63537/]www.pets-ural.ru/content/forum/viewtopic.php?p=63537#63537[/url]
[url=http://www.simplemachines.org/about/smf/stats.php/]www.simplemachines.org/about/smf/stats.php[/url]
[url=http://ya.2bb.ru/viewtopic.php?id=19040#p240217/]ya.2bb.ru/viewtopic.php?id=19040#p240217[/url]
[url=http://macadamlab.ru/wiki/index.php?title=пїЅпїЅпїЅпїЅпїЅпїЅ_пїЅпїЅпїЅпїЅпїЅпїЅ/]macadamlab.ru/wiki/index.php?title=пїЅпїЅпїЅпїЅпїЅпїЅ_пїЅпїЅпїЅпїЅпїЅпїЅ[/url]
ThomasVer
July 22, 2024 @ 8:19 am
http://ciprodelivery.pro/# ciprofloxacin mail online
MarikaJeX4460
July 22, 2024 @ 8:23 am
XEvil 6.0 automatically solve most kind of captchas,
Including such type of captchas: ReCaptcha v.2, ReCaptcha v.3, Hotmail (Microsoft), Google, Solve Media, BitcoinFaucet, Steam, Amazon, Twitter, Microsoft, Twitch, Outlook, +12k
+ hCaptcha, ArkoseLabs FunCaptcha, ReCaptcha Enterprize supported in new XEvil 6.0!
1.) Fast, easy, precisionly
XEvil is the fastest captcha killer in the world. Its has no solving limits, no threads number limits
you can solve even 1.000.000.000 captchas per day and it will cost 0 (ZERO) USD! Just buy license for 59 USD and all!
2.) Several APIs support
XEvil supports more than 6 different, worldwide known API: 2captcha.com, anti-captchas.com (antigate), RuCaptcha, DeathByCaptcha, etc.
just send your captcha via HTTP request, as you can send into any of that service – and XEvil will solve your captcha!
So, XEvil is compatible with hundreds of applications for SEO/SMM/password recovery/parsing/posting/clicking/cryptocurrency/etc.
3.) Useful support and manuals
After purchase, you got access to a private tech.support forum, Wiki, Skype/Telegram online support
Developers will train XEvil to your type of captcha for FREE and very fast – just send them examples
4.) How to get free trial use of XEvil full version?
– Try to search in Google “Home of XEvil”
– you will find IPs with opened port 80 of XEvil users (click on any IP to ensure)
– try to send your captcha via 2captcha API ino one of that IPs
– if you got BAD KEY error, just tru another IP
– enjoy! 🙂
– (its not work for hCaptcha!)
WARNING: Free XEvil DEMO does NOT support ReCaptcha, hCaptcha and most other types of captcha!
http://XEvil.Net/
Xazrisv
July 22, 2024 @ 8:26 am
Привет!
Как быстро и легально купить диплом в Москве
[url=http://arusak-diploms-srednee.ru/kupit-diplom-v-rostove-na-donu ]arusak-diploms-srednee.ru/kupit-diplom-v-rostove-na-donu [/url]
Iariorezu
July 22, 2024 @ 9:26 am
[u][b] Добрый день![/b][/u]
Заказать документ о получении высшего образования вы сможете у нас. Мы предлагаем документы об окончании любых ВУЗов России. Вы получите диплом по любой специальности, включая документы СССР. Даем 100% гарантию, что при проверке документа работодателями, никаких подозрений не появится.
[url=http://ingprint.ru/club/user/58725/forum/message/6938/203480//]ingprint.ru/club/user/58725/forum/message/6938/203480/[/url]
[url=http://forums.unigild.com/index.php?/topic/11161-настоящие-дипломы-легально-и-удобно//]forums.unigild.com/index.php?/topic/11161-настоящие-дипломы-легально-и-удобно/[/url]
[url=http://moust.lv/read-blog/227/]moust.lv/read-blog/227[/url]
[url=http://c90226sl.beget.tech/2024/07/07/kupit-diplom-v-moskve-legko-i-bezopasno//]c90226sl.beget.tech/2024/07/07/kupit-diplom-v-moskve-legko-i-bezopasno/[/url]
[url=http://video.listbb.ru/viewtopic.php?f=3&t=405/]video.listbb.ru/viewtopic.php?f=3&t=405[/url]
Sazrkbe
July 22, 2024 @ 9:49 am
[u][b] Здравствуйте![/b][/u]
[b]Пошаговая инструкция по официальной покупке диплома о высшем образовании [/b]
[url=http://vseogirls.ru/byistroe-oformlenie-diploma-lyuboy-spetsialnosti/]vseogirls.ru/byistroe-oformlenie-diploma-lyuboy-spetsialnosti[/url]
Рады помочь!
Oariorhua
July 22, 2024 @ 9:49 am
[u][b] Привет![/b][/u]
Купить диплом любого университета
[b]Мы предлагаем[/b] выгодно и быстро заказать диплом, который выполняется на бланке ГОЗНАКа и заверен печатями, штампами, подписями. Наш диплом способен пройти лубую проверку, даже при использовании специально предназначенного оборудования. Решите свои задачи быстро и просто с нашим сервисом.
[b]Где заказать диплом по необходимой специальности?[/b]
[url=http://ulmo.ukrbb.net/index.php/]ulmo.ukrbb.net/index.php[/url]
[url=http://gaeloxeparo.lucialpiazzale.com/sovety-cto-pozvolat-najti-nadeznyj-magazin-s-diplomami/]gaeloxeparo.lucialpiazzale.com/sovety-cto-pozvolat-najti-nadeznyj-magazin-s-diplomami[/url]
[url=http://backshowtime.ru/vash-nastoyashhiy-diplom-legalno-i-nadezhno/]backshowtime.ru/vash-nastoyashhiy-diplom-legalno-i-nadezhno[/url]
[url=http://sonnick84.bloguetechno.com/Советы-специалиста-что-помогут-быстро-купить-диплом-63719658/]sonnick84.bloguetechno.com/Советы-специалиста-что-помогут-быстро-купить-диплом-63719658[/url]
[url=http://www.bodybuilding.net/members/worksale.html/]www.bodybuilding.net/members/worksale.html[/url]
Diplomi_xvmi
July 22, 2024 @ 10:08 am
диплом о высшем образовании с занесением в реестр стоимость [url=https://ast-diploms.com/]ast-diploms.com[/url] .
Cazrdzq
July 22, 2024 @ 10:24 am
[u][b] Привет, друзья![/b][/u]
[b]Мы предлагаем документы техникумов[/b], расположенных в любом регионе России. Вы можете заказать качественно сделанный диплом от любого учебного заведения, за любой год, включая документы старого образца СССР. Документы выпускаются на бумаге высшего качества. Это позволяет делать настоящие дипломы, не отличимые от оригинала. Они заверяются всеми необходимыми печатями и подписями.
[url=http://belsprosshina.by/company/personal/user/127/forum/message/1571/5818//]belsprosshina.by/company/personal/user/127/forum/message/1571/5818/[/url]
Yrefafq
July 22, 2024 @ 10:33 am
[u][b] Привет![/b][/u]
[b]Заказать диплом университета.[/b]
[url=http://girlscools.ru/zakazhite-diplom-v-lyuboe-vremya/]girlscools.ru/zakazhite-diplom-v-lyuboe-vremya[/url]
[b]Успешной учебы![/b]
Diplomi_ttKt
July 22, 2024 @ 10:38 am
купить диплом о высшем образовании проверка [url=https://diplomasx.com/]diplomasx.com[/url] .
MarikaJeX7977
July 22, 2024 @ 11:14 am
XEvil 5.0 automatically solve most kind of captchas,
Including such type of captchas: ReCaptcha-2, ReCaptcha v.3, Hotmail (Microsoft), Google captcha, Solve Media, BitcoinFaucet, Steam, Amazon, Twitter, Microsoft, Twitch, Outlook, +12k
+ hCaptcha, ArkoseLabs FunCaptcha, ReCaptcha Enterprize supported in new XEvil 6.0!
1.) Fast, easy, precisionly
XEvil is the fastest captcha killer in the world. Its has no solving limits, no threads number limits
you can solve even 1.000.000.000 captchas per day and it will cost 0 (ZERO) USD! Just buy license for 59 USD and all!
2.) Several APIs support
XEvil supports more than 6 different, worldwide known API: 2captcha.com, anti-captcha (antigate), rucaptcha.com, death-by-captcha, etc.
just send your captcha via HTTP request, as you can send into any of that service – and XEvil will solve your captcha!
So, XEvil is compatible with hundreds of applications for SEO/SMM/password recovery/parsing/posting/clicking/cryptocurrency/etc.
3.) Useful support and manuals
After purchase, you got access to a private tech.support forum, Wiki, Skype/Telegram online support
Developers will train XEvil to your type of captcha for FREE and very fast – just send them examples
4.) How to get free trial use of XEvil full version?
– Try to search in Google “Home of XEvil”
– you will find IPs with opened port 80 of XEvil users (click on any IP to ensure)
– try to send your captcha via 2captcha API ino one of that IPs
– if you got BAD KEY error, just tru another IP
– enjoy! 🙂
– (its not work for hCaptcha!)
WARNING: Free XEvil DEMO does NOT support ReCaptcha, hCaptcha and most other types of captcha!
http://xrumersale.site/
Chrisgak
July 22, 2024 @ 11:25 am
After some difficult years in the late 2010s, Manchester United https://england.manchester-united-ar.com returned to greatness in English football by 2024.
EdwinCug
July 22, 2024 @ 11:26 am
The Formula One World Championship https://world-circuit-racing-championship.formula-1-ar.com, known as the Formula Championship in motor racing, is the highest tier of professional motor racing.
ThomasVer
July 22, 2024 @ 11:30 am
http://amoxildelivery.pro/# order amoxicillin online no prescription
Bryanquirl
July 22, 2024 @ 11:39 am
Michael Jordan https://chicago-bulls.michael-jordan-ar.com is one of the greatest basketball players of all time, whose career with the Chicago Bulls is legendary.
Stevenfaili
July 22, 2024 @ 11:39 am
дизайн интерьер полная версия дизайн интерьера квартиры
MyronMalry
July 22, 2024 @ 12:33 pm
https://paxloviddelivery.pro/# п»їpaxlovid
how to get clomid price [url=https://clomiddelivery.pro/#]can i get generic clomid without insurance[/url] buy clomid no prescription
Lazrgmb
July 22, 2024 @ 12:52 pm
[u][b] Здравствуйте![/b][/u]
[b]Мы предлагаем дипломы[/b] любой профессии по приятным ценам.
[url=http://laporteproperty.com/en/cb-profile/pluginclass/cbblogs?action=blogs&func=show&id=124/]laporteproperty.com/en/cb-profile/pluginclass/cbblogs?action=blogs&func=show&id=124[/url]
MarikaJeX7226
July 22, 2024 @ 2:00 pm
XEvil 5.0 automatically solve most kind of captchas,
Including such type of captchas: ReCaptcha-2, ReCaptcha v.3, Hotmail (Microsoft), Google, Solve Media, BitcoinFaucet, Steam, Amazon, Twitter, Microsoft, Twitch, Outlook, +12000
+ hCaptcha, ArkoseLabs FunCaptcha, ReCaptcha Enterprize supported in new XEvil 6.0!
1.) Fast, easy, precisionly
XEvil is the fastest captcha killer in the world. Its has no solving limits, no threads number limits
you can solve even 1.000.000.000 captchas per day and it will cost 0 (ZERO) USD! Just buy license for 59 USD and all!
2.) Several APIs support
XEvil supports more than 6 different, worldwide known API: 2captcha.com, anti-captchas.com (antigate), RuCaptcha, DeathByCaptcha, etc.
just send your captcha via HTTP request, as you can send into any of that service – and XEvil will solve your captcha!
So, XEvil is compatible with hundreds of applications for SEO/SMM/password recovery/parsing/posting/clicking/cryptocurrency/etc.
3.) Useful support and manuals
After purchase, you got access to a private tech.support forum, Wiki, Skype/Telegram online support
Developers will train XEvil to your type of captcha for FREE and very fast – just send them examples
4.) How to get free trial use of XEvil full version?
– Try to search in Google “Home of XEvil”
– you will find IPs with opened port 80 of XEvil users (click on any IP to ensure)
– try to send your captcha via 2captcha API ino one of that IPs
– if you got BAD KEY error, just tru another IP
– enjoy! 🙂
– (its not work for hCaptcha!)
WARNING: Free XEvil DEMO does NOT support ReCaptcha, hCaptcha and most other types of captcha!
http://XEvil.Net/
Lazrybn
July 22, 2024 @ 3:25 pm
[u][b] Привет, друзья![/b][/u]
[b]Мы можем предложить дипломы[/b] любых профессий по доступным тарифам.
[url=http://poltavagok.ru/poluchite-diplom-v-lyuboe-vremya//]poltavagok.ru/poluchite-diplom-v-lyuboe-vremya/[/url]
Jamesbug
July 22, 2024 @ 3:40 pm
buying amoxicillin online: amoxicillin medicine over the counter – amoxicillin generic
Williamlap
July 22, 2024 @ 4:04 pm
rental car bar Montenegro https://montenegro-car-rental-hire.com
MyronMalry
July 22, 2024 @ 4:07 pm
https://paxloviddelivery.pro/# Paxlovid buy online
paxlovid covid [url=http://paxloviddelivery.pro/#]paxlovid cost without insurance[/url] п»їpaxlovid
ThomasVer
July 22, 2024 @ 4:17 pm
http://ciprodelivery.pro/# ciprofloxacin mail online
MarikaJeX0769
July 22, 2024 @ 4:37 pm
XEvil 6.0 automatically solve most kind of captchas,
Including such type of captchas: ReCaptcha v.2, ReCaptcha-3, Hotmail (Microsoft), Google, Solve Media, BitcoinFaucet, Steam, Amazon, Twitter, Microsoft, Twitch, Outlook, +12k
+ hCaptcha, ArkoseLabs FunCaptcha, ReCaptcha Enterprize supported in new XEvil 6.0!
1.) Fast, easy, precisionly
XEvil is the fastest captcha killer in the world. Its has no solving limits, no threads number limits
you can solve even 1.000.000.000 captchas per day and it will cost 0 (ZERO) USD! Just buy license for 59 USD and all!
2.) Several APIs support
XEvil supports more than 6 different, worldwide known API: 2Captcha, anti-captchas.com (antigate), rucaptcha.com, DeathByCaptcha, etc.
just send your captcha via HTTP request, as you can send into any of that service – and XEvil will solve your captcha!
So, XEvil is compatible with hundreds of applications for SEO/SMM/password recovery/parsing/posting/clicking/cryptocurrency/etc.
3.) Useful support and manuals
After purchase, you got access to a private tech.support forum, Wiki, Skype/Telegram online support
Developers will train XEvil to your type of captcha for FREE and very fast – just send them examples
4.) How to get free trial use of XEvil full version?
– Try to search in Google “Home of XEvil”
– you will find IPs with opened port 80 of XEvil users (click on any IP to ensure)
– try to send your captcha via 2captcha API ino one of that IPs
– if you got BAD KEY error, just tru another IP
– enjoy! 🙂
– (its not work for hCaptcha!)
WARNING: Free XEvil DEMO does NOT support ReCaptcha, hCaptcha and most other types of captcha!
http://xrumersale.site/
KeithOrara
July 22, 2024 @ 4:42 pm
Mike Tyson https://american-boxer.mike-tyson-ar.com one of the most famous and influential boxers in history, was born on June 30, 1966 in Brooklyn, New York.
Herbertwef
July 22, 2024 @ 4:44 pm
Muhammad Ali https://american-boxer.muhammad-ali-ar.com is perhaps one of the most famous and greatest athletes in the history of boxing.
Jeffreybit
July 22, 2024 @ 4:50 pm
Manny Pacquiao https://filipino-boxer.manny-pacquiao-ar.com is one of the most prominent boxers in the history of the sport.
best hotel in hormoz island
July 22, 2024 @ 4:58 pm
clean and affordable residence in hormuz island
book a residence in red beach hotel
July 22, 2024 @ 5:00 pm
very gooooooooooooooood
Diplomi_xrSi
July 22, 2024 @ 5:12 pm
[u][b] Добрый день![/b][/u]
[b]Заказать документ[/b] о получении высшего образования можно у нас в Москве.
[url=http://ast-diplomas.com/kupit-diplom-perm /]ast-diplomas.com/kupit-diplom-perm [/url]
Vazrksc
July 22, 2024 @ 5:19 pm
[u][b] Здравствуйте![/b][/u]
Заказать документ университета вы сможете у нас в столице. Мы оказываем услуги по производству и продаже документов об окончании любых ВУЗов РФ.
[url=http://tucmas.fi/phpBB/viewtopic.php?f=4&t=214822/]tucmas.fi/phpBB/viewtopic.php?f=4&t=214822[/url]
[url=http://arbitrajniki.ru/forums/topic/kupit-diplom-cena-x545i//]arbitrajniki.ru/forums/topic/kupit-diplom-cena-x545i/[/url]
[url=http://forum.combat-arnis.ru/profile.php?mode=viewprofile&u=333543/]forum.combat-arnis.ru/profile.php?mode=viewprofile&u=333543[/url]
[url=http://nailpub.ru/forum/threads/hvordan-vinne-penger-pa-spilleautomater-pa-nettet-norske-casinobonuser.41199/#post-1151573/]nailpub.ru/forum/threads/hvordan-vinne-penger-pa-spilleautomater-pa-nettet-norske-casinobonuser.41199/#post-1151573[/url]
[url=http://bestnet.ru/support/forum/index.php?PAGE_NAME=profile_view&UID=137524/]bestnet.ru/support/forum/index.php?PAGE_NAME=profile_view&UID=137524[/url]
Iariorxxf
July 22, 2024 @ 6:04 pm
[u][b] Привет![/b][/u]
Купить документ ВУЗа вы имеете возможность у нас. Мы предлагаем документы об окончании любых университетов России. Вы сможете получить диплом по любым специальностям, любого года выпуска, в том числе документы Советского Союза. Даем гарантию, что в случае проверки документов работодателем, подозрений не появится.
[url=http://nsk.ukrbb.net/viewtopic.php?f=41&t=27376/]nsk.ukrbb.net/viewtopic.php?f=41&t=27376[/url]
[url=http://bestcoolfun.ru/vyisshee-obrazovanie-dlya-vas-legalno-i-byistro/]bestcoolfun.ru/vyisshee-obrazovanie-dlya-vas-legalno-i-byistro[/url]
[url=http://www.import-moto.com/users/88/]www.import-moto.com/users/88[/url]
[url=http://u2c.tv/member.php?u=30984/]u2c.tv/member.php?u=30984[/url]
[url=http://avtolux48.ru/people/user/367/blog/7868//]avtolux48.ru/people/user/367/blog/7868/[/url]
Dnrtdjo
July 22, 2024 @ 6:23 pm
[u][b] Привет![/b][/u]
[b]Заказать документ[/b] университета вы сможете в нашей компании в Москве.
[url=http://diplomasx24.ru/kupit-diplom-moskva/]diplomasx24.ru/kupit-diplom-moskva[/url]
[b]Успешной учебы![/b]
Oariordlo
July 22, 2024 @ 6:37 pm
[u][b] Здравствуйте![/b][/u]
Приобрести диплом университета
[b]Наша компания предлагает[/b] выгодно приобрести диплом, который выполняется на бланке ГОЗНАКа и заверен печатями, водяными знаками, подписями. Данный документ пройдет любые проверки, даже с использованием специальных приборов. Достигайте свои цели максимально быстро с нашей компанией.
[b]Где заказать диплом по необходимой специальности?[/b]
[url=http://school97.ru/vesti/view_profile.php?UID=210272/]school97.ru/vesti/view_profile.php?UID=210272[/url]
[url=http://forum.hi-def.ru/index.php?showtopic=29591/]forum.hi-def.ru/index.php?showtopic=29591[/url]
[url=http://o97765bq.beget.tech/2024/07/07/assortiment-i-varianty-dostavki-v-onlayn-magazine-pokupaem-attestat.html/]o97765bq.beget.tech/2024/07/07/assortiment-i-varianty-dostavki-v-onlayn-magazine-pokupaem-attestat.html[/url]
[url=http://www.zpu-journal.ru/forum/view_profile.php?UID=308472/]www.zpu-journal.ru/forum/view_profile.php?UID=308472[/url]
[url=http://galantclub.od.ua/member.php?u=15983/]galantclub.od.ua/member.php?u=15983[/url]
Jariordzp
July 22, 2024 @ 6:44 pm
[u][b] Здравствуйте![/b][/u]
Заказать диплом о высшем образовании
[b]Наша компания предлагает[/b] быстро купить диплом, который выполняется на оригинальной бумаге и заверен мокрыми печатями, водяными знаками, подписями. Наш диплом пройдет лубую проверку, даже при использовании профессиональных приборов. Решите свои задачи быстро и просто с нашей компанией.
[b]Где заказать диплом по актуальной специальности?[/b]
[url=http://astrcolcult.ru/?post_type=topic&p=34434/]astrcolcult.ru/?post_type=topic&p=34434[/url]
[url=http://autoprajs.ru/15204.html/]autoprajs.ru/15204.html[/url]
[url=http://arsenal.listbb.ru/viewtopic.php?f=14&t=419/]arsenal.listbb.ru/viewtopic.php?f=14&t=419[/url]
[url=http://promintern.listbb.ru/viewtopic.php?f=16&t=474/]promintern.listbb.ru/viewtopic.php?f=16&t=474[/url]
[url=http://burgasdent.listbb.ru/viewtopic.php?f=3&t=355/]burgasdent.listbb.ru/viewtopic.php?f=3&t=355[/url]
MarikaJeX5499
July 22, 2024 @ 7:17 pm
XEvil 5.0 automatically solve most kind of captchas,
Including such type of captchas: ReCaptcha v.2, ReCaptcha v.3, Hotmail, Google captcha, SolveMedia, BitcoinFaucet, Steam, Amazon, Twitter, Microsoft, Twitch, Outlook, +12k
+ hCaptcha, ArkoseLabs FunCaptcha, ReCaptcha Enterprize supported in new XEvil 6.0!
1.) Fast, easy, precisionly
XEvil is the fastest captcha killer in the world. Its has no solving limits, no threads number limits
you can solve even 1.000.000.000 captchas per day and it will cost 0 (ZERO) USD! Just buy license for 59 USD and all!
2.) Several APIs support
XEvil supports more than 6 different, worldwide known API: 2Captcha, anti-captchas.com (antigate), rucaptcha.com, DeathByCaptcha, etc.
just send your captcha via HTTP request, as you can send into any of that service – and XEvil will solve your captcha!
So, XEvil is compatible with hundreds of applications for SEO/SMM/password recovery/parsing/posting/clicking/cryptocurrency/etc.
3.) Useful support and manuals
After purchase, you got access to a private tech.support forum, Wiki, Skype/Telegram online support
Developers will train XEvil to your type of captcha for FREE and very fast – just send them examples
4.) How to get free trial use of XEvil full version?
– Try to search in Google “Home of XEvil”
– you will find IPs with opened port 80 of XEvil users (click on any IP to ensure)
– try to send your captcha via 2captcha API ino one of that IPs
– if you got BAD KEY error, just tru another IP
– enjoy! 🙂
– (its not work for hCaptcha!)
WARNING: Free XEvil DEMO does NOT support ReCaptcha, hCaptcha and most other types of captcha!
http://XEvil.Net/
ThomasVer
July 22, 2024 @ 7:18 pm
http://amoxildelivery.pro/# can you buy amoxicillin over the counter
Diplomi_jhSi
July 22, 2024 @ 7:49 pm
[u][b] Добрый день![/b][/u]
[b]Купить документ[/b] ВУЗа вы имеете возможность у нас в Москве.
[url=http://asxdiplomik.com/kupit-diplom-specialista /]asxdiplomik.com/kupit-diplom-specialista [/url]
Lazreqn
July 22, 2024 @ 8:16 pm
[u][b] Привет, друзья![/b][/u]
[b]Приобрести диплом любого ВУЗа.[/b]
[url=http://igpsclub.ru/social/read-blog/5452/]igpsclub.ru/social/read-blog/5452[/url]
Xazrhdf
July 22, 2024 @ 8:42 pm
[u][b] Привет, друзья![/b][/u]
Приобрести документ университета.
[url=http://etoprosto.ru/ru/forum/?category=5&action=topic/]etoprosto.ru/ru/forum/?category=5&action=topic[/url]
[url=http://dnd.listbb.ru/viewtopic.php?f=3&t=506/]dnd.listbb.ru/viewtopic.php?f=3&t=506[/url]
[url=http://musictech.gr/forum/viewtopic.php?f=8&t=41325/]musictech.gr/forum/viewtopic.php?f=8&t=41325[/url]
[url=http://ya.webtalk.ru/viewtopic.php?id=9716#p47364/]ya.webtalk.ru/viewtopic.php?id=9716#p47364[/url]
[url=http://graph.org/Diplomy-bez-ozhidaniya-i-slozhnostej-07-17/]graph.org/Diplomy-bez-ozhidaniya-i-slozhnostej-07-17[/url]
Dnrtytz
July 22, 2024 @ 8:55 pm
[u][b] Здравствуйте![/b][/u]
[b]Заказать документ[/b] о получении высшего образования можно в нашей компании в столице.
[url=http://diplomyx24.ru/kupit-diplom-voronezh/]diplomyx24.ru/kupit-diplom-voronezh[/url]
[b]Успехов в учебе![/b]
MyronMalry
July 22, 2024 @ 10:01 pm
https://paxloviddelivery.pro/# paxlovid buy
doxycycline india price [url=https://doxycyclinedelivery.pro/#]doxyhexal[/url] doxycycline 100mg online pharmacy
Uazrzie
July 22, 2024 @ 10:05 pm
[u][b] Привет, друзья![/b][/u]
[b]Приобрести диплом о высшем образовании.[/b]
[url=http://www.import-moto.com/users/88/]www.import-moto.com/users/88[/url]
MarikaJeX8291
July 22, 2024 @ 10:10 pm
XEvil 6.0 automatically solve most kind of captchas,
Including such type of captchas: ReCaptcha v.2, ReCaptcha-3, Hotmail, Google, Solve Media, BitcoinFaucet, Steam, Amazon, Twitter, Microsoft, Twitch, Outlook, +12k
+ hCaptcha, ArkoseLabs FunCaptcha, ReCaptcha Enterprize supported in new XEvil 6.0!
1.) Fast, easy, precisionly
XEvil is the fastest captcha killer in the world. Its has no solving limits, no threads number limits
you can solve even 1.000.000.000 captchas per day and it will cost 0 (ZERO) USD! Just buy license for 59 USD and all!
2.) Several APIs support
XEvil supports more than 6 different, worldwide known API: 2captcha.com, anti-captchas.com (antigate), RuCaptcha, death-by-captcha, etc.
just send your captcha via HTTP request, as you can send into any of that service – and XEvil will solve your captcha!
So, XEvil is compatible with hundreds of applications for SEO/SMM/password recovery/parsing/posting/clicking/cryptocurrency/etc.
3.) Useful support and manuals
After purchase, you got access to a private tech.support forum, Wiki, Skype/Telegram online support
Developers will train XEvil to your type of captcha for FREE and very fast – just send them examples
4.) How to get free trial use of XEvil full version?
– Try to search in Google “Home of XEvil”
– you will find IPs with opened port 80 of XEvil users (click on any IP to ensure)
– try to send your captcha via 2captcha API ino one of that IPs
– if you got BAD KEY error, just tru another IP
– enjoy! 🙂
– (its not work for hCaptcha!)
WARNING: Free XEvil DEMO does NOT support ReCaptcha, hCaptcha and most other types of captcha!
http://xrumersale.site/
Sazrzvp
July 22, 2024 @ 10:19 pm
[u][b] Здравствуйте![/b][/u]
[b]Купить диплом о среднем полном образовании, в чем подвох и как избежать обмана? [/b]
[url=http://mirnuy.forumieren.de/t8214-topic#13486/]mirnuy.forumieren.de/t8214-topic#13486[/url]
Всегда вам поможем!
Xazrhfi
July 22, 2024 @ 11:29 pm
[u][b] Привет, друзья![/b][/u]
Приобрести документ о получении высшего образования.
[url=http://deviva.ru/viewtopic.php?id=9128#p62064/]deviva.ru/viewtopic.php?id=9128#p62064[/url]
[url=http://visogo9427.ixbb.ru/viewtopic.php?id=118#p118/]visogo9427.ixbb.ru/viewtopic.php?id=118#p118[/url]
[url=http://msfo-soft.ru/msfo/forum/messages/forum24/topic10406/message293458/?result=new#message293458/]msfo-soft.ru/msfo/forum/messages/forum24/topic10406/message293458/?result=new#message293458[/url]
[url=http://rosen.moibb.ru/viewtopic.php?f=2&t=504/]rosen.moibb.ru/viewtopic.php?f=2&t=504[/url]
[url=http://anytalkworld.com/read-blog/6932/]anytalkworld.com/read-blog/6932[/url]
ThomasVer
July 23, 2024 @ 12:11 am
http://doxycyclinedelivery.pro/# cost doxycycline tablets
book a residence in red beach hotel
July 23, 2024 @ 12:42 am
goooooooooooooood
Sazradt
July 23, 2024 @ 12:59 am
[u][b] Добрый день![/b][/u]
[b]Возможно ли купить диплом стоматолога, и как это происходит [/b]
[url=http://fremontrp.listbb.ru/viewtopic.php?f=18&t=432/]fremontrp.listbb.ru/viewtopic.php?f=18&t=432[/url]
Поможем вам всегда!
MarikaJeX1033
July 23, 2024 @ 1:09 am
XEvil 5.0 automatically solve most kind of captchas,
Including such type of captchas: ReCaptcha v.2, ReCaptcha v.3, Hotmail (Microsoft), Google captcha, SolveMedia, BitcoinFaucet, Steam, Amazon, Twitter, Microsoft, Twitch, Outlook, +12k
+ hCaptcha, ArkoseLabs FunCaptcha, ReCaptcha Enterprize supported in new XEvil 6.0!
1.) Fast, easy, precisionly
XEvil is the fastest captcha killer in the world. Its has no solving limits, no threads number limits
you can solve even 1.000.000.000 captchas per day and it will cost 0 (ZERO) USD! Just buy license for 59 USD and all!
2.) Several APIs support
XEvil supports more than 6 different, worldwide known API: 2Captcha, anti-captchas.com (antigate), rucaptcha.com, DeathByCaptcha, etc.
just send your captcha via HTTP request, as you can send into any of that service – and XEvil will solve your captcha!
So, XEvil is compatible with hundreds of applications for SEO/SMM/password recovery/parsing/posting/clicking/cryptocurrency/etc.
3.) Useful support and manuals
After purchase, you got access to a private tech.support forum, Wiki, Skype/Telegram online support
Developers will train XEvil to your type of captcha for FREE and very fast – just send them examples
4.) How to get free trial use of XEvil full version?
– Try to search in Google “Home of XEvil”
– you will find IPs with opened port 80 of XEvil users (click on any IP to ensure)
– try to send your captcha via 2captcha API ino one of that IPs
– if you got BAD KEY error, just tru another IP
– enjoy! 🙂
– (its not work for hCaptcha!)
WARNING: Free XEvil DEMO does NOT support ReCaptcha, hCaptcha and most other types of captcha!
http://xrumersale.site/
MyronMalry
July 23, 2024 @ 1:18 am
https://paxloviddelivery.pro/# Paxlovid over the counter
amoxicillin tablets in india [url=https://amoxildelivery.pro/#]amoxicillin 500 mg purchase without prescription[/url] buy amoxicillin from canada
Cazrzso
July 23, 2024 @ 1:49 am
[u][b] Привет![/b][/u]
[b]Мы можем предложить документы ВУЗов[/b], которые находятся на территории всей РФ. Можно приобрести диплом от любого учебного заведения, за любой год, в том числе документы СССР. Документы печатаются на бумаге самого высшего качества. Это позволяет делать настоящие дипломы, которые невозможно отличить от оригинала. Документы заверяются всеми необходимыми печатями и штампами.
[url=http://hvacprotalk.com/read-blog/219/]hvacprotalk.com/read-blog/219[/url]
ThomasVer
July 23, 2024 @ 3:08 am
http://amoxildelivery.pro/# buying amoxicillin in mexico
Georgemor
July 23, 2024 @ 3:11 am
The golf https://arabic.golfclub-ar.com industry in the Arab world is growing rapidly, attracting players from all over the world.
DonaldNof
July 23, 2024 @ 3:32 am
The road to the Premier League https://english-championship.premier-league-ar.com begins long before a team gets promoted to the English Premier League for the first time
DavidciP
July 23, 2024 @ 3:36 am
The Italian football championship https://italian-championship.serie-a-ar.com known as Serie A, has seen an impressive revival in recent years.
Brettrem
July 23, 2024 @ 3:40 am
In the German football https://german-championship.bundesliga-football-ar.com championship known as the Bundesliga, rivalries between clubs have always been intense.
RobertAdods
July 23, 2024 @ 3:41 am
In recent years, the leading positions in the Spanish https://spanish-championship.laliga-ar.com championship have been firmly occupied by two major giants – Barcelona and Real Madrid.
MarikaJeX3396
July 23, 2024 @ 4:04 am
XEvil 5.0 automatically solve most kind of captchas,
Including such type of captchas: ReCaptcha v.2, ReCaptcha v.3, Hotmail (Microsoft), Google, Solve Media, BitcoinFaucet, Steam, Amazon, Twitter, Microsoft, Twitch, Outlook, +12k
+ hCaptcha, ArkoseLabs FunCaptcha, ReCaptcha Enterprize supported in new XEvil 6.0!
1.) Fast, easy, precisionly
XEvil is the fastest captcha killer in the world. Its has no solving limits, no threads number limits
you can solve even 1.000.000.000 captchas per day and it will cost 0 (ZERO) USD! Just buy license for 59 USD and all!
2.) Several APIs support
XEvil supports more than 6 different, worldwide known API: 2Captcha, anti-captchas.com (antigate), RuCaptcha, death-by-captcha, etc.
just send your captcha via HTTP request, as you can send into any of that service – and XEvil will solve your captcha!
So, XEvil is compatible with hundreds of applications for SEO/SMM/password recovery/parsing/posting/clicking/cryptocurrency/etc.
3.) Useful support and manuals
After purchase, you got access to a private tech.support forum, Wiki, Skype/Telegram online support
Developers will train XEvil to your type of captcha for FREE and very fast – just send them examples
4.) How to get free trial use of XEvil full version?
– Try to search in Google “Home of XEvil”
– you will find IPs with opened port 80 of XEvil users (click on any IP to ensure)
– try to send your captcha via 2captcha API ino one of that IPs
– if you got BAD KEY error, just tru another IP
– enjoy! 🙂
– (its not work for hCaptcha!)
WARNING: Free XEvil DEMO does NOT support ReCaptcha, hCaptcha and most other types of captcha!
http://XEvil.Net/
Vazrcqc
July 23, 2024 @ 4:07 am
[u][b] Привет, друзья![/b][/u]
Заказать документ о получении высшего образования можно у нас в Москве. Мы предлагаем документы об окончании любых университетов России.
[url=http://wiki.mysupp.ru/index.php?title=gosznacdiplomy/]wiki.mysupp.ru/index.php?title=gosznacdiplomy[/url]
[url=http://mos-it.ru/index.php?subaction=userinfo&user=uciga/]mos-it.ru/index.php?subaction=userinfo&user=uciga[/url]
[url=http://j013.koreawebcenter.com/bbs/board.php?bo_table=free&wr_id=668203/]j013.koreawebcenter.com/bbs/board.php?bo_table=free&wr_id=668203[/url]
[url=http://feki-php.8u.cz/profile.php?lookup=11161/]feki-php.8u.cz/profile.php?lookup=11161[/url]
[url=http://b24activities.ru/club/forum/messages/forum4/topic76/message131/?result=reply#message131/]b24activities.ru/club/forum/messages/forum4/topic76/message131/?result=reply#message131[/url]
Treftoq
July 23, 2024 @ 4:11 am
[u][b] Привет![/b][/u]
[b]Стоимость дипломов высшего и среднего образования и процесс их получения [/b]
[url=http://service-in.ru/forum/user/6215//]service-in.ru/forum/user/6215/[/url]
[u][b] Рады помочь![u][b].
Cazrpos
July 23, 2024 @ 4:15 am
[u][b] Привет, друзья![/b][/u]
[b]Мы предлагаем документы техникумов[/b], которые находятся в любом регионе России. Можно приобрести диплом от любого учебного заведения, за любой год, указав необходимую специальность и хорошие оценки за все дисциплины. Дипломы и аттестаты выпускаются на бумаге высшего качества. Это дает возможности делать настоящие дипломы, которые не отличить от оригинала. Они будут заверены необходимыми печатями и штампами.
[url=http://www.verdoos.com/read-blog/22128/]www.verdoos.com/read-blog/22128[/url]
book a residence in red beach hotel
July 23, 2024 @ 4:20 am
goooooooooooooooooooood
clean and affordable residence in hormuz island
هزینه سفر به جزیره هرمز
July 23, 2024 @ 4:21 am
great
Jamesbug
July 23, 2024 @ 4:23 am
amoxicillin 875 125 mg tab: price for amoxicillin 875 mg – amoxicillin online no prescription
Jamesbug
July 23, 2024 @ 5:25 am
buy cipro without rx: ciprofloxacin generic price – buy cipro no rx
Mazrvkg
July 23, 2024 @ 6:16 am
[u][b] Добрый день![/b][/u]
[b]Приобретение диплома ПТУ с сокращенной программой обучения в Москве[/b]
[url=http://newfinbiz.ru/kupit-diplomyi-dlya-vseh-spetsialnostey/]newfinbiz.ru/kupit-diplomyi-dlya-vseh-spetsialnostey[/url]
Sazrmqp
July 23, 2024 @ 6:17 am
[u][b] Здравствуйте![/b][/u]
Мы готовы предложить [b]дипломы[/b] любой профессии по приятным тарифам. [b]Цена[/b] будет зависеть от выбранной специальности, года выпуска и ВУЗа. Всегда стараемся поддерживать для покупателей адекватную ценовую политику. Для нас очень важно, чтобы документы были доступными для большого количества наших граждан.
[url=http://top100bookmark.com/story17159922/купить-диплом-сантехника/]top100bookmark.com/story17159922/купить-диплом-сантехника[/url]
MarikaJeX9157
July 23, 2024 @ 6:58 am
XEvil 5.0 automatically solve most kind of captchas,
Including such type of captchas: ReCaptcha-2, ReCaptcha-3, Hotmail (Microsoft), Google captcha, SolveMedia, BitcoinFaucet, Steam, Amazon, Twitter, Microsoft, Twitch, Outlook, +12000
+ hCaptcha, ArkoseLabs FunCaptcha, ReCaptcha Enterprize supported in new XEvil 6.0!
1.) Fast, easy, precisionly
XEvil is the fastest captcha killer in the world. Its has no solving limits, no threads number limits
you can solve even 1.000.000.000 captchas per day and it will cost 0 (ZERO) USD! Just buy license for 59 USD and all!
2.) Several APIs support
XEvil supports more than 6 different, worldwide known API: 2Captcha, anti-captchas.com (antigate), RuCaptcha, death-by-captcha, etc.
just send your captcha via HTTP request, as you can send into any of that service – and XEvil will solve your captcha!
So, XEvil is compatible with hundreds of applications for SEO/SMM/password recovery/parsing/posting/clicking/cryptocurrency/etc.
3.) Useful support and manuals
After purchase, you got access to a private tech.support forum, Wiki, Skype/Telegram online support
Developers will train XEvil to your type of captcha for FREE and very fast – just send them examples
4.) How to get free trial use of XEvil full version?
– Try to search in Google “Home of XEvil”
– you will find IPs with opened port 80 of XEvil users (click on any IP to ensure)
– try to send your captcha via 2captcha API ino one of that IPs
– if you got BAD KEY error, just tru another IP
– enjoy! 🙂
– (its not work for hCaptcha!)
WARNING: Free XEvil DEMO does NOT support ReCaptcha, hCaptcha and most other types of captcha!
http://xrumersale.site/
MyronMalry
July 23, 2024 @ 7:17 am
http://doxycyclinedelivery.pro/# odering doxycycline
purchase cipro [url=https://ciprodelivery.pro/#]ciprofloxacin[/url] where can i buy cipro online
Martinbeque
July 23, 2024 @ 7:25 am
In an era when many young footballers struggle to find their place at elite clubs, Javi’s https://barcelona.gavi-ar.com story at Barcelona stands out as an exceptional one.
RobertBruig
July 23, 2024 @ 7:26 am
The Saudi Football League https://saudi-arabian-championship.saudi-pro-league-ar.com known as the Saudi Professional League, is one of the most competitive and dynamic leagues in the world.
IsaiasBug
July 23, 2024 @ 7:29 am
Rodrigo Goes https://real-madrid.rodrygo-ar.com better known as Rodrigo, is one of the brightest young talents in modern football.
Kennethtauff
July 23, 2024 @ 7:29 am
https://euroavia24.com – Cheap flights, hotels and transfers around the world!
ThomasVer
July 23, 2024 @ 7:31 am
https://clomiddelivery.pro/# can i get cheap clomid pills
Oscarsnads
July 23, 2024 @ 7:53 am
Arsenal https://arsenal.mesut-ozil-ar.com made a high-profile signing in 2013, signing star midfielder Mesut Ozil from Real Madrid.
Dnrtvdg
July 23, 2024 @ 9:08 am
[u][b] Добрый день![/b][/u]
[b]Приобрести документ[/b] института вы имеете возможность в нашей компании.
[url=http://diploms-x.com/kupit-diplom-rostov-na-donu/]diploms-x.com/kupit-diplom-rostov-na-donu[/url]
[b]Успешной учебы![/b]
Uazrpcm
July 23, 2024 @ 9:14 am
[u][b] Привет, друзья![/b][/u]
[b]Приобрести диплом любого ВУЗа.[/b]
[url=http://sontopic.com/blogs/1909/Купить-диплом-без-риска/]sontopic.com/blogs/1909/Купить-диплом-без-риска[/url]
ThomasVer
July 23, 2024 @ 10:13 am
https://ciprodelivery.pro/# cipro online no prescription in the usa
sex video download hd says:
July 23, 2024 @ 10:32 am
Later, you will will see how Bing stacks up towards Google search in a
aspect-by-aspect comparison. This text gives you
a better have a look at a few of the options found in Bing and what it offers customers.
Lazrvls
July 23, 2024 @ 10:37 am
[u][b] Здравствуйте![/b][/u]
[b]Купить диплом университета.[/b]
[url=http://logan.in.ua/people/user/7276/blog/7929//]logan.in.ua/people/user/7276/blog/7929/[/url]
MyronMalry
July 23, 2024 @ 10:39 am
http://clomiddelivery.pro/# can i get cheap clomid no prescription
paxlovid india [url=http://paxloviddelivery.pro/#]Paxlovid over the counter[/url] paxlovid for sale
Davidpaf
July 23, 2024 @ 11:09 am
Thibaut Courtois https://real-madrid.thibaut-courtois-ar.com was born on May 11, 1992 in Belgium.
Mazryss
July 23, 2024 @ 11:12 am
[u][b] Привет![/b][/u]
[b]Как купить диплом о высшем образовании с минимальными рисками[/b]
[url=http://houseofnails.store/2024/06/30/диплом-самара-купить//]houseofnails.store/2024/06/30/диплом-самара-купить/[/url]
Victorrat
July 23, 2024 @ 11:17 am
Bayern Munich’s https://bayern.jamal-musiala-ar.com young midfielder, Jamal Musiala, has become one of the brightest talents in European football.
Trefxeo
July 23, 2024 @ 11:36 am
[u][b] Здравствуйте![/b][/u]
[b]Приобретение диплома ПТУ с сокращенной программой обучения в Москве [/b]
[url=http://vk.com/prymechatel?w=wall-147947644_398/]vk.com/prymechatel?w=wall-147947644_398[/url]
[u][b] Поможем вам всегда![u][b].
www.sites.google.com new
July 23, 2024 @ 11:47 am
This contribution from the fuel engine is limited to high-velocity eventualities, as throughout regular driving, there is no
such facilitation, and the car operates solely on electric energy.
High speed was elevated on the Volt, from the electronically restricted eighty
miles per hour (130 km/h) to one hundred and one miles
per hour (160 km/h). It’s because throughout this interval,
a lot of the UK’s top festivals are in full swing.
Dnrtgcs
July 23, 2024 @ 11:53 am
[u][b] Привет, друзья![/b][/u]
[b]Купить документ[/b] о получении высшего образования можно у нас.
[url=http://diploms-x24.ru/kupit-diplom-nizhnij-novgorod/]diploms-x24.ru/kupit-diplom-nizhnij-novgorod[/url]
[b]Успешной учебы![/b]
is any video downloader free?
July 23, 2024 @ 12:03 pm
Winnie the Pooh first appeared in short tales for magazines akin to “Vanity Honest.” Pooh was drawn by a number of artists in the 1920s, but it surely was political cartoonist E.
H. Shepard who scrawled the famous Pooh drawings for the Winnie the Pooh books.
Besides Winnie the Pooh, Christopher Robin had several other stuffed animals: Eeyore, Piglet, Tigger,
Kanga, and Roo.
Diplomi_nket
July 23, 2024 @ 12:09 pm
купить дипломы вуза срочно [url=https://diploms-x.com/]diploms-x.com[/url] .
LarryMog
July 23, 2024 @ 12:19 pm
завод подъемно транспортного оборудования вертикальный грузовой подъемник
Xazrvsf
July 23, 2024 @ 12:30 pm
[u][b] Привет, друзья![/b][/u]
Купить документ о получении высшего образования.
[url=http://entrainment.listbb.ru/viewtopic.php?f=96&t=24256/]entrainment.listbb.ru/viewtopic.php?f=96&t=24256[/url]
[url=http://brawlstarsacc.listbb.ru/viewtopic.php?f=3&t=492/]brawlstarsacc.listbb.ru/viewtopic.php?f=3&t=492[/url]
[url=http://newru.getbb.ru/viewtopic.php?f=29&t=1373/]newru.getbb.ru/viewtopic.php?f=29&t=1373[/url]
[url=http://poetzinc.com/create-blog//]poetzinc.com/create-blog/[/url]
[url=http://3drus.ru/forum/topic_34885/]3drus.ru/forum/topic_34885[/url]
Sazrsje
July 23, 2024 @ 2:17 pm
[u][b] Привет, друзья![/b][/u]
[b]Как получить диплом техникума официально и без лишних проблем [/b]
[url=http://myweektour.ru/oformlenie-diploma-lyuboy-spetsialnosti/]myweektour.ru/oformlenie-diploma-lyuboy-spetsialnosti[/url]
Окажем помощь!
ThomasVer
July 23, 2024 @ 2:38 pm
https://amoxildelivery.pro/# amoxicillin without a prescription
Danrlti
July 23, 2024 @ 3:17 pm
[u][b] Привет![/b][/u]
Мы изготавливаем дипломы любых профессий.
[url=http://www.2dommedical.com/2024/07/14/купить-заочный-диплом/]www.2dommedical.com/2024/07/14/купить-заочный-диплом/[/url]
Рады помочь!.
Xazrxzv
July 23, 2024 @ 3:46 pm
[u][b] Здравствуйте![/b][/u]
Приобрести документ ВУЗа.
[url=http://dopalnenie.listbb.ru/viewtopic.php?f=3&t=717/]dopalnenie.listbb.ru/viewtopic.php?f=3&t=717[/url]
[url=http://worksale777.blogspot.com/2024/07/blog-post_80.html/]worksale777.blogspot.com/2024/07/blog-post_80.html[/url]
[url=http://ironway.ru/forum/viewtopic.php?f=5&t=23801/]ironway.ru/forum/viewtopic.php?f=5&t=23801[/url]
[url=http://rrsclub.ru/showthread.php?p=45104#post45104/]rrsclub.ru/showthread.php?p=45104#post45104[/url]
[url=http://most.maxbb.ru/viewtopic.php?f=4&t=701/]most.maxbb.ru/viewtopic.php?f=4&t=701[/url]
Diplomi_odSi
July 23, 2024 @ 3:58 pm
[u][b] Привет![/b][/u]
[b]Приобрести документ[/b] университета вы имеете возможность у нас в столице.
[url=http://diplomasx.com/kupit-diplom-perm /]diplomasx.com/kupit-diplom-perm [/url]
Frankhof
July 23, 2024 @ 4:29 pm
Luis Suarez https://inter-miami.luis-suarez-ar.com the famous Uruguayan footballer, ended his brilliant career in European clubs and decided to try his hand at a new challenge – Major League Soccer.
BrianFlago
July 23, 2024 @ 4:45 pm
Al-Nasr https://saudi.al-nassr-ar.com is one of the most famous football teams in the Kingdom of Saudi Arabia.
MyronMalry
July 23, 2024 @ 4:48 pm
http://ciprodelivery.pro/# purchase cipro
paxlovid india [url=http://paxloviddelivery.pro/#]paxlovid for sale[/url] paxlovid covid
MichaelLop
July 23, 2024 @ 4:55 pm
Al-Ittihad https://saudi.al-ittihad-ar.com is one of the most famous football clubs in Saudi Arabia. Founded in 1927, the Saudi football giant has come a long way to the pinnacle of success.
Jamesbug
July 23, 2024 @ 5:04 pm
amoxicillin 775 mg: amoxicillin where to get – amoxicillin over the counter in canada
JosephErult
July 23, 2024 @ 5:06 pm
Al-Nasr Club https://saudi.al-hilal-ar.com from Riyadh has a rich history of success, but its growth has been particularly impressive in recent years.
Uazrtei
July 23, 2024 @ 5:34 pm
[u][b] Привет, друзья![/b][/u]
Где купить диплом по нужной специальности?
[url=http://themepacific.com/support/users/lutroduyda//]themepacific.com/support/users/lutroduyda/[/url]
Jamesbug
July 23, 2024 @ 6:04 pm
doxycycline over the counter south africa: how much is doxycycline cost – buy doxycycline for dogs
ThomasVer
July 23, 2024 @ 6:07 pm
http://clomiddelivery.pro/# can you get generic clomid pills
Cazrlrq
July 23, 2024 @ 6:09 pm
[u][b] Привет![/b][/u]
[b]Мы предлагаем документы ВУЗов[/b], которые расположены в любом регионе РФ. Можно купить диплом за любой год, указав необходимую специальность и хорошие оценки за все дисциплины. Дипломы и аттестаты выпускаются на бумаге высшего качества. Это позволяет делать государственные дипломы, не отличимые от оригиналов. Документы заверяются всеми необходимыми печатями и штампами.
[url=http://olive-penguin-h97mx0.mystrikingly.com/blog/61b0aa44b55/]olive-penguin-h97mx0.mystrikingly.com/blog/61b0aa44b55[/url]
Diplomi_trSi
July 23, 2024 @ 6:32 pm
[u][b] Привет, друзья![/b][/u]
[b]Купить документ[/b] о получении высшего образования вы имеете возможность в нашей компании.
[url=http://ast-diplomas.com/kupit-diplom-chelyabinsk /]ast-diplomas.com/kupit-diplom-chelyabinsk [/url]
Keithrarce
July 23, 2024 @ 6:38 pm
FC Barcelona https://spain.fc-barcelona-ar.com is undoubtedly one of the most famous and well-known football clubs in the world.
Yrefyzv
July 23, 2024 @ 7:08 pm
[u][b] Привет![/b][/u]
[b]Приобрести диплом о высшем образовании.[/b]
[url=http://www.musichunt.pro/blogs/view.htm?id=79464/]www.musichunt.pro/blogs/view.htm?id=79464[/url]
[b]Хорошей учебы![/b]
Razrvix
July 23, 2024 @ 7:21 pm
[u][b] Привет, друзья![/b][/u]
Приобрести диплом любого университета:
[url=http://hosting.show/profile/abacoc//]hosting.show/profile/abacoc/[/url]
[url=http://school5.p-fam.ru/users/apemov/]school5.p-fam.ru/users/apemov[/url]
[url=http://clindoeilinfo.com/a-peine-arrivee-parmi-les-reseaux-sociaux-lapplication-threads-serait-dans-le-collimateur-de-la-justice/#comment-109282/]clindoeilinfo.com/a-peine-arrivee-parmi-les-reseaux-sociaux-lapplication-threads-serait-dans-le-collimateur-de-la-justice/#comment-109282[/url]
[url=http://knx-fr.com/member.php?action=profile&uid=10128/]knx-fr.com/member.php?action=profile&uid=10128[/url]
[url=http://forumjizni.ru/member.php?u=121019/]forumjizni.ru/member.php?u=121019[/url]
Lariorqqw
July 23, 2024 @ 8:09 pm
Купить диплом о среднем полном образовании, в чем подвох и как избежать обмана?
[url=http://ast-diplomy.com/kupit-diplom-moskva/]ast-diplomy.com/kupit-diplom-moskva[/url]
Lazrtju
July 23, 2024 @ 8:14 pm
[b]Заказать диплом о высшем образовании.[/b]
[url=http://forum.vorchun.ru/posting.php?mode=post&f=2/]forum.vorchun.ru/posting.php?mode=post&f=2[/url]
[url=http://gromscream.80lvl.ru/viewtopic.php?f=3&t=2070/]gromscream.80lvl.ru/viewtopic.php?f=3&t=2070[/url]
[url=http://nitrnd.com/blogs/158705/Официальные-Дипломы-Гарантия-Качества-и-Подлинности/]nitrnd.com/blogs/158705/Официальные-Дипломы-Гарантия-Качества-и-Подлинности[/url]
[url=http://superforum.spybb.ru/post.php?action=post&fid=2/]superforum.spybb.ru/post.php?action=post&fid=2[/url]
[url=http://dskrnd.ru/forum/user/6379//]dskrnd.ru/forum/user/6379/[/url]
MyronMalry
July 23, 2024 @ 8:25 pm
http://paxloviddelivery.pro/# paxlovid india
buy paxlovid online [url=https://paxloviddelivery.pro/#]paxlovid cost without insurance[/url] paxlovid generic
Lazrliz
July 23, 2024 @ 8:36 pm
[u][b] Привет![/b][/u]
[b]Мы изготавливаем дипломы[/b] любой профессии по доступным тарифам.
[url=http://angelicaleyva.es/купить-диплом-тамбов//]angelicaleyva.es/купить-диплом-тамбов/[/url]
Williamraics
July 23, 2024 @ 8:44 pm
[u][b] Привет![/b][/u]
Мы готовы предложить дипломы.
[url=http://bazhenova.greatforum.ru/posting.php?mode=post&f=17&sid=97fe575b3fcc0fa37bdc337ea855e684/]bazhenova.greatforum.ru/posting.php?mode=post&f=17&sid=97fe575b3fcc0fa37bdc337ea855e684[/url]
[url=http://wiki.wonikrobotics.com/AllegroHandWiki/index.php/Special:AWCforum/st/id15466/]wiki.wonikrobotics.com/AllegroHandWiki/index.php/Special:AWCforum/st/id15466[/url]
[url=http://blogs.rufox.ru/~worksale/48159.htm/]blogs.rufox.ru/~worksale/48159.htm[/url]
[url=http://wiki.wonikrobotics.com/AllegroHandWiki/index.php/Special:AWCforum/st/id15473/]wiki.wonikrobotics.com/AllegroHandWiki/index.php/Special:AWCforum/st/id15473[/url]
[url=http://soad.msk.ru/forum/index.php?sid=762e4b94a9d4a2c9734fb98e4b8842ae/]soad.msk.ru/forum/index.php?sid=762e4b94a9d4a2c9734fb98e4b8842ae[/url]
ThomasVer
July 23, 2024 @ 11:03 pm
https://paxloviddelivery.pro/# paxlovid india
Diplomi_lret
July 23, 2024 @ 11:25 pm
купить диплом зоотехника [url=https://diploms-x.com/]diploms-x.com[/url] .
Manrpgt
July 24, 2024 @ 12:07 am
[u][b] Привет, друзья![/b][/u]
Заказать документ ВУЗа
[url=http://ast-diplomas.com/kupit-diplom-sankt-peterburg]ast-diplomas.com/kupit-diplom-sankt-peterburg[/url]
Dnrtvmt
July 24, 2024 @ 12:11 am
[u][b] Здравствуйте![/b][/u]
[b]Приобрести документ[/b] ВУЗа можно в нашем сервисе.
[url=http://ast-diplomas24.ru/kupit-diplom-omsk/]ast-diplomas24.ru/kupit-diplom-omsk[/url]
[b]Хорошей учебы![/b]
Xazrasy
July 24, 2024 @ 12:20 am
[b]Привет, друзья![/b]
Как официально купить диплом вуза с упрощенным обучением в Москве
[url=http://arusak-diploms-srednee.ru/diplom-sredne-spetcialnoe-obrazovanie В ]arusak-diploms-srednee.ru/diplom-sredne-spetcialnoe-obrazovanie В [/url]
ThomasVer
July 24, 2024 @ 2:22 am
http://paxloviddelivery.pro/# Paxlovid buy online
MyronMalry
July 24, 2024 @ 2:28 am
https://doxycyclinedelivery.pro/# doxycycline no prescription
buy clomid without prescription [url=https://clomiddelivery.pro/#]cost of cheap clomid tablets[/url] can i buy clomid
Dnrtsgp
July 24, 2024 @ 2:39 am
[u][b] Добрый день![/b][/u]
[b]Купить документ[/b] института вы имеете возможность у нас в Москве.
[url=http://ast-diplomas.com/kupit-diplom-vracha/]ast-diplomas.com/kupit-diplom-vracha[/url]
[b]Успешной учебы![/b]
Yrefaor
July 24, 2024 @ 3:39 am
[u][b] Привет![/b][/u]
[b]Приобрести диплом любого ВУЗа.[/b]
[url=http://укбихай.рф/forum/messages/forum1/topic837/message744/?result=new#message744/]укбихай.рф/forum/messages/forum1/topic837/message744/?result=new#message744[/url]
[b]Хорошей учебы![/b]
CarrollKet
July 24, 2024 @ 3:53 am
boat hire Kotor https://boat-hire-in-montenegro.com
MarionPhace
July 24, 2024 @ 3:58 am
Montenegro Greece https://rent-a-yacht-montenegro.com
Lazrsez
July 24, 2024 @ 4:17 am
[u][b] Привет, друзья![/b][/u]
[b]Купить диплом о высшем образовании.[/b]
[url=http://igpsclub.ru/social/read-blog/5452/]igpsclub.ru/social/read-blog/5452[/url]
MichaelSmavy
July 24, 2024 @ 4:21 am
FC Bayern Munich (Munich) https://germany.bayern-munchen-ar.com is one of the most famous and recognized football clubs in Germany and Europe
Randalldyday
July 24, 2024 @ 4:24 am
Arsenal https://england.arsenal-ar.com is one of the most famous and successful football clubs in the history of English football.
TimothySep
July 24, 2024 @ 4:24 am
Real Madrid’s https://spain.real-madrid-ar.com history goes back more than a century. The club was founded in 1902 by a group of football enthusiasts led by Juan Padilla
Uazrigi
July 24, 2024 @ 5:01 am
[u][b] Добрый день![/b][/u]
Где заказать диплом специалиста?
[url=http://vitroshop.ru/index.html/]vitroshop.ru/index.html[/url]
MyronMalry
July 24, 2024 @ 5:53 am
https://paxloviddelivery.pro/# paxlovid india
п»їpaxlovid [url=https://paxloviddelivery.pro/#]paxlovid buy[/url] Paxlovid over the counter
Razrogh
July 24, 2024 @ 6:07 am
[u][b] Здравствуйте![/b][/u]
Заказать диплом о высшем образовании:
[url=http://forum.dalmatin-club.ru/topic/49757-купить-диплом-j18b//]forum.dalmatin-club.ru/topic/49757-купить-диплом-j18b/[/url]
[url=http://forum.stpku.ru/viewtopic.php?f=2&t=49716/]forum.stpku.ru/viewtopic.php?f=2&t=49716[/url]
[url=http://propusk-spb.ru/index.php?subaction=userinfo&user=isare/]propusk-spb.ru/index.php?subaction=userinfo&user=isare[/url]
[url=http://kvartirker.ru/profile/elusyzy//]kvartirker.ru/profile/elusyzy/[/url]
[url=http://акп33.рф/users/ohumiwu/]акп33.рф/users/ohumiwu[/url]
Jamesbug
July 24, 2024 @ 6:54 am
how to get generic clomid: cheap clomid without dr prescription – get cheap clomid pills
Lariorpal
July 24, 2024 @ 7:02 am
Сколько стоит диплом высшего и среднего образования и как его получить?
[url=http://diplomyx.com/kupit-diplom-ekaterinbur/]diplomyx.com/kupit-diplom-ekaterinbur[/url]
Xazrtpc
July 24, 2024 @ 7:04 am
[u][b] Привет![/b][/u]
Заказать документ о получении высшего образования.
[url=http://sz.80lvl.ru/viewtopic.php?f=9&t=1929/]sz.80lvl.ru/viewtopic.php?f=9&t=1929[/url]
[url=http://rodina.listbb.ru/viewtopic.php?f=3&t=636/]rodina.listbb.ru/viewtopic.php?f=3&t=636[/url]
[url=http://wiki.intradition.ru/index.php/Закажите_диплом_прямо_сейчас_и_начните_новую_карьеру/]wiki.intradition.ru/index.php/Закажите_диплом_прямо_сейчас_и_начните_новую_карьеру[/url]
[url=http://wo.linyway.com/read-blog/4129/]wo.linyway.com/read-blog/4129[/url]
[url=http://doctorsforum.ru/viewtopic.php?f=193&t=39110/]doctorsforum.ru/viewtopic.php?f=193&t=39110[/url]
Lazrjbe
July 24, 2024 @ 7:06 am
[b]Приобрести диплом любого университета.[/b]
[url=http://bezone.ru/node/339527/]bezone.ru/node/339527[/url]
[url=http://komfort.rusff.me/viewtopic.php?id=13894#p40941/]komfort.rusff.me/viewtopic.php?id=13894#p40941[/url]
[url=http://office.listbb.ru/posting.php?mode=post&f=2&sid=cf6c27f9202d08a5b20e3a50e4e5c0ac/]office.listbb.ru/posting.php?mode=post&f=2&sid=cf6c27f9202d08a5b20e3a50e4e5c0ac[/url]
[url=http://visacart.80lvl.ru/ucp.php?mode=login&sid=cc10ef0b7077243777c6230b38576b24/]visacart.80lvl.ru/ucp.php?mode=login&sid=cc10ef0b7077243777c6230b38576b24[/url]
[url=http://mestovstrechi.flybb.ru/viewtopic.php?f=4&t=1273/]mestovstrechi.flybb.ru/viewtopic.php?f=4&t=1273[/url]
ThomasVer
July 24, 2024 @ 7:16 am
https://amoxildelivery.pro/# where to buy amoxicillin 500mg without prescription
Iariorejy
July 24, 2024 @ 7:29 am
[u][b] Привет, друзья![/b][/u]
Заказать документ института можно в нашей компании. Мы оказываем услуги по продаже документов об окончании любых ВУЗов РФ. Вы сможете получить диплом по любым специальностям, включая документы Советского Союза. Гарантируем, что в случае проверки документов работодателями, подозрений не возникнет.
[url=http://kurkure.ru/index.php/cb-profile/pluginclass/cbblogs?action=blogs&func=show&id=148/]kurkure.ru/index.php/cb-profile/pluginclass/cbblogs?action=blogs&func=show&id=148[/url]
[url=http://nagaevo.ekafe.ru/viewtopic.php?f=86&t=2512/]nagaevo.ekafe.ru/viewtopic.php?f=86&t=2512[/url]
[url=http://verkehrsknoten.de/index.php/forum/sozialvorschriften/91107#92071/]verkehrsknoten.de/index.php/forum/sozialvorschriften/91107#92071[/url]
[url=http://aquarium.zone/read-blog/1055/]aquarium.zone/read-blog/1055[/url]
[url=http://galantclub.od.ua/member.php?u=15983/]galantclub.od.ua/member.php?u=15983[/url]
Oariorswj
July 24, 2024 @ 7:40 am
[u][b] Здравствуйте![/b][/u]
Приобрести диплом о высшем образовании
[b]Наши специалисты предлагают[/b] быстро и выгодно приобрести диплом, который выполняется на оригинальном бланке и заверен мокрыми печатями, штампами, подписями официальных лиц. Диплом способен пройти любые проверки, даже при использовании специального оборудования. Достигайте своих целей быстро и просто с нашей компанией.
[b]Где заказать диплом специалиста?[/b]
[url=http://anatoliyrud.ekafe.ru/viewforum.php?f=29/]anatoliyrud.ekafe.ru/viewforum.php?f=29[/url]
[url=http://datasphere.ru/club/user/15/blog//]datasphere.ru/club/user/15/blog/[/url]
[url=http://gti-club.ru/forum/member.php?u=23001/]gti-club.ru/forum/member.php?u=23001[/url]
[url=http://animesocial.su/blogs/3037/FAQ-и-служба-поддержка-Приобретаем-документы-в-сети-интернет/]animesocial.su/blogs/3037/FAQ-и-служба-поддержка-Приобретаем-документы-в-сети-интернет[/url]
[url=http://www.shtfsocial.com/blogs/182706/Что-готов-сейчас-предложить-интернет-магазин-с-дипломами/]www.shtfsocial.com/blogs/182706/Что-готов-сейчас-предложить-интернет-магазин-с-дипломами[/url]
Williamraics
July 24, 2024 @ 7:44 am
[u][b] Привет![/b][/u]
Мы изготавливаем дипломы.
[url=http://1776reloaded.us/blogs/48/ упить-?иплом-Ѕыстро-и-Ћегально/]1776reloaded.us/blogs/48/ упить-?иплом-Ѕыстро-и-Ћегально[/url]
[url=http://sovetushka.forum2x2.ru/login/]sovetushka.forum2x2.ru/login[/url]
[url=http://blogs.rufox.ru/~worksale/48138.htm/]blogs.rufox.ru/~worksale/48138.htm[/url]
[url=http://allukrnews.ru/nastoyashhie-diplomyi-byistro-nadezhno-legalno//]allukrnews.ru/nastoyashhie-diplomyi-byistro-nadezhno-legalno/[/url]
[url=http://3drus.ru/index.php?do=forum&act=add_topic&subaction=1/]3drus.ru/index.php?do=forum&act=add_topic&subaction=1[/url]
Stevenfes
July 24, 2024 @ 8:10 am
Thai Company Directory https://thaicorporates.com List of companies and business information.
ThomasCRURO
July 24, 2024 @ 8:22 am
печать наклеек краснодар https://salavat-rik.ru
Charlesbub
July 24, 2024 @ 8:27 am
Ремонт плоской кровли https://remontiruem-krovly.ru в Москве, цена работы за 1 м?. Прайс лист на работы под ключ, отзывы и фото.
MichaelMaw
July 24, 2024 @ 8:51 am
AC Milan https://italy.milan-ar.com is one of the most successful and decorated football clubs in the world.
Jariorisx
July 24, 2024 @ 11:19 am
[u][b] Здравствуйте![/b][/u]
Приобрести диплом любого университета
[b]Мы предлагаем[/b] быстро купить диплом, который выполнен на бланке ГОЗНАКа и заверен печатями, штампами, подписями официальных лиц. Документ пройдет лубую проверку, даже с использованием специфических приборов. Достигайте цели быстро и просто с нашими дипломами.
[b]Где купить диплом по нужной специальности?[/b]
[url=http://slliver.getbb.ru/viewtopic.php?f=62&t=826/]slliver.getbb.ru/viewtopic.php?f=62&t=826[/url]
[url=http://malispa.ru/users/122/]malispa.ru/users/122[/url]
[url=http://alik.forumrpg.ru/viewtopic.php?id=6639#p275762/]alik.forumrpg.ru/viewtopic.php?id=6639#p275762[/url]
[url=http://forum.qwas.ru/kupit-diplom-t19852.html/]forum.qwas.ru/kupit-diplom-t19852.html[/url]
[url=http://ya.2bb.ru/viewtopic.php?id=19040#p240217/]ya.2bb.ru/viewtopic.php?id=19040#p240217[/url]
Manrlzu
July 24, 2024 @ 11:32 am
[u][b] Привет![/b][/u]
Купить документ о получении высшего образования
[url=http://diplomyx.com/kupit-diplom-magistra]diplomyx.com/kupit-diplom-magistra[/url]
Xazrjov
July 24, 2024 @ 11:55 am
[b]Здравствуйте![/b]
Диплом техникума купить официально с упрощенным обучением в Москве
[url=http://arusak-diploms-srednee.ru/diplom-sredne-spetcialnoe-obrazovanie В ]arusak-diploms-srednee.ru/diplom-sredne-spetcialnoe-obrazovanie В [/url]
Jamestyday
July 24, 2024 @ 12:39 pm
Galatasaray https://turkey.galatasaray-ar.com is one of the most famous football clubs in Turkiye, with a glorious and eventful history.
MichaelDup
July 24, 2024 @ 12:42 pm
The fascinating story of Ja Morant’s https://spain.atletico-madrid-ar.com meteoric rise, from status from rookie to leader of the Memphis Grizzlies and rising NBA superstar.
Georgebub
July 24, 2024 @ 12:45 pm
In the world of football, Atletico Madrid https://spain.atletico-madrid-ar.com has long been considered the second most important club in Spain after the dominant, Real Madrid.
Cazrvrp
July 24, 2024 @ 1:42 pm
[u][b] Привет![/b][/u]
Мы готовы предложить документы ВУЗов
[url=http://argayash.flybb.ru/viewtopic.php?f=9&t=1164/]argayash.flybb.ru/viewtopic.php?f=9&t=1164[/url]
[url=http://wmrp.listbb.ru/viewtopic.php?f=51&t=692/]wmrp.listbb.ru/viewtopic.php?f=51&t=692[/url]
[url=http://kostromag.ru/forum/science/9044.aspx/]kostromag.ru/forum/science/9044.aspx[/url]
[url=http://most.maxbb.ru/viewtopic.php?f=4&t=706/]most.maxbb.ru/viewtopic.php?f=4&t=706[/url]
[url=http://modern-constructions.org/blogs/66575/дипломы-о-высшем-образовании-от-аккредитованных-вузов/]modern-constructions.org/blogs/66575/дипломы-о-высшем-образовании-от-аккредитованных-вузов[/url]
DavidSquag
July 24, 2024 @ 2:23 pm
The future football star Shabab Al-Ahly https://dubai.shabab-al-ahli-ar.com was born in Dubai in 2000. From a young age, he showed exceptional football abilities and joined the youth academy of one of the UAE’s leading clubs, Shabab Al-Ahly.
Diplomi_fmSi
July 24, 2024 @ 2:37 pm
[u][b] Привет![/b][/u]
[b]Заказать документ[/b] института вы сможете в нашей компании в Москве.
[url=http://diplomyx.com/kupit-diplom-kazan /]diplomyx.com/kupit-diplom-kazan [/url]
Sazrcbj
July 24, 2024 @ 3:27 pm
[u][b] Привет, друзья![/b][/u]
Приобрести документ о получении высшего образования
[url=http://alivechrist.com/read-blog/205/]alivechrist.com/read-blog/205[/url]
[url=http://sonnick84.tinyblogging.com/Подробное-описание-покупки-аттестата-в-известном-интернет-магазине-72055738/]sonnick84.tinyblogging.com/Подробное-описание-покупки-аттестата-в-известном-интернет-магазине-72055738[/url]
[url=http://whatson.plus/blogs/16346/С-умом-покупаем-диплом-в-сети-экспертный-обзор/]whatson.plus/blogs/16346/С-умом-покупаем-диплом-в-сети-экспертный-обзор[/url]
[url=http://samovod.ru/content/articles/65991//]samovod.ru/content/articles/65991/[/url]
[url=http://priusforum.ru/forums/index.php?autocom=gallery&req=si&img=5715/]priusforum.ru/forums/index.php?autocom=gallery&req=si&img=5715[/url]
Dnrtyue
July 24, 2024 @ 3:32 pm
[u][b] Привет, друзья![/b][/u]
[b]Приобрести документ[/b] о получении высшего образования можно в нашей компании в Москве.
[url=http://diploms-x.com/kupit-diplom-nizhnij-novgorod/]diploms-x.com/kupit-diplom-nizhnij-novgorod[/url]
[b]Удачи![/b]
Trefwsl
July 24, 2024 @ 4:08 pm
[u][b] Привет, друзья![/b][/u]
[b]Как приобрести диплом техникума с минимальными рисками [/b]
[url=http://www.pandora.ukrbb.net/viewtopic.php?f=2&t=8550/]www.pandora.ukrbb.net/viewtopic.php?f=2&t=8550[/url]
[u][b] Будем рады вам помочь![u][b].
Terrymeexy
July 24, 2024 @ 4:12 pm
купить новостройку от застройщика https://kvartiranew43.ru
TimothyLex
July 24, 2024 @ 4:14 pm
купить 1 комнатную новостройку https://kvartira-new43.ru
GregorygibiA
July 24, 2024 @ 4:19 pm
Indibet is a premier online casino offering a wide array of games including slots, table games, and live dealer options. Renowned for its user-friendly interface and robust security measures, Indibet ensures a top-notch gaming experience with exciting bonuses and 24/7 customer support.
RobertMox
July 24, 2024 @ 4:19 pm
купить 2 комнатную квартиру в новостройке https://kvartiru-kupit43.ru
Cazrysa
July 24, 2024 @ 4:32 pm
[u][b] Привет![/b][/u]
Мы можем предложить документы техникумов
[url=http://dog-ola.ru/viewtopic.php?f=28&t=7509/]dog-ola.ru/viewtopic.php?f=28&t=7509[/url]
[url=http://ulmo.ukrbb.net/index.php/]ulmo.ukrbb.net/index.php[/url]
[url=http://kai87la.flybb.ru/viewtopic.php?f=5&t=432/]kai87la.flybb.ru/viewtopic.php?f=5&t=432[/url]
[url=http://linkthere.club/read-blog/8929/]linkthere.club/read-blog/8929[/url]
[url=http://mnogozip.ru/forum/user/23882//]mnogozip.ru/forum/user/23882/[/url]
Lazrlda
July 24, 2024 @ 4:59 pm
[u][b] Привет, друзья![/b][/u]
[b]Купить диплом любого ВУЗа.[/b]
[url=http://blogs.rufox.ru/~sonnick84/48075.htm/]blogs.rufox.ru/~sonnick84/48075.htm[/url]
Diplomi_fgSi
July 24, 2024 @ 5:23 pm
[u][b] Привет, друзья![/b][/u]
[b]Купить документ[/b] о получении высшего образования вы сможете в нашей компании в Москве.
[url=http://diploms-x.com/kupit-diplom-o-vysshem-obrazovanii /]diploms-x.com/kupit-diplom-o-vysshem-obrazovanii [/url]
Iariorysu
July 24, 2024 @ 6:08 pm
[u][b] Привет![/b][/u]
Заказать документ университета можно в нашей компании. Мы оказываем услуги по продаже документов об окончании любых университетов РФ. Вы сможете получить необходимый диплом по любой специальности, любого года выпуска, включая документы старого образца. Гарантируем, что при проверке документа работодателями, подозрений не появится.
[url=http://ya.0bb.ru/viewtopic.php?id=11909#p31755/]ya.0bb.ru/viewtopic.php?id=11909#p31755[/url]
[url=http://fforum.ixbb.ru/viewtopic.php?id=186#p4064/]fforum.ixbb.ru/viewtopic.php?id=186#p4064[/url]
[url=http://driveme.rusff.me/viewtopic.php?id=2167#p105223/]driveme.rusff.me/viewtopic.php?id=2167#p105223[/url]
[url=http://followmylive.com/read-blog/969/]followmylive.com/read-blog/969[/url]
[url=http://followgrown.com/read-blog/2594/]followgrown.com/read-blog/2594[/url]
Oariortmb
July 24, 2024 @ 6:09 pm
[u][b] Привет, друзья![/b][/u]
Купить диплом университета
[b]Мы предлагаем[/b] выгодно и быстро приобрести диплом, который выполнен на оригинальном бланке и заверен мокрыми печатями, водяными знаками, подписями. Данный документ пройдет любые проверки, даже при использовании специального оборудования. Достигайте цели быстро и просто с нашими дипломами.
[b]Где приобрести диплом по нужной специальности?[/b]
[url=http://newspromworld.ru/kupit-diplom-bez-riska/]newspromworld.ru/kupit-diplom-bez-riska[/url]
[url=http://finttech.ru/garantiya-podlinnosti-pokupka-diplomov-legalno/]finttech.ru/garantiya-podlinnosti-pokupka-diplomov-legalno[/url]
[url=http://iloveyouclub.net/blogs/1893/Что-сможет-в-наше-время-предоставить-магазин-с-документами/]iloveyouclub.net/blogs/1893/Что-сможет-в-наше-время-предоставить-магазин-с-документами[/url]
[url=http://sonnick84.bloguetechno.com/Правильно-заказываем-документы-в-сети-интернет-обзор-63708705/]sonnick84.bloguetechno.com/Правильно-заказываем-документы-в-сети-интернет-обзор-63708705[/url]
[url=http://www.nasseej.net/blogs/148009/Находим-надежный-онлайн-магазин-для-приобретения-документов/]www.nasseej.net/blogs/148009/Находим-надежный-онлайн-магазин-для-приобретения-документов[/url]
Dnrtagi
July 24, 2024 @ 6:21 pm
[u][b] Здравствуйте![/b][/u]
[b]Заказать документ[/b] о получении высшего образования можно в нашей компании в столице.
[url=http://ast-diplomy.com/kupit-diplom-o-srednem-obrazovanii/]ast-diplomy.com/kupit-diplom-o-srednem-obrazovanii[/url]
[b]Удачи![/b]
Jamesbug
July 24, 2024 @ 6:29 pm
paxlovid pill: paxlovid price – paxlovid for sale
Jamesbug
July 24, 2024 @ 7:27 pm
doxycycline australia: doxycycline gel in india – doxycycline discount
Lazrylo
July 24, 2024 @ 7:30 pm
[u][b] Здравствуйте![/b][/u]
[b]Заказать диплом ВУЗа.[/b]
[url=http://medbereg.ru/club/user/15/blog/4123//]medbereg.ru/club/user/15/blog/4123/[/url]
Xazraei
July 24, 2024 @ 8:11 pm
[u][b] Добрый день![/b][/u]
Заказать документ университета.
[url=http://rosen.moibb.ru/viewtopic.php?f=2&t=505/]rosen.moibb.ru/viewtopic.php?f=2&t=505[/url]
[url=http://russiajoy.ru/realizuyte-svoi-ambitsii-s-nashim-diplomom/]russiajoy.ru/realizuyte-svoi-ambitsii-s-nashim-diplomom[/url]
[url=http://sadcr.listbb.ru/viewtopic.php?f=53&t=393/]sadcr.listbb.ru/viewtopic.php?f=53&t=393[/url]
[url=http://carolinahurricanesclub.com/create-blog/]carolinahurricanesclub.com/create-blog[/url]
[url=http://cittaviva.net/read-blog/73/]cittaviva.net/read-blog/73[/url]
Qazramg
July 24, 2024 @ 8:35 pm
[u][b] Здравствуйте![/b][/u]
[b]Приобрести диплом университета.[/b]
[url=http://pdapal.com/read-blog/880/]pdapal.com/read-blog/880[/url]
[url=http://devilz.ro/events/event/43-дипломы-по-выгодным-расценкам-в-популярном-онлайн-магазине//]devilz.ro/events/event/43-дипломы-по-выгодным-расценкам-в-популярном-онлайн-магазине/[/url]
[url=http://bakinsky-dvorik.ru/club/user/127071/blog/4743//]bakinsky-dvorik.ru/club/user/127071/blog/4743/[/url]
[url=http://meisterbook.com/read-blog/15544/]meisterbook.com/read-blog/15544[/url]
[url=http://sonnick84.blogadvize.com/34775342/Планируете-заказать-аттестат-у-проверенного-производителя-Заходите/]sonnick84.blogadvize.com/34775342/Планируете-заказать-аттестат-у-проверенного-производителя-Заходите[/url]
Vazrnkg
July 24, 2024 @ 9:24 pm
[u][b] Здравствуйте![/b][/u]
Приобрести документ ВУЗа вы можете в нашей компании в Москве. Мы предлагаем документы об окончании любых ВУЗов Российской Федерации.
[url=http://bestnet.ru/support/forum/index.php?PAGE_NAME=profile_view&UID=137524/]bestnet.ru/support/forum/index.php?PAGE_NAME=profile_view&UID=137524[/url]
[url=http://dianov.bget.ru/forum/thread57492.html#1281061/]dianov.bget.ru/forum/thread57492.html#1281061[/url]
[url=http://www.dxzw.com/member/index.php?uid=acuxusu/]www.dxzw.com/member/index.php?uid=acuxusu[/url]
[url=http://avestalaw.ru/index.php?subaction=userinfo&user=ynijyhu/]avestalaw.ru/index.php?subaction=userinfo&user=ynijyhu[/url]
[url=http://pedolog-pro.ru/forums/topic/новая-ссылка-на-мегу-2/page/501/#post-330598/]pedolog-pro.ru/forums/topic/новая-ссылка-на-мегу-2/page/501/#post-330598[/url]
Xazrtdd
July 24, 2024 @ 10:57 pm
[u][b] Здравствуйте![/b][/u]
Заказать документ университета.
[url=http://o91707v5.beget.tech/2024/07/15/diplomy-s-garantiey-oficialnosti.html/]o91707v5.beget.tech/2024/07/15/diplomy-s-garantiey-oficialnosti.html[/url]
[url=http://panther.80lvl.ru/posting.php?mode=post&f=19/]panther.80lvl.ru/posting.php?mode=post&f=19[/url]
[url=http://epistles.ru/blogs/1566/Получите-диплом-без-лишних-забот-и-усилий/]epistles.ru/blogs/1566/Получите-диплом-без-лишних-забот-и-усилий[/url]
[url=http://kurgetrp.listbb.ru/posting.php?mode=post&f=11&sid=6ca59cd4d714f92dc95dbeda5784d023/]kurgetrp.listbb.ru/posting.php?mode=post&f=11&sid=6ca59cd4d714f92dc95dbeda5784d023[/url]
[url=http://mastergrad.getbb.ru/viewtopic.php?f=12&t=2286/]mastergrad.getbb.ru/viewtopic.php?f=12&t=2286[/url]
Trefmyi
July 24, 2024 @ 11:56 pm
[u][b] Привет, друзья![/b][/u]
[b]Сколько стоит получить диплом высшего и среднего образования легально? [/b]
[url=http://autoschool-progress.kz/index.php?subaction=userinfo&user=usefizaq/]autoschool-progress.kz/index.php?subaction=userinfo&user=usefizaq[/url]
[u][b] Поможем вам всегда![u][b].
Lazrpix
July 24, 2024 @ 11:58 pm
[u][b] Добрый день![/b][/u]
[b]Мы изготавливаем дипломы[/b] любых профессий по невысоким ценам.
[url=http://pdbsoftware.com/2024/06/23/купить-диплом-в-новочеркасске//]pdbsoftware.com/2024/06/23/купить-диплом-в-новочеркасске/[/url]
Sazrxev
July 25, 2024 @ 12:32 am
[u][b] Добрый день![/b][/u]
Мы изготавливаем [b]дипломы[/b] психологов, юристов, экономистов и прочих профессий по выгодным ценам. [b]Цена[/b] зависит от определенной специальности, года получения и образовательного учреждения. Всегда стараемся поддерживать для заказчиков адекватную ценовую политику. Для нас очень важно, чтобы дипломы были доступными для подавляющей массы граждан.
[url=http://7bookmarks.com/story16996838/купить-диплом-бакалавра/]7bookmarks.com/story16996838/купить-диплом-бакалавра[/url]
Lazrzlh
July 25, 2024 @ 2:27 am
[u][b] Привет, друзья![/b][/u]
[b]Мы изготавливаем дипломы[/b] психологов, юристов, экономистов и прочих профессий по доступным ценам.
[url=http://weekofsport.ru/vash-diplom-kachestvo-i-autentichnost/]weekofsport.ru/vash-diplom-kachestvo-i-autentichnost[/url]
Jamesunsum
July 25, 2024 @ 3:40 am
купить двухкомнатную квартиру https://novye-kvartiryspb.ru
Jasoncob
July 25, 2024 @ 3:43 am
купить квартиру в новостройке цены https://kvartirukupit43.ru
ThomasCaX
July 25, 2024 @ 3:44 am
квартиры от застройщика жк квартиры от застройщика жк
Williamgat
July 25, 2024 @ 3:48 am
купить квартиру новостройке застройщика цены https://newkvartiry-spb.ru
Sazrjfa
July 25, 2024 @ 3:56 am
[u][b] Здравствуйте![/b][/u]
Приобрести документ о получении высшего образования
[url=http://twanty2.com/read-blog/181/]twanty2.com/read-blog/181[/url]
[url=http://ethiopianregister.com/read-blog/1477/]ethiopianregister.com/read-blog/1477[/url]
[url=http://antrem.ru/cb-profile/pluginclass/cbblogs?action=blogs&func=show&id=282/]antrem.ru/cb-profile/pluginclass/cbblogs?action=blogs&func=show&id=282[/url]
[url=http://tras.ru/cb-profile/pluginclass/cbblogs.html?action=blogs&func=show&id=125/]tras.ru/cb-profile/pluginclass/cbblogs.html?action=blogs&func=show&id=125[/url]
[url=http://theigbos.com/blogs/3595/Документы-по-выгодным-ценам-в-известном-онлайн-магазине/]theigbos.com/blogs/3595/Документы-по-выгодным-ценам-в-известном-онлайн-магазине[/url]
MatthewSor
July 25, 2024 @ 4:21 am
купить квартиру новостройка застройщика отделкой https://novyekvartiry2.ru
Cazrqau
July 25, 2024 @ 6:29 am
[u][b] Привет, друзья![/b][/u]
Мы предлагаем документы техникумов
[url=http://irishpal.com/read-blog/1234/]irishpal.com/read-blog/1234[/url]
[url=http://ya.webtalk.ru/viewtopic.php?id=9740#p47411/]ya.webtalk.ru/viewtopic.php?id=9740#p47411[/url]
[url=http://telesat.gtaserv.ru/posting.php?mode=post&f=251&sid=a607bfe8ff57dd5bdf90c88cfe0a81a4/]telesat.gtaserv.ru/posting.php?mode=post&f=251&sid=a607bfe8ff57dd5bdf90c88cfe0a81a4[/url]
[url=http://forumkasino.bestff.ru/viewtopic.php?id=4292#p9735/]forumkasino.bestff.ru/viewtopic.php?id=4292#p9735[/url]
[url=http://pokemon-go.onl/users/6146/]pokemon-go.onl/users/6146[/url]
Sazrvhm
July 25, 2024 @ 6:43 am
[u][b] Привет, друзья![/b][/u]
Заказать документ института
[url=http://bestchristian.com/read-blog/1877/]bestchristian.com/read-blog/1877[/url]
[url=http://sonnick84.qowap.com/87843716/Проверенный-магазин-с-широким-каталогом-дипломов/]sonnick84.qowap.com/87843716/Проверенный-магазин-с-широким-каталогом-дипломов[/url]
[url=http://kopoluzhona.image-perth.org/zakazyvaem-dokumenty-v-nadeznom-onlajn-magazine-sovety-eksperta/]kopoluzhona.image-perth.org/zakazyvaem-dokumenty-v-nadeznom-onlajn-magazine-sovety-eksperta[/url]
[url=http://forum.enchald.com/files/file/9-вы-хотите-приобрести-диплом-переходите-в-наш-магазин//]forum.enchald.com/files/file/9-вы-хотите-приобрести-диплом-переходите-в-наш-магазин/[/url]
[url=http://zdravamir.ru/index.php?option=com_myblog&show=faq-n-o–on-n-n-n-.html&Itemid=/]zdravamir.ru/index.php?option=com_myblog&show=faq-n-o–on-n-n-n-.html&Itemid=[/url]
Dnrtfdx
July 25, 2024 @ 6:51 am
[u][b] Здравствуйте![/b][/u]
[b]Заказать документ[/b] ВУЗа вы можете у нас в Москве.
[url=http://ast-diplomy24.ru/kupit-diplom-rostov-na-donu/]ast-diplomy24.ru/kupit-diplom-rostov-na-donu[/url]
[b]Успехов в учебе![/b]
肩こり 辛い
July 25, 2024 @ 7:19 am
I visited many web pages however the audio quality for audio songs present
at this site is in fact fabulous.
あまちにゃん
July 25, 2024 @ 7:20 am
Hi there this is kinda of off topic but I was wanting to know if blogs use WYSIWYG editors or if you have
to manually code with HTML. I’m starting a blog soon but have no coding know-how so I wanted to
get advice from someone with experience. Any help would
be enormously appreciated!
AllenCom
July 25, 2024 @ 7:33 am
квартира в новостройке от застройщика недвижимость новостройки купить
Danielthush
July 25, 2024 @ 7:35 am
недвижимость купить квартиру купить квартиру недорого
WilliamMapse
July 25, 2024 @ 7:39 am
купить двухкомнатную квартиру в новостройке https://kvartiranovostroi.ru
Vazrkcs
July 25, 2024 @ 8:27 am
[u][b] Добрый день![/b][/u]
Приобрести документ университета можно у нас в Москве. Мы оказываем услуги по продаже документов об окончании любых университетов РФ.
[url=http://rf-isolation.ru/index.php?/topic/736-предложения/#comment-4548/]rf-isolation.ru/index.php?/topic/736-предложения/#comment-4548[/url]
[url=http://forum.teens4greece.com/main3/viewtopic.php?f=31&t=2&p=1709#p1709/]forum.teens4greece.com/main3/viewtopic.php?f=31&t=2&p=1709#p1709[/url]
[url=http://d.mwoff.org/index.php?/topic/50-купить-диплом-i590z//]d.mwoff.org/index.php?/topic/50-купить-диплом-i590z/[/url]
[url=http://certificate.winko.net/bbs/board.php?bo_table=free&wr_id=292870/]certificate.winko.net/bbs/board.php?bo_table=free&wr_id=292870[/url]
[url=http://fxprimer.ru/index.php?topic=432785.new#new/]fxprimer.ru/index.php?topic=432785.new#new[/url]
TommieRerve
July 25, 2024 @ 9:12 am
купить новую квартиру застройщик https://kvartira-novostroi.ru
RaymondSmozy
July 25, 2024 @ 9:24 am
купить однокомнатную квартиру в новостройке купить квартиру новостройка застройщика отделкой
Cazrecc
July 25, 2024 @ 9:24 am
[u][b] Привет, друзья![/b][/u]
Мы можем предложить документы техникумов
[url=http://mireait.listbb.ru/viewtopic.php?f=2&t=488/]mireait.listbb.ru/viewtopic.php?f=2&t=488[/url]
[url=http://boltushka.flybb.ru/viewtopic.php?f=14&t=1537&sid=b469566d37804aae103e8065e31b81c9/]boltushka.flybb.ru/viewtopic.php?f=14&t=1537&sid=b469566d37804aae103e8065e31b81c9[/url]
[url=http://vbocharov-and-friends.ru/viewtopic.php?f=21&t=3933&sid=cb38fdf353995f974789fb422fd54f5f/]vbocharov-and-friends.ru/viewtopic.php?f=21&t=3933&sid=cb38fdf353995f974789fb422fd54f5f[/url]
[url=http://domovou.3nx.ru/viewtopic.php?p=7050#7050/]domovou.3nx.ru/viewtopic.php?p=7050#7050[/url]
[url=http://mymotospeed.ru/dostupnoe-oformlenie-diplomov/]mymotospeed.ru/dostupnoe-oformlenie-diplomov[/url]
Dnrtsjr
July 25, 2024 @ 9:35 am
[u][b] Привет, друзья![/b][/u]
[b]Заказать документ[/b] ВУЗа можно в нашей компании в Москве.
[url=http://ast-diplom24.ru/kupit-diplom-voronezh/]ast-diplom24.ru/kupit-diplom-voronezh[/url]
[b]Успешной учебы![/b]
Sazryan
July 25, 2024 @ 11:36 am
[u][b] Здравствуйте![/b][/u]
Мы изготавливаем [b]дипломы[/b] любой профессии по невысоким тарифам. [b]Цена[/b] может зависеть от выбранной специальности, года выпуска и университета. Всегда стараемся поддерживать для покупателей адекватную ценовую политику. Важно, чтобы документы были доступны для подавляющей массы наших граждан.
[url=http://bookmarktiger.com/story17073205/купить-диплом-Саратов/]bookmarktiger.com/story17073205/купить-диплом-Саратов[/url]
ThomasMAICT
July 25, 2024 @ 11:44 am
Помощь в решении задач https://zadachireshaem-online.ru. Опытные авторы с профессиональной подготовкой окажут консультацию в решении задач на заказ недорого, быстро, качественно
RogerWot
July 25, 2024 @ 11:45 am
купить 1 квартиру в новостройке https://kvartira-novostroyka2.ru
Johnnygraky
July 25, 2024 @ 11:49 am
Заказать контрольную работу https://kontrolnye-reshim.ru, недорого, цены. Решение контрольных работ на заказ срочно.
Xazrjba
July 25, 2024 @ 12:13 pm
[u][b] Привет![/b][/u]
Заказать документ о получении высшего образования.
[url=http://x91392sl.beget.tech/2024/07/13/diplom-lyuboy-specialnosti-za-korotkiy-srok.html/]x91392sl.beget.tech/2024/07/13/diplom-lyuboy-specialnosti-za-korotkiy-srok.html[/url]
[url=http://newsofmebel.ru/vash-karernyiy-rost-nachinaetsya-s-diploma-poluchite-ego-byistro/]newsofmebel.ru/vash-karernyiy-rost-nachinaetsya-s-diploma-poluchite-ego-byistro[/url]
[url=http://iqtorg.ru/forum/messages/forum2/topic630/message704/?result=new#message704/]iqtorg.ru/forum/messages/forum2/topic630/message704/?result=new#message704[/url]
[url=http://polegasm.net/index.php/forum/welcome-mat/151613/]polegasm.net/index.php/forum/welcome-mat/151613[/url]
[url=http://politictoday.ru/byistraya-dostavka-diplomov/]politictoday.ru/byistraya-dostavka-diplomov[/url]
DavidPreof
July 25, 2024 @ 1:58 pm
Заказать дипломную работу https://diplomzakazat-oline.ru недорого. Дипломные работы на заказ с гарантией.
CharlesDef
July 25, 2024 @ 2:04 pm
Заказать курсовую работу https://kursovye-napishem.ru в Москве: цены на написание и выполнение, недорого
骨格 矯正 保険 適用
July 25, 2024 @ 2:17 pm
J.P. Roth, The logistics of Roman military at battle
(264 B.C. Ayrault Dodge Theodore, Hannibal: A History of the Art of Warfare
Among the many Carthaginians and Romans All the way down to the Battle of Pydna, 168 BC.
Diplomi_fcSi
July 25, 2024 @ 3:10 pm
[u][b] Добрый день![/b][/u]
[b]Приобрести документ[/b] института можно у нас.
[url=http://ast-diplomy.com/kupit-diplom-s-registraciej /]ast-diplomy.com/kupit-diplom-s-registraciej [/url]
ArthurEsoft
July 25, 2024 @ 3:33 pm
Accessibility Team Meeting Notes https://make.wordpress.org/accessibility/2021/06/11/accessibility-team-meeting-notes-june-11-2021
Richardvah
July 25, 2024 @ 3:50 pm
Красивая музыка https://melodia.space для души слушать онлайн.
Lazrlhj
July 25, 2024 @ 3:54 pm
[u][b] Привет, друзья![/b][/u]
[b]Заказать диплом любого ВУЗа.[/b]
[url=http://itach-soft.com/club/user/6/blog/999//]itach-soft.com/club/user/6/blog/999/[/url]
Xazrgby
July 25, 2024 @ 4:23 pm
[u][b] Добрый день![/b][/u]
Заказать документ ВУЗа.
[url=http://vv.flybb.ru/viewtopic.php?f=23&t=2251/]vv.flybb.ru/viewtopic.php?f=23&t=2251[/url]
[url=http://worksale.jofo.me/2279463.html?_ga=2.212027927.2025584804.1721122678-746589305.1721122678/]worksale.jofo.me/2279463.html?_ga=2.212027927.2025584804.1721122678-746589305.1721122678[/url]
[url=http://graph.org/Poluchite-diplomy-za-korotkij-srok-07-16/]graph.org/Poluchite-diplomy-za-korotkij-srok-07-16[/url]
[url=http://gazeta.ekafe.ru/posting.php?mode=reply&f=48&t=32117&sid=ef3a9d82983d6c6bfd12c1492bcce01a/]gazeta.ekafe.ru/posting.php?mode=reply&f=48&t=32117&sid=ef3a9d82983d6c6bfd12c1492bcce01a[/url]
[url=http://o91746bp.beget.tech/content/add/topic/]o91746bp.beget.tech/content/add/topic[/url]
Mazrnly
July 25, 2024 @ 5:00 pm
[u][b] Привет![/b][/u]
[b]Можно ли быстро купить диплом старого образца и в чем подвох?[/b]
[url=http://newsbig.raidersfanteamshop.com/poleznye-sovety-po-priobreteniu-diploma-v-internete//]newsbig.raidersfanteamshop.com/poleznye-sovety-po-priobreteniu-diploma-v-internete/[/url]
Diplomi_yxSi
July 25, 2024 @ 5:52 pm
[u][b] Здравствуйте![/b][/u]
[b]Купить документ[/b] ВУЗа вы можете в нашей компании в столице.
[url=http://ast-diplomas.com/kupit-diplom-kandidata-nauk /]ast-diplomas.com/kupit-diplom-kandidata-nauk [/url]
StephenFrith
July 25, 2024 @ 6:01 pm
Помощь студентам в выполнении рефератов https://referatkupit-oline.ru. Низкие цены и быстрое написание рефератов!
Danielsnoni
July 25, 2024 @ 6:05 pm
создание сайтов seo https://seoraskrutka43.ru
Stevendox
July 25, 2024 @ 6:11 pm
продвижение сайта в поисковых системах https://seo-raskrutka43.ru
Lazripv
July 25, 2024 @ 6:57 pm
[u][b] Привет, друзья![/b][/u]
[b]Мы предлагаем дипломы[/b] любой профессии по приятным тарифам.
[url=http://blackmastercomic.com/blog/2024/06/22/диплом-в-три-клика//]blackmastercomic.com/blog/2024/06/22/диплом-в-три-клика/[/url]
Cazrzsl
July 25, 2024 @ 7:00 pm
[u][b] Здравствуйте![/b][/u]
Купить диплом академии
[url=http://feelosum.com/blogs/1859/дипломы-без-лишних-вопросов/]feelosum.com/blogs/1859/дипломы-без-лишних-вопросов[/url]
[url=http://wmrp.listbb.ru/viewtopic.php?f=51&t=692/]wmrp.listbb.ru/viewtopic.php?f=51&t=692[/url]
[url=http://moskovskij.getbb.ru/viewtopic.php?f=12&t=3092/]moskovskij.getbb.ru/viewtopic.php?f=12&t=3092[/url]
[url=http://human.forumieren.de/t317-topic#387/]human.forumieren.de/t317-topic#387[/url]
[url=http://oren.kabb.ru/viewtopic.php?f=50&t=34246/]oren.kabb.ru/viewtopic.php?f=50&t=34246[/url]
Sazrxwl
July 25, 2024 @ 8:36 pm
[u][b] Здравствуйте![/b][/u]
Приобрести документ университета
[url=http://terios2.ru/forums/index.php?autocom=gallery&req=si&img=2983/]terios2.ru/forums/index.php?autocom=gallery&req=si&img=2983[/url]
[url=http://80.87.194.225/blogs/1/otlichnye-tseny-i-vysokoe-kachestvo-pokupay-dokumenty-onlayn.php/]80.87.194.225/blogs/1/otlichnye-tseny-i-vysokoe-kachestvo-pokupay-dokumenty-onlayn.php[/url]
[url=http://chipokids.ru/company/personal/user/4275/forum/message/1660/1719//]chipokids.ru/company/personal/user/4275/forum/message/1660/1719/[/url]
[url=http://telegra.ph/Kakie-onlajn-magaziny-s-dokumentami-est-na-segodnyashnij-moment-07-07/]telegra.ph/Kakie-onlajn-magaziny-s-dokumentami-est-na-segodnyashnij-moment-07-07[/url]
[url=http://balacixynu.trexgame.net/glavnye-zatraty-proizvoditela-diplomov-avtorskij-obzor/]balacixynu.trexgame.net/glavnye-zatraty-proizvoditela-diplomov-avtorskij-obzor[/url]
Mazrttc
July 25, 2024 @ 9:43 pm
[u][b] Привет![/b][/u]
[b]Как получить диплом техникума с упрощенным обучением в Москве официально[/b]
[url=http://wegug.in/blogs/752/ упите-?ипломы-Ѕыстро-и-Ќадежно/]wegug.in/blogs/752/ упите-?ипломы-Ѕыстро-и-Ќадежно[/url]
Sazriyh
July 25, 2024 @ 11:18 pm
[u][b] Добрый день![/b][/u]
Заказать документ о получении высшего образования
[url=http://chillibell.com/read-blog/35/]chillibell.com/read-blog/35[/url]
[url=http://dixbe.com/blogs/242/Подробное-описание-приобретения-диплома-в-онлайн-магазине/]dixbe.com/blogs/242/Подробное-описание-приобретения-диплома-в-онлайн-магазине[/url]
[url=http://jointsnmotion-hub.mn.co/posts/61679297/]jointsnmotion-hub.mn.co/posts/61679297[/url]
[url=http://rusere.ru/communication/forum/messages/forum11/message3684/2971-chetyre-glavnykh-gruppy-onlayn_magazinov_-chto-realizuyut-dokumenty?result=new/]rusere.ru/communication/forum/messages/forum11/message3684/2971-chetyre-glavnykh-gruppy-onlayn_magazinov_-chto-realizuyut-dokumenty?result=new[/url]
[url=http://payhip.com/BestNews/blog/news/kak-najti-nadezhnyj-internet-magazin-kotoryj-realizuet-dokumenty/]payhip.com/BestNews/blog/news/kak-najti-nadezhnyj-internet-magazin-kotoryj-realizuet-dokumenty[/url]
Dnrtcot
July 25, 2024 @ 11:32 pm
[u][b] Добрый день![/b][/u]
[b]Заказать документ[/b] о получении высшего образования вы сможете у нас.
[url=http://asxdiplomik24.ru/kupit-diplom-moskva/]asxdiplomik24.ru/kupit-diplom-moskva[/url]
[b]Успехов в учебе![/b]
Cazrnsi
July 26, 2024 @ 12:56 am
[u][b] Добрый день![/b][/u]
Мы можем предложить документы техникумов
[url=http://crewties.moibb.ru/viewtopic.php?f=2&t=434/]crewties.moibb.ru/viewtopic.php?f=2&t=434[/url]
[url=http://etoprosto.ru/ru/forum/?category=5&action=topic/]etoprosto.ru/ru/forum/?category=5&action=topic[/url]
[url=http://oren.kabb.ru/viewtopic.php?f=50&t=34348/]oren.kabb.ru/viewtopic.php?f=50&t=34348[/url]
[url=http://lubov.listbb.ru/viewtopic.php?f=2&t=498/]lubov.listbb.ru/viewtopic.php?f=2&t=498[/url]
[url=http://jeepgarage.ru/forum/topic.php?forum=24&topic=771/]jeepgarage.ru/forum/topic.php?forum=24&topic=771[/url]
Uazrhug
July 26, 2024 @ 1:29 am
[u][b] Привет![/b][/u]
Где приобрести диплом специалиста?
[url=http://www.leenkup.com/read-blog/16408/]www.leenkup.com/read-blog/16408[/url]
Dnrtuib
July 26, 2024 @ 2:13 am
[u][b] Добрый день![/b][/u]
[b]Приобрести документ[/b] о получении высшего образования можно в нашем сервисе.
[url=http://ast-diplomy24.ru/kupit-diplom-krasnoyarsk/]ast-diplomy24.ru/kupit-diplom-krasnoyarsk[/url]
[b]Успехов в учебе![/b]
Williamraics
July 26, 2024 @ 3:26 am
[u][b] Добрый день![/b][/u]
Мы изготавливаем дипломы.
[url=http://rpinnovative.1stbb.ru/posting.php?mode=post&f=8&sid=1c15344b9efd67a14a72d7f76e5dbf36/]rpinnovative.1stbb.ru/posting.php?mode=post&f=8&sid=1c15344b9efd67a14a72d7f76e5dbf36[/url]
[url=http://yexanin202.ixbb.ru/viewtopic.php?id=109#p109/]yexanin202.ixbb.ru/viewtopic.php?id=109#p109[/url]
[url=http://pokemon-go.onl/users/6146/]pokemon-go.onl/users/6146[/url]
[url=http://freedomrp.getbb.ru/viewtopic.php?f=110&t=789/]freedomrp.getbb.ru/viewtopic.php?f=110&t=789[/url]
[url=http://s-s-o.ru/forum.php?PAGE_NAME=profile_view&UID=52661/]s-s-o.ru/forum.php?PAGE_NAME=profile_view&UID=52661[/url]
Cazrbug
July 26, 2024 @ 4:13 am
[u][b] Здравствуйте![/b][/u]
Мы можем предложить документы техникумов
[url=http://mir.4admins.ru/viewtopic.php?f=10&t=13047/]mir.4admins.ru/viewtopic.php?f=10&t=13047[/url]
[url=http://academy4elements.mybb.ru/viewtopic.php?id=963#p2097/]academy4elements.mybb.ru/viewtopic.php?id=963#p2097[/url]
[url=http://kvartal8b.getbb.ru/viewtopic.php?f=19&t=21990/]kvartal8b.getbb.ru/viewtopic.php?f=19&t=21990[/url]
[url=http://nachosking.com/read-blog/1131/]nachosking.com/read-blog/1131[/url]
[url=http://gazeta.ekafe.ru/viewforum.php?f=48/]gazeta.ekafe.ru/viewforum.php?f=48[/url]
WilliamHehon
July 26, 2024 @ 4:15 am
seo продвижение корпоративных сайтов в казани раскрутка сайта цена казань
Brandonpen
July 26, 2024 @ 4:16 am
купить продвижение сайта https://seo-optimizatsia.ru
Jamesskego
July 26, 2024 @ 4:28 am
Останні новини України https://gromrady.org.ua сьогодні онлайн – головні події світу
HarryPrign
July 26, 2024 @ 4:29 am
Новинний ресурс https://actualnews.kyiv.ua про всі важливі події в Україні та світі.
RobertCaurf
July 26, 2024 @ 4:32 am
Новини сьогодні https://gau.org.ua останні новини України та світу онлайн
Sazrrxc
July 26, 2024 @ 4:51 am
[u][b] Привет, друзья![/b][/u]
Диплом специалиста
[url=http://pika.listbb.ru/posting.php?mode=post&f=9/]pika.listbb.ru/posting.php?mode=post&f=9[/url]
[url=http://elfae.ruhelp.com/viewtopic.php?id=17289#p39567/]elfae.ruhelp.com/viewtopic.php?id=17289#p39567[/url]
[url=http://nsibirsk.ru/forum/obyavleniya/newtopic/#google_vignette/]nsibirsk.ru/forum/obyavleniya/newtopic/#google_vignette[/url]
[url=http://industrial.getbb.ru/viewtopic.php?f=4&t=3107/]industrial.getbb.ru/viewtopic.php?f=4&t=3107[/url]
[url=http://mf.getbb.ru/viewtopic.php?f=28&t=664/]mf.getbb.ru/viewtopic.php?f=28&t=664[/url]
Cazrmyu
July 26, 2024 @ 4:52 am
[u][b] Здравствуйте![/b][/u]
Получить диплом академии
[url=http://webnewsrealty.ru/dostupnyie-diplomyi-vseh-urovney/]webnewsrealty.ru/dostupnyie-diplomyi-vseh-urovney[/url]
[url=http://menaharia.com/create-blog//]menaharia.com/create-blog/[/url]
[url=http://allnewstroy.ru/poluchite-ofitsialnyiy-diplom-byistro-i-legko/]allnewstroy.ru/poluchite-ofitsialnyiy-diplom-byistro-i-legko[/url]
[url=http://boltushka.flybb.ru/viewtopic.php?f=14&t=1537&sid=b469566d37804aae103e8065e31b81c9/]boltushka.flybb.ru/viewtopic.php?f=14&t=1537&sid=b469566d37804aae103e8065e31b81c9[/url]
[url=http://worldavtonew.ru/oformlenie-diplomov-bez-ozhidaniya/]worldavtonew.ru/oformlenie-diplomov-bez-ozhidaniya[/url]
chastnoeporno.top
July 26, 2024 @ 5:51 am
A round of applause for your article. Much thanks again.
My website: anilingus.tv
Xazrdro
July 26, 2024 @ 6:32 am
[u][b] Привет, друзья![/b][/u]
Купить документ о получении высшего образования.
[url=http://boltushka.flybb.ru/viewtopic.php?f=14&t=1517/]boltushka.flybb.ru/viewtopic.php?f=14&t=1517[/url]
[url=http://mockwanasvyazi.getbb.ru/viewtopic.php?f=12&t=1531/]mockwanasvyazi.getbb.ru/viewtopic.php?f=12&t=1531[/url]
[url=http://socialnetwork.cloudyzx.com/read-blog/13987/]socialnetwork.cloudyzx.com/read-blog/13987[/url]
[url=http://eurodelo.ru/poluchite-diplom-o-kotorom-vsegda-mechtali/]eurodelo.ru/poluchite-diplom-o-kotorom-vsegda-mechtali[/url]
[url=http://mastergrad.getbb.ru/viewtopic.php?f=12&t=2371/]mastergrad.getbb.ru/viewtopic.php?f=12&t=2371[/url]
Lazrsri
July 26, 2024 @ 7:50 am
[u][b] Добрый день![/b][/u]
[b]Мы предлагаем дипломы[/b] психологов, юристов, экономистов и любых других профессий по невысоким тарифам.
[url=http://tailblog.com/wp/2024/06/30/купить-диплом-в-анапе//]tailblog.com/wp/2024/06/30/купить-диплом-в-анапе/[/url]
EddieVor
July 26, 2024 @ 7:55 am
Новини України https://kiev-online.com.ua останні події в Україні та світі сьогодні, новини України за минулий день онлайн
StephenApose
July 26, 2024 @ 7:55 am
Популярные репортажи https://infotolium.com в больших фотографиях, новости, события в мире
Kennethmut
July 26, 2024 @ 7:57 am
Україна свіжі новини https://kiev-pravda.kiev.ua останні події на сьогодні
DavidZit
July 26, 2024 @ 8:28 am
Свіжі новини України https://lenta.kyiv.ua останні новини з-за кордону, новини політики, економіки, спорту, культури.
Manrcat
July 26, 2024 @ 8:39 am
[u][b] Здравствуйте![/b][/u]
Заказать документ университета
[url=http://ast-diplomy.com/kupit-diplom-moskva]ast-diplomy.com/kupit-diplom-moskva[/url]
Xazrfcq
July 26, 2024 @ 8:54 am
[b]Здравствуйте![/b]
Полезные советы по безопасной покупке диплома о высшем образовании
[url=http://arusak-diploms-srednee.ru/kupit-diplom-magistra В ]arusak-diploms-srednee.ru/kupit-diplom-magistra В [/url]
JoshuaElith
July 26, 2024 @ 9:24 am
Україна останні новини https://lentanews.kyiv.ua головні новини та останні події
Xazrxms
July 26, 2024 @ 9:27 am
[u][b] Привет![/b][/u]
Купить документ ВУЗа.
[url=http://munchkin.flybb.ru/viewtopic.php?f=7&t=4189/]munchkin.flybb.ru/viewtopic.php?f=7&t=4189[/url]
[url=http://kinopuk.ru/viewtopic.php?id=9085#p56666/]kinopuk.ru/viewtopic.php?id=9085#p56666[/url]
[url=http://newsbizlife.ru/prostoe-i-byistroe-oformlenie-diploma/]newsbizlife.ru/prostoe-i-byistroe-oformlenie-diploma[/url]
[url=http://minimoo.eu/index.php/en/forum/welcome-mat/666566/]minimoo.eu/index.php/en/forum/welcome-mat/666566[/url]
[url=http://art-gymnastics.ru/users/54/]art-gymnastics.ru/users/54[/url]
Lazraek
July 26, 2024 @ 10:34 am
[u][b] Привет, друзья![/b][/u]
[b]Мы можем предложить дипломы[/b] любой профессии по невысоким ценам.
[url=http://houseofnails.store/2024/06/30/диплом-самара-купить//]houseofnails.store/2024/06/30/диплом-самара-купить/[/url]
Iariorhmv
July 26, 2024 @ 11:23 am
[u][b] Привет![/b][/u]
Купить документ о получении высшего образования вы имеете возможность у нас в столице. Мы предлагаем документы об окончании любых ВУЗов Российской Федерации. Вы получите диплом по любым специальностям, любого года выпуска, включая документы Советского Союза. Гарантируем, что в случае проверки документов работодателями, никаких подозрений не появится.
[url=http://horordark.ru/kak-poluchit-diplom-byistro-i-bez-problem/]horordark.ru/kak-poluchit-diplom-byistro-i-bez-problem[/url]
[url=http://hanaverse.top/blogs/918/Основные-расходы-современного-производителя-документов-обзор/]hanaverse.top/blogs/918/Основные-расходы-современного-производителя-документов-обзор[/url]
[url=http://nwkqnvz5.ixbb.ru/viewtopic.php?id=107#p107/]nwkqnvz5.ixbb.ru/viewtopic.php?id=107#p107[/url]
[url=http://familyportal.forumrom.com/viewtopic.php?id=23177#p60214/]familyportal.forumrom.com/viewtopic.php?id=23177#p60214[/url]
[url=http://farm86.com/blogs/324/Как-приобрести-недорого-качественный-диплом-в-сети/]farm86.com/blogs/324/Как-приобрести-недорого-качественный-диплом-в-сети[/url]
Razrsql
July 26, 2024 @ 11:32 am
[u][b] Привет![/b][/u]
Приобрести диплом любого университета:
[url=http://nis.com.tn/index.php?option=com_kunena&view=topic&catid=2&id=6776&Itemid=151/]nis.com.tn/index.php?option=com_kunena&view=topic&catid=2&id=6776&Itemid=151[/url]
[url=http://forum.stpku.ru/viewtopic.php?f=2&t=49711/]forum.stpku.ru/viewtopic.php?f=2&t=49711[/url]
[url=http://roblox-studio-scripts.ru/topic/19607-kupit-diplom-m493f//]roblox-studio-scripts.ru/topic/19607-kupit-diplom-m493f/[/url]
[url=http://cmsthemefinder.com/userinfo.php?uid=35792#/]cmsthemefinder.com/userinfo.php?uid=35792#[/url]
[url=http://nailpub.ru/forum/threads/norske-mobil-casino-eller-norske-betting-sider.41827/#post-1152463/]nailpub.ru/forum/threads/norske-mobil-casino-eller-norske-betting-sider.41827/#post-1152463[/url]
Richardjew
July 26, 2024 @ 12:36 pm
Головні новини https://pto-kyiv.com.ua України та світу
JeromeTut
July 26, 2024 @ 12:50 pm
Головні новини https://mediashare.com.ua про регіон України. Будьте в курсі останніх новин
DerekGaups
July 26, 2024 @ 12:50 pm
Новини та аналітика https://newsportal.kyiv.ua ситуація в Україні.
Shawnevelo
July 26, 2024 @ 12:53 pm
Новини України https://sensus.org.ua та світу сьогодні. Головні та останні новини дня
Sazrxxq
July 26, 2024 @ 1:02 pm
[u][b] Привет![/b][/u]
Заказать документ университета
[url=http://yobasket.es/es/perfil-usuario/pluginclass/cbblogs.html?action=blogs&func=show&id=120/]yobasket.es/es/perfil-usuario/pluginclass/cbblogs.html?action=blogs&func=show&id=120[/url]
[url=http://pora-valit.com/wiki/index.php?title=В_случае_если_потребовались_клапана,_задвижки,_манометры_-_заходите!/]pora-valit.com/wiki/index.php?title=В_случае_если_потребовались_клапана,_задвижки,_манометры_-_заходите![/url]
[url=http://friend24.in/blogs/2843/Ищем-надежный-и-проверенный-интернет-магазин-с-дипломами/]friend24.in/blogs/2843/Ищем-надежный-и-проверенный-интернет-магазин-с-дипломами[/url]
[url=http://sonnick84.blogstival.com/50719489/Советы-профессионала-которые-помогут-быстро-заказать-диплом/]sonnick84.blogstival.com/50719489/Советы-профессионала-которые-помогут-быстро-заказать-диплом[/url]
[url=http://brand-the-change.mn.co/posts/61638953/]brand-the-change.mn.co/posts/61638953[/url]
HenryNerge
July 26, 2024 @ 1:09 pm
Новини, останні події https://prp.org.ua в Україні та світі, новини політики, бізнесу та економіки, законодавства
Lazrjwb
July 26, 2024 @ 1:45 pm
[b]Купить диплом любого ВУЗа.[/b]
[url=http://inteam.maxbb.ru/viewtopic.php?f=1&t=1534&sid=85a2e5d3a3c5b95c7c1b54d218b274c4/]inteam.maxbb.ru/viewtopic.php?f=1&t=1534&sid=85a2e5d3a3c5b95c7c1b54d218b274c4[/url]
[url=http://ladymystery.ru/forum/topic.asp?TOPIC_ID=9490/]ladymystery.ru/forum/topic.asp?TOPIC_ID=9490[/url]
[url=http://galantclub.od.ua/member.php?u=15983/]galantclub.od.ua/member.php?u=15983[/url]
[url=http://rulezrp.listbb.ru/viewtopic.php?f=2&t=599/]rulezrp.listbb.ru/viewtopic.php?f=2&t=599[/url]
[url=http://vetreriameliante.it/index.php/kforum/jm-slideshow-module/9813-img-https#9813/]vetreriameliante.it/index.php/kforum/jm-slideshow-module/9813-img-https#9813[/url]
Diplomi_ffSi
July 26, 2024 @ 2:21 pm
[u][b] Привет![/b][/u]
[b]Приобрести документ[/b] о получении высшего образования можно в нашей компании.
[url=http://diplomasx.com/kupit-diplom-krasnodar /]diplomasx.com/kupit-diplom-krasnodar [/url]
Qazrkqs
July 26, 2024 @ 2:41 pm
[u][b] Привет, друзья![/b][/u]
[b]Приобрести диплом ВУЗа.[/b]
[url=http://socialmoby.com/read-blog/379/]socialmoby.com/read-blog/379[/url]
[url=http://cbr.by/club/user/1977/blog/326//]cbr.by/club/user/1977/blog/326/[/url]
[url=http://huarenjiaohui.com/index.php?/gallery/image/122-вы-хотите-приобрести-диплом-заходите-в-наш-онлайн-магазин//]huarenjiaohui.com/index.php?/gallery/image/122-вы-хотите-приобрести-диплом-заходите-в-наш-онлайн-магазин/[/url]
[url=http://forumblogger.my.id/blogs/1916/Как-найти-проверенный-магазин-что-продает-документы/]forumblogger.my.id/blogs/1916/Как-найти-проверенный-магазин-что-продает-документы[/url]
[url=http://igpsclub.ru/social/read-blog/5450/]igpsclub.ru/social/read-blog/5450[/url]
Williamraics
July 26, 2024 @ 2:45 pm
[u][b] Привет, друзья![/b][/u]
Мы готовы предложить дипломы.
[url=http://forummsk.getbb.ru/viewtopic.php?f=12&t=2582/]forummsk.getbb.ru/viewtopic.php?f=12&t=2582[/url]
[url=http://most.maxbb.ru/posting.php?mode=post&f=4/]most.maxbb.ru/posting.php?mode=post&f=4[/url]
[url=http://israelafrica.mn.co/posts/61678496/]israelafrica.mn.co/posts/61678496[/url]
[url=http://vv.flybb.ru/viewtopic.php?f=23&t=2232/]vv.flybb.ru/viewtopic.php?f=23&t=2232[/url]
[url=http://3drus.ru/index.php?do=forum&act=add_topic&subaction=1/]3drus.ru/index.php?do=forum&act=add_topic&subaction=1[/url]
Dnrtvdc
July 26, 2024 @ 3:27 pm
[u][b] Здравствуйте![/b][/u]
[b]Заказать документ[/b] о получении высшего образования вы сможете у нас.
[url=http://ast-diplom.com/kupit-diplom-rostov-na-donu/]ast-diplom.com/kupit-diplom-rostov-na-donu[/url]
[b]Успешной учебы![/b]
Kevinwhiff
July 26, 2024 @ 3:58 pm
Корисні та цікаві статті https://sevsovet.com.ua про здоров’я, дозвілля, кар’єру.
Sazrnui
July 26, 2024 @ 4:15 pm
[u][b] Добрый день![/b][/u]
Приобрести документ о получении высшего образования
[url=http://affiliatemetro.com/read-blog/819/]affiliatemetro.com/read-blog/819[/url]
[url=http://ayema.ng/blogs/33240/Детальное-описание-покупки-аттестата-в-известном-интернет-магазине/]ayema.ng/blogs/33240/Детальное-описание-покупки-аттестата-в-известном-интернет-магазине[/url]
[url=http://sonnick84.aioblogs.com/81933752/Что-потребуется-из-техники-для-создания-диплома/]sonnick84.aioblogs.com/81933752/Что-потребуется-из-техники-для-создания-диплома[/url]
[url=http://homes-for-homeless-children.mn.co/posts/61748147/]homes-for-homeless-children.mn.co/posts/61748147[/url]
[url=http://gta-balkan.com/events/event/41-вы-хотите-купить-диплом-переходите-в-наш-онлайн-магазин//]gta-balkan.com/events/event/41-вы-хотите-купить-диплом-переходите-в-наш-онлайн-магазин/[/url]
DavidPew
July 26, 2024 @ 4:17 pm
Головні новини https://status.net.ua сьогодні, найсвіжіші та останні новини України онлайн
TerryZes
July 26, 2024 @ 4:21 pm
Останні новини https://thingshistory.com зовнішньої та внутрішньої політики в країні та світі.
Danielpap
July 26, 2024 @ 4:39 pm
Останні новини світу https://uamc.com.ua про Україну від порталу новин Ukraine Today
Diplomi_xgSi
July 26, 2024 @ 5:02 pm
[u][b] Здравствуйте![/b][/u]
[b]Приобрести документ[/b] о получении высшего образования вы сможете в нашей компании.
[url=http://diplomyx.com/kupit-diplom-krasnodar /]diplomyx.com/kupit-diplom-krasnodar [/url]
Yrefzxp
July 26, 2024 @ 5:10 pm
[u][b] Здравствуйте![/b][/u]
[b]Заказать диплом о высшем образовании.[/b]
[url=http://clubalameda.com/index.php/cb-profile/pluginclass/cbblogs?action=blogs&func=show&id=121//]clubalameda.com/index.php/cb-profile/pluginclass/cbblogs?action=blogs&func=show&id=121/[/url]
[url=http://traveltravelforum.com/showthread.php/32859-Купить-диплом-Ваш-путь-к-успеху?p=67847/]traveltravelforum.com/showthread.php/32859-Купить-диплом-Ваш-путь-к-успеху?p=67847[/url]
[url=http://fidelize.site/blogs/95/Что-понадобится-для-создания-неотличимого-от-оригинала-диплома/]fidelize.site/blogs/95/Что-понадобится-для-создания-неотличимого-от-оригинала-диплома[/url]
[url=http://mykinotime.ru/page/4//]mykinotime.ru/page/4/[/url]
[url=http://malaysiasteelinstitute.com/купить-диплом//]malaysiasteelinstitute.com/купить-диплом/[/url]
Dnrttew
July 26, 2024 @ 6:13 pm
[u][b] Привет![/b][/u]
[b]Заказать документ[/b] университета можно в нашей компании в столице.
[url=http://ast-diplomas24.ru/kupit-diplom-rostov-na-donu/]ast-diplomas24.ru/kupit-diplom-rostov-na-donu[/url]
[b]Успешной учебы![/b]
Trefjxg
July 26, 2024 @ 6:56 pm
[u][b] Добрый день![/b][/u]
Как избежать рисков при покупке диплома колледжа или ПТУ в России
[url=http://nigerianelections2019.com/купить-диплом-проверенные-способы//]nigerianelections2019.com/купить-диплом-проверенные-способы/[/url]
Рады оказаться полезными!.
Lariorbul
July 26, 2024 @ 7:43 pm
Покупка диплома о среднем полном образовании: как избежать мошенничества?
[url=http://bharatpage.com/read-blog/587/]bharatpage.com/read-blog/587[/url]
[url=http://forum.aigames.su/index.php?/gallery/image/183-благодаря-чему-онлайн-магазины-с-дипломами-сегодня-так-популярны//]forum.aigames.su/index.php?/gallery/image/183-благодаря-чему-онлайн-магазины-с-дипломами-сегодня-так-популярны/[/url]
[url=http://freedomrp.getbb.ru/viewtopic.php?f=110&t=793/]freedomrp.getbb.ru/viewtopic.php?f=110&t=793[/url]
[url=http://exoltech.us/blogs/207844/Официальный-диплом-без-учебы-и-экзаменов/]exoltech.us/blogs/207844/Официальный-диплом-без-учебы-и-экзаменов[/url]
[url=http://msfo-soft.ru/msfo/forum/messages/forum24/topic10394/message292502/?result=new#message292502/]msfo-soft.ru/msfo/forum/messages/forum24/topic10394/message292502/?result=new#message292502[/url]
Manrhjh
July 26, 2024 @ 8:00 pm
[u][b] Здравствуйте![/b][/u]
Купить документ о получении высшего образования
[url=http://ast-diploms24.ru/kupit-diplom-nizhnij-novgorod]ast-diploms24.ru/kupit-diplom-nizhnij-novgorod[/url]
Xazrjaj
July 26, 2024 @ 8:22 pm
[b]Здравствуйте![/b]
Сколько стоит диплом высшего и среднего образования и как его получить?
[url=http://arusak-diploms-srednee.ru/kupit-diplom-v-nizhnem-novgorodeВ ]arusak-diploms-srednee.ru/kupit-diplom-v-nizhnem-novgorodeВ [/url]
Cazrfyx
July 26, 2024 @ 9:18 pm
[u][b] Привет, друзья![/b][/u]
Мы предлагаем документы ВУЗов
[url=http://abs70.ru/member.php?u=1918/]abs70.ru/member.php?u=1918[/url]
[url=http://paladiny.ru/forummess.dwar.php?TopicID=27683/]paladiny.ru/forummess.dwar.php?TopicID=27683[/url]
[url=http://robotaforum.2bb.ru/post.php?fid=40/]robotaforum.2bb.ru/post.php?fid=40[/url]
[url=http://impressiverp.listbb.ru/viewtopic.php?f=16&t=965/]impressiverp.listbb.ru/viewtopic.php?f=16&t=965[/url]
[url=http://cardinalparkmld.listbb.ru/viewtopic.php?f=3&t=486/]cardinalparkmld.listbb.ru/viewtopic.php?f=3&t=486[/url]
Mazrpwo
July 26, 2024 @ 9:49 pm
[u][b] Привет![/b][/u]
[b]Как официально приобрести аттестат 11 класса с минимальными затратами времени[/b]
[url=http://lucsa.id/Купить-Проверенный-Диплом//]lucsa.id/Купить-Проверенный-Диплом/[/url]
Xazrtfr
July 26, 2024 @ 10:53 pm
[u][b] Здравствуйте![/b][/u]
Заказать документ о получении высшего образования.
[url=http://visacart.80lvl.ru/viewtopic.php?f=1&t=783/]visacart.80lvl.ru/viewtopic.php?f=1&t=783[/url]
[url=http://poznanie.gtaserv.ru/memberlist.php/]poznanie.gtaserv.ru/memberlist.php[/url]
[url=http://forums.worldsamba.org/showthread.php?tid=1048928/]forums.worldsamba.org/showthread.php?tid=1048928[/url]
[url=http://heroes.app/blogs/428782/Ваш-путь-к-успеху-начинается-с-диплома/]heroes.app/blogs/428782/Ваш-путь-к-успеху-начинается-с-диплома[/url]
[url=http://veneraroleplay.listbb.ru/posting.php?mode=post&f=4/]veneraroleplay.listbb.ru/posting.php?mode=post&f=4[/url]
Cazrnbi
July 27, 2024 @ 12:02 am
[u][b] Добрый день![/b][/u]
Мы можем предложить документы ВУЗов
[url=http://jeepgarage.ru/forum/topic.php?forum=24&topic=780/]jeepgarage.ru/forum/topic.php?forum=24&topic=780[/url]
[url=http://sllife.ru/social/user/4216/forum/message/2443/7155/#message7155/]sllife.ru/social/user/4216/forum/message/2443/7155/#message7155[/url]
[url=http://gamemotors.ru/diplomyi-s-garantiey-ofitsialnosti/]gamemotors.ru/diplomyi-s-garantiey-ofitsialnosti[/url]
[url=http://machineintelligence.mn.co/members/26295417/]machineintelligence.mn.co/members/26295417[/url]
[url=http://korrespondentweek.ru/vash-diplom-vash-uspeh//]korrespondentweek.ru/vash-diplom-vash-uspeh/[/url]
Lazrgtl
July 27, 2024 @ 1:43 am
[u][b] Добрый день![/b][/u]
[b]Мы готовы предложить дипломы[/b] любой профессии по доступным ценам.
[url=http://learn4fun.vn/купить-диплом-игитм-или-любого-вуза-ро//]learn4fun.vn/купить-диплом-игитм-или-любого-вуза-ро/[/url]
Vazrglt
July 27, 2024 @ 3:19 am
[u][b] Привет![/b][/u]
Заказать документ ВУЗа вы можете у нас в Москве. Мы оказываем услуги по производству и продаже документов об окончании любых университетов Российской Федерации.
[url=http://rf-unicron.ru/index.php?/topic/446-купить-диплом-a881v//]rf-unicron.ru/index.php?/topic/446-купить-диплом-a881v/[/url]
[url=http://mtpkrskstate.ru/forum/user/199846//]mtpkrskstate.ru/forum/user/199846/[/url]
[url=http://”www.myvibor.ru/forum/index.php?action=post;topic=2803.0;last_msg=110269″/]”www.myvibor.ru/forum/index.php?action=post;topic=2803.0;last_msg=110269″[/url]
[url=http://ssd-astra.ru/forum/viewtopic.php?f=14&t=7009/]ssd-astra.ru/forum/viewtopic.php?f=14&t=7009[/url]
[url=http://zakupki.motilek.ru/index.php?name=account&op=info&uname=iromeni/]zakupki.motilek.ru/index.php?name=account&op=info&uname=iromeni[/url]
Robertlense
July 27, 2024 @ 3:51 am
can i get clomid pill: where can i get cheap clomid without dr prescription – how can i get clomid
Robertlense
July 27, 2024 @ 4:23 am
amoxicillin 500 mg capsule: how to get amoxicillin – amoxicillin generic brand
Sazrrsq
July 27, 2024 @ 5:03 am
[u][b] Привет![/b][/u]
Заказать документ о получении высшего образования
[url=http://myteana.ru/forums/index.php?autocom=gallery&req=si&img=5106/]myteana.ru/forums/index.php?autocom=gallery&req=si&img=5106[/url]
[url=http://ou812chat.com/read-blog/1168/]ou812chat.com/read-blog/1168[/url]
[url=http://saopaulofansclub.com/read-blog/2718/]saopaulofansclub.com/read-blog/2718[/url]
[url=http://talkiachat.com/blogs/27/Легко-приобретаем-документы-в-онлайн-магазине-Russian-Diplom/]talkiachat.com/blogs/27/Легко-приобретаем-документы-в-онлайн-магазине-Russian-Diplom[/url]
[url=http://igpsclub.ru/social/read-blog/5461/]igpsclub.ru/social/read-blog/5461[/url]
Trefpju
July 27, 2024 @ 6:09 am
[u][b] Привет, друзья![/b][/u]
Купить диплом о среднем полном образовании, в чем подвох и как избежать обмана?
[url=http://tanhashop.com/category/uncategorized/page/2//]tanhashop.com/category/uncategorized/page/2/[/url]
Рады помочь!.
Jariordwc
July 27, 2024 @ 6:22 am
[u][b] Добрый день![/b][/u]
Заказать диплом ВУЗа
[url=http://roving.club/read-blog/1453/]roving.club/read-blog/1453[/url]
[url=http://vsetutonline.com/forum/member.php?u=76509/]vsetutonline.com/forum/member.php?u=76509[/url]
[url=http://ya.6bb.ru/viewtopic.php?id=3201#p5134/]ya.6bb.ru/viewtopic.php?id=3201#p5134[/url]
[url=http://fauna.0pk.me/viewtopic.php?id=1336#p6250/]fauna.0pk.me/viewtopic.php?id=1336#p6250[/url]
[url=http://myanimalgram.com/read-blog/352/]myanimalgram.com/read-blog/352[/url]
Larioryhq
July 27, 2024 @ 6:34 am
Как оказалось, купить диплом кандидата наук не так уж и сложно
[url=http://usa.life/read-blog/63422/]usa.life/read-blog/63422[/url]
[url=http://olive-penguin-h97mx0.mystrikingly.com/blog/dec09af525d/]olive-penguin-h97mx0.mystrikingly.com/blog/dec09af525d[/url]
[url=http://rabotavinternete.forum2x2.ru/t55636-topic#138412/]rabotavinternete.forum2x2.ru/t55636-topic#138412[/url]
[url=http://gerosland.com/read-blog/165/]gerosland.com/read-blog/165[/url]
[url=http://forums.unigild.com/index.php?/topic/15237-диплом-в-кратчайшие-сроки-ваш-шаг-к-новой-жизни//]forums.unigild.com/index.php?/topic/15237-диплом-в-кратчайшие-сроки-ваш-шаг-к-новой-жизни/[/url]
Craighot
July 27, 2024 @ 7:09 am
Mixing Reinvented ChipMixer For Your Privacy
Melvinker
July 27, 2024 @ 7:20 am
Газоблоки https://gasoblok.ru являются самым популярным материалом для строительства домов из газобетона, они стали эффективной, надежной и недорогой технологией. Качественные газобетонные блоки производятся промышленным способом и обрабатываются в специальных автоклавах.
Arthurwhort
July 27, 2024 @ 7:25 am
Строительство заборов из металлического штакетника под ключ в Санкт-Петербурге https://trudolubov.com/product/zabory-pod-klyuch/zabor-metallicheskiy-shtaketnik/. Цены на сайте.
RandyEloni
July 27, 2024 @ 8:04 am
car rental Montenegro tripadvisor rental cars Montenegro
Dnrtcyk
July 27, 2024 @ 8:34 am
[u][b] Добрый день![/b][/u]
[b]Приобрести документ[/b] университета вы можете у нас в Москве.
[url=http://ast-diplomy.com/otzyvy/]ast-diplomy.com/otzyvy[/url]
[b]Успехов в учебе![/b]
Dnrtptp
July 27, 2024 @ 11:57 am
[u][b] Привет, друзья![/b][/u]
[b]Заказать документ[/b] университета можно у нас.
[url=http://diplomasx24.ru/kupit-diplom-magistra/]diplomasx24.ru/kupit-diplom-magistra[/url]
[b]Успехов в учебе![/b]
Uazryco
July 27, 2024 @ 12:12 pm
[u][b] Привет![/b][/u]
[b]Купить диплом о высшем образовании.[/b]
[url=http://student-news.ru/kupit-diplom-doveryayte-professionalam/]student-news.ru/kupit-diplom-doveryayte-professionalam[/url]
岡山では何が有名ですか?
July 27, 2024 @ 12:52 pm
The PS3 makes use of the Blu-ray format, whereas the Xbox 360 uses a typical DVD9.
In April 2011, SCEA once more requested that the case be
dismissed and made claims that the plaintiffs refiled declare was inadequate and that they
had been hackers and wanted to violate Sony’s intellectual property and asked the judge to grant search rights on their PS3 methods.
MichaelNob
July 27, 2024 @ 1:52 pm
Мазь для суставов https://sustalits.ru заказать онлайн.
Vazrxtc
July 27, 2024 @ 2:17 pm
[u][b] Здравствуйте![/b][/u]
Приобрести документ университета вы сможете в нашей компании в столице. Мы оказываем услуги по продаже документов об окончании любых ВУЗов РФ.
[url=http://otpusk.ru/user/index.php?backurl=/forum/topic/add/forum2//]otpusk.ru/user/index.php?backurl=/forum/topic/add/forum2/[/url]
[url=http://artem-energo.ru/forums.php?m=posts&q=18303&n=last#bottom/]artem-energo.ru/forums.php?m=posts&q=18303&n=last#bottom[/url]
[url=http://master39.net/forum/messages/forum1/topic42/message28722/?result=reply#message28722/]master39.net/forum/messages/forum1/topic42/message28722/?result=reply#message28722[/url]
[url=http://www.fromrus.su/index.php?/topic/13511-купить-диплом-цена-n116y//]www.fromrus.su/index.php?/topic/13511-купить-диплом-цена-n116y/[/url]
[url=http://forum.computest.ru/post/678927/#p678927/]forum.computest.ru/post/678927/#p678927[/url]
Lazrfss
July 27, 2024 @ 2:28 pm
[u][b] Добрый день![/b][/u]
[b]Мы изготавливаем дипломы[/b] психологов, юристов, экономистов и прочих профессий по приятным ценам.
[url=http://russiajoy.ru/oformlenie-diploma-onlayn-za-neskolko-shagov/]russiajoy.ru/oformlenie-diploma-onlayn-za-neskolko-shagov[/url]
Cazruld
July 27, 2024 @ 2:36 pm
[u][b] Привет![/b][/u]
Мы можем предложить документы техникумов
[url=http://moskvic.actieforum.com/t3750-topic#7042/]moskvic.actieforum.com/t3750-topic#7042[/url]
[url=http://draiv.flybb.ru/viewtopic.php?f=2&t=757/]draiv.flybb.ru/viewtopic.php?f=2&t=757[/url]
[url=http://russiajoy.ru/kupite-diplom-po-vyigodnoy-tsene/]russiajoy.ru/kupite-diplom-po-vyigodnoy-tsene[/url]
[url=http://everyday.flyboard.ru/viewtopic.php?f=6&t=2171/]everyday.flyboard.ru/viewtopic.php?f=6&t=2171[/url]
[url=http://rakisochi.ru/users/20/]rakisochi.ru/users/20[/url]
Diplomi_puSi
July 27, 2024 @ 2:55 pm
[u][b] Добрый день![/b][/u]
[b]Приобрести документ[/b] университета можно у нас в Москве.
[url=http://asxdiplomik.com/kupit-diplom-bakalavra-ili-specialista /]asxdiplomik.com/kupit-diplom-bakalavra-ili-specialista [/url]
Lazrfbs
July 27, 2024 @ 3:12 pm
[u][b] Добрый день![/b][/u]
[b]Приобрести диплом любого университета.[/b]
[url=http://mediamemorial.ru/club/user/69594/forum/message/3756/13238//]mediamemorial.ru/club/user/69594/forum/message/3756/13238/[/url]
Xazreey
July 27, 2024 @ 3:16 pm
[u][b] Привет![/b][/u]
Купить документ о получении высшего образования.
[url=http://newsofmebel.ru/vash-karernyiy-rost-nachinaetsya-s-diploma-poluchite-ego-byistro/]newsofmebel.ru/vash-karernyiy-rost-nachinaetsya-s-diploma-poluchite-ego-byistro[/url]
[url=http://recept-food.ru/diplomyi-priznannyie-rabotodatelyami/]recept-food.ru/diplomyi-priznannyie-rabotodatelyami[/url]
[url=http://woman.build2.ru/viewtopic.php?id=14575#p44702/]woman.build2.ru/viewtopic.php?id=14575#p44702[/url]
[url=http://subscribe.ru/member/quick/]subscribe.ru/member/quick[/url]
[url=http://bestcoolfun.ru/poluchite-diplom-v-kratchayshie-sroki/]bestcoolfun.ru/poluchite-diplom-v-kratchayshie-sroki[/url]
Jimmiesap
July 27, 2024 @ 4:47 pm
Автомобільний портал https://autodream.com.ua новини та огляди новинок авторинку.
JasonPiefe
July 27, 2024 @ 4:57 pm
Новини автомобільного ринку https://autoguide.kyiv.ua та автопромисловості
Anthonysar
July 27, 2024 @ 4:58 pm
Автомобільний портал https://allauto.kyiv.ua який захоплює своїми тест-драйвами та новими новинами автосвіту
Cazrylu
July 27, 2024 @ 5:26 pm
[u][b] Привет, друзья![/b][/u]
Мы готовы предложить документы ВУЗов
[url=http://ulmo.ukrbb.net/index.php/]ulmo.ukrbb.net/index.php[/url]
[url=http://forumkasino.bestff.ru/viewtopic.php?id=4292#p9735/]forumkasino.bestff.ru/viewtopic.php?id=4292#p9735[/url]
[url=http://myfootballday.ru/diplomyi-s-dostavkoy-na-dom/]myfootballday.ru/diplomyi-s-dostavkoy-na-dom[/url]
[url=http://human.forumieren.de/t316-topic#386/]human.forumieren.de/t316-topic#386[/url]
[url=http://dachaweek.ru/diplomyi-dlya-vseh-spetsialnostey/]dachaweek.ru/diplomyi-dlya-vseh-spetsialnostey[/url]
Xazrqsp
July 27, 2024 @ 6:00 pm
[u][b] Привет![/b][/u]
Заказать документ университета.
[url=http://rosen.moibb.ru/viewtopic.php?f=2&t=505/]rosen.moibb.ru/viewtopic.php?f=2&t=505[/url]
[url=http://nitrnd.com/blogs/162079/Дипломы-для-успешной-карьеры/]nitrnd.com/blogs/162079/Дипломы-для-успешной-карьеры[/url]
[url=http://true.pahom.su/content/add/topic/]true.pahom.su/content/add/topic[/url]
[url=http://miku-crdb.ru/forum/messages/forum1/topic348/message377/?result=new#message377/]miku-crdb.ru/forum/messages/forum1/topic348/message377/?result=new#message377[/url]
[url=http://mirfinrealty.ru/byistroe-poluchenie-diploma-nadezhno-i-konfidentsialno/]mirfinrealty.ru/byistroe-poluchenie-diploma-nadezhno-i-konfidentsialno[/url]
Diplomi_dzSi
July 27, 2024 @ 6:10 pm
[u][b] Привет![/b][/u]
[b]Купить документ[/b] института вы можете у нас в столице.
[url=http://diplomasx.com/kupit-diplom-bakalavra-ili-specialista /]diplomasx.com/kupit-diplom-bakalavra-ili-specialista [/url]
JosephSuema
July 27, 2024 @ 6:19 pm
best company to rent a car in Montenegro Montenegro car rental
Sazrrzt
July 27, 2024 @ 9:08 pm
[u][b] Привет, друзья![/b][/u]
Диплом по специальности на выбор
[url=http://newsofgames.ru/obrazovanie-mechtyi-diplomyi-vedushhih-vuzov/]newsofgames.ru/obrazovanie-mechtyi-diplomyi-vedushhih-vuzov[/url]
[url=http://pnevmokzn.80lvl.ru/search.php?search_id=egosearch/]pnevmokzn.80lvl.ru/search.php?search_id=egosearch[/url]
[url=http://kvitka.ukrbb.net/viewtopic.php?f=58&t=23712/]kvitka.ukrbb.net/viewtopic.php?f=58&t=23712[/url]
[url=http://ya.5bb.ru/viewtopic.php?id=20002#p60473/]ya.5bb.ru/viewtopic.php?id=20002#p60473[/url]
[url=http://wiki.why42.ru/wiki/дипломы_ведущих_вузов_для_успешного_будущего/]wiki.why42.ru/wiki/дипломы_ведущих_вузов_для_успешного_будущего[/url]
Cazrmdn
July 27, 2024 @ 9:10 pm
[u][b] Привет, друзья![/b][/u]
Приобрести диплом о высшем образовании
[url=http://torica.4adm.ru/viewtopic.php?f=6&t=2228/]torica.4adm.ru/viewtopic.php?f=6&t=2228[/url]
[url=http://mamapoltava.listbb.ru/viewtopic.php?f=39&t=5438/]mamapoltava.listbb.ru/viewtopic.php?f=39&t=5438[/url]
[url=http://forum-ahangaran2007.flybb.ru/viewtopic.php?f=5&t=727/]forum-ahangaran2007.flybb.ru/viewtopic.php?f=5&t=727[/url]
[url=http://crewties.moibb.ru/viewtopic.php?f=2&t=433/]crewties.moibb.ru/viewtopic.php?f=2&t=433[/url]
[url=http://borovsk-tract.maxbb.ru/posting.php?mode=post&f=1/]borovsk-tract.maxbb.ru/posting.php?mode=post&f=1[/url]
Sazriko
July 27, 2024 @ 11:02 pm
[u][b] Привет, друзья![/b][/u]
Заказать документ института
[url=http://fighter71.com/read-blog/1020/]fighter71.com/read-blog/1020[/url]
[url=http://network-91053.mn.co/posts/61837969/]network-91053.mn.co/posts/61837969[/url]
[url=http://imforma.it/cb-profile/pluginclass/cbblogs.html?action=blogs&func=show&id=121/]imforma.it/cb-profile/pluginclass/cbblogs.html?action=blogs&func=show&id=121[/url]
[url=http://cms-blog-news.de/modules.php?name=Journal&file=display&jid=209/]cms-blog-news.de/modules.php?name=Journal&file=display&jid=209[/url]
[url=http://baetezhylefy.timeforchangecounselling.com/glavnye-zatraty-izgotovitela-dokumentov-avtorskij-obzor/]baetezhylefy.timeforchangecounselling.com/glavnye-zatraty-izgotovitela-dokumentov-avtorskij-obzor[/url]
Uazrcck
July 27, 2024 @ 11:25 pm
[u][b] Здравствуйте![/b][/u]
[b]Заказать диплом университета.[/b]
[url=http://rucollectshoeclub.com/read-blog/1839/]rucollectshoeclub.com/read-blog/1839[/url]
Dnrtkvk
July 28, 2024 @ 2:44 am
[u][b] Добрый день![/b][/u]
[b]Приобрести документ[/b] о получении высшего образования можно в нашем сервисе.
[url=http://ast-diplomy24.ru/kupit-diplom-voronezh/]ast-diplomy24.ru/kupit-diplom-voronezh[/url]
[b]Успехов в учебе![/b]
Lazrjmr
July 28, 2024 @ 3:55 am
[u][b] Здравствуйте![/b][/u]
[b]Заказать диплом любого ВУЗа.[/b]
[url=http://midoma.ru/node/shirokiy-vybor-dokumentov-v-izvestnom-interne/]midoma.ru/node/shirokiy-vybor-dokumentov-v-izvestnom-interne[/url]
ジャパンマテリアルの業種は?
July 28, 2024 @ 4:59 am
I do not even know how I ended up here, but I thought this post was great.
I do not know who you are but definitely you are going
to a famous blogger if you are not already 😉 Cheers!
Razrfxn
July 28, 2024 @ 5:03 am
[u][b] Привет![/b][/u]
Заказать диплом о высшем образовании:
[url=http://legislador.art.br/forum/viewtopic.php?p=101210#p101210/]legislador.art.br/forum/viewtopic.php?p=101210#p101210[/url]
[url=http://daongil.com/bbs/board.php?bo_table=free&wr_id=297936/]daongil.com/bbs/board.php?bo_table=free&wr_id=297936[/url]
[url=http://дагсервис.рф/content/landsdiplomy/]дагсервис.рф/content/landsdiplomy[/url]
[url=http://netl.free.fr/index.php?file=Members&op=detail&autor=eganofa/]netl.free.fr/index.php?file=Members&op=detail&autor=eganofa[/url]
[url=http://bank-garantiya.ru/index.php?subaction=userinfo&user=ociratep/]bank-garantiya.ru/index.php?subaction=userinfo&user=ociratep[/url]
アイリスオーヤマの株主優待はありますか?
July 28, 2024 @ 5:10 am
Hi friends, pleasant article and good arguments commented here, I am really enjoying by these.
Roberttiz
July 28, 2024 @ 5:18 am
Официальный сайт по продаже оригинальных кроссовок изи буст https://yeezy-boost-shop.ru в Москве. Мы продаем yeezy boost оригинал с доставкой по всей России. В нашей линейке есть такие модели Adidas yeezy 350, yeezy 500, yeezy slide.
CecilOvams
July 28, 2024 @ 5:34 am
В нашем интернет магазине https://shop-uggs.ru представлен широкий ассортимент оригинальных женских, мужских и детских UGG Australia. Вы можете купить угги у нас в Москве, а так же с доставкой по России без предоплаты. Мы привозим 2 пары обуви на примерку, вы сможете сначала примерить уги и только потом оплачивать те угги которые вам подошли.
JamesCat
July 28, 2024 @ 5:39 am
Сериал “911: Служба спасения” https://911-sluzhba-spaseniya.watch рассказывает о работе сотрудников экстренных служб Лос-Анджелеса, включая пожарных, медиков и операторов 911. Они ежедневно сталкиваются с опасными ситуациями, спасая жизни и решая сложные личные проблемы. Смотрите онлайн в хорошем качестве HD, бесплатно, все сезоны.
Dnrtead
July 28, 2024 @ 5:58 am
[u][b] Добрый день![/b][/u]
[b]Приобрести документ[/b] института можно у нас в Москве.
[url=http://diploms-x.com/kupit-diplom-voronezh/]diploms-x.com/kupit-diplom-voronezh[/url]
[b]Удачи![/b]
GerardoTat
July 28, 2024 @ 6:02 am
Щоденні новини https://autoinfo.kyiv.ua із автомобільного середовища. Поради автоаматорам. Тест-драйви
Cazrrtv
July 28, 2024 @ 7:17 am
[u][b] Привет![/b][/u]
Купить диплом о высшем образовании
[url=http://pytalovo.4admins.ru/viewtopic.php?f=15&t=2374/]pytalovo.4admins.ru/viewtopic.php?f=15&t=2374[/url]
[url=http://vsetutonline.com/forum/member.php?u=76509/]vsetutonline.com/forum/member.php?u=76509[/url]
[url=http://restoranica.ru/forum/user/4872//]restoranica.ru/forum/user/4872/[/url]
[url=http://veneraroleplay.listbb.ru/viewtopic.php?f=4&t=328/]veneraroleplay.listbb.ru/viewtopic.php?f=4&t=328[/url]
[url=http://paintball-keller-lev.de/viewtopic.php?f=3&t=100163/]paintball-keller-lev.de/viewtopic.php?f=3&t=100163[/url]
Lazrbqx
July 28, 2024 @ 7:34 am
[b]Заказать диплом любого ВУЗа.[/b]
[url=http://skvortsy.listbb.ru/viewtopic.php?f=31&t=1800/]skvortsy.listbb.ru/viewtopic.php?f=31&t=1800[/url]
[url=http://avtolux48.ru/people/user/374/blog/7864//]avtolux48.ru/people/user/374/blog/7864/[/url]
[url=http://vseogirls.ru/diplomyi-na-zakaz-byistro-i-nadezhno/]vseogirls.ru/diplomyi-na-zakaz-byistro-i-nadezhno[/url]
[url=http://goalissimo.org/forum/viewtopic.php?f=4&t=695403/]goalissimo.org/forum/viewtopic.php?f=4&t=695403[/url]
[url=http://avtovideotest.ru/vash-nastoyashhiy-diplom-byistro-i-konfidentsialno/]avtovideotest.ru/vash-nastoyashhiy-diplom-byistro-i-konfidentsialno[/url]
Xazrqge
July 28, 2024 @ 7:38 am
[u][b] Здравствуйте![/b][/u]
Заказать документ о получении высшего образования.
[url=http://ya.0bb.ru/viewtopic.php?id=11999#p32011/]ya.0bb.ru/viewtopic.php?id=11999#p32011[/url]
[url=http://argayash.flybb.ru/viewtopic.php?f=9&t=1153/]argayash.flybb.ru/viewtopic.php?f=9&t=1153[/url]
[url=http://shopwheel.ru/club/user/52/blog/?b24statAction=addLogEntry/]shopwheel.ru/club/user/52/blog/?b24statAction=addLogEntry[/url]
[url=http://saumalkol.com/forum/разное-2/3592-легальный-диплом-без-лишних-хлопот-узнайте-как.html/]saumalkol.com/forum/разное-2/3592-легальный-диплом-без-лишних-хлопот-узнайте-как.html[/url]
[url=http://smd.mybb.ru/viewtopic.php?id=4109#p23191/]smd.mybb.ru/viewtopic.php?id=4109#p23191[/url]
Cazroor
July 28, 2024 @ 7:50 am
[u][b] Привет![/b][/u]
Мы предлагаем документы техникумов
[url=http://mafiaclans.ru/viewtopic.php?f=43&t=7667/]mafiaclans.ru/viewtopic.php?f=43&t=7667[/url]
[url=http://wecanchat.mn.co/spaces/9349852/feed/]wecanchat.mn.co/spaces/9349852/feed[/url]
[url=http://ya.9bb.ru/post.php?fid=2/]ya.9bb.ru/post.php?fid=2[/url]
[url=http://school97.ru/vesti20/index.php?PAGE_NAME=profile_view&UID=210605/]school97.ru/vesti20/index.php?PAGE_NAME=profile_view&UID=210605[/url]
[url=http://ya.bestbb.ru/viewtopic.php?id=3034#p6702/]ya.bestbb.ru/viewtopic.php?id=3034#p6702[/url]
Uazrpba
July 28, 2024 @ 8:26 am
[u][b] Привет, друзья![/b][/u]
Где заказать диплом по нужной специальности?
[url=http://swayycases.com/диплом-университета-купить-выгодные//]swayycases.com/диплом-университета-купить-выгодные/[/url]
marc jacobs outlet
July 28, 2024 @ 8:40 am
Enjoy premium Marc Jacobs products for less at the marc jacobs outlet.
Herbertageve
July 28, 2024 @ 8:42 am
Авто статті https://automobile.kyiv.ua з порадами з ремонту та обслуговування, авто блог з професійними порадами, огляди новинок
DouglasKib
July 28, 2024 @ 8:50 am
Слимофор https://slymofor.ru инновационное средство для быстрого похудения.
Williamraics
July 28, 2024 @ 8:56 am
[u][b] Привет![/b][/u]
Мы можем предложить дипломы.
[url=http://drovokol.ru/forum/user/12134//]drovokol.ru/forum/user/12134/[/url]
[url=http://rossensor.ru/forum/?PAGE_NAME=profile_view&UID=43403/]rossensor.ru/forum/?PAGE_NAME=profile_view&UID=43403[/url]
[url=http://avtolux48.ru/people/user/374/blog//]avtolux48.ru/people/user/374/blog/[/url]
[url=http://kh.txi.ru/forum/index.php?/]kh.txi.ru/forum/index.php?[/url]
[url=http://feelosum.com/blogs/1677/Гарантия-Подлинности-Покупка-Дипломов-Легально/]feelosum.com/blogs/1677/Гарантия-Подлинности-Покупка-Дипломов-Легально[/url]
種類株式の配当は普通株式に優先する?
July 28, 2024 @ 9:24 am
I just like the valuable info you supply in your articles.
I will bookmark your blog and check again right here frequently.
I am moderately certain I’ll be informed lots of
new stuff proper right here! Best of luck for the
next!
迷惑LINEが来たらどうすればいいですか?
July 28, 2024 @ 9:37 am
This paragraph will help the internet people for building up new website or even a blog from start to
end.
湘南美容クリニックの運営会社は?
July 28, 2024 @ 10:02 am
Triage is a system of prioritizing patients once
they enter the ER, no matter who obtained there first.
We’re all acquainted with depictions of heart patients within the ER.
Emergency room experts know the right way to deal with dire cardiac situations, but much more common than coronary
heart assaults are chest pains, which is likely to be precursors or signs of
different well being problems.
ボンビーガールはなぜ終了したのですか?
July 28, 2024 @ 10:24 am
Hi there, i read your blog from time to time and i own a similar one and i was just wondering if you get a lot of spam responses?
If so how do you protect against it, any plugin or anything you
can advise? I get so much lately it’s driving me crazy
so any help is very much appreciated.
Xazrowt
July 28, 2024 @ 10:35 am
[u][b] Привет![/b][/u]
Заказать документ института.
[url=http://kurgetrp.listbb.ru/viewtopic.php?f=11&t=456/]kurgetrp.listbb.ru/viewtopic.php?f=11&t=456[/url]
[url=http://besconeshnocte.flybb.ru/viewtopic.php?f=2&t=953/]besconeshnocte.flybb.ru/viewtopic.php?f=2&t=953[/url]
[url=http://simplemachines.org/about/smf/stats.php/]simplemachines.org/about/smf/stats.php[/url]
[url=http://lubov.listbb.ru/ucp.php?mode=login&sid=c1b711d4af4ca6eb78846ad2341b7374/]lubov.listbb.ru/ucp.php?mode=login&sid=c1b711d4af4ca6eb78846ad2341b7374[/url]
[url=http://hb9lc.org/wiki/index.php/Дипломы_любой_сложности_быстро_и_без_проблем/]hb9lc.org/wiki/index.php/Дипломы_любой_сложности_быстро_и_без_проблем[/url]
Cazrnyk
July 28, 2024 @ 10:43 am
[u][b] Привет, друзья![/b][/u]
Мы готовы предложить документы ВУЗов
[url=http://jeepgarage.ru/forum/topic.php?forum=24&topic=779/]jeepgarage.ru/forum/topic.php?forum=24&topic=779[/url]
[url=http://sklad-slabov.ru/forum/user/8415//]sklad-slabov.ru/forum/user/8415/[/url]
[url=http://wayworld.listbb.ru/viewtopic.php?f=2&t=24826/]wayworld.listbb.ru/viewtopic.php?f=2&t=24826[/url]
[url=http://elizabethjohns.ru/forum/user/6051//]elizabethjohns.ru/forum/user/6051/[/url]
[url=http://dogovor.top/wiki/болучите_диплом_о_высшем_образовании_без_лишних_хлопот/]dogovor.top/wiki/болучите_диплом_о_высшем_образовании_без_лишних_хлопот[/url]
TrentonVULLY
July 28, 2024 @ 11:23 am
Авто статті https://bestauto.kyiv.ua з порадами з ремонту та обслуговування, авто блог з професійними порадами, огляди новинок
Bernardlal
July 28, 2024 @ 11:28 am
Автомобільні новини https://autonovosti.kyiv.ua України та світу, тест-драйви нових авто, поради експертів
Phillipglity
July 28, 2024 @ 11:28 am
Автоновини України https://avtomobilist.kyiv.ua огляди машин та новини для автомобілістів
Thomasoredo
July 28, 2024 @ 11:36 am
Автомобільні новини https://avtonews.kyiv.ua України. Все для автовласника.
Lazrgmr
July 28, 2024 @ 12:30 pm
[u][b] Привет, друзья![/b][/u]
[b]Мы можем предложить дипломы[/b] психологов, юристов, экономистов и прочих профессий по приятным тарифам.
[url=http://diplomman.ru/]diplomman.ru[/url]
Sazrpvf
July 28, 2024 @ 3:04 pm
[u][b] Привет![/b][/u]
Приобрести документ института
[url=http://portugues.ru/forum/album.php?albumid=91&attachmentid=3900&commentid=1664/]portugues.ru/forum/album.php?albumid=91&attachmentid=3900&commentid=1664[/url]
[url=http://flerus-shop.hcp.dilhost.ru/club/user/3/forum/message/876/872//]flerus-shop.hcp.dilhost.ru/club/user/3/forum/message/876/872/[/url]
[url=http://tricityfriends.com/read-blog/578/]tricityfriends.com/read-blog/578[/url]
[url=http://network-2295034.mn.co/posts/61679299/]network-2295034.mn.co/posts/61679299[/url]
[url=http://eyawei-networks.mn.co/posts/61679446/]eyawei-networks.mn.co/posts/61679446[/url]
Lazrtfx
July 28, 2024 @ 3:13 pm
[u][b] Добрый день![/b][/u]
[b]Мы можем предложить дипломы[/b] любой профессии по доступным тарифам.
[url=http://mans-diplomas.ru/]mans-diplomas.ru[/url]
Jamesabary
July 28, 2024 @ 3:29 pm
Авто статті https://black-star.com.ua з порадами з ремонту та обслуговування
Jamespyday
July 28, 2024 @ 3:34 pm
DMV Test на русском языке https://papadmv.com тесты с ответами ПДД США 2024. Тренировочные dmv test на русском для сдачи на права, изучите правила дорожного движения США для разных штатов.
Xazrbgo
July 28, 2024 @ 3:40 pm
[b]Привет![/b]
Как официально купить аттестат 11 класса с упрощенным обучением в Москве
[url=http://arusak-diploms-srednee.ru/kupit-diplom-kandidata-nauk В ]arusak-diploms-srednee.ru/kupit-diplom-kandidata-nauk В [/url]
Manrehs
July 28, 2024 @ 3:41 pm
[u][b] Привет, друзья![/b][/u]
Приобрести документ о получении высшего образования
[url=http://diplomyx.com/kupit-diplom-nizhnij-novgorod]diplomyx.com/kupit-diplom-nizhnij-novgorod[/url]
Razrbkv
July 28, 2024 @ 4:06 pm
[u][b] Привет![/b][/u]
Приобрести диплом любого ВУЗа:
[url=http://pax.nichost.ru/forum/view_profile.php?UID=133467/]pax.nichost.ru/forum/view_profile.php?UID=133467[/url]
[url=http://b24activities.ru/club/forum/messages/forum4/topic76/message135/?result=reply#message135/]b24activities.ru/club/forum/messages/forum4/topic76/message135/?result=reply#message135[/url]
[url=http://sskyn.com/home-uid-83477.html/]sskyn.com/home-uid-83477.html[/url]
[url=http://led119.ru/forum/user/107639//]led119.ru/forum/user/107639/[/url]
[url=http://bizon15.ru/index.php?subaction=userinfo&user=abapaxo/]bizon15.ru/index.php?subaction=userinfo&user=abapaxo[/url]
LarryApope
July 28, 2024 @ 4:28 pm
Undress AI & Bulk Nude AI Generator nudify online. Make AI nudes and bulk generate undress AI photos of any girl for almost free!
ThomasCyday
July 28, 2024 @ 6:24 pm
Щоденні новини https://k-moto.com.ua із автомобільного середовища. Поради автоаматорам. Тест-драйви автомобілів з пробігом та огляди новинок
Diplomi_zjSi
July 28, 2024 @ 6:33 pm
Привет!
Заказать документ о получении высшего образования вы сможете в нашей компании в Москве.
[url=http://ast-diplom.com/kupit-diplom-s-registraciej/]ast-diplom.com/kupit-diplom-s-registraciej[/url]
Lazrmax
July 28, 2024 @ 6:34 pm
[b]Купить диплом любого ВУЗа.[/b]
[url=http://fish-sea-products.ru/forum/user/5826//]fish-sea-products.ru/forum/user/5826/[/url]
[url=http://bn.tobase.ru/viewtopic.php?f=30&t=3343/]bn.tobase.ru/viewtopic.php?f=30&t=3343[/url]
[url=http://polosedan-club.com/search/search/]polosedan-club.com/search/search[/url]
[url=http://sfpdsa.listbb.ru/posting.php?mode=post&f=2/]sfpdsa.listbb.ru/posting.php?mode=post&f=2[/url]
[url=http://elizabethjohns.ru/forum/user/6036//]elizabethjohns.ru/forum/user/6036/[/url]
BillyTix
July 28, 2024 @ 7:00 pm
Найбільший автомобільний портал https://mirauto.kyiv.ua України
Lazrtyn
July 28, 2024 @ 7:13 pm
[u][b] Привет, друзья![/b][/u]
[b]Приобрести диплом любого ВУЗа.[/b]
[url=http://livestockkenya.com/index.php/cb-profile/pluginclass/cbblogs?action=blogs&func=show&id=244/]livestockkenya.com/index.php/cb-profile/pluginclass/cbblogs?action=blogs&func=show&id=244[/url]
Roberttum
July 28, 2024 @ 7:23 pm
PrestigeAvto https://prestige-avto.com.ua України автомобільний журнал
Uazrxto
July 28, 2024 @ 7:43 pm
[u][b] Привет![/b][/u]
Где купить диплом специалиста?
[url=http://www.mizmiz.de/read-blog/23918/]www.mizmiz.de/read-blog/23918[/url]
Williamraics
July 28, 2024 @ 7:55 pm
[u][b] Привет, друзья![/b][/u]
Мы изготавливаем дипломы.
[url=http://fabnews.ru/forum/showthread.php?p=79754#post79754/]fabnews.ru/forum/showthread.php?p=79754#post79754[/url]
[url=http://demo.4admins.ru/posting.php?mode=post&f=10&sid=5c319a4b18b2bf2335b52b43d1fb9b6f/]demo.4admins.ru/posting.php?mode=post&f=10&sid=5c319a4b18b2bf2335b52b43d1fb9b6f[/url]
[url=http://domovou.3nx.ru/viewtopic.php?p=6845#6845/]domovou.3nx.ru/viewtopic.php?p=6845#6845[/url]
[url=http://vetstate.ru/forum/?PAGE_NAME=profile_view&UID=125341/]vetstate.ru/forum/?PAGE_NAME=profile_view&UID=125341[/url]
[url=http://dolgoprudni.rusff.me/viewtopic.php?id=2296#p5154/]dolgoprudni.rusff.me/viewtopic.php?id=2296#p5154[/url]
Mazrzvx
July 28, 2024 @ 9:27 pm
[u][b] Добрый день![/b][/u]
[b]Рекомендации по безопасной покупке диплома о высшем образовании[/b]
[url=http://diploms-man.ru/]diploms-man.ru[/url]
Eanrrgj
July 28, 2024 @ 9:27 pm
[u][b] Привет![/b][/u]
[b]Мы можем предложить дипломы[/b] любой профессии по выгодным ценам.
[url=http://rucollectshoeclub.com/read-blog/1841/]rucollectshoeclub.com/read-blog/1841[/url]
[url=http://ozelim.org/sosyal/blogs/382/Выбираем-проверенный-онлайн-магазин-с-дипломами-и-аттестатами/]ozelim.org/sosyal/blogs/382/Выбираем-проверенный-онлайн-магазин-с-дипломами-и-аттестатами[/url]
[url=http://farm86.com/blogs/332/Главные-расходы-российского-производителя-документов-обзор/]farm86.com/blogs/332/Главные-расходы-российского-производителя-документов-обзор[/url]
[url=http://wakelet.com/wake/fNFEDznBMphL_kDaIfyqk/]wakelet.com/wake/fNFEDznBMphL_kDaIfyqk[/url]
[url=http://chillibell.com/read-blog/36/]chillibell.com/read-blog/36[/url]
Diplomi_zjSi
July 28, 2024 @ 9:44 pm
Добрый день!
Купить документ о получении высшего образования можно в нашей компании в столице.
[url=http://asxdiplomik.com/kupit-diplom-tehnikuma-kolledzhaВ /]asxdiplomik.com/kupit-diplom-tehnikuma-kolledzhaВ [/url]
Yrefzvo
July 28, 2024 @ 10:17 pm
[u][b] Добрый день![/b][/u]
[b]Приобрести диплом ВУЗа.[/b]
[url=http://sharaepewixu.iamarrows.com/cto-neobhodimo-iz-tehniki-dla-proizvodstva-diploma//]sharaepewixu.iamarrows.com/cto-neobhodimo-iz-tehniki-dla-proizvodstva-diploma/[/url]
[url=http://lada-xray.net/member.php?tab=visitor_messaging&u=2434&page=2/]lada-xray.net/member.php?tab=visitor_messaging&u=2434&page=2[/url]
[url=http://sciens.ru/page/blwdr//]sciens.ru/page/blwdr/[/url]
[url=http://friend24.in/blogs/2853/Что-именно-требуется-из-техники-для-создания-диплома/]friend24.in/blogs/2853/Что-именно-требуется-из-техники-для-создания-диплома[/url]
[url=http://newsato.ru/page/2/]newsato.ru/page/2[/url]
Jariorjkb
July 28, 2024 @ 10:53 pm
[u][b] Привет, друзья![/b][/u]
Приобрести диплом университета
[url=http://ou812chat.com/read-blog/1237/]ou812chat.com/read-blog/1237[/url]
[url=http://dogovor.top/wiki/Оформление_диплома_любой_специальности/]dogovor.top/wiki/Оформление_диплома_любой_специальности[/url]
[url=http://ulmo.ukrbb.net/index.php/]ulmo.ukrbb.net/index.php[/url]
[url=http://firstamendment.tv/read-blog/50348/]firstamendment.tv/read-blog/50348[/url]
[url=http://breyerhorses.ru/personal/profile/?register=yes/]breyerhorses.ru/personal/profile/?register=yes[/url]
Larioreve
July 28, 2024 @ 11:28 pm
Как быстро получить диплом магистра? Легальные способы
[url=http://u90517ol.beget.tech/2024/07/11/oformlenie-diploma-pod-klyuch-bezopasno-i-legalno.html/]u90517ol.beget.tech/2024/07/11/oformlenie-diploma-pod-klyuch-bezopasno-i-legalno.html[/url]
[url=http://ya3bbru.bbok.ru/viewtopic.php?id=4042#p7095/]ya3bbru.bbok.ru/viewtopic.php?id=4042#p7095[/url]
[url=http://feelosum.com/blogs/1728/Ваш-новый-диплом-уже-сегодня/]feelosum.com/blogs/1728/Ваш-новый-диплом-уже-сегодня[/url]
[url=http://podsliving.in/forums/topic/купить-диплом-в-москве-бытовые-услови//]podsliving.in/forums/topic/купить-диплом-в-москве-бытовые-услови/[/url]
[url=http://astroroleplay.getbb.ru/viewtopic.php?f=6&t=488/]astroroleplay.getbb.ru/viewtopic.php?f=6&t=488[/url]
Xazrmzy
July 29, 2024 @ 12:12 am
[u][b] Добрый день![/b][/u]
Купить документ института.
[url=http://exoltech.ps/blogs/post/new/]exoltech.ps/blogs/post/new[/url]
[url=http://teplichnaya.ru/viewtopic.php?f=18&t=641602/]teplichnaya.ru/viewtopic.php?f=18&t=641602[/url]
[url=http://fsmi.wiki/index.php?title=Получите_диплом_в_кратчайшие_сроки/]fsmi.wiki/index.php?title=Получите_диплом_в_кратчайшие_сроки[/url]
[url=http://wiki.3cdr.ru/wiki/index.php/Ваш_диплом_за_короткое_время:_качественно_и_безопасно/]wiki.3cdr.ru/wiki/index.php/Ваш_диплом_за_короткое_время:_качественно_и_безопасно[/url]
[url=http://israelafrica.mn.co/posts/62321047/]israelafrica.mn.co/posts/62321047[/url]
Cazrlze
July 29, 2024 @ 1:12 am
[u][b] Добрый день![/b][/u]
Мы можем предложить документы техникумов
[url=http://1wum.ru/forum/?PAGE_NAME=profile_view&UID=40009&MUL_MODE=/]1wum.ru/forum/?PAGE_NAME=profile_view&UID=40009&MUL_MODE=[/url]
[url=http://financeokey.ru/poluchite-diplom-o-vyisshem-obrazovanii-bez-lishnih-hlopot/]financeokey.ru/poluchite-diplom-o-vyisshem-obrazovanii-bez-lishnih-hlopot[/url]
[url=http://elizabethjohns.ru/forum/user/6051//]elizabethjohns.ru/forum/user/6051/[/url]
[url=http://magiclife.ukraine7.com/t6383-topic#15965/]magiclife.ukraine7.com/t6383-topic#15965[/url]
[url=http://pika.listbb.ru/posting.php?mode=post&f=9/]pika.listbb.ru/posting.php?mode=post&f=9[/url]
Oariorjqd
July 29, 2024 @ 1:27 am
[u][b] Здравствуйте![/b][/u]
Купить диплом о высшем образовании
[b]Наши специалисты предлагают[/b] выгодно заказать диплом, который выполнен на бланке ГОЗНАКа и заверен печатями, штампами, подписями. Наш диплом пройдет любые проверки, даже с применением профессиональных приборов. Решите свои задачи максимально быстро с нашей компанией.
[b]Где заказать диплом специалиста?[/b]
[url=http://famenest.com/read-blog/7940/]famenest.com/read-blog/7940[/url]
[url=http://lubercy.ixbb.ru/viewtopic.php?id=7221#p15723/]lubercy.ixbb.ru/viewtopic.php?id=7221#p15723[/url]
[url=http://avtomobil1980.ixbb.ru/viewtopic.php?id=112#p112/]avtomobil1980.ixbb.ru/viewtopic.php?id=112#p112[/url]
[url=http://kingasteroid.com/blogs/284/Быстрое-Оформление-Дипломов-Надежно-и-Легально/]kingasteroid.com/blogs/284/Быстрое-Оформление-Дипломов-Надежно-и-Легально[/url]
[url=http://sonnick84.pointblog.net/Что-понадобится-для-создания-качественного-диплома-ВУЗа-69447016/]sonnick84.pointblog.net/Что-понадобится-для-создания-качественного-диплома-ВУЗа-69447016[/url]
Mazrgfw
July 29, 2024 @ 2:39 am
[u][b] Добрый день![/b][/u]
[b]Рекомендации по безопасной покупке диплома о высшем образовании[/b]
[url=http://mandiplomikx.ru/]mandiplomikx.ru[/url]
RonaldFoumb
July 29, 2024 @ 3:03 am
Автомобільні новини https://sedan.kyiv.ua України та світу, тест-драйви автомобілів, автоспорт
Manrknt
July 29, 2024 @ 3:05 am
[u][b] Добрый день![/b][/u]
Приобрести документ о получении высшего образования
[url=http://diplomasx24.ru/kupit-diplom-vracha]diplomasx24.ru/kupit-diplom-vracha[/url]
Xazribt
July 29, 2024 @ 3:07 am
[u][b] Привет![/b][/u]
Заказать документ ВУЗа.
[url=http://bazhenova.greatforum.ru/viewtopic.php?f=17&t=5076/]bazhenova.greatforum.ru/viewtopic.php?f=17&t=5076[/url]
[url=http://crystalroleplay.listbb.ru/viewtopic.php?f=14&t=2499/]crystalroleplay.listbb.ru/viewtopic.php?f=14&t=2499[/url]
[url=http://flashtest.80lvl.ru/viewtopic.php?f=3&t=1919/]flashtest.80lvl.ru/viewtopic.php?f=3&t=1919[/url]
[url=http://yourealtynews.ru/diplomyi-dlya-uspeshnoy-kareryi/]yourealtynews.ru/diplomyi-dlya-uspeshnoy-kareryi[/url]
[url=http://veneraroleplay.listbb.ru/viewtopic.php?f=4&t=320/]veneraroleplay.listbb.ru/viewtopic.php?f=4&t=320[/url]
Xazrhwz
July 29, 2024 @ 3:16 am
[b]Привет, друзья![/b]
Всё, что нужно знать о покупке аттестата о среднем образовании без рисков
[url=http://landik-diploms-srednee.ru/svidetelstvo-o-razvode В ]landik-diploms-srednee.ru/svidetelstvo-o-razvode В [/url]
MarioGom
July 29, 2024 @ 3:20 am
Авто статті https://road.kyiv.ua з порадами з ремонту та обслуговування. Авто блог з професійними порадами.
WilberSnume
July 29, 2024 @ 3:30 am
The fascinating story of Ja Morant’s https://grizzlies-de-memphis.ja-morant-fr.com meteoric rise, from status from rookie to leader of the Memphis Grizzlies and rising NBA superstar.
Michaellit
July 29, 2024 @ 3:32 am
Свежие новости https://diesel.kyiv.ua автомобильного рынка, новинки автопрома
業界分析で何をするのでしょうか?
July 29, 2024 @ 3:33 am
Excellent beat ! I wish to apprentice at the same time as you amend
your website, how could i subscribe for a weblog website?
The account helped me a applicable deal. I have been tiny bit familiar of this your broadcast offered vibrant clear concept
Iariorcbq
July 29, 2024 @ 3:49 am
[u][b] Добрый день![/b][/u]
Заказать документ института можно в нашей компании. Мы оказываем услуги по продаже документов об окончании любых университетов России. Вы получите диплом по любой специальности, включая документы старого образца. Даем 100% гарантию, что при проверке документа работодателями, подозрений не появится.
[url=http://gamerszona.ru/index.php/]gamerszona.ru/index.php[/url]
[url=http://ja360.fun/read-blog/23/]ja360.fun/read-blog/23[/url]
[url=http://contraband.ch/read-blog/4228/]contraband.ch/read-blog/4228[/url]
[url=http://lidertude.com/blogs/1191/Что-необходимо-из-оборудования-для-создания-документов/]lidertude.com/blogs/1191/Что-необходимо-из-оборудования-для-создания-документов[/url]
[url=http://ck2zm153m.anchor-blog.com/8552292/Как-подыскать-проверенный-магазин-с-документами-Обзор-от-эксперта/]ck2zm153m.anchor-blog.com/8552292/Как-подыскать-проверенный-магазин-с-документами-Обзор-от-эксперта[/url]
Cazrnfw
July 29, 2024 @ 3:55 am
[u][b] Привет![/b][/u]
Мы готовы предложить документы ВУЗов
[url=http://galantclub.od.ua/member.php?u=15983/]galantclub.od.ua/member.php?u=15983[/url]
[url=http://visacart.80lvl.ru/viewtopic.php?f=1&t=797/]visacart.80lvl.ru/viewtopic.php?f=1&t=797[/url]
[url=http://zdshi-tula.ru/forum/?PAGE_NAME=message&FID=1&TID=234&MID=38997&result=new#message38997/]zdshi-tula.ru/forum/?PAGE_NAME=message&FID=1&TID=234&MID=38997&result=new#message38997[/url]
[url=http://girlscools.ru/zakazhite-diplom-pryamo-seychas/]girlscools.ru/zakazhite-diplom-pryamo-seychas[/url]
[url=http://besconeshnocte.flybb.ru/viewtopic.php?f=2&t=959/]besconeshnocte.flybb.ru/viewtopic.php?f=2&t=959[/url]
Lazrdxl
July 29, 2024 @ 4:12 am
[u][b] Привет![/b][/u]
[b]Мы изготавливаем дипломы[/b] любых профессий по невысоким ценам.
[url=http://mans-diplomasxx.ru/]mans-diplomasxx.ru[/url]
Qazrosa
July 29, 2024 @ 4:56 am
[u][b] Добрый день![/b][/u]
[b]Заказать диплом университета.[/b]
[url=http://doomelang.com/read-blog/6524/]doomelang.com/read-blog/6524[/url]
[url=http://lifethelife.com/read-blog/292/]lifethelife.com/read-blog/292[/url]
[url=http://micchat.online/read-blog/1479/]micchat.online/read-blog/1479[/url]
[url=http://fbadult.com/blogs/2838/Правильно-заказываем-документы-в-интернете-обзор/]fbadult.com/blogs/2838/Правильно-заказываем-документы-в-интернете-обзор[/url]
[url=http://forosmex.com/index.php?/gallery/image/543-4-е-ключевых-группы-интернет-магазинов-что-реализуют-документы//]forosmex.com/index.php?/gallery/image/543-4-е-ключевых-группы-интернет-магазинов-что-реализуют-документы/[/url]
NTTの民営化を断行したのは誰ですか?
July 29, 2024 @ 4:58 am
Over shorter time periods, such as one year, Jensen’s alpha
is a useful performance metric, adjusting returns for market beta risk.
Should the enterprise pick a growth strategy in line with market opportunity, and aim to grow sales, increase profits and broaden market share,
they would consider vertical and horizontal integration; and, also
diversification.
Trefjjt
July 29, 2024 @ 6:49 am
[u][b] Здравствуйте![/b][/u]
Быстрое обучение и получение диплома магистра – возможно ли это?
[url=http://ayurastroyoga.com/настоящий-диплом-как-и-где-купить//]ayurastroyoga.com/настоящий-диплом-как-и-где-купить/[/url]
Рады оказаться полезными!.
Lazrqgy
July 29, 2024 @ 7:09 am
[u][b] Привет, друзья![/b][/u]
[b]Мы изготавливаем дипломы[/b] психологов, юристов, экономистов и любых других профессий по выгодным ценам.
[url=http://mans-diploms.ru/]mans-diploms.ru[/url]
KennethReevy
July 29, 2024 @ 7:35 am
download free screen capture for windows https://screen-capture-free.com
Lloydthype
July 29, 2024 @ 7:51 am
Indibet https://indibeti.in is a premier online casino offering a wide array of games including slots, table games, and live dealer options. Renowned for its user-friendly interface and robust security measures, Indibet ensures a top-notch gaming experience with exciting bonuses and 24/7 customer support.
Doylebophy
July 29, 2024 @ 7:55 am
L’histoire epique du FC Barcelone https://espagne.barcelona-fr.com 120 ans de triomphes et de tribulations.
Ronaldruink
July 29, 2024 @ 8:07 am
Rodri https://manchester-city.rodrigo-hernandez.com le maestro du milieu de terrain de Manchester City
HarryBob
July 29, 2024 @ 9:13 am
Immerse yourself in the charm of France https://france.life-in-france.net a land of fine cuisine, impressive architecture and picturesque landscapes. An unrivaled lifestyle.
Dnrtutr
July 29, 2024 @ 9:18 am
[u][b] Здравствуйте![/b][/u]
[b]Приобрести документ[/b] о получении высшего образования вы можете у нас в Москве.
[url=http://diplomasx24.ru/kupit-diplom-krasnoyarsk/]diplomasx24.ru/kupit-diplom-krasnoyarsk[/url]
[b]Хорошей учебы![/b]
Eanrtry
July 29, 2024 @ 10:01 am
[u][b] Привет, друзья![/b][/u]
[b]Мы изготавливаем дипломы[/b] любых профессий по приятным ценам.
[url=http://alex-neil.ru/index.php/cb-profile/pluginclass/cbblogs?action=blogs&func=show&id=232/]alex-neil.ru/index.php/cb-profile/pluginclass/cbblogs?action=blogs&func=show&id=232[/url]
[url=http://naya.social/read-blog/154/]naya.social/read-blog/154[/url]
[url=http://verdoos.com/read-blog/21962/]verdoos.com/read-blog/21962[/url]
[url=http://indungii.ro/forum/index.php?/gallery/image/9-faq-и-тех-поддержка-заказываем-диплом-в-сети//]indungii.ro/forum/index.php?/gallery/image/9-faq-и-тех-поддержка-заказываем-диплом-в-сети/[/url]
[url=http://taxecure.mn.co/posts/61838293/]taxecure.mn.co/posts/61838293[/url]
Vazrlhz
July 29, 2024 @ 10:03 am
[u][b] Привет![/b][/u]
Приобрести документ ВУЗа вы можете у нас. Мы предлагаем документы об окончании любых ВУЗов Российской Федерации.
[url=http://silabgarza.net/forums/showthread.php?tid=268324/]silabgarza.net/forums/showthread.php?tid=268324[/url]
[url=http://classicalmusicmp3freedownload.com/ja/index.php?title=diplomandoci/]classicalmusicmp3freedownload.com/ja/index.php?title=diplomandoci[/url]
[url=http://автосервис-ковров.рф/index.php?subaction=userinfo&user=yhufokimo/]автосервис-ковров.рф/index.php?subaction=userinfo&user=yhufokimo[/url]
[url=http://”www.inline.spb.ru/in/index.php?option=com_fireboard&Itemid=15&func=view&catid=7&id=4924#4924″/]”www.inline.spb.ru/in/index.php?option=com_fireboard&Itemid=15&func=view&catid=7&id=4924#4924″[/url]
[url=http://lodowisko.pszow.pl/forum/viewtopic.php?f=21&t=18714&p=53313#p53313/]lodowisko.pszow.pl/forum/viewtopic.php?f=21&t=18714&p=53313#p53313[/url]
Lazrxjo
July 29, 2024 @ 11:12 am
[u][b] Привет![/b][/u]
[b]Купить диплом ВУЗа.[/b]
[url=http://islider.ru/read-blog/564/]islider.ru/read-blog/564[/url]
Dominictix
July 29, 2024 @ 11:15 am
buying from online mexican pharmacy: purple pharmacy mexico price list – buying prescription drugs in mexico
Edwardhen
July 29, 2024 @ 11:31 am
Deep Fake Nudes undress AI with Undress AI and Deepnude
Dominictix
July 29, 2024 @ 11:49 am
mexico drug stores pharmacies: medication from mexico pharmacy – medication from mexico pharmacy
RichardHew
July 29, 2024 @ 11:50 am
Частная платная клиника https://mypsyhealth.ru психиатрии, психологии, психотерапии и наркологии анонимно в Москве.
Lariorgox
July 29, 2024 @ 12:14 pm
Процесс получения диплома стоматолога: реально ли это сделать быстро?
[url=http://sovetushka.forum2x2.ru/t28885-topic#63656/]sovetushka.forum2x2.ru/t28885-topic#63656[/url]
[url=http://wiki.mdomtv.net/index.php?title=Купить_диплом:_Экономьте_время_и_деньги/]wiki.mdomtv.net/index.php?title=Купить_диплом:_Экономьте_время_и_деньги[/url]
[url=http://onlinekinospace.ru/byistroe-oformlenie-diploma-nadezhno-i-konfidentsialno/]onlinekinospace.ru/byistroe-oformlenie-diploma-nadezhno-i-konfidentsialno[/url]
[url=http://tweecampus.com/read-blog/81813/]tweecampus.com/read-blog/81813[/url]
[url=http://moscowurban.getbb.ru/viewtopic.php?f=12&t=1663/]moscowurban.getbb.ru/viewtopic.php?f=12&t=1663[/url]
Dnrtxxq
July 29, 2024 @ 12:19 pm
[u][b] Привет, друзья![/b][/u]
[b]Заказать документ[/b] о получении высшего образования вы сможете у нас в столице.
[url=http://diplomasx.com/kupit-diplom-sankt-peterburg/]diplomasx.com/kupit-diplom-sankt-peterburg[/url]
[b]Хорошей учебы![/b]
Charlessah
July 29, 2024 @ 4:41 pm
The Boston Celtics https://celtics-de-boston.bill-russel.com are one of the most successful teams in the history of the National Basketball Association (NBA).
JamesEvoli
July 29, 2024 @ 4:45 pm
Discover Kyrie Irving’s https://mavericks-de-dallas.kyrie-irving-fr.com journey with the Dallas Mavericks, from his early days to forming a power duo with Luka Doncic and competing for a championship.
Ronaldskala
July 29, 2024 @ 4:46 pm
Learn about Jayson Tatum’s https://celtics-de-boston.jayson-tatum.com rise from young rookie to key leader of the Boston Celtics in the NBA, his impact on the team and his success on the court.
ThomasSlono
July 29, 2024 @ 4:49 pm
The story of Michael Phelps https://amerique.michael-phelps.com how he became the greatest swimmer of all time, overcoming adversity, setting records and inspiring the world.
KevinSkync
July 29, 2024 @ 5:57 pm
Mavericks rising superstar Luka Doncic https://mavericks-de-dallas.lukadoncic-fr.com continues to amaze the basketball world with his game.
Dominictix
July 29, 2024 @ 6:07 pm
pharmacies in mexico that ship to usa: mexican border pharmacies shipping to usa – mexican rx online
Cazripq
July 29, 2024 @ 7:20 pm
[u][b] Привет![/b][/u]
Мы готовы предложить документы техникумов
[url=http://pitomec.ru/forum/post/13176/]pitomec.ru/forum/post/13176[/url]
[url=http://grp.7olimp.ru/viewforum.php?f=1/]grp.7olimp.ru/viewforum.php?f=1[/url]
[url=http://metalfans.maxbb.ru/viewtopic.php?f=7&t=708/]metalfans.maxbb.ru/viewtopic.php?f=7&t=708[/url]
[url=http://panther.80lvl.ru/index.php/]panther.80lvl.ru/index.php[/url]
[url=http://net4women.ru/blogs/4018/Купите-диплом-для-новой-карьеры/]net4women.ru/blogs/4018/Купите-диплом-для-новой-карьеры[/url]
Sazrsdt
July 29, 2024 @ 8:54 pm
[u][b] Здравствуйте![/b][/u]
Заказать документ о получении высшего образования
[url=http://leenkup.com/read-blog/16519/]leenkup.com/read-blog/16519[/url]
[url=http://sharemeglobal.com/blogs/903/На-что-именно-обращать-внимание-при-заказе-диплома-в-сети/]sharemeglobal.com/blogs/903/На-что-именно-обращать-внимание-при-заказе-диплома-в-сети[/url]
[url=http://sonnick84.glifeblog.com/27739762/Выбираем-надежный-интернет-магазин-для-покупки-документов/]sonnick84.glifeblog.com/27739762/Выбираем-надежный-интернет-магазин-для-покупки-документов[/url]
[url=http://hrv-club.ru/forums/index.php?autocom=gallery&req=si&img=6055/]hrv-club.ru/forums/index.php?autocom=gallery&req=si&img=6055[/url]
[url=http://respectmyopinion.com/blogs/26/Проверенный-и-надежный-магазин-с-обширным-ассортиментом-документов/]respectmyopinion.com/blogs/26/Проверенный-и-надежный-магазин-с-обширным-ассортиментом-документов[/url]
Cazropn
July 29, 2024 @ 10:15 pm
[u][b] Здравствуйте![/b][/u]
Мы можем предложить документы техникумов
[url=http://allnewstroy.ru/oformite-diplom-pryamo-seychas/]allnewstroy.ru/oformite-diplom-pryamo-seychas[/url]
[url=http://moneycool.bestbb.ru/search.php?action=search&keywords=&author=&forum=-1&search_in=all&sort_by=0&sort_dir=DESC&show_as=topics&search=%CE%F2%EF%F0%E0%E2%E8%F2%FC/]moneycool.bestbb.ru/search.php?action=search&keywords=&author=&forum=-1&search_in=all&sort_by=0&sort_dir=DESC&show_as=topics&search=%CE%F2%EF%F0%E0%E2%E8%F2%FC[/url]
[url=http://rrsclub.ru/member.php?u=1164/]rrsclub.ru/member.php?u=1164[/url]
[url=http://mnogozip.ru/forum/user/23882//]mnogozip.ru/forum/user/23882/[/url]
[url=http://testforum.1stbb.ru/viewtopic.php?f=3&t=442/]testforum.1stbb.ru/viewtopic.php?f=3&t=442[/url]
Diplomi_iwSi
July 29, 2024 @ 11:42 pm
Привет, друзья!
Приобрести документ университета вы сможете в нашем сервисе.
[url=http://ast-diplomas.com/kupit-diplom-novosibirsk/]ast-diplomas.com/kupit-diplom-novosibirsk[/url]
Lazresu
July 29, 2024 @ 11:44 pm
[u][b] Здравствуйте![/b][/u]
[b]Мы готовы предложить дипломы[/b] любой профессии по доступным ценам.
[url=http://lighttur.ru/ofitsialnyie-diplomyi-podlinnost-i-nadezhnost/]lighttur.ru/ofitsialnyie-diplomyi-podlinnost-i-nadezhnost[/url]
Lazrfoc
July 30, 2024 @ 12:00 am
[u][b] Привет![/b][/u]
[b]Заказать диплом университета.[/b]
[url=http://antrem.ru/cb-profile/pluginclass/cbblogs?action=blogs&func=show&id=283/]antrem.ru/cb-profile/pluginclass/cbblogs?action=blogs&func=show&id=283[/url]
Xazricb
July 30, 2024 @ 12:21 am
[u][b] Добрый день![/b][/u]
Приобрести документ о получении высшего образования.
[url=http://polosedan-club.com/search/search/]polosedan-club.com/search/search[/url]
[url=http://study.flybb.ru/viewtopic.php?f=4&t=419/]study.flybb.ru/viewtopic.php?f=4&t=419[/url]
[url=http://ukrevent.ru/diplom-dlya-karernogo-rosta-prosto-i-udobno/]ukrevent.ru/diplom-dlya-karernogo-rosta-prosto-i-udobno[/url]
[url=http://subscribe.ru/member/quick/]subscribe.ru/member/quick[/url]
[url=http://ukrom.in.ua/users/327/]ukrom.in.ua/users/327[/url]
Vazrwdm
July 30, 2024 @ 12:50 am
[u][b] Добрый день![/b][/u]
Заказать документ о получении высшего образования можно у нас. Мы предлагаем документы об окончании любых университетов РФ.
[url=http://zanasite.free.fr/laripostedumardi/doku.php?id=russianydiplomix/]zanasite.free.fr/laripostedumardi/doku.php?id=russianydiplomix[/url]
[url=http://school5.p-fam.ru/users/uwitari/]school5.p-fam.ru/users/uwitari[/url]
[url=http://www.detskiy-mir.net/index.php?subaction=userinfo&user=uxofapas/]www.detskiy-mir.net/index.php?subaction=userinfo&user=uxofapas[/url]
[url=http://wiki.serialkiller.adele.im/doku.php?id=landsdiplomy/]wiki.serialkiller.adele.im/doku.php?id=landsdiplomy[/url]
[url=http://clindoeilinfo.com/a-peine-arrivee-parmi-les-reseaux-sociaux-lapplication-threads-serait-dans-le-collimateur-de-la-justice/#comment-109282/]clindoeilinfo.com/a-peine-arrivee-parmi-les-reseaux-sociaux-lapplication-threads-serait-dans-le-collimateur-de-la-justice/#comment-109282[/url]
Yrefsqk
July 30, 2024 @ 1:08 am
[u][b] Здравствуйте![/b][/u]
[b]Купить диплом о высшем образовании.[/b]
[url=http://topnewsgadget.ru/page/2/]topnewsgadget.ru/page/2[/url]
[url=http://ip.webmasterhome.cn/?ip=frees-diplom.com/]ip.webmasterhome.cn/?ip=frees-diplom.com[/url]
[url=http://hellsms.com/cb-profile/pluginclass/cbblogs?action=blogs&func=show&id=6637/]hellsms.com/cb-profile/pluginclass/cbblogs?action=blogs&func=show&id=6637[/url]
[url=http://pakians.com/blogs/97805/Что-именно-требуется-из-оборудования-для-изготовления-диплома/]pakians.com/blogs/97805/Что-именно-требуется-из-оборудования-для-изготовления-диплома[/url]
[url=http://onlinekinospace.ru/page/2/]onlinekinospace.ru/page/2[/url]
Dnrtwnu
July 30, 2024 @ 1:53 am
[u][b] Привет![/b][/u]
[b]Купить документ[/b] ВУЗа вы можете в нашей компании в столице.
[url=http://diplomasx24.ru/kupit-diplom-krasnoyarsk/]diplomasx24.ru/kupit-diplom-krasnoyarsk[/url]
[b]Успешной учебы![/b]
Lazrgsm
July 30, 2024 @ 2:20 am
[u][b] Добрый день![/b][/u]
[b]Мы готовы предложить дипломы[/b] психологов, юристов, экономистов и любых других профессий по доступным ценам.
[url=http://aboutallfinance.ru/kupite-diplom-bez-ozhidaniya-i-slozhnostey/]aboutallfinance.ru/kupite-diplom-bez-ozhidaniya-i-slozhnostey[/url]
Mazrkpz
July 30, 2024 @ 2:20 am
[u][b] Здравствуйте![/b][/u]
Приобрести документ института можно у нас. Мы оказываем услуги по продаже документов об окончании любых университетов России. Вы получите необходимый диплом по любым специальностям, включая документы СССР. Гарантируем, что при проверке документов работодателем, подозрений не появится.
[b]Мы можем предложить дипломы[/b] психологов, юристов, экономистов и любых других профессий по выгодным ценам. Цена может зависеть от выбранной специальности, года выпуска и образовательного учреждения. Всегда стараемся поддерживать для клиентов адекватную политику тарифов. Важно, чтобы документы были доступными для большого количества наших граждан.
[url=http://comedyforme.ru/diplom-bez-zatrudneniy-variantyi-dlya-zanyatyih-lyudey/]comedyforme.ru/diplom-bez-zatrudneniy-variantyi-dlya-zanyatyih-lyudey[/url]
[b]Всегда вам поможем![/b]
Diplomi_ljSi
July 30, 2024 @ 2:29 am
Привет, друзья!
Заказать документ института можно у нас в столице.
[url=http://ast-diploms.com/kupit-diplom-kandidata-nauk/]ast-diploms.com/kupit-diplom-kandidata-nauk[/url]
Trefayv
July 30, 2024 @ 2:30 am
[u][b] Привет![/b][/u]
Как официально купить диплом вуза с упрощенным обучением в Москве
[url=http://remotebillpay.com/купить-диплом-о-высшем-образовании-ре/]remotebillpay.com/купить-диплом-о-высшем-образовании-ре[/url]
Рады оказать помощь!.
Xazrvwk
July 30, 2024 @ 3:10 am
[u][b] Добрый день![/b][/u]
Купить документ о получении высшего образования.
[url=http://figarohair.ru/conf/viewtopic.php?f=11&t=12656&sid=b855b67a79a79da9457b5d595541f640/]figarohair.ru/conf/viewtopic.php?f=11&t=12656&sid=b855b67a79a79da9457b5d595541f640[/url]
[url=http://forum.vorchun.ru/viewtopic.php?f=2&t=253067/]forum.vorchun.ru/viewtopic.php?f=2&t=253067[/url]
[url=http://newmedtime.ru/ofitsialnyie-diplomyi-po-dostupnyim-tsenam/]newmedtime.ru/ofitsialnyie-diplomyi-po-dostupnyim-tsenam[/url]
[url=http://blogsfere.com/viewtopic.php?t=181808/]blogsfere.com/viewtopic.php?t=181808[/url]
[url=http://mamapoltava.listbb.ru/viewtopic.php?f=39&t=5321/]mamapoltava.listbb.ru/viewtopic.php?f=39&t=5321[/url]
Dnrtqrb
July 30, 2024 @ 4:39 am
[u][b] Привет![/b][/u]
[b]Заказать документ[/b] института можно у нас.
[url=http://ast-diplomas.com/otzyvy/]ast-diplomas.com/otzyvy[/url]
[b]Хорошей учебы![/b]
Mazrghe
July 30, 2024 @ 5:01 am
[u][b] Здравствуйте![/b][/u]
[b]Узнайте, как приобрести диплом о высшем образовании без рисков[/b]
[url=http://mandiplomikx.ru/]mandiplomikx.ru[/url]
Mazrjvd
July 30, 2024 @ 10:20 am
[u][b] Привет![/b][/u]
[b]Сколько стоит диплом высшего и среднего образования и как это происходит?[/b]
[url=http://diploms-man.ru/]diploms-man.ru[/url]
Uazrssd
July 30, 2024 @ 10:45 am
[u][b] Привет![/b][/u]
[b]Приобрести диплом о высшем образовании.[/b]
[url=http://trum.flybb.ru/viewtopic.php?f=15&t=4688&sid=87dec77769dda0a19b59f24b871611b0/]trum.flybb.ru/viewtopic.php?f=15&t=4688&sid=87dec77769dda0a19b59f24b871611b0[/url]
Edwardbug
July 30, 2024 @ 11:53 am
Синергель https://sinergels.ru состав препарата и инструкция по применению. Показания и противопоказания.
PercyHex
July 30, 2024 @ 2:37 pm
AI Nude Generator https://ainudegenerator.app Transform Photos with AI Nude App.
Treflhl
July 30, 2024 @ 3:17 pm
[u][b] Привет![/b][/u]
Реально ли приобрести диплом стоматолога? Основные этапы
[url=http://higherranker.com/купить-диплом-проверенные-способы/]higherranker.com/купить-диплом-проверенные-способы[/url]
Рады помочь!.
Razrsto
July 30, 2024 @ 3:58 pm
[u][b] Здравствуйте![/b][/u]
Приобрести диплом университета:
[url=http://mpsamp.com/bbs/board.php?bo_table=free&wr_id=245178/]mpsamp.com/bbs/board.php?bo_table=free&wr_id=245178[/url]
[url=http://опт-кирпич.рф/index.php?subaction=userinfo&user=ubitaxos/]опт-кирпич.рф/index.php?subaction=userinfo&user=ubitaxos[/url]
[url=http://sborkakuhni.com/users/ibyfid/]sborkakuhni.com/users/ibyfid[/url]
[url=http://kirpich-opt-265.1gb.ru/index.php?subaction=userinfo&user=uluhaxur/]kirpich-opt-265.1gb.ru/index.php?subaction=userinfo&user=uluhaxur[/url]
[url=http://daongil.com/bbs/board.php?bo_table=free&wr_id=298085/]daongil.com/bbs/board.php?bo_table=free&wr_id=298085[/url]
Yrefhgb
July 30, 2024 @ 4:23 pm
[u][b] Здравствуйте![/b][/u]
[b]Заказать диплом любого университета.[/b]
[url=http://sdflex.kr/bbs/board.php?bo_table=faq&wr_id=19048&page=1/]sdflex.kr/bbs/board.php?bo_table=faq&wr_id=19048&page=1[/url]
[url=http://khelafat.com/blogs/4390/Ценообразование-документов-авторский-обзор-специалистов/]khelafat.com/blogs/4390/Ценообразование-документов-авторский-обзор-специалистов[/url]
[url=http://medik-look.ru/page/2/]medik-look.ru/page/2[/url]
[url=http://hyteratelsizleri.com/urundetay/hytera-pd355/]hyteratelsizleri.com/urundetay/hytera-pd355[/url]
[url=http://forum.unrivaled.ro/index.php?/gallery/image/532-аттестаты-дипломы-и-сертификаты-в-знаменитом-онлайн-магазине/]forum.unrivaled.ro/index.php?/gallery/image/532-аттестаты-дипломы-и-сертификаты-в-знаменитом-онлайн-магазине[/url]
Xazrcqh
July 30, 2024 @ 4:47 pm
[u][b] Здравствуйте![/b][/u]
Заказать документ о получении высшего образования.
[url=http://agrofly.ch/Официальные_дипломы_без_лишних_хлопот/]agrofly.ch/Официальные_дипломы_без_лишних_хлопот[/url]
[url=http://cartagena.activeboard.com/forum.spark/]cartagena.activeboard.com/forum.spark[/url]
[url=http://lolipopnews.ru/diplomyi-lyuboy-slozhnosti-byistro-i-bez-problem/]lolipopnews.ru/diplomyi-lyuboy-slozhnosti-byistro-i-bez-problem[/url]
[url=http://benedeek.com/blogs/91884/Подлинные-дипломы-от-ведущих-вузов/]benedeek.com/blogs/91884/Подлинные-дипломы-от-ведущих-вузов[/url]
[url=http://forexrassia.ru/diplomyi-lyuboy-slozhnosti-byistro-i-bez-problem/]forexrassia.ru/diplomyi-lyuboy-slozhnosti-byistro-i-bez-problem[/url]
Dnrtogr
July 30, 2024 @ 5:33 pm
[u][b] Привет, друзья![/b][/u]
[b]Купить документ[/b] о получении высшего образования можно в нашем сервисе.
[url=http://diplomasx.com/kupit-diplom-rostov-na-donu/]diplomasx.com/kupit-diplom-rostov-na-donu[/url]
[b]Удачи![/b]
Trefzgb
July 30, 2024 @ 5:51 pm
[u][b] Привет, друзья![/b][/u]
Приобретение диплома ПТУ с сокращенной программой обучения в Москве
[url=http://remotebillpay.com/купить-диплом-о-высшем-образовании-ре/]remotebillpay.com/купить-диплом-о-высшем-образовании-ре[/url]
Рады помочь!.
Dominictix
July 30, 2024 @ 5:56 pm
mexican rx online: mexico pharmacies prescription drugs – medication from mexico pharmacy
Dominictix
July 30, 2024 @ 6:35 pm
best online pharmacies in mexico: mexican pharmaceuticals online – medicine in mexico pharmacies
Dnrtpfv
July 30, 2024 @ 8:20 pm
[u][b] Здравствуйте![/b][/u]
[b]Приобрести документ[/b] ВУЗа можно в нашем сервисе.
[url=http://ast-diploms24.ru/kupit-diplom-omsk/]ast-diploms24.ru/kupit-diplom-omsk[/url]
[b]Хорошей учебы![/b]
Uazrrhk
July 30, 2024 @ 8:22 pm
[u][b] Здравствуйте![/b][/u]
Где заказать диплом специалиста?
[url=http://themepacific.com/support/users/lutroduyda//]themepacific.com/support/users/lutroduyda/[/url]
Diplomi_tzSi
July 30, 2024 @ 10:38 pm
Привет, друзья!
Заказать документ института можно у нас в Москве.
[url=http://diplomyx.com/kupit-diplom-s-registraciej/]diplomyx.com/kupit-diplom-s-registraciej[/url]
Danrakv
July 30, 2024 @ 11:31 pm
[u][b] Привет, друзья![/b][/u]
Мы предлагаем дипломы любой профессии.
[url=http://conectta2.com/read-blog/814_ishem-proverennyj-magazin-s-diplomami-i-attestatami]conectta2.com/read-blog/814_ishem-proverennyj-magazin-s-diplomami-i-attestatami[/url]
Поможем вам всегда!.
NelsonbiC
July 30, 2024 @ 11:55 pm
buying from online mexican pharmacy: mexican rx online – reputable mexican pharmacies online
Cazrupg
July 31, 2024 @ 12:03 am
[u][b] Привет, друзья![/b][/u]
Купить диплом о высшем образовании
[url=http://club2108.ru/forum/post.php?fid=57/]club2108.ru/forum/post.php?fid=57[/url]
[url=http://realkeys.flybb.ru/viewtopic.php?f=1&t=459/]realkeys.flybb.ru/viewtopic.php?f=1&t=459[/url]
[url=http://generalarminius.com/viewtopic.php?t=156570/]generalarminius.com/viewtopic.php?t=156570[/url]
[url=http://pnevmokzn.80lvl.ru/ucp.php?mode=register&sid=4bb286368ef29b1bf276b15c81477724/]pnevmokzn.80lvl.ru/ucp.php?mode=register&sid=4bb286368ef29b1bf276b15c81477724[/url]
[url=http://ya3bbru.bbok.ru/viewtopic.php?id=4092#p7202/]ya3bbru.bbok.ru/viewtopic.php?id=4092#p7202[/url]
Sazraqi
July 31, 2024 @ 12:04 am
[u][b] Добрый день![/b][/u]
Диплом бакалавра
[url=http://u2c.tv/member.php?u=30984/]u2c.tv/member.php?u=30984[/url]
[url=http://ya3bbru.bbok.ru/viewtopic.php?id=4083#p7183/]ya3bbru.bbok.ru/viewtopic.php?id=4083#p7183[/url]
[url=http://forum.kam.su/member.php?u=5734/]forum.kam.su/member.php?u=5734[/url]
[url=http://sankt-peterburg.forum2x2.ru/t20701-topic#77611/]sankt-peterburg.forum2x2.ru/t20701-topic#77611[/url]
[url=http://peaceofficial.5nx.ru/posting.php?mode=post&f=111&sid=d4866bda1a098dc4a544e09cc8148f4e/]peaceofficial.5nx.ru/posting.php?mode=post&f=111&sid=d4866bda1a098dc4a544e09cc8148f4e[/url]
Diplomi_csSi
July 31, 2024 @ 1:21 am
Здравствуйте!
Приобрести документ о получении высшего образования можно в нашей компании.
[url=http://ast-diplom.com/kupit-diplom-s-registraciej/]ast-diplom.com/kupit-diplom-s-registraciej[/url]
Iarioryre
July 31, 2024 @ 1:33 am
[u][b] Привет, друзья![/b][/u]
Приобрести документ о получении высшего образования вы имеете возможность в нашей компании. Мы оказываем услуги по производству и продаже документов об окончании любых ВУЗов России. Вы сможете получить диплом по любым специальностям, любого года выпуска, включая документы старого образца. Гарантируем, что при проверке документов работодателями, подозрений не возникнет.
[url=http://wowonder.xyz/read-blog/78394/]wowonder.xyz/read-blog/78394[/url]
[url=http://meco.eeconme.com/read-blog/3907/]meco.eeconme.com/read-blog/3907[/url]
[url=http://app-s.ru/blog/pochemu_onlajn_magaziny_s_diplomami_segodnja_tak_populjarny/2024-07-10-1904/]app-s.ru/blog/pochemu_onlajn_magaziny_s_diplomami_segodnja_tak_populjarny/2024-07-10-1904[/url]
[url=http://forum.safe-animals.ru/index.php/]forum.safe-animals.ru/index.php[/url]
[url=http://fijianvibe.com/blogs/783/Прочитайте-наш-материал-если-нужно-купить-диплом-в-интернете/]fijianvibe.com/blogs/783/Прочитайте-наш-материал-если-нужно-купить-диплом-в-интернете[/url]
Razrgao
July 31, 2024 @ 2:39 am
[u][b] Привет![/b][/u]
Заказать диплом любого ВУЗа:
[url=http://dachamania.ru/photo_gallrey/profile.php?uid=57180/]dachamania.ru/photo_gallrey/profile.php?uid=57180[/url]
[url=http://rytof.ru/node/16245/]rytof.ru/node/16245[/url]
[url=http://rivertravel.net/viewtopic.php?f=21&t=8495/]rivertravel.net/viewtopic.php?f=21&t=8495[/url]
[url=http://payhotel.ru/forum/messages/forum2/topic7566/message7822/?result=reply#message7822/]payhotel.ru/forum/messages/forum2/topic7566/message7822/?result=reply#message7822[/url]
[url=http://”cf58051.tmweb.ru/index.php?action=post;topic=2257.0;last_msg=132488″/]”cf58051.tmweb.ru/index.php?action=post;topic=2257.0;last_msg=132488″[/url]
Xazrcgh
July 31, 2024 @ 2:54 am
[b]Добрый день![/b]
Как купить аттестат 11 класса с официальным упрощенным обучением в Москве
[url=http://landik-diploms-srednee.ru/kupit-diplom-v-novosibirske]landik-diploms-srednee.ru/kupit-diplom-v-novosibirske[/url]
Manruqt
July 31, 2024 @ 2:55 am
[u][b] Добрый день![/b][/u]
Заказать документ о получении высшего образования
[url=http://diploms-x24.ru/kupit-diplom-vracha]diploms-x24.ru/kupit-diplom-vracha[/url]
Lazrxjm
July 31, 2024 @ 3:04 am
[b]Заказать диплом любого ВУЗа.[/b]
[url=http://fotomaniya.getbb.ru/viewtopic.php?f=16&t=11163/]fotomaniya.getbb.ru/viewtopic.php?f=16&t=11163[/url]
[url=http://soad.msk.ru/forum/posting.php?mode=newtopic&f=16/]soad.msk.ru/forum/posting.php?mode=newtopic&f=16[/url]
[url=http://goup.hashnode.dev/kupit-nastoyashij-diplom-prostoe-reshenie/]goup.hashnode.dev/kupit-nastoyashij-diplom-prostoe-reshenie[/url]
[url=http://etoprosto.ru/ru/forum/?category=5&action=topic/]etoprosto.ru/ru/forum/?category=5&action=topic[/url]
[url=http://igrosoft.getbb.ru/viewtopic.php?f=11&t=1332/]igrosoft.getbb.ru/viewtopic.php?f=11&t=1332[/url]
Williamraics
July 31, 2024 @ 3:07 am
[u][b] Добрый день![/b][/u]
Мы изготавливаем дипломы.
[url=http://elitemagyaritasok.info/e107_plugins/forum/forum_post.php?f=nt&id=21#/]elitemagyaritasok.info/e107_plugins/forum/forum_post.php?f=nt&id=21#[/url]
[url=http://altasugar.it/new/index.php?option=com_kunena&view=topic&catid=3&id=118950&Itemid=151/]altasugar.it/new/index.php?option=com_kunena&view=topic&catid=3&id=118950&Itemid=151[/url]
[url=http://a63.flybb.ru/viewtopic.php?f=16&t=3173/]a63.flybb.ru/viewtopic.php?f=16&t=3173[/url]
[url=http://allseoreg.wordpress.com/2024/07/08/легальна€-покупка-дипломов-онлайн/]allseoreg.wordpress.com/2024/07/08/легальна€-покупка-дипломов-онлайн[/url]
[url=http://moskvic.actieforum.com/t3720-topic#7009/]moskvic.actieforum.com/t3720-topic#7009[/url]
web site
July 31, 2024 @ 3:39 am
Hmm is anyone else encountering problems with the images on this
blog loading? I’m trying to find out if its a problem on my end or
if it’s the blog. Any feed-back would be greatly appreciated.
Eanrade
July 31, 2024 @ 3:42 am
[u][b] Здравствуйте![/b][/u]
[b]Мы готовы предложить дипломы[/b] любых профессий по доступным ценам.
[url=http://xpertmapping.com/как-можно-будет-недорого-приобрести-а/]xpertmapping.com/как-можно-будет-недорого-приобрести-а[/url]
[url=http://sonnick84.thezenweb.com/Документы-по-комфортным-ценам-в-знаменитом-интернет-магазине-66055539/]sonnick84.thezenweb.com/Документы-по-комфортным-ценам-в-знаменитом-интернет-магазине-66055539[/url]
[url=http://zekond.com/read-blog/53359/]zekond.com/read-blog/53359[/url]
[url=http://woorichat.com/read-blog/829/]woorichat.com/read-blog/829[/url]
[url=http://ffxiv.one/blogs/143/На-что-именно-обратить-свое-внимание-при-покупки-диплома-в/]ffxiv.one/blogs/143/На-что-именно-обратить-свое-внимание-при-покупки-диплома-в[/url]
Lazrevj
July 31, 2024 @ 5:57 am
[u][b] Привет, друзья![/b][/u]
[b]Мы изготавливаем дипломы[/b] любых профессий по доступным тарифам.
[url=http://thecareerer.com/wordpress/2024/06/30/купить-диплом-продавца-2/]thecareerer.com/wordpress/2024/06/30/купить-диплом-продавца-2[/url]
Treflzh
July 31, 2024 @ 6:16 am
[u][b] Привет, друзья![/b][/u]
Официальное получение диплома техникума с упрощенным обучением в Москве
[url=http://dadiler.com/купить-диплом-советы-и-рекомендации/]dadiler.com/купить-диплом-советы-и-рекомендации[/url]
Рады помочь!.
Uazrtkt
July 31, 2024 @ 7:47 am
[u][b] Привет, друзья![/b][/u]
Где приобрести диплом специалиста?
[url=http://buro-alfa.ru/alfa/2022/04/22/zachem-neobhodimo-poluchenie-diploma-onlayn.html/]buro-alfa.ru/alfa/2022/04/22/zachem-neobhodimo-poluchenie-diploma-onlayn.html[/url]
Trefyfx
July 31, 2024 @ 8:50 am
[u][b] Здравствуйте![/b][/u]
Как оказалось, купить диплом кандидата наук не так уж и сложно
[url=http://sex8.zone/home.php?mod=space&uid=9145316&do=profile/]sex8.zone/home.php?mod=space&uid=9145316&do=profile[/url]
Поможем вам всегда!.
Xazruce
July 31, 2024 @ 8:52 am
[u][b] Здравствуйте![/b][/u]
Заказать документ ВУЗа.
[url=http://altersoftonline.maxbb.ru/posting.php?mode=post&f=11/]altersoftonline.maxbb.ru/posting.php?mode=post&f=11[/url]
[url=http://newsato.ru/vash-shans-na-uspeh-s-nashim-diplomom/]newsato.ru/vash-shans-na-uspeh-s-nashim-diplomom[/url]
[url=http://derivsocial.org/read-blog/2619/]derivsocial.org/read-blog/2619[/url]
[url=http://woman.build2.ru/viewtopic.php?id=14556#p44661/]woman.build2.ru/viewtopic.php?id=14556#p44661[/url]
[url=http://vnuci.listbb.ru/viewtopic.php?f=2&t=544/]vnuci.listbb.ru/viewtopic.php?f=2&t=544[/url]
Lazrjnb
July 31, 2024 @ 9:32 am
[u][b] Здравствуйте![/b][/u]
[b]Заказать диплом о высшем образовании.[/b]
[url=http://rugladyseminars.com/cb-profile/pluginclass/cbblogs?action=blogs&func=show&id=399/]rugladyseminars.com/cb-profile/pluginclass/cbblogs?action=blogs&func=show&id=399[/url]
Yrefcre
July 31, 2024 @ 9:52 am
[u][b] Добрый день![/b][/u]
[b]Купить диплом о высшем образовании.[/b]
[url=http://qyshikisoxa.lowescouponn.com/cto-potrebuetsa-iz-oborudovania-dla-izgotovlenia-diploma/]qyshikisoxa.lowescouponn.com/cto-potrebuetsa-iz-oborudovania-dla-izgotovlenia-diploma[/url]
[url=http://filmbaaz.in/blogs/942/Что-понадобится-для-производства-качественного-диплома-университета/]filmbaaz.in/blogs/942/Что-понадобится-для-производства-качественного-диплома-университета[/url]
[url=http://madesports.net/купить-диплом-в-бульгуме-анонимно-и-по/]madesports.net/купить-диплом-в-бульгуме-анонимно-и-по[/url]
[url=http://gaeloxeparo.lucialpiazzale.com/cena-dokumentov-sovety-i-obzor-ekspertov/]gaeloxeparo.lucialpiazzale.com/cena-dokumentov-sovety-i-obzor-ekspertov[/url]
[url=http://madesports.net/купить-диплом-в-бульгуме-анонимно-и-по/]madesports.net/купить-диплом-в-бульгуме-анонимно-и-по[/url]
Sazrtyr
July 31, 2024 @ 10:03 am
[u][b] Добрый день![/b][/u]
Диплом любой специальности
[url=http://silich.ru/forum/member.php?u=4810/]silich.ru/forum/member.php?u=4810[/url]
[url=http://stroimsa.forum2x2.ru/t1838-topic#4746/]stroimsa.forum2x2.ru/t1838-topic#4746[/url]
[url=http://netcallvoip.com/wiki/index.php/дипломы_для_успешных_людей:_начните_новую_главу_жизни/]netcallvoip.com/wiki/index.php/дипломы_для_успешных_людей:_начните_новую_главу_жизни[/url]
[url=http://burgasdent.listbb.ru/viewtopic.php?f=3&t=409/]burgasdent.listbb.ru/viewtopic.php?f=3&t=409[/url]
[url=http://liquidationrama.com/read-blog/1135/]liquidationrama.com/read-blog/1135[/url]
Cazrwdn
July 31, 2024 @ 10:08 am
[u][b] Добрый день![/b][/u]
Приобрести диплом института
[url=http://monstaluck.mn.co/posts/62527858/]monstaluck.mn.co/posts/62527858[/url]
[url=http://alternativaprofi.com/forum/viewthread.php?thread_id=33707/]alternativaprofi.com/forum/viewthread.php?thread_id=33707[/url]
[url=http://cslregister.com/forum/member.php?u=6225/]cslregister.com/forum/member.php?u=6225[/url]
[url=http://vip.rolevaya.info/viewtopic.php?id=5053#p8679/]vip.rolevaya.info/viewtopic.php?id=5053#p8679[/url]
[url=http://semeyka.listbb.ru/viewtopic.php?f=20&t=790/]semeyka.listbb.ru/viewtopic.php?f=20&t=790[/url]
Cazrwok
July 31, 2024 @ 10:26 am
[u][b] Добрый день![/b][/u]
Мы готовы предложить документы техникумов
[url=http://buldingnews.ru/poluchite-diplom-i-nachnite-novuyu-zhizn/]buldingnews.ru/poluchite-diplom-i-nachnite-novuyu-zhizn[/url]
[url=http://kolhos.listbb.ru/viewtopic.php?f=2&t=637/]kolhos.listbb.ru/viewtopic.php?f=2&t=637[/url]
[url=http://seaman.com.ua/forum/index.php?threads/diplomy-dlja-vashego-uspexa.37862//]seaman.com.ua/forum/index.php?threads/diplomy-dlja-vashego-uspexa.37862/[/url]
[url=http://video.listbb.ru/viewtopic.php?f=3&t=419/]video.listbb.ru/viewtopic.php?f=3&t=419[/url]
[url=http://sadcr.listbb.ru/viewtopic.php?f=53&t=398/]sadcr.listbb.ru/viewtopic.php?f=53&t=398[/url]
Sazredi
July 31, 2024 @ 10:46 am
[u][b] Привет![/b][/u]
Приобрести документ о получении высшего образования
[url=http://shopy.partners/read-blog/1547/]shopy.partners/read-blog/1547[/url]
[url=http://naya.social/read-blog/99/]naya.social/read-blog/99[/url]
[url=http://upuge.com/read-blog/31643_vybiraem-proverennyj-internet-magazin-dlya-priobreteniya-diploma.html/]upuge.com/read-blog/31643_vybiraem-proverennyj-internet-magazin-dlya-priobreteniya-diploma.html[/url]
[url=http://gamerszona.ru/index.php?/blogs/entry/6-какие-онлайн-магазины-с-документами-на-сегодняшний-момент-есть//]gamerszona.ru/index.php?/blogs/entry/6-какие-онлайн-магазины-с-документами-на-сегодняшний-момент-есть/[/url]
[url=http://kaiztech.net/IPB/index.php?/gallery/image/424-каталог-и-варианты-доставки-в-онлайн-магазине-заказываем-аттестат//]kaiztech.net/IPB/index.php?/gallery/image/424-каталог-и-варианты-доставки-в-онлайн-магазине-заказываем-аттестат/[/url]
Mazrwlj
July 31, 2024 @ 11:38 am
[u][b] Привет, друзья![/b][/u]
[b]Полезная информация как официально купить диплом о высшем образовании[/b]
[url=http://diplomyx-man.ru/]diplomyx-man.ru[/url]
Xazrxyv
July 31, 2024 @ 11:49 am
[u][b] Привет![/b][/u]
Заказать документ института.
[url=http://polegasm.net/index.php/forum/welcome-mat/151619/]polegasm.net/index.php/forum/welcome-mat/151619[/url]
[url=http://amengram.com/blogs/401/Легальный-диплом-без-лишних-хлопот-узнайте-как/]amengram.com/blogs/401/Легальный-диплом-без-лишних-хлопот-узнайте-как[/url]
[url=http://productinn.mn.co/posts/62245206/]productinn.mn.co/posts/62245206[/url]
[url=http://vatrusha.maxbb.ru/viewtopic.php?f=2&t=701/]vatrusha.maxbb.ru/viewtopic.php?f=2&t=701[/url]
[url=http://oldforum.citysakh.ru/?talkid=26720/]oldforum.citysakh.ru/?talkid=26720[/url]
Larioryjy
July 31, 2024 @ 12:50 pm
Как официально купить диплом вуза с упрощенным обучением в Москве
[url=http://avtolux48.ru/people/user/367/blog/7897/]avtolux48.ru/people/user/367/blog/7897[/url]
[url=http://les-ailes-chalaisiennes.com/forum/topic/диплом-любой-специальности-качестве/#postid-19484/]les-ailes-chalaisiennes.com/forum/topic/диплом-любой-специальности-качестве/#postid-19484[/url]
[url=http://master39.net/forum/user/4854/]master39.net/forum/user/4854[/url]
[url=http://toplentanews.ru/gde-kupit-diplom-polnoe-rukovodstvo/]toplentanews.ru/gde-kupit-diplom-polnoe-rukovodstvo[/url]
[url=http://niceroleplay.getbb.ru/viewtopic.php?f=1&t=1223/]niceroleplay.getbb.ru/viewtopic.php?f=1&t=1223[/url]
Xazrguc
July 31, 2024 @ 2:01 pm
[b]Здравствуйте![/b]
Официальная покупка диплома вуза с сокращенной программой обучения в Москве
[url=http://arusak-diploms-srednee.ru/kupit-diplom-v-rostove-na-donu]arusak-diploms-srednee.ru/kupit-diplom-v-rostove-na-donu[/url]
Manrizu
July 31, 2024 @ 2:04 pm
[u][b] Добрый день![/b][/u]
Купить документ о получении высшего образования
[url=http://diplomyx24.ru/kupit-diplom-o-srednem-obrazovanii]diplomyx24.ru/kupit-diplom-o-srednem-obrazovanii[/url]
Williamraics
July 31, 2024 @ 2:21 pm
[u][b] Привет, друзья![/b][/u]
Мы можем предложить дипломы.
[url=http://4x4zubry.by/forum/messages/forum1/topic2034/message202266/?result=new#message202266/]4x4zubry.by/forum/messages/forum1/topic2034/message202266/?result=new#message202266[/url]
[url=http://bn.tobase.ru/viewtopic.php?f=30&t=3346/]bn.tobase.ru/viewtopic.php?f=30&t=3346[/url]
[url=http://desteg.getbb.ru/viewtopic.php?f=6&t=801/]desteg.getbb.ru/viewtopic.php?f=6&t=801[/url]
[url=http://sfera100.ru/forum/user/2581/]sfera100.ru/forum/user/2581[/url]
[url=http://forexrassia.ru/nastoyashhie-diplomyi-bez-ozhidaniya-i-problem/]forexrassia.ru/nastoyashhie-diplomyi-bez-ozhidaniya-i-problem[/url]
Lazrovh
July 31, 2024 @ 2:21 pm
[b]Купить диплом любого университета.[/b]
[url=http://elektrofahrrad-tests.de/forums/newthread.php?fid=2&processed=1/]elektrofahrrad-tests.de/forums/newthread.php?fid=2&processed=1[/url]
[url=http://balans.kz/bonus/]balans.kz/bonus[/url]
[url=http://mybaltika.info/ru/blogs/587/10253/]mybaltika.info/ru/blogs/587/10253[/url]
[url=http://obrezanie05.ru/users/15/]obrezanie05.ru/users/15[/url]
[url=http://ya.10bb.ru/viewtopic.php?id=3955#p7051/]ya.10bb.ru/viewtopic.php?id=3955#p7051[/url]
Eanrzey
July 31, 2024 @ 2:41 pm
[u][b] Здравствуйте![/b][/u]
[b]Мы готовы предложить дипломы[/b] психологов, юристов, экономистов и других профессий по доступным ценам.
[url=http://network-66643.mn.co/posts/61724716/]network-66643.mn.co/posts/61724716[/url]
[url=http://sonnick84.anchor-blog.com/8509136/Как-заказать-сейчас-качественный-диплом-в-сети/]sonnick84.anchor-blog.com/8509136/Как-заказать-сейчас-качественный-диплом-в-сети[/url]
[url=http://ecoheal.ru/index.php/jomsocial/groups/viewbulletin/296-bystryj-vybor-magazina-realizuyushchego-diplomy?groupid=1/]ecoheal.ru/index.php/jomsocial/groups/viewbulletin/296-bystryj-vybor-magazina-realizuyushchego-diplomy?groupid=1[/url]
[url=http://contraband.ch/read-blog/3846/]contraband.ch/read-blog/3846[/url]
[url=http://transfer-tur.ru/ru/cb-profile/pluginclass/cbblogs?action=blogs&func=show&id=165/]transfer-tur.ru/ru/cb-profile/pluginclass/cbblogs?action=blogs&func=show&id=165[/url]
Mazrkpm
July 31, 2024 @ 4:54 pm
[u][b] Привет, друзья![/b][/u]
[b]Официальная покупка диплома вуза с упрощенной программой обучения[/b]
[url=http://diploms-man.ru/]diploms-man.ru[/url]
Vazrfcm
July 31, 2024 @ 6:11 pm
[u][b] Привет![/b][/u]
Заказать документ о получении высшего образования вы можете в нашей компании в Москве. Мы оказываем услуги по продаже документов об окончании любых ВУЗов РФ.
[url=http://axioma-estate.ru/index.php?subaction=userinfo&user=ilojoxod/]axioma-estate.ru/index.php?subaction=userinfo&user=ilojoxod[/url]
[url=http://gektorstroi.ru/forum/?PAGE_NAME=profile_view&UID=181570/]gektorstroi.ru/forum/?PAGE_NAME=profile_view&UID=181570[/url]
[url=http://forum.dalmatin-club.ru/topic/49653-купить-диплом-цена-j794a//]forum.dalmatin-club.ru/topic/49653-купить-диплом-цена-j794a/[/url]
[url=http://kuvandyk.ru/users.php?m=details&id=56318/]kuvandyk.ru/users.php?m=details&id=56318[/url]
[url=http://gdz-fizika.ru/index.php?subaction=userinfo&user=ijedyxa/]gdz-fizika.ru/index.php?subaction=userinfo&user=ijedyxa[/url]
Dnrtcnk
July 31, 2024 @ 6:35 pm
[b]Заказать документ[/b] о получении высшего образования вы сможете в нашем сервисе.
[url=http://ilovebookmarking.com/story17652594/diplom/]ilovebookmarking.com/story17652594/diplom[/url]
[url=http://meshbookmarks.com/story17701519/diplom/]meshbookmarks.com/story17701519/diplom[/url]
[url=http://tadalive.com/blog/337167/diplom-kupit-848855xw/]tadalive.com/blog/337167/diplom-kupit-848855xw[/url]
[url=http://brettswebsite.com/diplom-949722kuqdo/]brettswebsite.com/diplom-949722kuqdo[/url]
[url=http://paltosuzca.com/diplom-kupit-89995ps/]paltosuzca.com/diplom-kupit-89995ps[/url]
Trefqqd
July 31, 2024 @ 9:13 pm
[u][b] Привет, друзья![/b][/u]
Где и как купить диплом о высшем образовании без лишних рисков
[url=http://remotebillpay.com/купить-диплом-о-высшем-образовании-ре/]remotebillpay.com/купить-диплом-о-высшем-образовании-ре[/url]
Будем рады вам помочь!.
Dnrtxzr
July 31, 2024 @ 9:19 pm
[b]Приобрести документ[/b] о получении высшего образования вы можете в нашей компании в столице.
[url=http://tadalive.com/blog/338145/diplom-kupit-126277el/]tadalive.com/blog/338145/diplom-kupit-126277el[/url]
[url=http://tigaedu.com/blog/index.php?entryid=328997/]tigaedu.com/blog/index.php?entryid=328997[/url]
[url=http://bruthindustan.com/diplom-kupit-666651mf/]bruthindustan.com/diplom-kupit-666651mf[/url]
[url=http://seolistlinks.com/story18942414/diplom/]seolistlinks.com/story18942414/diplom[/url]
[url=http://digitalmagazine.xyz/diplom-920313tpflr/]digitalmagazine.xyz/diplom-920313tpflr[/url]
Cazrenf
July 31, 2024 @ 9:23 pm
[u][b] Здравствуйте![/b][/u]
Мы готовы предложить документы техникумов
[url=http://dachaweek.ru/kupite-diplom-byistro-i-legalno/]dachaweek.ru/kupite-diplom-byistro-i-legalno[/url]
[url=http://ic-info.ru/forum/user/166501//]ic-info.ru/forum/user/166501/[/url]
[url=http://dailygram.com/blog/1307115/легальное-получение-диплома-в-короткие-сроки//]dailygram.com/blog/1307115/легальное-получение-диплома-в-короткие-сроки/[/url]
[url=http://newfinbiz.ru/kachestvennyie-diplomyi-bez-posrednikov/]newfinbiz.ru/kachestvennyie-diplomyi-bez-posrednikov[/url]
[url=http://vip.rolevaya.info/viewtopic.php?id=5053#p8679/]vip.rolevaya.info/viewtopic.php?id=5053#p8679[/url]
Diplomi_eoSi
July 31, 2024 @ 9:37 pm
Добрый день!
Приобрести документ о получении высшего образования вы имеете возможность в нашей компании в Москве.
[url=http://ast-diplomas.com/kupit-diplom-kazan/]ast-diplomas.com/kupit-diplom-kazan[/url]
Trefkmh
July 31, 2024 @ 11:41 pm
[u][b] Привет, друзья![/b][/u]
Пошаговая инструкция по официальной покупке диплома о высшем образовании
[url=http://lampcanvas.com/купить-диплом-о-высшем-образовании-со/]lampcanvas.com/купить-диплом-о-высшем-образовании-со[/url]
Рады оказаться полезными!.
Lazrwuo
July 31, 2024 @ 11:45 pm
[u][b] Здравствуйте![/b][/u]
[b]Мы изготавливаем дипломы[/b] любой профессии по приятным тарифам.
[url=http://montrealpal.com/read-blog/815_kak-najti-nadezhnyj-magazin-prodayushij-diplomy/]montrealpal.com/read-blog/815_kak-najti-nadezhnyj-magazin-prodayushij-diplomy[/url]
Diplomi_uaSi
August 1, 2024 @ 12:19 am
Привет!
Заказать документ ВУЗа можно в нашей компании в Москве.
[url=http://asxdiplomik.com/kupit-diplom-krasnodar/]asxdiplomik.com/kupit-diplom-krasnodar[/url]
Xazrima
August 1, 2024 @ 1:17 am
[u][b] Привет![/b][/u]
Купить документ о получении высшего образования.
[url=http://revolverp.forumex.ru/viewtopic.php?f=11&t=317/]revolverp.forumex.ru/viewtopic.php?f=11&t=317[/url]
[url=http://plut-uchukov.flybb.ru/viewtopic.php?f=1&t=507/]plut-uchukov.flybb.ru/viewtopic.php?f=1&t=507[/url]
[url=http://mosfo.getbb.ru/viewtopic.php?f=12&t=922/]mosfo.getbb.ru/viewtopic.php?f=12&t=922[/url]
[url=http://kalininabad.flyboard.ru/viewtopic.php?f=11&t=2829/]kalininabad.flyboard.ru/viewtopic.php?f=11&t=2829[/url]
[url=http://biznas.com/Biz-postsm335972_Default.aspx#post335972/]biznas.com/Biz-postsm335972_Default.aspx#post335972[/url]
Dominictix
August 1, 2024 @ 1:31 am
mexican online pharmacies prescription drugs: medication from mexico pharmacy – reputable mexican pharmacies online
Xazrbih
August 1, 2024 @ 4:12 am
[u][b] Добрый день![/b][/u]
Приобрести документ университета.
[url=http://pdlspd.listbb.ru/viewtopic.php?f=3&t=471/]pdlspd.listbb.ru/viewtopic.php?f=3&t=471[/url]
[url=http://dyado.listbb.ru/viewtopic.php?f=2&t=462/]dyado.listbb.ru/viewtopic.php?f=2&t=462[/url]
[url=http://diigo.com/item/note/8p2f0/toi5?k=2c284abd9e67279ec0e70971a73240bb/]diigo.com/item/note/8p2f0/toi5?k=2c284abd9e67279ec0e70971a73240bb[/url]
[url=http://dp.getbb.ru/viewtopic.php?f=3&t=744/]dp.getbb.ru/viewtopic.php?f=3&t=744[/url]
[url=http://testimonyforgod.com/read-blog/5738/]testimonyforgod.com/read-blog/5738[/url]
Vazrdnr
August 1, 2024 @ 4:59 am
[u][b] Привет, друзья![/b][/u]
Приобрести документ о получении высшего образования вы можете в нашей компании в Москве. Мы предлагаем документы об окончании любых университетов Российской Федерации.
[url=http://uralremput.ru/index.php?subaction=userinfo&user=ijifi/]uralremput.ru/index.php?subaction=userinfo&user=ijifi[/url]
[url=http://rf-isolation.ru/index.php?/topic/726-event огненное-магическое-оружие-6-20к-gp/#comment-4486/]rf-isolation.ru/index.php?/topic/726-event огненное-магическое-оружие-6-20к-gp/#comment-4486[/url]
[url=http://mtm-2000.ru/users/izezavas/]mtm-2000.ru/users/izezavas[/url]
[url=http://whirlpowertool.ru/index.php?subaction=userinfo&user=ikacyrul/]whirlpowertool.ru/index.php?subaction=userinfo&user=ikacyrul[/url]
[url=http://printpoligraf.kiev.ua/index.php?subaction=userinfo&user=ikewag/]printpoligraf.kiev.ua/index.php?subaction=userinfo&user=ikewag[/url]
accounting homework help
August 1, 2024 @ 9:01 am
Accounting is necessary for students to analyse and evaluate financial data, which is a process that can be time-consuming and requires a high degree of ability. For this reason, a significant number of students look for accounting homework help on the internet, hoping to find someone who can assist them in improving their scores on accounting assignments. Are you in need of assistance with your accounting class? There is no need for you to be concerned at all since the team of experts is ready to provide you the highest possible level of online accounting homework help within the specified time frame. When it comes to assignments, quality should be the first priority, and students always want assistance that is of high quality and dependable in order to attain the marks they seek in their assignments. Consequently, the specialists are always working to improve the quality of the accounting solutions they provide.
Dnrtyer
August 1, 2024 @ 10:24 am
[b]Приобрести документ[/b] о получении высшего образования можно у нас в Москве.
[url=http://bookmarksusa.com/story17676650/diplom/]bookmarksusa.com/story17676650/diplom[/url]
[url=http://bookmarksurl.com/story3006864/diplom/]bookmarksurl.com/story3006864/diplom[/url]
[url=http://royalbookmarking.com/story17666351/diplom/]royalbookmarking.com/story17666351/diplom[/url]
[url=http://singnalsocial.com/story2935171/diplom/]singnalsocial.com/story2935171/diplom[/url]
[url=http://tigaedu.com/blog/index.php?entryid=325170/]tigaedu.com/blog/index.php?entryid=325170[/url]
Trefmqx
August 1, 2024 @ 12:15 pm
[u][b] Привет, друзья![/b][/u]
Сколько стоит диплом высшего и среднего образования и как его получить?
[url=http://higherranker.com/купить-диплом-проверенные-способы/]higherranker.com/купить-диплом-проверенные-способы[/url]
Рады оказать помощь!.
Lazrrzq
August 1, 2024 @ 12:42 pm
[u][b] Привет, друзья![/b][/u]
[b]Мы изготавливаем дипломы[/b] любой профессии по приятным тарифам.
[url=http://newsbig.fotosdefrases.com/rekomendacii-po-priobreteniu-dokumenta-v-internete/]newsbig.fotosdefrases.com/rekomendacii-po-priobreteniu-dokumenta-v-internete[/url]
Dnrtpym
August 1, 2024 @ 1:15 pm
[b]Приобрести документ[/b] о получении высшего образования можно в нашем сервисе.
[url=http://letusbookmark.com/story19169220/diplom/]letusbookmark.com/story19169220/diplom[/url]
[url=http://utahsyardsale.com/author/alyssaainsw/]utahsyardsale.com/author/alyssaainsw[/url]
[url=http://bookmarklogin.com/story17759664/diplom/]bookmarklogin.com/story17759664/diplom[/url]
[url=http://pirooztak.ir/?option=com_k2&view=itemlist&task=user&id=1358403/]pirooztak.ir/?option=com_k2&view=itemlist&task=user&id=1358403[/url]
[url=http://managerhotels.com/business-sales/diplom-112785oamig/]managerhotels.com/business-sales/diplom-112785oamig[/url]
Yrefwzs
August 1, 2024 @ 2:02 pm
[u][b] Добрый день![/b][/u]
[b]Заказать диплом университета.[/b]
[url=http://toyota-porte.ru/forums/index.php?autocom=gallery&req=si&img=2223/]toyota-porte.ru/forums/index.php?autocom=gallery&req=si&img=2223[/url]
[url=http://arikkeu.com/g5/bbs/board.php?bo_table=arikkeu1234_&wr_id=82313/]arikkeu.com/g5/bbs/board.php?bo_table=arikkeu1234_&wr_id=82313[/url]
[url=http://kotanusantara.cloud/index.php?/events/event/13-быстрый-выбор-онлайн-магазина-что-реализует-дипломы-и-аттестаты/]kotanusantara.cloud/index.php?/events/event/13-быстрый-выбор-онлайн-магазина-что-реализует-дипломы-и-аттестаты[/url]
[url=http://find-topdeals.com/blogs/133404/Где-купить-диплом-с-гарантией-качества/]find-topdeals.com/blogs/133404/Где-купить-диплом-с-гарантией-качества[/url]
[url=http://weekinato.ru/page/3/]weekinato.ru/page/3[/url]
Trefdqk
August 1, 2024 @ 3:00 pm
[u][b] Здравствуйте![/b][/u]
Полезные советы по безопасной покупке диплома о высшем образовании
[url=http://csleague.ca/диплом-о-среднем-образовании-купить-н/]csleague.ca/диплом-о-среднем-образовании-купить-н[/url]
Рады оказаться полезными!.
ScottieEffex
August 1, 2024 @ 3:00 pm
RuNet https://gallerix.asia ?????????????????? ??4?????,?????????????????
journey to wellness
August 1, 2024 @ 3:08 pm
Starting a journey to wellness is an exciting adventure that promises to enhance your life in every aspect—physical, mental, emotional, and spiritual. Whether your goal is to improve your health, achieve a greater sense of balance, or feel better in your daily life, adopting a holistic approach can bring profound benefits. This journey is about more than just exercising or eating right; it’s about making a comprehensive change that touches every part of your being.
Steveblurb
August 1, 2024 @ 3:42 pm
Центр сертификации https://www.rospromtest.ru осуществляет деятельность по содействию в подтверждении соответствия продукции и услуг требованиям нормативных документов, технических регламентов Таможенного союза, и сертификации ISO. Мы оказываем полный комплекс услуг в сфере сертификации.
Sazrgqx
August 1, 2024 @ 3:44 pm
[u][b] Привет![/b][/u]
Мы готовы предложить дипломы психологов, юристов, экономистов и любых других профессий.
[url=http://offmarketbusinessforsale.com/kupit-diplom-544589cuefs/]offmarketbusinessforsale.com/kupit-diplom-544589cuefs[/url]
[url=http://bookmarkforest.com/story17605765/kupit-diplom/]bookmarkforest.com/story17605765/kupit-diplom[/url]
[url=http://fam2fam.com/groups/kupit-diplom-721876npoww/]fam2fam.com/groups/kupit-diplom-721876npoww[/url]
[url=http://wisesocialsmedia.com/story2957134/kupit-diplom/]wisesocialsmedia.com/story2957134/kupit-diplom[/url]
[url=http://segisocial.com/kupit-diplom-521450vifpy/]segisocial.com/kupit-diplom-521450vifpy[/url]
Mazrqop
August 1, 2024 @ 3:50 pm
[b]Купить документ о получении высшего образования.[/b]
[url=http://enrollbookmarks.com/story17610146/kupit-diplom/]enrollbookmarks.com/story17610146/kupit-diplom[/url]
[url=http://bookmarkity.com/story17722154/kupit-diplom/]bookmarkity.com/story17722154/kupit-diplom[/url]
[url=http://bookmarkpath.com/story17614552/kupit-diplom/]bookmarkpath.com/story17614552/kupit-diplom[/url]
[url=http://bookmarksden.com/story17813900/kupit-diplom/]bookmarksden.com/story17813900/kupit-diplom[/url]
[url=http://thekiwisocial.com/story3019387/kupit-diplom/]thekiwisocial.com/story3019387/kupit-diplom[/url]
Arnoldstota
August 1, 2024 @ 4:12 pm
medication from mexico pharmacy [url=http://mexicandeliverypharma.com/#]mexican drugstore online[/url] mexican mail order pharmacies
Xazrsxj
August 1, 2024 @ 5:59 pm
[u][b] Здравствуйте![/b][/u]
Купить документ о получении высшего образования.
[url=http://forexrassia.ru/oformlenie-diplomov-s-minimalnyimi-zatratami/]forexrassia.ru/oformlenie-diplomov-s-minimalnyimi-zatratami[/url]
[url=http://grot.getbb.ru/posting.php?mode=post&f=6&sid=46598a45c144d2d30d432aa01834fa32/]grot.getbb.ru/posting.php?mode=post&f=6&sid=46598a45c144d2d30d432aa01834fa32[/url]
[url=http://chromerp.listbb.ru/viewtopic.php?f=13&t=510/]chromerp.listbb.ru/viewtopic.php?f=13&t=510[/url]
[url=http://myworldavto.ru/legkiy-sposob-poluchit-diplom-lyuboy-spetsialnosti/]myworldavto.ru/legkiy-sposob-poluchit-diplom-lyuboy-spetsialnosti[/url]
[url=http://cardinalparkmld.listbb.ru/viewtopic.php?f=3&t=468/]cardinalparkmld.listbb.ru/viewtopic.php?f=3&t=468[/url]
Uazrucz
August 1, 2024 @ 6:03 pm
[b]Где приобрести диплом по нужной специальности?[/b]
[url=http://top10bookmark.com/story17539591/diplom/]top10bookmark.com/story17539591/diplom[/url]
[url=http://localtrusted.co.uk/wiki/index.php?title=Diplom_660323lPlWX/]localtrusted.co.uk/wiki/index.php?title=Diplom_660323lPlWX[/url]
[url=http://youwantech.com/xe/board/412263/]youwantech.com/xe/board/412263[/url]
[url=http://youlangue.lu/blog/index.php?entryid=87296/]youlangue.lu/blog/index.php?entryid=87296[/url]
[url=http://comfortrent.ru/2024/07/27/diplom-177737swopr/]comfortrent.ru/2024/07/27/diplom-177737swopr[/url]
Edwardbub
August 1, 2024 @ 6:28 pm
order balloons to your home https://buy-balloons-dubai.com
Mazrwsg
August 1, 2024 @ 6:30 pm
[u][b] Привет, друзья![/b][/u]
[b]Всё, что нужно знать о покупке аттестата о среднем образовании[/b]
[url=http://mans-diplom.ru/]mans-diplom.ru[/url]
Razrkyi
August 1, 2024 @ 7:34 pm
[u][b] Добрый день![/b][/u]
Заказать диплом о высшем образовании:
[url=http://b24activities.ru/club/forum/messages/forum4/topic76/message138/?result=reply#message138/]b24activities.ru/club/forum/messages/forum4/topic76/message138/?result=reply#message138[/url]
[url=http://e-audit.gefest.me/index.php?subaction=userinfo&user=uvituf/]e-audit.gefest.me/index.php?subaction=userinfo&user=uvituf[/url]
[url=http://cntu-vek.ru/forum/user/13073//]cntu-vek.ru/forum/user/13073/[/url]
[url=http://mpsamp.com/bbs/board.php?bo_table=free&wr_id=245178/]mpsamp.com/bbs/board.php?bo_table=free&wr_id=245178[/url]
[url=http://geolan-ksl.ru/forum/user/85791//]geolan-ksl.ru/forum/user/85791/[/url]
Xazrovj
August 1, 2024 @ 8:54 pm
[u][b] Привет, друзья![/b][/u]
Купить документ ВУЗа.
[url=http://dragonlivers.ixbb.ru/viewtopic.php?id=114#p114/]dragonlivers.ixbb.ru/viewtopic.php?id=114#p114[/url]
[url=http://ashinova.ru/category-5/t-1737#1737/]ashinova.ru/category-5/t-1737#1737[/url]
[url=http://coup.forum2x2.ru/t3797-topic#10048/]coup.forum2x2.ru/t3797-topic#10048[/url]
[url=http://myweektour.ru/vash-nadezhnyiy-pomoshhnik-v-poluchenii-diploma/]myweektour.ru/vash-nadezhnyiy-pomoshhnik-v-poluchenii-diploma[/url]
[url=http://ford-talks.ru/viewtopic.php?f=12&t=5090/]ford-talks.ru/viewtopic.php?f=12&t=5090[/url]
Diplomi_zhSi
August 1, 2024 @ 9:26 pm
Здравствуйте!
Приобрести документ института можно у нас в Москве.
[url=http://ast-diploms.com/kupit-diplom-tehnikuma-kolledzha/]ast-diploms.com/kupit-diplom-tehnikuma-kolledzha[/url]
Mazroeg
August 1, 2024 @ 11:59 pm
[u][b] Здравствуйте![/b][/u]
[b]Аттестат школы купить официально с упрощенным обучением в Москве[/b]
[url=http://diploms-man.ru/]diploms-man.ru[/url]
Diplomi_xzSi
August 2, 2024 @ 12:20 am
Здравствуйте!
Купить документ института можно у нас.
[url=http://ast-diplom.com/kupit-diplom-kandidata-nauk /]ast-diplom.com/kupit-diplom-kandidata-nauk [/url]
Cazronb
August 2, 2024 @ 1:28 am
[u][b] Добрый день![/b][/u]
Получить диплом университета
[url=http://bvf.ru/forum/showthread.php?p=25250301#post25250301/]bvf.ru/forum/showthread.php?p=25250301#post25250301[/url]
[url=http://diigo.com/item/note/8p2f0/ouhw?k=1f797285cdcc57109d315eeccc1b6075/]diigo.com/item/note/8p2f0/ouhw?k=1f797285cdcc57109d315eeccc1b6075[/url]
[url=http://boyara-analia.flybb.ru/viewtopic.php?f=7&t=710/]boyara-analia.flybb.ru/viewtopic.php?f=7&t=710[/url]
[url=http://club-kia.com/forum/thread30848/]club-kia.com/forum/thread30848[/url]
[url=http://darknews.ru/kupite-diplom-i-nachnite-novuyu-zhizn/]darknews.ru/kupite-diplom-i-nachnite-novuyu-zhizn[/url]
Sazrqyt
August 2, 2024 @ 1:34 am
[u][b] Добрый день![/b][/u]
Диплом бакалавра
[url=http://dposhop.ru/forum/user/2936/]dposhop.ru/forum/user/2936[/url]
[url=http://bintarotrojan.com/blogs/11480/Быстрый-путь-к-высшему-образованию-и-новым-возможностям/]bintarotrojan.com/blogs/11480/Быстрый-путь-к-высшему-образованию-и-новым-возможностям[/url]
[url=http://datasphere.ru/club/user/15/blog/]datasphere.ru/club/user/15/blog[/url]
[url=http://kids-news.ru/byistroe-poluchenie-diplomov-vseh-urovney/]kids-news.ru/byistroe-poluchenie-diplomov-vseh-urovney[/url]
[url=http://boyara-analia.flybb.ru/viewtopic.php?f=7&t=707/]boyara-analia.flybb.ru/viewtopic.php?f=7&t=707[/url]
Dnrtkac
August 2, 2024 @ 2:10 am
[b]Приобрести документ[/b] ВУЗа можно в нашей компании в столице.
[url=http://medexmd.com/community/profile/jeffryrehfisch/]medexmd.com/community/profile/jeffryrehfisch[/url]
[url=http://get-social-now.com/story2910780/diplom/]get-social-now.com/story2910780/diplom[/url]
[url=http://investicos.com/uncategorized/diplom-709333ynjoc/]investicos.com/uncategorized/diplom-709333ynjoc[/url]
[url=http://yourbuyersguide.co.uk/diplom-227383ulbap/]yourbuyersguide.co.uk/diplom-227383ulbap[/url]
[url=http://tadalive.com/blog/336492/diplom-kupit-986320pi/]tadalive.com/blog/336492/diplom-kupit-986320pi[/url]
Oariorgqa
August 2, 2024 @ 2:17 am
[u][b] Привет, друзья![/b][/u]
Купить диплом любого университета
[b]Мы предлагаем[/b] выгодно заказать диплом, который выполняется на оригинальном бланке и заверен печатями, водяными знаками, подписями должностных лиц. Наш диплом способен пройти любые проверки, даже с использованием специально предназначенного оборудования. Достигайте своих целей быстро с нашим сервисом.
[b]Где купить диплом по необходимой специальности?[/b]
[url=http://dragonlivers.ixbb.ru/viewtopic.php?id=106#p106/]dragonlivers.ixbb.ru/viewtopic.php?id=106#p106[/url]
[url=http://vipka.mybb.ru/viewtopic.php?id=2881#p5045/]vipka.mybb.ru/viewtopic.php?id=2881#p5045[/url]
[url=http://invertebratenetwork.com/blogs/245/Что-может-сегодня-предоставить-интернет-магазин-с-дипломами/]invertebratenetwork.com/blogs/245/Что-может-сегодня-предоставить-интернет-магазин-с-дипломами[/url]
[url=http://biocenter.pro/club/blogs/sonnick84-blog-bc/faq-i-tekhpodderzhka-pokupaem-diplom-v-seti//]biocenter.pro/club/blogs/sonnick84-blog-bc/faq-i-tekhpodderzhka-pokupaem-diplom-v-seti/[/url]
[url=http://azat.on.kg/blogs/1046/Благодаря-чему-онлайн-магазины-с-дипломами-столь-популярны/]azat.on.kg/blogs/1046/Благодаря-чему-онлайн-магазины-с-дипломами-столь-популярны[/url]
Sazrcqc
August 2, 2024 @ 2:36 am
[u][b] Добрый день![/b][/u]
Мы готовы предложить дипломы любой профессии.
[url=http://thekiwisocial.com/story3020264/kupit-diplom/]thekiwisocial.com/story3020264/kupit-diplom[/url]
[url=http://isocialfans.com/story3021058/kupit-diplom/]isocialfans.com/story3021058/kupit-diplom[/url]
[url=http://penteadistadesucesso.com/kupit-diplom-640789foebi/]penteadistadesucesso.com/kupit-diplom-640789foebi[/url]
[url=http://nerdsmaster.com/groups/kupit-diplom-463592nmbdl/]nerdsmaster.com/groups/kupit-diplom-463592nmbdl[/url]
[url=http://bookmark-media.com/story17739373/kupit-diplom/]bookmark-media.com/story17739373/kupit-diplom[/url]
Mazrgnd
August 2, 2024 @ 2:49 am
[b]Заказать документ о получении высшего образования.[/b]
[url=http://businessbookmark.com/story3006486/kupit-diplom/]businessbookmark.com/story3006486/kupit-diplom[/url]
[url=http://bookmarkingfeed.com/story17601278/kupit-diplom/]bookmarkingfeed.com/story17601278/kupit-diplom[/url]
[url=http://tbookmark.com/story17573782/kupit-diplom/]tbookmark.com/story17573782/kupit-diplom[/url]
[url=http://hariharparagovernmentiti.com/2024/07/27/kupit-diplom-609843kmjjt/]hariharparagovernmentiti.com/2024/07/27/kupit-diplom-609843kmjjt[/url]
[url=http://socialmediaentry.com/story2983929/kupit-diplom/]socialmediaentry.com/story2983929/kupit-diplom[/url]
Trefyfs
August 2, 2024 @ 3:02 am
[u][b] Здравствуйте![/b][/u]
Как купить аттестат 11 класса с официальным упрощенным обучением в Москве
[url=http://tanhashop.com/category/uncategorized/page/2/]tanhashop.com/category/uncategorized/page/2[/url]
Поможем вам всегда!.
Lazrpxm
August 2, 2024 @ 3:55 am
[u][b] Привет, друзья![/b][/u]
[b]Мы предлагаем дипломы[/b] любой профессии по невысоким ценам.
[url=http://techhackly.xyz/2024/06/30/купить-дипломы-университета/]techhackly.xyz/2024/06/30/купить-дипломы-университета[/url]
Dnrtqjk
August 2, 2024 @ 5:03 am
[b]Купить документ[/b] о получении высшего образования можно в нашей компании.
[url=http://thegmariecollection.com/diplom-725038pfhik/]thegmariecollection.com/diplom-725038pfhik[/url]
[url=http://socialwoot.com/story19183071/diplom/]socialwoot.com/story19183071/diplom[/url]
[url=http://tiannaxander.com/diplom-703513munsd/]tiannaxander.com/diplom-703513munsd[/url]
[url=http://webworlddesigners.com/diplom-999976cpora/]webworlddesigners.com/diplom-999976cpora[/url]
[url=http://ottawapianomovingspecialist.ca/diplom-593454iqwfn/]ottawapianomovingspecialist.ca/diplom-593454iqwfn[/url]
Lazrvvc
August 2, 2024 @ 5:34 am
[u][b] Привет![/b][/u]
[b]Приобрести диплом любого университета.[/b]
[url=http://cyclopede.com/cb-profile/pluginclass/cbblogs?action=blogs&func=show&id=270/]cyclopede.com/cb-profile/pluginclass/cbblogs?action=blogs&func=show&id=270[/url]
Uazrcwp
August 2, 2024 @ 5:37 am
[b]Где купить диплом по актуальной специальности?[/b]
[url=http://reech.life/index.php/blog/4931/diplom-kupit-10043lt/]reech.life/index.php/blog/4931/diplom-kupit-10043lt[/url]
[url=http://kankerala.org/uncategorized/diplom-154092ybwxh/]kankerala.org/uncategorized/diplom-154092ybwxh[/url]
[url=http://freeflashgamesnow.com/profile/2771936/ShastaGarst/]freeflashgamesnow.com/profile/2771936/ShastaGarst[/url]
[url=http://selfhackathon.com/diplom-kupit-764261th/]selfhackathon.com/diplom-kupit-764261th[/url]
[url=http://yourbuyersguide.co.uk/diplom-kupit-900655va/]yourbuyersguide.co.uk/diplom-kupit-900655va[/url]
Trefhmt
August 2, 2024 @ 5:50 am
[u][b] Здравствуйте![/b][/u]
Как официально купить аттестат 11 класса с упрощенным обучением в Москве
[url=http://dadiler.com/купить-диплом-советы-и-рекомендации/]dadiler.com/купить-диплом-советы-и-рекомендации[/url]
Будем рады вам помочь!.
pay someone to take my online accounting class
August 2, 2024 @ 5:56 am
The majority of the ideas that are linked with statistics are numerical in nature, which necessitates good calculating abilities. This is one of the biggest challenges that students face when doing their statistics assignments, among the many challenges that are related with various courses and areas. The heavy demands of the course frequently make it difficult for students to find the time and energy to participate in online statistics programmes. In online classrooms, it is necessary for students to complete their homework in its entirety and reply to all statistical issues. This is because homework plays a significant part in ensuring that students absorb the subject. You may be wondering, “Do my online stats class for me?” if you are interested in receiving help. When it comes to online classes and assignments, the answer is a resounding yes, because engaging the services of a third party is an excellent potential choice.
Jariorluu
August 2, 2024 @ 6:26 am
[u][b] Здравствуйте![/b][/u]
Купить диплом любого ВУЗа
[url=http://forum-pmr.net/member.php?u=42748/]forum-pmr.net/member.php?u=42748[/url]
[url=http://larashare.net/events/event/49-огромная-популярность-магазинов-что-продают-дипломы/]larashare.net/events/event/49-огромная-популярность-магазинов-что-продают-дипломы[/url]
[url=http://asepbook.com/read-blog/4002/]asepbook.com/read-blog/4002[/url]
[url=http://klinlife.ru/forums/index.php?/topic/39943-kak-zakazat-diplom-onlain-prostie-shagi/]klinlife.ru/forums/index.php?/topic/39943-kak-zakazat-diplom-onlain-prostie-shagi[/url]
[url=http://forum.drustvogil-galad.si/index.php/topic,14420.0/]forum.drustvogil-galad.si/index.php/topic,14420.0[/url]
Qazrdxz
August 2, 2024 @ 6:28 am
[u][b] Привет, друзья![/b][/u]
[b]Заказать диплом о высшем образовании.[/b]
[url=http://productinn.mn.co/posts/61679264/]productinn.mn.co/posts/61679264[/url]
[url=http://uconnect.ae/read-blog/78595/]uconnect.ae/read-blog/78595[/url]
[url=http://sonnick84.blogofchange.com/28737644/Как-можно-будет-приобрести-недорого-качественный-диплом-в-сети/]sonnick84.blogofchange.com/28737644/Как-можно-будет-приобрести-недорого-качественный-диплом-в-сети[/url]
[url=http://machineintelligence.mn.co/posts/61679253/]machineintelligence.mn.co/posts/61679253[/url]
[url=http://wiki.mysupp.ru/index.php?title=Ассортимент_и_способы_доставки_в_онлайн-магазине._Покупаем_аттестат/]wiki.mysupp.ru/index.php?title=Ассортимент_и_способы_доставки_в_онлайн-магазине._Покупаем_аттестат[/url]
Lariormjx
August 2, 2024 @ 6:28 am
Как быстро и легально купить аттестат 11 класса в Москве
[url=http://aboutallfinance.ru/kupite-diplom-i-poluchite-novyie-vozmozhnosti/]aboutallfinance.ru/kupite-diplom-i-poluchite-novyie-vozmozhnosti[/url]
[url=http://akbmpq9ux.bloggosite.com/34731505/Как-подобрать-надежный-магазин-с-документами-Обзор-от-специалиста/]akbmpq9ux.bloggosite.com/34731505/Как-подобрать-надежный-магазин-с-документами-Обзор-от-специалиста[/url]
[url=http://pajarojaguar.org/mediawiki01/index.php/Диплом_любой_профессии:_качественно_и_надежно/]pajarojaguar.org/mediawiki01/index.php/Диплом_любой_профессии:_качественно_и_надежно[/url]
[url=http://alik.forumrpg.ru/viewtopic.php?id=6883#p277428/]alik.forumrpg.ru/viewtopic.php?id=6883#p277428[/url]
[url=http://forum.7x.ru/member.php?u=16498/]forum.7x.ru/member.php?u=16498[/url]
Lazrobb
August 2, 2024 @ 6:38 am
[u][b] Здравствуйте![/b][/u]
[b]Мы можем предложить дипломы[/b] любых профессий по приятным тарифам.
[url=http://phugiabetong.vn/купить-диплом-в-березниках/]phugiabetong.vn/купить-диплом-в-березниках[/url]
Lazrznn
August 2, 2024 @ 8:13 am
[u][b] Здравствуйте![/b][/u]
[b]Купить диплом университета.[/b]
[url=http://deris.by/news/blogs/1/obshirnyy-vybor-dokumentov-v-populyarnom-internet-magazine.php/]deris.by/news/blogs/1/obshirnyy-vybor-dokumentov-v-populyarnom-internet-magazine.php[/url]
Xazrgvi
August 2, 2024 @ 8:33 am
[b]Привет![/b]
Сколько стоит диплом высшего и среднего образования и как его получить?
[url=http://landik-diploms-srednee.ru/kupit-diplom-v-ekaterinburge]landik-diploms-srednee.ru/kupit-diplom-v-ekaterinburge[/url]
Manreiv
August 2, 2024 @ 8:55 am
[u][b] Здравствуйте![/b][/u]
Купить документ института
[url=http://asxdiplomik24.ru/kupit-diplom-rostov-na-donu]asxdiplomik24.ru/kupit-diplom-rostov-na-donu[/url]
Lazrqai
August 2, 2024 @ 9:21 am
[b]Купить диплом ВУЗа.[/b]
[url=http://corsica.forhikers.com/forum/p/127339/]corsica.forhikers.com/forum/p/127339[/url]
[url=http://msfo-soft.ru/msfo/forum/messages/forum24/topic10323/message288506/?result=new#message288506/]msfo-soft.ru/msfo/forum/messages/forum24/topic10323/message288506/?result=new#message288506[/url]
[url=http://coastalplainplants.org/wiki/index.php/Высшее_Образование_для_Вас:_Легально_и_Быстро/]coastalplainplants.org/wiki/index.php/Высшее_Образование_для_Вас:_Легально_и_Быстро[/url]
[url=http://orthograf.getbb.ru/viewtopic.php?f=2&t=465/]orthograf.getbb.ru/viewtopic.php?f=2&t=465[/url]
[url=http://naturetour.ru/club/user/186/blog/1616/]naturetour.ru/club/user/186/blog/1616[/url]
Williamraics
August 2, 2024 @ 9:25 am
[u][b] Добрый день![/b][/u]
Мы изготавливаем дипломы.
[url=http://seaman.com.ua/forum/index.php?threads/kupit-diplom-ot-licenzirovannyx-uchrezhdenij.37472/]seaman.com.ua/forum/index.php?threads/kupit-diplom-ot-licenzirovannyx-uchrezhdenij.37472[/url]
[url=http://telegra.ph/Kupit-Oficialnyj-Diplom-Bystro-i-Legalno-07-09/]telegra.ph/Kupit-Oficialnyj-Diplom-Bystro-i-Legalno-07-09[/url]
[url=http://alik.forumrpg.ru/viewtopic.php?id=6853#p276083/]alik.forumrpg.ru/viewtopic.php?id=6853#p276083[/url]
[url=http://mastercode.ru/forum/?PAGE_NAME=profile_view&UID=4746/]mastercode.ru/forum/?PAGE_NAME=profile_view&UID=4746[/url]
[url=http://yur-dom.ru/communication/forum/messages/forum3/message247/290-kupit-diplom-bystro-i-legalno?result=new#message247/]yur-dom.ru/communication/forum/messages/forum3/message247/290-kupit-diplom-bystro-i-legalno?result=new#message247[/url]
Xazrwyd
August 2, 2024 @ 10:17 am
[u][b] Здравствуйте![/b][/u]
Приобрести документ университета.
[url=http://sensemi.getbb.ru/viewtopic.php?f=6&t=607/]sensemi.getbb.ru/viewtopic.php?f=6&t=607[/url]
[url=http://vared.flyboard.ru/viewtopic.php?f=1&t=547/]vared.flyboard.ru/viewtopic.php?f=1&t=547[/url]
[url=http://openmotonews.ru/oformlenie-diplomov-s-garantiey-kachestva/]openmotonews.ru/oformlenie-diplomov-s-garantiey-kachestva[/url]
[url=http://gamerplayz2k.mn.co/posts/62086690/]gamerplayz2k.mn.co/posts/62086690[/url]
[url=http://ilovehabbo.bbon.ru/viewtopic.php?id=1560#p28977/]ilovehabbo.bbon.ru/viewtopic.php?id=1560#p28977[/url]
Cazryhq
August 2, 2024 @ 11:31 am
[u][b] Привет![/b][/u]
Купить диплом о высшем образовании
[url=http://anoreksja.org.pl/posting.php?mode=post&f=13/]anoreksja.org.pl/posting.php?mode=post&f=13[/url]
[url=http://kolba.com.ua/index.php?topic=141018.new#new/]kolba.com.ua/index.php?topic=141018.new#new[/url]
[url=http://sadcr.listbb.ru/viewtopic.php?f=53&t=398/]sadcr.listbb.ru/viewtopic.php?f=53&t=398[/url]
[url=http://toursoul.ru/?p=19243/]toursoul.ru/?p=19243[/url]
[url=http://magnat-matras.ru/forum/user/50906/]magnat-matras.ru/forum/user/50906[/url]
Sazrzls
August 2, 2024 @ 11:36 am
[u][b] Привет![/b][/u]
Диплом экономиста
[url=http://burgasdent.listbb.ru/viewtopic.php?f=3&t=408/]burgasdent.listbb.ru/viewtopic.php?f=3&t=408[/url]
[url=http://ai-db.science/wiki/Оформите_диплом_за_пару_дней/]ai-db.science/wiki/Оформите_диплом_за_пару_дней[/url]
[url=http://vetreriameliante.it/index.php/kforum/jm-slideshow-module/9865#9865/]vetreriameliante.it/index.php/kforum/jm-slideshow-module/9865#9865[/url]
[url=http://foro.rune-nifelheim.com/general-ragnarok-m/vysokokachestvennye-diplomy-na-zakaz/]foro.rune-nifelheim.com/general-ragnarok-m/vysokokachestvennye-diplomy-na-zakaz[/url]
[url=http://koxma.4adm.ru/viewtopic.php?f=298&t=5534&view=next&sid=05f6e58705ea31cb4f7032cc409091a8/]koxma.4adm.ru/viewtopic.php?f=298&t=5534&view=next&sid=05f6e58705ea31cb4f7032cc409091a8[/url]
Yrefkhm
August 2, 2024 @ 12:08 pm
[u][b] Привет![/b][/u]
[b]Заказать диплом любого университета.[/b]
[url=http://bbs.boway.net/home.php?mod=space&uid=910685/]bbs.boway.net/home.php?mod=space&uid=910685[/url]
[url=http://boosterblog.com/vote-864130-668401?adresse=frees-diplom.com/]boosterblog.com/vote-864130-668401?adresse=frees-diplom.com[/url]
[url=http://thenet.work/gallery/image/481-быстрый-выбор-онлайн-магазина-реализующего-дипломы-и-аттестаты/]thenet.work/gallery/image/481-быстрый-выбор-онлайн-магазина-реализующего-дипломы-и-аттестаты[/url]
[url=http://led-electro.ru/bitrix/rk.php?goto=frees-diplom.com/]led-electro.ru/bitrix/rk.php?goto=frees-diplom.com[/url]
[url=http://jejucordelia.com/eng/bbs/board.php?bo_table=review_e&wr_id=186722/]jejucordelia.com/eng/bbs/board.php?bo_table=review_e&wr_id=186722[/url]
Sazrelq
August 2, 2024 @ 2:14 pm
[u][b] Привет![/b][/u]
Приобрести документ о получении высшего образования
[url=http://leenkup.com/read-blog/16623/]leenkup.com/read-blog/16623[/url]
[url=http://islgaming.community/read-blog/86/]islgaming.community/read-blog/86[/url]
[url=http://refine.live/events/event/15-быстрый-поиск-магазина-что-реализует-дипломы-и-аттестаты/]refine.live/events/event/15-быстрый-поиск-магазина-что-реализует-дипломы-и-аттестаты[/url]
[url=http://vutruao.vn/read-blog/40/]vutruao.vn/read-blog/40[/url]
[url=http://ccde-cambodia.com/cb-profile/pluginclass/cbblogs?action=blogs&func=show&id=117/]ccde-cambodia.com/cb-profile/pluginclass/cbblogs?action=blogs&func=show&id=117[/url]
Yrefjpj
August 2, 2024 @ 2:42 pm
[u][b] Привет![/b][/u]
[b]Купить диплом университета.[/b]
[url=http://facesocial.demo3.dedicatedhost247.com/read-blog/14635/]facesocial.demo3.dedicatedhost247.com/read-blog/14635[/url]
[url=http://walkner.cc/page3.php?messagePage=28398/]walkner.cc/page3.php?messagePage=28398[/url]
[url=http://mybaltika.info/ru/blogs/587/10348/]mybaltika.info/ru/blogs/587/10348[/url]
[url=http://financeokey.ru/page/2/]financeokey.ru/page/2[/url]
[url=http://hspk.co.kr/g5/bbs/board.php?bo_table=trz7pxdei6&wr_id=5498/]hspk.co.kr/g5/bbs/board.php?bo_table=trz7pxdei6&wr_id=5498[/url]
Iariornwg
August 2, 2024 @ 3:19 pm
[u][b] Добрый день![/b][/u]
Купить документ университета вы сможете у нас в столице. Мы предлагаем документы об окончании любых университетов РФ. Вы получите необходимый диплом по любым специальностям, любого года выпуска, в том числе документы Советского Союза. Даем 100% гарантию, что при проверке документа работодателем, подозрений не возникнет.
[url=http://hip-hop.ru/forum/id298234-worksale/]hip-hop.ru/forum/id298234-worksale[/url]
[url=http://upkisb.ru/cb-profile/pluginclass/cbblogs?action=blogs&func=show&id=124/]upkisb.ru/cb-profile/pluginclass/cbblogs?action=blogs&func=show&id=124[/url]
[url=http://avtolux48.ru/people/]avtolux48.ru/people[/url]
[url=http://rst.adk.audio/company/personal/]rst.adk.audio/company/personal[/url]
[url=http://ackeer.com/read-blog/477/]ackeer.com/read-blog/477[/url]
JamesNix
August 2, 2024 @ 4:05 pm
Федеральный закон https://zakonobosago.ru “Об обязательном страховании гражданской ответственности владельцев транспортных средств” (ОСАГО)
Danrldb
August 2, 2024 @ 4:49 pm
[u][b] Привет, друзья![/b][/u]
Мы изготавливаем дипломы любой профессии.
[url=http://bbs.heyshell.com/forum.php?mod=viewthread&tid=28283]bbs.heyshell.com/forum.php?mod=viewthread&tid=28283[/url]
Будем рады вам помочь!.
Cazrefl
August 2, 2024 @ 5:01 pm
[u][b] Привет![/b][/u]
Мы предлагаем документы ВУЗов
[url=http://asg-amt.phorum.pl/viewtopic.php?p=32745#32745/]asg-amt.phorum.pl/viewtopic.php?p=32745#32745[/url]
[url=http://grot.getbb.ru/viewtopic.php?f=6&t=1080/]grot.getbb.ru/viewtopic.php?f=6&t=1080[/url]
[url=http://bs.listbb.ru/viewtopic.php?f=3&t=644/]bs.listbb.ru/viewtopic.php?f=3&t=644[/url]
[url=http://topnewsgadget.ru/vyisokokachestvennyie-diplomyi-bez-lishnih-hlopot/]topnewsgadget.ru/vyisokokachestvennyie-diplomyi-bez-lishnih-hlopot[/url]
[url=http://terios2.ru/forums/index.php?autocom=gallery&req=si&img=3081/]terios2.ru/forums/index.php?autocom=gallery&req=si&img=3081[/url]
Qazrfoc
August 2, 2024 @ 5:22 pm
[u][b] Добрый день![/b][/u]
[b]Приобрести диплом университета.[/b]
[url=http://pumasoccershoesfans.com/read-blog/2126/]pumasoccershoesfans.com/read-blog/2126[/url]
[url=http://online-casino-australia.mn.co/posts/61724753/]online-casino-australia.mn.co/posts/61724753[/url]
[url=http://gamingforum.se/index.php?/gallery/image/123-покупаем-диплом-в-проверенном-интернет-магазине-по-комфортной-цене/]gamingforum.se/index.php?/gallery/image/123-покупаем-диплом-в-проверенном-интернет-магазине-по-комфортной-цене[/url]
[url=http://derpyshare.mn.co/posts/61679397/]derpyshare.mn.co/posts/61679397[/url]
[url=http://encontrocentral.com/read-blog/2287/]encontrocentral.com/read-blog/2287[/url]
Jariorexc
August 2, 2024 @ 5:28 pm
[u][b] Привет, друзья![/b][/u]
Приобрести диплом о высшем образовании
[url=http://roving.club/read-blog/1453/]roving.club/read-blog/1453[/url]
[url=http://telearchaeology.org/TAWiki/index.php/Оформление_диплома_под_ключ:_безопасно_и_легально/]telearchaeology.org/TAWiki/index.php/Оформление_диплома_под_ключ:_безопасно_и_легально[/url]
[url=http://bvf.ru/forum/showthread.php?p=25249084#post25249084/]bvf.ru/forum/showthread.php?p=25249084#post25249084[/url]
[url=http://jaitun.com/read-blog/391/]jaitun.com/read-blog/391[/url]
[url=http://x70795vj.beget.tech/2024/07/10/blagodarya-chemu-onlayn-magaziny-s-dokumentami-segodnya-tak-populyarny/]x70795vj.beget.tech/2024/07/10/blagodarya-chemu-onlayn-magaziny-s-dokumentami-segodnya-tak-populyarny[/url]
Larioreoz
August 2, 2024 @ 5:37 pm
Как получить диплом о среднем образовании в Москве и других городах
[url=http://nedvigimost.bbok.ru/viewtopic.php?id=27448#p46954/]nedvigimost.bbok.ru/viewtopic.php?id=27448#p46954[/url]
[url=http://telodosocial.it/blogs/755/Как-найти-надежный-магазин-с-документами-Обзор-от-специалиста/]telodosocial.it/blogs/755/Как-найти-надежный-магазин-с-документами-Обзор-от-специалиста[/url]
[url=http://podsliving.in/forums/topic/купить-диплом-в-москве-бытовые-услови/]podsliving.in/forums/topic/купить-диплом-в-москве-бытовые-услови[/url]
[url=http://followgrown.com/read-blog/2702/]followgrown.com/read-blog/2702[/url]
[url=http://forum-pmr.net/member.php?u=42748/]forum-pmr.net/member.php?u=42748[/url]
Dnrtpgv
August 2, 2024 @ 5:40 pm
[b]Заказать документ[/b] университета можно в нашем сервисе.
[url=http://taondinternational.rudraserver.com/blog/index.php?entryid=134634/]taondinternational.rudraserver.com/blog/index.php?entryid=134634[/url]
[url=http://tigaedu.com/blog/index.php?entryid=335070/]tigaedu.com/blog/index.php?entryid=335070[/url]
[url=http://tigaedu.com/blog/index.php?entryid=334163/]tigaedu.com/blog/index.php?entryid=334163[/url]
[url=http://localtrusted.co.uk/wiki/index.php?title=Diplom_869614qrCpm/]localtrusted.co.uk/wiki/index.php?title=Diplom_869614qrCpm[/url]
[url=http://investicos.com/uncategorized/diplom-49323iclhh/]investicos.com/uncategorized/diplom-49323iclhh[/url]
Trefknw
August 2, 2024 @ 6:10 pm
[u][b] Привет![/b][/u]
Как не попасть впросак при покупке диплома колледжа или ПТУ в России
[url=http://theplaygamepicks.com/диплом-техникума-где-и-как-купить/]theplaygamepicks.com/диплом-техникума-где-и-как-купить[/url]
Поможем вам всегда!.
Xazrwaw
August 2, 2024 @ 6:13 pm
[u][b] Привет, друзья![/b][/u]
Заказать документ о получении высшего образования.
[url=http://club2108.ru/forum/post.php?fid=57/]club2108.ru/forum/post.php?fid=57[/url]
[url=http://transexit.g-talk.ru/viewtopic.php?f=10&t=2120/]transexit.g-talk.ru/viewtopic.php?f=10&t=2120[/url]
[url=http://veneraroleplay.listbb.ru/viewtopic.php?f=4&t=323/]veneraroleplay.listbb.ru/viewtopic.php?f=4&t=323[/url]
[url=http://kome.maxbb.ru/viewtopic.php?f=22&t=2245/]kome.maxbb.ru/viewtopic.php?f=22&t=2245[/url]
[url=http://deanonnic.listbb.ru/viewtopic.php?f=5&t=457/]deanonnic.listbb.ru/viewtopic.php?f=5&t=457[/url]
Lazronr
August 2, 2024 @ 7:00 pm
[u][b] Привет, друзья![/b][/u]
[b]Мы можем предложить дипломы[/b] любой профессии по выгодным ценам.
[url=http://make-coin.ru/legkiy-sposob-poluchit-diplom-onlayn/]make-coin.ru/legkiy-sposob-poluchit-diplom-onlayn[/url]
Dnrtbbf
August 2, 2024 @ 8:22 pm
[b]Купить документ[/b] о получении высшего образования вы можете в нашей компании в столице.
[url=http://ranatourandtravels.com/diplom-791904dsxhy/]ranatourandtravels.com/diplom-791904dsxhy[/url]
[url=http://mysocialport.com/story2999067/diplom/]mysocialport.com/story2999067/diplom[/url]
[url=http://alonegocio.net.br/author/maricelapri/]alonegocio.net.br/author/maricelapri[/url]
[url=http://aplamaharashtra.com/archives/1846/]aplamaharashtra.com/archives/1846[/url]
[url=http://welnesbiolabs.com/diplom-794144bpjsh/]welnesbiolabs.com/diplom-794144bpjsh[/url]
Lazrmid
August 2, 2024 @ 8:37 pm
[b]Приобрести диплом о высшем образовании.[/b]
[url=http://rodme.ru/posting.php?mode=post&f=6/]rodme.ru/posting.php?mode=post&f=6[/url]
[url=http://a63.flybb.ru/viewtopic.php?f=16&t=3193/]a63.flybb.ru/viewtopic.php?f=16&t=3193[/url]
[url=http://aviapoisk.getbb.ru/posting.php?mode=post&f=27&sid=770996810a1494e5f6926adef577ad08/]aviapoisk.getbb.ru/posting.php?mode=post&f=27&sid=770996810a1494e5f6926adef577ad08[/url]
[url=http://allukrnews.ru/kupit-diplom-bez-posrednikov-legko-i-byistro/]allukrnews.ru/kupit-diplom-bez-posrednikov-legko-i-byistro[/url]
[url=http://network-6063768.mn.co/posts/61899861/]network-6063768.mn.co/posts/61899861[/url]
Williamraics
August 2, 2024 @ 8:43 pm
[u][b] Здравствуйте![/b][/u]
Мы изготавливаем дипломы.
[url=http://vared.flyboard.ru/viewtopic.php?f=1&t=534/]vared.flyboard.ru/viewtopic.php?f=1&t=534[/url]
[url=http://kalininabad.flyboard.ru/viewtopic.php?f=11&t=2809/]kalininabad.flyboard.ru/viewtopic.php?f=11&t=2809[/url]
[url=http://yur-dom.ru/communication/forum/messages/forum3/message257/300-kupit-diplom-bystro-i-nadezhno?result=new#message257/]yur-dom.ru/communication/forum/messages/forum3/message257/300-kupit-diplom-bystro-i-nadezhno?result=new#message257[/url]
[url=http://o91746bp.beget.tech/2024/07/08/vash-diplom-legalno-nadezhno-i-udobno/]o91746bp.beget.tech/2024/07/08/vash-diplom-legalno-nadezhno-i-udobno[/url]
[url=http://wap.fortboyard2012forum.4adm.ru/viewtopic.php?f=27&t=4152/]wap.fortboyard2012forum.4adm.ru/viewtopic.php?f=27&t=4152[/url]
Diplomi_kgSi
August 2, 2024 @ 8:53 pm
Добрый день!
Заказать документ ВУЗа можно в нашей компании в столице.
[url=http://ast-diploms.com/kupit-diplom-tehnikuma-kolledzha/]ast-diploms.com/kupit-diplom-tehnikuma-kolledzha[/url]
Vazrapp
August 2, 2024 @ 9:32 pm
[u][b] Добрый день![/b][/u]
Купить документ о получении высшего образования вы можете в нашей компании. Мы оказываем услуги по продаже документов об окончании любых университетов России.
[url=http://fishtrade3000.ru/forum/user/1332/]fishtrade3000.ru/forum/user/1332[/url]
[url=http://sborkakuhni.com/users/ulogyjan/]sborkakuhni.com/users/ulogyjan[/url]
[url=http://akb-video.ru/profile.php?u=uriqunel/]akb-video.ru/profile.php?u=uriqunel[/url]
[url=http://clindoeilinfo.com/a-peine-arrivee-parmi-les-reseaux-sociaux-lapplication-threads-serait-dans-le-collimateur-de-la-justice/#comment-79454/]clindoeilinfo.com/a-peine-arrivee-parmi-les-reseaux-sociaux-lapplication-threads-serait-dans-le-collimateur-de-la-justice/#comment-79454[/url]
[url=http://новодвинцы.рф/forum/messages/forum4/topic840/message264507/?result=reply#message264507/]новодвинцы.рф/forum/messages/forum4/topic840/message264507/?result=reply#message264507[/url]
Diplomi_jfSi
August 2, 2024 @ 11:32 pm
Добрый день!
Купить документ института можно у нас в Москве.
[url=http://ast-diplomy.com/kupit-diplom-kandidata-nauk/]ast-diplomy.com/kupit-diplom-kandidata-nauk[/url]
Mazrehu
August 3, 2024 @ 1:18 am
[u][b] Здравствуйте![/b][/u]
[b]Купить диплом магистра оказалось возможно, быстрое обучение и диплом на руки[/b]
[url=http://mandiplomiks.ru/]mandiplomiks.ru[/url]
pay someone to take my online nursing class
August 3, 2024 @ 3:26 am
Since the beginning of their existence, they have been giving students competent answers to their tasks, and they have always delivered excellent results. Over time, they have refined their strategies and approaches to connecting with students, writers, and members of the team in order to give access around the clock, reduce the amount of time that delays occur, and finish their assignments before the deadline. If you do not have a lot of time to delegate your responsibilities, you can still rely on them because the transactions and business activities they do are smooth and trouble-free. It is possible to rely on their assistance regardless of whether you need to fulfill the requirements of your assignment or care for other duties. Additionally, if you are short on time, you might want to think about pay someone to take my online English class. This will guarantee that you fulfill all of the standards that are necessary for your academic course.
Cazrube
August 3, 2024 @ 3:55 am
[u][b] Добрый день![/b][/u]
Мы можем предложить документы техникумов
[url=http://aurorahcs.com/forum/viewtopic.php?f=8&t=685504/]aurorahcs.com/forum/viewtopic.php?f=8&t=685504[/url]
[url=http://karasteamfulldmroleplay.getbb.ru/viewtopic.php?f=62&t=945/]karasteamfulldmroleplay.getbb.ru/viewtopic.php?f=62&t=945[/url]
[url=http://collieforum.ru/posting.php?mode=post&f=17/]collieforum.ru/posting.php?mode=post&f=17[/url]
[url=http://cardinalparkmld.listbb.ru/viewtopic.php?f=3&t=480/]cardinalparkmld.listbb.ru/viewtopic.php?f=3&t=480[/url]
[url=http://caribbeantown.listbb.ru/viewtopic.php?f=5&t=512/]caribbeantown.listbb.ru/viewtopic.php?f=5&t=512[/url]
Mazrtps
August 3, 2024 @ 6:35 am
[u][b] Здравствуйте![/b][/u]
[b]Процесс получения диплома стоматолога: реально ли это сделать быстро?[/b]
[url=http://diploms-man.ru/]diploms-man.ru[/url]
NelsonbiC
August 3, 2024 @ 7:25 am
medication from mexico pharmacy: mexican border pharmacies shipping to usa – medication from mexico pharmacy
Vazrwjr
August 3, 2024 @ 8:08 am
[u][b] Здравствуйте![/b][/u]
Купить документ ВУЗа можно в нашей компании в Москве. Мы предлагаем документы об окончании любых ВУЗов Российской Федерации.
[url=http://forum.stpku.ru/viewtopic.php?f=2&t=49723/]forum.stpku.ru/viewtopic.php?f=2&t=49723[/url]
[url=http://web.symbol.rs/forum/member.php?action=profile&uid=667970/]web.symbol.rs/forum/member.php?action=profile&uid=667970[/url]
[url=http://карачаевский-район.рф/forum/forum1/topic1/]карачаевский-район.рф/forum/forum1/topic1[/url]
[url=http://dayz.tselik.ru/profile.php?action=show&member=10398/]dayz.tselik.ru/profile.php?action=show&member=10398[/url]
[url=http://kochtagebuch.org/doku.php?id=gosznacdiplomy/]kochtagebuch.org/doku.php?id=gosznacdiplomy[/url]
Trefhur
August 3, 2024 @ 8:14 am
[u][b] Добрый день![/b][/u]
Как приобрести диплом техникума с минимальными рисками
[url=http://jubileetrip.com/2024/07/12/купить-диплом-проверенные-способы/]jubileetrip.com/2024/07/12/купить-диплом-проверенные-способы[/url]
Рады оказать помощь!.
Dnrtukp
August 3, 2024 @ 8:51 am
[b]Заказать документ[/b] университета вы сможете у нас.
[url=http://bookmarkangaroo.com/story17758718/diplom/]bookmarkangaroo.com/story17758718/diplom[/url]
[url=http://localtrusted.co.uk/wiki/index.php?title=Diplom_366989bPxBe/]localtrusted.co.uk/wiki/index.php?title=Diplom_366989bPxBe[/url]
[url=http://youwantech.com/xe/board/460422/]youwantech.com/xe/board/460422[/url]
[url=http://utahsyardsale.com/author/kieranceden/]utahsyardsale.com/author/kieranceden[/url]
[url=http://dmozbookmark.com/story17704598/diplom/]dmozbookmark.com/story17704598/diplom[/url]
Razrfmp
August 3, 2024 @ 9:59 am
[u][b] Привет![/b][/u]
Купить диплом любого ВУЗа:
[url=http://sborkakuhni.com/users/usolovo/]sborkakuhni.com/users/usolovo[/url]
[url=http://altaiklad.ru/viewtopic.php?f=15&t=5343/]altaiklad.ru/viewtopic.php?f=15&t=5343[/url]
[url=http://nnklad.ru/viewtopic.php?f=4&t=1708/]nnklad.ru/viewtopic.php?f=4&t=1708[/url]
[url=http://krasrec.ru/forum/user/199860/]krasrec.ru/forum/user/199860[/url]
[url=http://fire-team.ru/forum/showthread.php?p=35344#post35344/]fire-team.ru/forum/showthread.php?p=35344#post35344[/url]
Uazryrk
August 3, 2024 @ 10:20 am
[u][b] Привет![/b][/u]
[b]Заказать диплом ВУЗа.[/b]
[url=http://amalgama-forum.com/album.php?albumid=95&attachmentid=1990&commentid=1380/]amalgama-forum.com/album.php?albumid=95&attachmentid=1990&commentid=1380[/url]
Trefdxi
August 3, 2024 @ 10:52 am
[u][b] Привет![/b][/u]
Вопросы и ответы: можно ли быстро купить диплом старого образца?
[url=http://caretrip.net/купить-диплом-вуза-быстро-и-легко/]caretrip.net/купить-диплом-вуза-быстро-и-легко[/url]
Поможем вам всегда!.
Dnrtlrn
August 3, 2024 @ 11:34 am
[b]Заказать документ[/b] ВУЗа можно у нас.
[url=http://bookmarkick.com/story17692343/diplom/]bookmarkick.com/story17692343/diplom[/url]
[url=http://starryjeju.com/qna/5650524/]starryjeju.com/qna/5650524[/url]
[url=http://zbookmarkhub.com/story17770159/diplom/]zbookmarkhub.com/story17770159/diplom[/url]
[url=http://samnetworkth.com/diplom-793784mybpn/]samnetworkth.com/diplom-793784mybpn[/url]
[url=http://sttimothysignal.org/groups/diplom-kupit-439120lu/]sttimothysignal.org/groups/diplom-kupit-439120lu[/url]
Dennisteaft
August 3, 2024 @ 12:52 pm
проститутки москвы https://saratov.prostitutki.sex
Yrefijf
August 3, 2024 @ 12:55 pm
[u][b] Привет, друзья![/b][/u]
[b]Заказать диплом любого ВУЗа.[/b]
[url=http://leenkup.com/read-blog/16570/]leenkup.com/read-blog/16570[/url]
[url=http://ourfathersfamily.com/blogs/2393/Главные-затраты-производителя-дипломов-обзор/]ourfathersfamily.com/blogs/2393/Главные-затраты-производителя-дипломов-обзор[/url]
[url=http://pickuptruckindubai.com/2024/06/10/купить-диплом-в-рязани-о-высшем-образо/]pickuptruckindubai.com/2024/06/10/купить-диплом-в-рязани-о-высшем-образо[/url]
[url=http://lidertude.com/blogs/1191/Что-необходимо-из-оборудования-для-создания-документов/]lidertude.com/blogs/1191/Что-необходимо-из-оборудования-для-создания-документов[/url]
[url=http://jbnucri.com/bbs/board.php?bo_table=companylist&wr_id=6824/]jbnucri.com/bbs/board.php?bo_table=companylist&wr_id=6824[/url]
TimothyBug
August 3, 2024 @ 12:58 pm
проститутки метро щелковская https://perm.prostitutki.sex
JavierPraps
August 3, 2024 @ 1:25 pm
проститутки азиатки проститутки у метро
Marcelorek
August 3, 2024 @ 1:31 pm
проститутки коломна https://rnd.prostitutki.sex
Arnoldstota
August 3, 2024 @ 1:45 pm
reputable mexican pharmacies online [url=https://mexicandeliverypharma.online/#]mexican border pharmacies shipping to usa[/url] buying prescription drugs in mexico
Yrefwwy
August 3, 2024 @ 3:34 pm
[u][b] Привет, друзья![/b][/u]
[b]Заказать диплом о высшем образовании.[/b]
[url=http://poltavagok.ru/page/4/]poltavagok.ru/page/4[/url]
[url=http://s-s-o.ru/forum.php?PAGE_NAME=profile_view&UID=52697/]s-s-o.ru/forum.php?PAGE_NAME=profile_view&UID=52697[/url]
[url=http://weekofsport.ru/page/3/]weekofsport.ru/page/3[/url]
[url=http://gamerlaunch.com/community/users/blog/6443244/2286460/расходы-любого-/?gid=535/]gamerlaunch.com/community/users/blog/6443244/2286460/расходы-любого-/?gid=535[/url]
[url=http://redpages.net/index.php?page=user&action=pub_profile&id=8802/]redpages.net/index.php?page=user&action=pub_profile&id=8802[/url]
Sazrdqo
August 3, 2024 @ 5:24 pm
[u][b] Здравствуйте![/b][/u]
Мы изготавливаем дипломы любой профессии.
[url=http://fridayad.in/user/profile/2635283/]fridayad.in/user/profile/2635283[/url]
[url=http://laviehub.com/blog/kupit-diplom-623393xhnyi/]laviehub.com/blog/kupit-diplom-623393xhnyi[/url]
[url=http://indexedbookmarks.com/story17617294/kupit-diplom/]indexedbookmarks.com/story17617294/kupit-diplom[/url]
[url=http://livebookmarking.com/story17634547/kupit-diplom/]livebookmarking.com/story17634547/kupit-diplom[/url]
[url=http://mykindadoctor.com/kupit-diplom-631425erjrv/]mykindadoctor.com/kupit-diplom-631425erjrv[/url]
Mazrflb
August 3, 2024 @ 5:32 pm
[b]Заказать документ о получении высшего образования.[/b]
[url=http://toprankdesign.co.uk/wiki/index.php?title=Kupit_Diplom_177706WuXfy/]toprankdesign.co.uk/wiki/index.php?title=Kupit_Diplom_177706WuXfy[/url]
[url=http://amongus.begandigital.com/kupit-diplom-964852txlsl/]amongus.begandigital.com/kupit-diplom-964852txlsl[/url]
[url=http://laviehub.com/blog/kupit-diplom-734614aksyf/]laviehub.com/blog/kupit-diplom-734614aksyf[/url]
[url=http://getsocialnetwork.com/story3038826/kupit-diplom/]getsocialnetwork.com/story3038826/kupit-diplom[/url]
[url=http://linkedbookmarker.com/story3023785/kupit-diplom/]linkedbookmarker.com/story3023785/kupit-diplom[/url]
Manuelgef
August 3, 2024 @ 5:39 pm
проститутки в химках https://vlg.prostitutki.sex
Lazrxdo
August 3, 2024 @ 5:53 pm
[u][b] Добрый день![/b][/u]
[b]Заказать диплом о высшем образовании.[/b]
[url=http://blogs.rufox.ru/~sonnick84/48075.htm/]blogs.rufox.ru/~sonnick84/48075.htm[/url]
Donaldcit
August 3, 2024 @ 5:58 pm
шлюхи балашиха проститутки рязанский проспект
Diplomi_xqSi
August 3, 2024 @ 7:48 pm
Привет!
Приобрести документ ВУЗа можно у нас.
[url=http://ast-diplomas.com/kupit-diplom-tehnikuma-kolledzha/]ast-diplomas.com/kupit-diplom-tehnikuma-kolledzha[/url]
Razrhol
August 3, 2024 @ 8:36 pm
[u][b] Привет![/b][/u]
Купить диплом ВУЗа:
[url=http://payhotel.ru/forum/messages/forum2/topic7566/message7822/?result=reply#message7822/]payhotel.ru/forum/messages/forum2/topic7566/message7822/?result=reply#message7822[/url]
[url=http://academijacrimea.ru/index.php?subaction=userinfo&user=epuci/]academijacrimea.ru/index.php?subaction=userinfo&user=epuci[/url]
[url=http://tamklad.ru/viewtopic.php?f=8&t=8774/]tamklad.ru/viewtopic.php?f=8&t=8774[/url]
[url=http://testowik.ru/index.php/forum/user/52911-asisa/]testowik.ru/index.php/forum/user/52911-asisa[/url]
[url=http://test.krestikom.net/forums/topic/946-купить-диплом-z971s/]test.krestikom.net/forums/topic/946-купить-диплом-z971s[/url]
Diplomi_bmSi
August 3, 2024 @ 10:30 pm
Добрый день!
Приобрести документ института вы имеете возможность у нас в столице.
[url=http://ast-diploms.com/kupit-diplom-specialista/]ast-diploms.com/kupit-diplom-specialista[/url]
Trefdeo
August 3, 2024 @ 10:42 pm
[u][b] Привет, друзья![/b][/u]
Официальная покупка аттестата о среднем образовании в Москве и других городах
[url=http://property25.org/где-купить-диплом-о-высшем-образовани/]property25.org/где-купить-диплом-о-высшем-образовани[/url]
Поможем вам всегда!.
Cazrzer
August 3, 2024 @ 11:17 pm
[u][b] Привет, друзья![/b][/u]
Получить диплом о высшем образовании
[url=http://forum.vorchun.ru/viewtopic.php?f=2&t=253166/]forum.vorchun.ru/viewtopic.php?f=2&t=253166[/url]
[url=http://school97.ru/vesti20/index.php?PAGE_NAME=profile_view&UID=210677/]school97.ru/vesti20/index.php?PAGE_NAME=profile_view&UID=210677[/url]
[url=http://local-urban-eats.mn.co/posts/62669843/]local-urban-eats.mn.co/posts/62669843[/url]
[url=http://3drus.ru/index.php?do=forum&act=add_topic&subaction=1/]3drus.ru/index.php?do=forum&act=add_topic&subaction=1[/url]
[url=http://lezt.ixbb.ru/viewtopic.php?id=122#p122/]lezt.ixbb.ru/viewtopic.php?id=122#p122[/url]
Uazrvjh
August 3, 2024 @ 11:50 pm
[b]Где заказать диплом по необходимой специальности?[/b]
[url=http://taondinternational.rudraserver.com/blog/index.php?entryid=133092/]taondinternational.rudraserver.com/blog/index.php?entryid=133092[/url]
[url=http://ottawapianomovingspecialist.ca/diplom-985797ghihg/]ottawapianomovingspecialist.ca/diplom-985797ghihg[/url]
[url=http://edux.izvrsna.hr/blog/index.php?entryid=266/]edux.izvrsna.hr/blog/index.php?entryid=266[/url]
[url=http://ashwinihydropneumatics.com/diplom-413589bzfkg/]ashwinihydropneumatics.com/diplom-413589bzfkg[/url]
[url=http://offmarketbusinessforsale.com/diplom-146734uucoc/]offmarketbusinessforsale.com/diplom-146734uucoc[/url]
Dnrtghy
August 4, 2024 @ 12:13 am
[b]Купить документ[/b] о получении высшего образования можно в нашей компании в Москве.
[url=http://isocialfans.com/story3020931/diplom/]isocialfans.com/story3020931/diplom[/url]
[url=http://jadegouvea.com/diplom-660460rafky/]jadegouvea.com/diplom-660460rafky[/url]
[url=http://vibesvietnam.com/diplom-547919bxzwm/]vibesvietnam.com/diplom-547919bxzwm[/url]
[url=http://echobookmarks.com/story17625423/diplom/]echobookmarks.com/story17625423/diplom[/url]
[url=http://itdongnam.com/diplom-799379yntop/]itdongnam.com/diplom-799379yntop[/url]
Lazrlmv
August 4, 2024 @ 12:38 am
[u][b] Добрый день![/b][/u]
[b]Мы изготавливаем дипломы[/b] психологов, юристов, экономистов и прочих профессий по приятным ценам.
[url=http://kikwit.alobi.cd/2024/06/30/купить-диплом-в-ессентуки/]kikwit.alobi.cd/2024/06/30/купить-диплом-в-ессентуки[/url]
Trefddm
August 4, 2024 @ 12:59 am
[u][b] Привет![/b][/u]
Купить диплом старого образца, можно ли это сделать по быстрой схеме?
[url=http://protectorakanaan.com/купить-диплом-проверенные-способы/]protectorakanaan.com/купить-диплом-проверенные-способы[/url]
Рады оказаться полезными!.
Xazregq
August 4, 2024 @ 1:34 am
[b]Привет![/b]
Приобрести документ о получении высшего образования
[url=http://arusak-diploms-srednee.ru/kupit-diplom-v-krasnodare]arusak-diploms-srednee.ru/kupit-diplom-v-krasnodare[/url]
tablet
August 4, 2024 @ 1:55 am
[url=https://avermox.com/]vermox drug[/url]
RobertKeync
August 4, 2024 @ 2:26 am
cytotec online [url=https://cytotecbestprice.pro/#]buy cytotec over the counter[/url] Misoprostol 200 mg buy online
Matthewadact
August 4, 2024 @ 2:33 am
60 mg prednisone daily: 25 mg prednisone – purchase prednisone canada
Patrickirowl
August 4, 2024 @ 2:38 am
https://cytotecbestprice.pro/# cytotec buy online usa
Dnrtvrs
August 4, 2024 @ 2:48 am
[b]Приобрести документ[/b] о получении высшего образования вы имеете возможность у нас.
[url=http://webworlddesigners.com/diplom-204603qvfft/]webworlddesigners.com/diplom-204603qvfft[/url]
[url=http://bookmarkpressure.com/story17612531/diplom/]bookmarkpressure.com/story17612531/diplom[/url]
[url=http://tourxperts.com/diplom-kupit-427593ed/]tourxperts.com/diplom-kupit-427593ed[/url]
[url=http://ada.waaron.org/blog/index.php?entryid=6074/]ada.waaron.org/blog/index.php?entryid=6074[/url]
[url=http://fatallisto.com/story7333631/diplom/]fatallisto.com/story7333631/diplom[/url]
Lewiscoata
August 4, 2024 @ 3:03 am
http://cytotecbestprice.pro/# Misoprostol 200 mg buy online
Matthewadact
August 4, 2024 @ 3:07 am
cytotec abortion pill: purchase cytotec – buy cytotec online
Patrickirowl
August 4, 2024 @ 3:12 am
https://zithromaxbestprice.pro/# zithromax for sale us
Sazrodt
August 4, 2024 @ 3:22 am
[u][b] Здравствуйте![/b][/u]
Мы изготавливаем дипломы любой профессии.
[url=http://smallbizblog.net/2024/07/28/kupit-diplom-124401knfez/]smallbizblog.net/2024/07/28/kupit-diplom-124401knfez[/url]
[url=http://kankerala.org/uncategorized/kupit-diplom-55331ytxxc/]kankerala.org/uncategorized/kupit-diplom-55331ytxxc[/url]
[url=http://bookmarkjourney.com/story17701993/kupit-diplom/]bookmarkjourney.com/story17701993/kupit-diplom[/url]
[url=http://arandaasesoria.com/kupit-diplom-515530vywbk/]arandaasesoria.com/kupit-diplom-515530vywbk[/url]
[url=http://itdongnam.com/kupit-diplom-245154uknzf/]itdongnam.com/kupit-diplom-245154uknzf[/url]
Mazrldb
August 4, 2024 @ 3:31 am
[b]Заказать документ университета.[/b]
[url=http://sttimothysignal.org/groups/kupit-diplom-638740sodon/]sttimothysignal.org/groups/kupit-diplom-638740sodon[/url]
[url=http://bookmarksparkle.com/story17767760/kupit-diplom/]bookmarksparkle.com/story17767760/kupit-diplom[/url]
[url=http://deadbeathomeowner.com/community/profile/terrellbey2078/]deadbeathomeowner.com/community/profile/terrellbey2078[/url]
[url=http://koolegardi.ir/2024/07/28/kupit-diplom-50167wipfq/]koolegardi.ir/2024/07/28/kupit-diplom-50167wipfq[/url]
[url=http://apollobookmarks.com/story17606182/kupit-diplom/]apollobookmarks.com/story17606182/kupit-diplom[/url]
Lewiscoata
August 4, 2024 @ 3:43 am
https://propeciabestprice.pro/# order cheap propecia
Lazrhdq
August 4, 2024 @ 4:25 am
[u][b] Привет![/b][/u]
[b]Купить диплом любого университета.[/b]
[url=http://imforma.it/cb-profile/pluginclass/cbblogs?action=blogs&func=show&id=118/]imforma.it/cb-profile/pluginclass/cbblogs?action=blogs&func=show&id=118[/url]
Oarioradr
August 4, 2024 @ 4:33 am
[u][b] Привет![/b][/u]
Приобрести диплом о высшем образовании
[b]Наша компания предлагает[/b] максимально быстро купить диплом, который выполнен на оригинальном бланке и заверен мокрыми печатями, водяными знаками, подписями. Наш документ пройдет лубую проверку, даже при помощи специфических приборов. Решайте свои задачи быстро и просто с нашей компанией.
[b]Где купить диплом специалиста?[/b]
[url=http://woman.build2.ru/viewtopic.php?id=14514#p44579/]woman.build2.ru/viewtopic.php?id=14514#p44579[/url]
[url=http://xiudda2iz.blogsumer.com/27945098/Вы-хотите-выяснить-как-заказать-диплом-в-сети-недорого-Ждем-Вас/]xiudda2iz.blogsumer.com/27945098/Вы-хотите-выяснить-как-заказать-диплом-в-сети-недорого-Ждем-Вас[/url]
[url=http://wiki.odessanews.biz/index.php/4_главных_группы_интернет_магазинов,_продающих_дипломы/]wiki.odessanews.biz/index.php/4_главных_группы_интернет_магазинов,_продающих_дипломы[/url]
[url=http://dbuniverse.net/forum/index.php?/gallery/image/250-за-счет-чего-магазины-с-дипломами-сегодня-так-популярны//]dbuniverse.net/forum/index.php?/gallery/image/250-за-счет-чего-магазины-с-дипломами-сегодня-так-популярны/[/url]
[url=http://iwasocial.com/read-blog/284/]iwasocial.com/read-blog/284[/url]
Iariorjpp
August 4, 2024 @ 7:20 am
[u][b] Добрый день![/b][/u]
Купить документ университета можно в нашем сервисе. Мы предлагаем документы об окончании любых университетов Российской Федерации. Вы получите необходимый диплом по любым специальностям, любого года выпуска, включая документы образца СССР. Даем 100% гарантию, что при проверке документа работодателем, никаких подозрений не появится.
[url=http://technoevents.ru/nastoyashhie-diplomyi-bez-ozhidaniya-i-problem/]technoevents.ru/nastoyashhie-diplomyi-bez-ozhidaniya-i-problem[/url]
[url=http://lubercy.ixbb.ru/index.php/]lubercy.ixbb.ru/index.php[/url]
[url=http://obozrevatelevents.ru/legalnaya-pokupka-diplomov-konfidentsialno-i-nadezhno/]obozrevatelevents.ru/legalnaya-pokupka-diplomov-konfidentsialno-i-nadezhno[/url]
[url=http://testsajtet.net/read-blog/215/]testsajtet.net/read-blog/215[/url]
[url=http://californiarpn2.listbb.ru/index.php/]californiarpn2.listbb.ru/index.php[/url]
Mazrard
August 4, 2024 @ 7:40 am
[u][b] Здравствуйте![/b][/u]
[b]Пошаговая инструкция по официальной покупке диплома о высшем образовании[/b]
[url=http://diploms-man.ru/]diploms-man.ru[/url]
Cazrnpj
August 4, 2024 @ 9:30 am
[u][b] Привет![/b][/u]
Заказать диплом о высшем образовании
[url=http://29ru.listbb.ru/viewtopic.php?f=26&t=1960/]29ru.listbb.ru/viewtopic.php?f=26&t=1960[/url]
[url=http://elizabethjohns.ru/forum/user/6053/]elizabethjohns.ru/forum/user/6053[/url]
[url=http://seaman.com.ua/forum/index.php?forums/morskoe-obrazovanie.12/create-thread/]seaman.com.ua/forum/index.php?forums/morskoe-obrazovanie.12/create-thread[/url]
[url=http://hitman.getbb.ru/viewtopic.php?f=16&t=962/]hitman.getbb.ru/viewtopic.php?f=16&t=962[/url]
[url=http://moskvic.actieforum.com/t3749-topic#7040/]moskvic.actieforum.com/t3749-topic#7040[/url]
Matthewadact
August 4, 2024 @ 9:35 am
buy cytotec pills online cheap: cytotec pills buy online – buy cytotec online fast delivery
Sazrnsx
August 4, 2024 @ 9:37 am
[u][b] Здравствуйте![/b][/u]
Диплом для вас
[url=http://raussga.flybb.ru/viewtopic.php?f=12&t=2219/]raussga.flybb.ru/viewtopic.php?f=12&t=2219[/url]
Patrickirowl
August 4, 2024 @ 9:49 am
http://nolvadexbestprice.pro/# tamoxifen cyp2d6
Matthewadact
August 4, 2024 @ 10:08 am
generic propecia no prescription: cost cheap propecia without prescription – buy propecia without dr prescription
Patrickirowl
August 4, 2024 @ 10:23 am
http://prednisonebestprice.pro/# prednisone tablets 2.5 mg
Qazrune
August 4, 2024 @ 10:26 am
[u][b] Привет, друзья![/b][/u]
[b]Приобрести диплом любого ВУЗа.[/b]
[url=http://thehubbcommunitynetwork.mn.co/posts/61837766/]thehubbcommunitynetwork.mn.co/posts/61837766[/url]
[url=http://sonnick84.ampblogs.com/Дипломы-справки-а-кроме-этого-свидетельства-низкие-цены-65066569/]sonnick84.ampblogs.com/Дипломы-справки-а-кроме-этого-свидетельства-низкие-цены-65066569[/url]
[url=http://poetzinc.com/read-blog/11601/]poetzinc.com/read-blog/11601[/url]
[url=http://sonnick84.idblogz.com/28686013/Надежный-и-проверенный-онлайн-магазин-с-большим-выбором-документов/]sonnick84.idblogz.com/28686013/Надежный-и-проверенный-онлайн-магазин-с-большим-выбором-документов[/url]
[url=http://darkgate.pl/events/event/24-дипломы-аттестаты-и-свидетельства-в-известном-интернет-магазине/]darkgate.pl/events/event/24-дипломы-аттестаты-и-свидетельства-в-известном-интернет-магазине[/url]
Jariorugl
August 4, 2024 @ 10:33 am
[u][b] Добрый день![/b][/u]
Купить диплом ВУЗа
[url=http://ic-info.ru/forum/user/166288/]ic-info.ru/forum/user/166288[/url]
[url=http://craneflower.net/blogs/168/Хотите-узнать-как-купить-документы-в-интернете-Ждем-Вас/]craneflower.net/blogs/168/Хотите-узнать-как-купить-документы-в-интернете-Ждем-Вас[/url]
[url=http://rogerfilms.com/index.php?title=Быстрое_оформление_диплома_любой_специальности/]rogerfilms.com/index.php?title=Быстрое_оформление_диплома_любой_специальности[/url]
[url=http://face2face.club/blogs/21/Изучите-наш-материал-в-случае-если-надо-заказать-диплом-в/]face2face.club/blogs/21/Изучите-наш-материал-в-случае-если-надо-заказать-диплом-в[/url]
[url=http://slliver.getbb.ru/viewtopic.php?f=62&t=922/]slliver.getbb.ru/viewtopic.php?f=62&t=922[/url]
Lariorzew
August 4, 2024 @ 11:00 am
Сколько стоит диплом высшего и среднего образования и как это происходит?
[url=http://naijamatta.com/read-blog/20895/]naijamatta.com/read-blog/20895[/url]
[url=http://loveyou.az/read-blog/191/]loveyou.az/read-blog/191[/url]
[url=http://forum.freaks.ro/index.php?/calendar/event/10-почему-онлайн-магазины-с-документами-столь-востребованы-и-популярны/]forum.freaks.ro/index.php?/calendar/event/10-почему-онлайн-магазины-с-документами-столь-востребованы-и-популярны[/url]
[url=http://vxengine.ru/blogs/556/Благодаря-чему-онлайн-магазины-с-документами-так-популярны/]vxengine.ru/blogs/556/Благодаря-чему-онлайн-магазины-с-документами-так-популярны[/url]
[url=http://komfort.rusff.me/viewtopic.php?id=13952#p41121/]komfort.rusff.me/viewtopic.php?id=13952#p41121[/url]
Uazrera
August 4, 2024 @ 11:02 am
[b]Где заказать диплом специалиста?[/b]
[url=http://worldlistpro.com/story19311298/diplom/]worldlistpro.com/story19311298/diplom[/url]
[url=http://tayarut.bkerem.org.il/diplom-529158agiay/]tayarut.bkerem.org.il/diplom-529158agiay[/url]
[url=http://moodjhomedia.com/story1913584/diplom/]moodjhomedia.com/story1913584/diplom[/url]
[url=http://youwantech.com/xe/board/408699/]youwantech.com/xe/board/408699[/url]
[url=http://mysocialname.com/story3018259/diplom/]mysocialname.com/story3018259/diplom[/url]
Lewiscoata
August 4, 2024 @ 11:28 am
http://cytotecbestprice.pro/# buy cytotec
Xazrhql
August 4, 2024 @ 11:58 am
[u][b] Добрый день![/b][/u]
Заказать документ института.
[url=http://mirstalkera.4admins.ru/viewtopic.php?f=50&t=2121/]mirstalkera.4admins.ru/viewtopic.php?f=50&t=2121[/url]
[url=http://derivsocial.org/read-blog/2604/]derivsocial.org/read-blog/2604[/url]
[url=http://kuvandyk.ru/message.php?msg=151/]kuvandyk.ru/message.php?msg=151[/url]
[url=http://myweektour.ru/realnyie-diplomyi-dlya-realnyih-professionalov/]myweektour.ru/realnyie-diplomyi-dlya-realnyih-professionalov[/url]
[url=http://ya.7bb.ru/viewtopic.php?id=12256#p34900/]ya.7bb.ru/viewtopic.php?id=12256#p34900[/url]
Lewiscoata
August 4, 2024 @ 12:10 pm
http://cytotecbestprice.pro/# cytotec online
Xazrfao
August 4, 2024 @ 12:49 pm
[b]Добрый день![/b]
Заказать документ о получении высшего образования
[url=http://arusak-diploms-srednee.ru/kupit-diplom-kandidata-nauk]arusak-diploms-srednee.ru/kupit-diplom-kandidata-nauk[/url]
Manrake
August 4, 2024 @ 12:50 pm
[u][b] Привет, друзья![/b][/u]
Заказать документ о получении высшего образования
[url=http://bookmarkstumble.com/story19215171/kupit-diplom]bookmarkstumble.com/story19215171/kupit-diplom[/url]
[url=http://maroonbookmarks.com/story17559438/kupit-diplom]maroonbookmarks.com/story17559438/kupit-diplom[/url]
[url=http://fam2fam.com/groups/kupit-diplom-345880uibbd]fam2fam.com/groups/kupit-diplom-345880uibbd[/url]
[url=http://thebookmarkking.com/story17647344/kupit-diplom]thebookmarkking.com/story17647344/kupit-diplom[/url]
[url=http://laviehub.com/blog/kupit-diplom-231476hsbyo]laviehub.com/blog/kupit-diplom-231476hsbyo[/url]
Mazrikm
August 4, 2024 @ 1:02 pm
[u][b] Привет![/b][/u]
[b]Всё о покупке аттестата о среднем образовании: полезные советы[/b]
[url=http://mans-diplomy.ru/]mans-diplomy.ru[/url]
Yrefrbu
August 4, 2024 @ 2:18 pm
[u][b] Добрый день![/b][/u]
[b]Заказать диплом о высшем образовании.[/b]
[url=http://ismayilovform.com/blog/page,1,26,16-qarin-eritmeyin-en-effektiv-yollari/]ismayilovform.com/blog/page,1,26,16-qarin-eritmeyin-en-effektiv-yollari[/url]
[url=http://4yo.us/blogs/140998/Цена-дипломов-авторский-обзор-специалистов/]4yo.us/blogs/140998/Цена-дипломов-авторский-обзор-специалистов[/url]
[url=http://sngine.codiantprod.com/blogs/223/Стоимость-документов-советы-и-авторский-обзор-специалистов?lang=ar_sa/]sngine.codiantprod.com/blogs/223/Стоимость-документов-советы-и-авторский-обзор-специалистов?lang=ar_sa[/url]
[url=http://fisketavling.nu/cb-profile/pluginclass/cbblogs?action=blogs&func=show&id=341/]fisketavling.nu/cb-profile/pluginclass/cbblogs?action=blogs&func=show&id=341[/url]
[url=http://thenet.work/gallery/image/481-быстрый-выбор-онлайн-магазина-реализующего-дипломы-и-аттестаты/]thenet.work/gallery/image/481-быстрый-выбор-онлайн-магазина-реализующего-дипломы-и-аттестаты[/url]
Eanrixh
August 4, 2024 @ 2:52 pm
[u][b] Привет![/b][/u]
[b]Мы готовы предложить дипломы[/b] любой профессии по приятным ценам.
[url=http://l67697qa.beget.tech/2024/07/04/zachem-v-nashe-vremya-pokupayut-diplomy-v-internete/]l67697qa.beget.tech/2024/07/04/zachem-v-nashe-vremya-pokupayut-diplomy-v-internete[/url]
[url=http://thesocialgroup.co.uk/blogs/521/Надежный-и-проверенный-интернет-магазин-с-широким-выбором-документов/]thesocialgroup.co.uk/blogs/521/Надежный-и-проверенный-интернет-магазин-с-широким-выбором-документов[/url]
[url=http://create-lifestyle.mn.co/posts/61724492/]create-lifestyle.mn.co/posts/61724492[/url]
[url=http://keros-mod.ru/index.php?/gallery/image/12-низкие-расценки-и-высокое-качество-документы-онлайн/]keros-mod.ru/index.php?/gallery/image/12-низкие-расценки-и-высокое-качество-документы-онлайн[/url]
[url=http://forum.freaks.ro/index.php?/calendar/event/5-вы-хотите-заказать-диплом-переходите-в-наш-онлайн-магазин/]forum.freaks.ro/index.php?/calendar/event/5-вы-хотите-заказать-диплом-переходите-в-наш-онлайн-магазин[/url]
Lazrohx
August 4, 2024 @ 2:58 pm
[b]Приобрести диплом ВУЗа.[/b]
[url=http://pnevmokzn.80lvl.ru/search.php?search_id=egosearch/]pnevmokzn.80lvl.ru/search.php?search_id=egosearch[/url]
[url=http://poboltaem.foroactivo.com/post/]poboltaem.foroactivo.com/post[/url]
[url=http://sz.80lvl.ru/posting.php?mode=post&f=9&sid=0cb9034ec4bd4042da5b850a985f842d/]sz.80lvl.ru/posting.php?mode=post&f=9&sid=0cb9034ec4bd4042da5b850a985f842d[/url]
[url=http://krasnodarforum.ru/member.php?u=2812/]krasnodarforum.ru/member.php?u=2812[/url]
[url=http://rolevikionline.g-talk.ru/ucp.php?mode=privacy&sid=9dfc6ff3de758b13f76f846d9cf80be8/]rolevikionline.g-talk.ru/ucp.php?mode=privacy&sid=9dfc6ff3de758b13f76f846d9cf80be8[/url]
Trefpox
August 4, 2024 @ 3:09 pm
[u][b] Здравствуйте![/b][/u]
Как не стать жертвой мошенников при покупке диплома о среднем полном образовании
[url=http://gaigoimoinhat.xyz/купить-диплом-о-высшем-образовании-со/]gaigoimoinhat.xyz/купить-диплом-о-высшем-образовании-со[/url]
Будем рады вам помочь!.
Dnrtnqx
August 4, 2024 @ 3:14 pm
[b]Приобрести документ[/b] института можно у нас в Москве.
[url=http://tigaedu.com/blog/index.php?entryid=325496/]tigaedu.com/blog/index.php?entryid=325496[/url]
[url=http://arandaasesoria.com/diplom-kupit-848841nt/]arandaasesoria.com/diplom-kupit-848841nt[/url]
[url=http://syg-us.com/entrenamiento-virtual/blog/index.php?entryid=46667/]syg-us.com/entrenamiento-virtual/blog/index.php?entryid=46667[/url]
[url=http://axisgraf.com/2024/07/28/diplom-794663qhgtl/]axisgraf.com/2024/07/28/diplom-794663qhgtl[/url]
[url=http://managerhotels.com/business-customer-service/diplom-660903bydcz/]managerhotels.com/business-customer-service/diplom-660903bydcz[/url]
Williamraics
August 4, 2024 @ 3:36 pm
[u][b] Здравствуйте![/b][/u]
Мы предлагаем дипломы.
[url=http://fighter71.com/read-blog/1345/]fighter71.com/read-blog/1345[/url]
[url=http://registraciavsaita.listbb.ru/ucp.php?mode=login&sid=3d0f9063e3a49419644371ddb0b93701/]registraciavsaita.listbb.ru/ucp.php?mode=login&sid=3d0f9063e3a49419644371ddb0b93701[/url]
[url=http://epistles.ru/blogs/1529/Купить-диплом-быстро-и-надежно/]epistles.ru/blogs/1529/Купить-диплом-быстро-и-надежно[/url]
[url=http://ostome.ru/forumbb/posting.php?mode=post&f=12/]ostome.ru/forumbb/posting.php?mode=post&f=12[/url]
[url=http://rossensor.ru/forum/?PAGE_NAME=message&FID=1&TID=16550&TITLE_SEO=16550-kupit-legalnyy-diplom_-bystro-i-konfidentsialno&MID=861008&result=new#message861008/]rossensor.ru/forum/?PAGE_NAME=message&FID=1&TID=16550&TITLE_SEO=16550-kupit-legalnyy-diplom_-bystro-i-konfidentsialno&MID=861008&result=new#message861008[/url]
RobertKeync
August 4, 2024 @ 3:37 pm
where can i buy prednisone [url=https://prednisonebestprice.pro/#]prednisone otc price[/url] prednisone purchase canada
Matthewadact
August 4, 2024 @ 4:39 pm
buy cheap propecia without insurance: get generic propecia prices – order propecia for sale
Yrefoup
August 4, 2024 @ 4:54 pm
[u][b] Добрый день![/b][/u]
[b]Приобрести диплом университета.[/b]
[url=http://stscrap.kr/gb5/bbs/board.php?bo_table=consult_20240205&wr_id=1375/]stscrap.kr/gb5/bbs/board.php?bo_table=consult_20240205&wr_id=1375[/url]
[url=http://thesocialmusic.co.uk/blogs/7254/Основные-затраты-российского-производителя-дипломов-обзор/]thesocialmusic.co.uk/blogs/7254/Основные-затраты-российского-производителя-дипломов-обзор[/url]
[url=http://watucowalu.cavandoragh.org/cto-trebuetsa-dla-proizvodstva-kacestvennogo-diploma-universiteta/]watucowalu.cavandoragh.org/cto-trebuetsa-dla-proizvodstva-kacestvennogo-diploma-universiteta[/url]
[url=http://fijianvibe.com/blogs/779/Что-именно-нужно-для-создания-неотличимого-от-оригинала-диплома?lang=en_us/]fijianvibe.com/blogs/779/Что-именно-нужно-для-создания-неотличимого-от-оригинала-диплома?lang=en_us[/url]
[url=http://isig.ac.cd/alumni/blogs/37709/Основные-затраты-российского-изготовителя-дипломов-обзор/]isig.ac.cd/alumni/blogs/37709/Основные-затраты-российского-изготовителя-дипломов-обзор[/url]
Patrickirowl
August 4, 2024 @ 4:57 pm
http://cytotecbestprice.pro/# Misoprostol 200 mg buy online
Matthewadact
August 4, 2024 @ 5:12 pm
cytotec buy online usa: buy cytotec in usa – cytotec abortion pill
Sazrarq
August 4, 2024 @ 5:17 pm
[u][b] Привет, друзья![/b][/u]
Купить документ о получении высшего образования
[url=http://uconnect.ae/read-blog/79550/]uconnect.ae/read-blog/79550[/url]
[url=http://naya.social/read-blog/120/]naya.social/read-blog/120[/url]
[url=http://xect2ppa5.blogdeazar.com/28386633/С-легкостью-заказываем-аттестат-в-известном-магазине-russian-diplom/]xect2ppa5.blogdeazar.com/28386633/С-легкостью-заказываем-аттестат-в-известном-магазине-russian-diplom[/url]
[url=http://humped.life/read-blog/535/]humped.life/read-blog/535[/url]
[url=http://miss2010.nuclear.ru/site_py/blogs/1/tekhpodderzhka-i-faq-priobretaem-diplom-v-seti.php/]miss2010.nuclear.ru/site_py/blogs/1/tekhpodderzhka-i-faq-priobretaem-diplom-v-seti.php[/url]
Dnrtege
August 4, 2024 @ 5:56 pm
[b]Купить документ[/b] института вы имеете возможность в нашей компании.
[url=http://hariharparagovernmentiti.com/2024/07/26/diplom-708108lipbb/]hariharparagovernmentiti.com/2024/07/26/diplom-708108lipbb[/url]
[url=http://comfortrent.ru/2024/07/28/diplom-104431hmdde/]comfortrent.ru/2024/07/28/diplom-104431hmdde[/url]
[url=http://localtrusted.co.uk/wiki/index.php?title=Diplom_578801rvmft/]localtrusted.co.uk/wiki/index.php?title=Diplom_578801rvmft[/url]
[url=http://gtj.kr/board_KtRj53/373645/]gtj.kr/board_KtRj53/373645[/url]
[url=http://tigaedu.com/blog/index.php?entryid=325467/]tigaedu.com/blog/index.php?entryid=325467[/url]
Diplomi_hxSi
August 4, 2024 @ 6:19 pm
Добрый день!
Купить документ университета вы сможете в нашей компании в Москве.
[url=http://diploms-x.com/kupit-diplom-specialista/]diploms-x.com/kupit-diplom-specialista[/url]
Lewiscoata
August 4, 2024 @ 7:35 pm
https://prednisonebestprice.pro/# prednisone 20 mg purchase
Lewiscoata
August 4, 2024 @ 8:11 pm
http://prednisonebestprice.pro/# prednisone 20mg by mail order
Qazrmrr
August 4, 2024 @ 8:59 pm
[u][b] Привет, друзья![/b][/u]
[b]Купить диплом университета.[/b]
[url=http://cms-blog-news.de/modules.php?name=Journal&file=display&jid=202/]cms-blog-news.de/modules.php?name=Journal&file=display&jid=202[/url]
[url=http://laporteproperty.com/en/cb-profile/pluginclass/cbblogs?action=blogs&func=show&id=127/]laporteproperty.com/en/cb-profile/pluginclass/cbblogs?action=blogs&func=show&id=127[/url]
[url=http://chumcity.xyz/read-blog/576/]chumcity.xyz/read-blog/576[/url]
[url=http://anamaliya.ru/otzyvy/kakie-onlayn-magaziny-s-diplomami-na-tekushchiy-den-imeyutsya/]anamaliya.ru/otzyvy/kakie-onlayn-magaziny-s-diplomami-na-tekushchiy-den-imeyutsya[/url]
[url=http://sonnick84.articlesblogger.com/51285236/Советы-специалиста-что-помогут-легко-приобрести-документы/]sonnick84.articlesblogger.com/51285236/Советы-специалиста-что-помогут-легко-приобрести-документы[/url]
Diplomi_bnSi
August 4, 2024 @ 9:00 pm
Добрый день!
Заказать документ ВУЗа можно в нашей компании в Москве.
[url=http://diplomasx.com/kupit-diplom-specialista /]diplomasx.com/kupit-diplom-specialista [/url]
Larioryer
August 4, 2024 @ 10:15 pm
Полезные советы по безопасной покупке диплома о высшем образовании
[url=http://technoevents.ru/diplom-lyuboy-stepeni-i-spetsialnosti-dostupno-kazhdomu/]technoevents.ru/diplom-lyuboy-stepeni-i-spetsialnosti-dostupno-kazhdomu[/url]
[url=http://forum.7x.ru/member.php?u=16498/]forum.7x.ru/member.php?u=16498[/url]
[url=http://theprome.com/read-blog/13736/]theprome.com/read-blog/13736[/url]
[url=http://respectmyopinion.com/blogs/32/Советы-что-позволят-найти-проверенный-магазин-с-дипломами/]respectmyopinion.com/blogs/32/Советы-что-позволят-найти-проверенный-магазин-с-дипломами[/url]
[url=http://u90517ol.beget.tech/2024/07/11/oformlenie-diploma-pod-klyuch-bezopasno-i-legalno/]u90517ol.beget.tech/2024/07/11/oformlenie-diploma-pod-klyuch-bezopasno-i-legalno[/url]
Xazrkpn
August 4, 2024 @ 10:59 pm
[u][b] Привет, друзья![/b][/u]
Приобрести документ ВУЗа.
[url=http://maminmir.getbb.ru/viewtopic.php?f=1&t=1128/]maminmir.getbb.ru/viewtopic.php?f=1&t=1128[/url]
[url=http://bmc.ukrbb.net/posting.php?mode=post&f=8/]bmc.ukrbb.net/posting.php?mode=post&f=8[/url]
[url=http://wap.fortboyard2012forum.4adm.ru/viewtopic.php?f=27&t=4159/]wap.fortboyard2012forum.4adm.ru/viewtopic.php?f=27&t=4159[/url]
[url=http://sovetushka.forum2x2.ru/t28893-topic#63665/]sovetushka.forum2x2.ru/t28893-topic#63665[/url]
[url=http://miku-crdb.ru/forum/messages/forum1/topic343/message372/?result=new#message372/]miku-crdb.ru/forum/messages/forum1/topic343/message372/?result=new#message372[/url]
Information Security Analysis
August 4, 2024 @ 11:37 pm
Every student needs to spend a lot of time getting better at academic writing. A lot of students say they have trouble with the steps they need to take to complete a writing task. At times, it might seem hard for them to combine their studies, careers, and fun activities. Many students choose to use academic writing services instead of doing their own work, even though most of them have the skills and knowledge to do it themselves. These services do most of the work, so students only have to finish polishing their work and turn it in. Every year, online class help helps a huge number of students with their homework, giving them more time to do other things besides study. You can now do an online class in a way that will get you great grades without wasting any of your time. At the moment, you can use services to pay someone to take my online class. There is a good chance that you will finish all of your homework well ahead of time.
Matthewadact
August 5, 2024 @ 12:09 am
where can i buy prednisone online without a prescription: prednisone 60 mg – prednisone prescription for sale
Vazrqgv
August 5, 2024 @ 12:09 am
[u][b] Привет, друзья![/b][/u]
Заказать документ о получении высшего образования вы сможете в нашей компании в столице. Мы предлагаем документы об окончании любых университетов России.
[url=http://adgoods.ru/user/profile/4810/]adgoods.ru/user/profile/4810[/url]
[url=http://японецавто.рф/forum/user/51624/]японецавто.рф/forum/user/51624[/url]
[url=http://speicher-photovoltaik.de/profile/?u=30952/]speicher-photovoltaik.de/profile/?u=30952[/url]
[url=http://to-kaz.mediatex-n.kz/index.php?subaction=userinfo&user=ilude/]to-kaz.mediatex-n.kz/index.php?subaction=userinfo&user=ilude[/url]
[url=http://mydnepr.pp.ua/forum/topic.php?forum=7&topic=941/]mydnepr.pp.ua/forum/topic.php?forum=7&topic=941[/url]
Matthewadact
August 5, 2024 @ 12:54 am
buying generic propecia tablets: buying cheap propecia without dr prescription – cost of propecia pills
Mazrxqc
August 5, 2024 @ 2:08 am
[u][b] Здравствуйте![/b][/u]
[b]Официальная покупка диплома вуза с сокращенной программой в Москве[/b]
[url=http://rushkadiplomik.ru/]rushkadiplomik.ru[/url]
Lazrfbr
August 5, 2024 @ 2:20 am
[b]Купить диплом о высшем образовании.[/b]
[url=http://wo.linyway.com/read-blog/3408/]wo.linyway.com/read-blog/3408[/url]
[url=http://vira-ss.flybb.ru/viewtopic.php?f=9&t=430/]vira-ss.flybb.ru/viewtopic.php?f=9&t=430[/url]
[url=http://mockwanasvyazi.getbb.ru/viewtopic.php?f=12&t=1459/]mockwanasvyazi.getbb.ru/viewtopic.php?f=12&t=1459[/url]
[url=http://spasibo.kz/bonus.php/]spasibo.kz/bonus.php[/url]
[url=http://mockwanasvyazi.getbb.ru/viewtopic.php?f=12&t=1481/]mockwanasvyazi.getbb.ru/viewtopic.php?f=12&t=1481[/url]
Treffrq
August 5, 2024 @ 2:55 am
[u][b] Здравствуйте![/b][/u]
Стоимость дипломов высшего и среднего образования и как избежать подделок
[url=http://csleague.ca/диплом-о-среднем-образовании-купить-н/]csleague.ca/диплом-о-среднем-образовании-купить-н[/url]
Будем рады вам помочь!.
Williamraics
August 5, 2024 @ 2:58 am
[u][b] Добрый день![/b][/u]
Мы изготавливаем дипломы.
[url=http://anoreksja.org.pl/posting.php?mode=post&f=13/]anoreksja.org.pl/posting.php?mode=post&f=13[/url]
[url=http://gromscream.80lvl.ru/viewtopic.php?f=3&t=2070/]gromscream.80lvl.ru/viewtopic.php?f=3&t=2070[/url]
[url=http://fsmi.wiki/index.php?title=Настоящие_Дипломы:_Доступно_и_Безопасно/]fsmi.wiki/index.php?title=Настоящие_Дипломы:_Доступно_и_Безопасно[/url]
[url=http://paulikipedia.ru/index.php/Легальная_Покупка_Дипломов:_Конфиденциально_и_Быстро/]paulikipedia.ru/index.php/Легальная_Покупка_Дипломов:_Конфиденциально_и_Быстро[/url]
[url=http://silton.ru/forum/user/5002/]silton.ru/forum/user/5002[/url]
Mazrgxg
August 5, 2024 @ 5:12 am
[u][b] Здравствуйте![/b][/u]
[b]Как правильно купить диплом колледжа и пту в России, подводные камни[/b]
[url=http://rushkadiplomik.ru/]rushkadiplomik.ru[/url]
Yrefjtv
August 5, 2024 @ 5:22 am
[u][b] Здравствуйте![/b][/u]
[b]Приобрести диплом ВУЗа.[/b]
[url=http://allnewstroy.ru/page/3/]allnewstroy.ru/page/3[/url]
[url=http://djjmeets.com/blogs/179064/Главные-расходы-российского-изготовителя-дипломов-авторский-обзор/]djjmeets.com/blogs/179064/Главные-расходы-российского-изготовителя-дипломов-авторский-обзор[/url]
[url=http://scientistsufo.ru/page/3/]scientistsufo.ru/page/3[/url]
[url=http://myliveroom.com/blogs/1057/Что-именно-понадобится-для-создания-качественного-документа/]myliveroom.com/blogs/1057/Что-именно-понадобится-для-создания-качественного-документа[/url]
[url=http://buzzakoo.com/blogs/772/Ключевые-затраты-производителя-дипломов-авторский-обзор/]buzzakoo.com/blogs/772/Ключевые-затраты-производителя-дипломов-авторский-обзор[/url]
Trefjww
August 5, 2024 @ 5:37 am
[u][b] Привет![/b][/u]
Как избежать рисков при покупке диплома колледжа или ПТУ в России
[url=http://protectorakanaan.com/купить-диплом-проверенные-способы/]protectorakanaan.com/купить-диплом-проверенные-способы[/url]
Рады оказаться полезными!.
Sazrtom
August 5, 2024 @ 6:00 am
[u][b] Здравствуйте![/b][/u]
Купить документ ВУЗа
[url=http://gutscheine-247.de/modules.php?name=Journal&file=display&jid=209/]gutscheine-247.de/modules.php?name=Journal&file=display&jid=209[/url]
[url=http://together-19.com/read-blog/26885/]together-19.com/read-blog/26885[/url]
[url=http://hanaverse.top/blogs/917/Как-можно-купить-сегодня-качественный-диплом-в-интернете/]hanaverse.top/blogs/917/Как-можно-купить-сегодня-качественный-диплом-в-интернете[/url]
[url=http://sonnick84.bloginder.com/28793133/Документы-по-низким-ценам-в-знаменитом-магазине/]sonnick84.bloginder.com/28793133/Документы-по-низким-ценам-в-знаменитом-магазине[/url]
[url=http://ja360.fun/read-blog/18/]ja360.fun/read-blog/18[/url]
Dnrtbvw
August 5, 2024 @ 7:23 am
[b]Купить документ[/b] университета можно у нас в столице.
[url=http://luxuriousrentz.com/diplom-kupit-688716gk/]luxuriousrentz.com/diplom-kupit-688716gk[/url]
[url=http://directlynews.com/diplom-585878vrmun/]directlynews.com/diplom-585878vrmun[/url]
[url=http://ada.waaron.org/blog/index.php?entryid=5408/]ada.waaron.org/blog/index.php?entryid=5408[/url]
[url=http://offmarketbusinessforsale.com/diplom-254766ojzcc/]offmarketbusinessforsale.com/diplom-254766ojzcc[/url]
[url=http://ez-bookmarking.com/story17628933/diplom/]ez-bookmarking.com/story17628933/diplom[/url]
Matthewadact
August 5, 2024 @ 7:46 am
by prednisone w not prescription: prednisone for dogs – generic over the counter prednisone
Matthewadact
August 5, 2024 @ 8:20 am
cost of generic propecia without rx: generic propecia – cost of cheap propecia pill
Yreftgy
August 5, 2024 @ 8:41 am
[u][b] Привет![/b][/u]
[b]Приобрести диплом любого университета.[/b]
[url=http://investorcartel.com/community/profile/alexandriaseymo/]investorcartel.com/community/profile/alexandriaseymo[/url]
[url=http://ismayilovform.com/blog/page,1,26,16-qarin-eritmeyin-en-effektiv-yollari/]ismayilovform.com/blog/page,1,26,16-qarin-eritmeyin-en-effektiv-yollari[/url]
[url=http://find-topdeals.com/blogs/133404/Где-купить-диплом-с-гарантией-качества/]find-topdeals.com/blogs/133404/Где-купить-диплом-с-гарантией-качества[/url]
[url=http://stscrap.kr/gb5/bbs/board.php?bo_table=consult_20240205&wr_id=1375/]stscrap.kr/gb5/bbs/board.php?bo_table=consult_20240205&wr_id=1375[/url]
[url=http://fromrus.su/index.php?/gallery/image/331-аттестаты-дипломы-и-справки-в-знаменитом-онлайн-магазине/]fromrus.su/index.php?/gallery/image/331-аттестаты-дипломы-и-справки-в-знаменитом-онлайн-магазине[/url]
Dnrtogh
August 5, 2024 @ 10:22 am
[b]Заказать документ[/b] университета можно в нашей компании в Москве.
[url=http://taondinternational.rudraserver.com/blog/index.php?entryid=133666/]taondinternational.rudraserver.com/blog/index.php?entryid=133666[/url]
[url=http://thesocialvibes.com/story3033118/diplom/]thesocialvibes.com/story3033118/diplom[/url]
[url=http://parathajoint.com/2024/07/28/diplom-kupit-292090kr/]parathajoint.com/2024/07/28/diplom-kupit-292090kr[/url]
[url=http://pwi2.dragonicgames.com/diplom-189759kytyz/]pwi2.dragonicgames.com/diplom-189759kytyz[/url]
[url=http://toplistar.com/story19386385/diplom/]toplistar.com/story19386385/diplom[/url]
Cazrqqp
August 5, 2024 @ 10:33 am
[u][b] Привет![/b][/u]
Мы предлагаем документы ВУЗов
[url=http://severka.flybb.ru/viewtopic.php?f=6&t=380/]severka.flybb.ru/viewtopic.php?f=6&t=380[/url]
[url=http://dskrnd.ru/forum/user/6709/]dskrnd.ru/forum/user/6709[/url]
[url=http://rodina.listbb.ru/ucp.php?mode=login&sid=605915d2bfe4419b5e3a3d3372250813/]rodina.listbb.ru/ucp.php?mode=login&sid=605915d2bfe4419b5e3a3d3372250813[/url]
[url=http://monstaluck.mn.co/posts/62527858/]monstaluck.mn.co/posts/62527858[/url]
[url=http://1776reloaded.us/blogs/173/Легальное-получение-диплома-в-короткие-сроки/]1776reloaded.us/blogs/173/Легальное-получение-диплома-в-короткие-сроки[/url]
Vazrbdk
August 5, 2024 @ 10:51 am
[u][b] Добрый день![/b][/u]
Купить документ о получении высшего образования можно в нашей компании в Москве. Мы оказываем услуги по продаже документов об окончании любых ВУЗов Российской Федерации.
[url=http://rf-4fun.ru/index.php?/topic/324-удача-или-упорство/page/2/#comment-3118/]rf-4fun.ru/index.php?/topic/324-удача-или-упорство/page/2/#comment-3118[/url]
[url=http://jm-eng16.com/bbs/board.php?bo_table=free&wr_id=373036/]jm-eng16.com/bbs/board.php?bo_table=free&wr_id=373036[/url]
[url=http://support.ipron.com/communication/forum/index.php?PAGE_NAME=profile_view&UID=17543/]support.ipron.com/communication/forum/index.php?PAGE_NAME=profile_view&UID=17543[/url]
[url=http://k-special.com/bbs/board.php?bo_table=free&wr_id=1094438/]k-special.com/bbs/board.php?bo_table=free&wr_id=1094438[/url]
[url=http://85sucai.com/member/index.php?uid=yhaxyn/]85sucai.com/member/index.php?uid=yhaxyn[/url]
Lewiscoata
August 5, 2024 @ 10:57 am
https://nolvadexbestprice.pro/# tamoxifen hip pain
Lewiscoata
August 5, 2024 @ 12:21 pm
http://zithromaxbestprice.pro/# zithromax tablets for sale
Matthewadact
August 5, 2024 @ 2:59 pm
buy cytotec over the counter: Cytotec 200mcg price – buy misoprostol over the counter
Matthewadact
August 5, 2024 @ 3:34 pm
tamoxifen 20 mg tablet: natural alternatives to tamoxifen – aromatase inhibitor tamoxifen
Roscoe Davey
August 5, 2024 @ 7:00 pm
Dear missouri.edu webmaster, You always provide great resources and references.
Diplomi_nbSi
August 5, 2024 @ 7:43 pm
Привет!
Купить документ о получении высшего образования вы можете в нашей компании в столице.
[url=http://diploms-rushkas.ru/]diploms-rushkas.ru[/url]
Mazrwuu
August 5, 2024 @ 7:49 pm
[u][b] Добрый день![/b][/u]
[b]Как официально купить диплом вуза с упрощенным обучением в Москве[/b]
[url=http://rushkadiplomik.ru/]rushkadiplomik.ru[/url]
Mazrzto
August 5, 2024 @ 7:54 pm
[b]Купить документ о получении высшего образования.[/b]
[url=http://get-social-now.com/story2915437/kupit-diplom/]get-social-now.com/story2915437/kupit-diplom[/url]
[url=http://getsocialnetwork.com/story3054247/kupit-diplom/]getsocialnetwork.com/story3054247/kupit-diplom[/url]
[url=http://edux.izvrsna.hr/blog/index.php?entryid=107/]edux.izvrsna.hr/blog/index.php?entryid=107[/url]
[url=http://userbookmark.com/story17644385/kupit-diplom/]userbookmark.com/story17644385/kupit-diplom[/url]
[url=http://investicos.com/uncategorized/kupit-diplom-936007oloew/]investicos.com/uncategorized/kupit-diplom-936007oloew[/url]
Matthewadact
August 5, 2024 @ 10:11 pm
get generic propecia without dr prescription: cheap propecia price – get propecia online
Diplomi_xzSi
August 5, 2024 @ 10:41 pm
Добрый день!
Заказать документ о получении высшего образования вы имеете возможность в нашей компании.
[url=http://rushkadiplomikx.ru/]rushkadiplomikx.ru[/url]
Matthewadact
August 5, 2024 @ 10:46 pm
buy propecia online: cost of generic propecia online – buying generic propecia without dr prescription
Mazryob
August 5, 2024 @ 10:54 pm
[u][b] Здравствуйте![/b][/u]
[b]Диплом пту купить официально с упрощенным обучением в Москве[/b]
[url=http://rushkadiplomik.ru/]rushkadiplomik.ru[/url]
Razrzcf
August 5, 2024 @ 11:32 pm
[u][b] Добрый день![/b][/u]
Заказать диплом любого университета:
[url=http://doktorpishet.ru/index.php?subaction=userinfo&user=oniliti/]doktorpishet.ru/index.php?subaction=userinfo&user=oniliti[/url]
[url=http://heimur.ru/index.php?/topic/132-купить-диплом-о-среднем-образовании-r266p/]heimur.ru/index.php?/topic/132-купить-диплом-о-среднем-образовании-r266p[/url]
[url=http://lamoonvideo.ru/profile.php?u=ojukyjab/]lamoonvideo.ru/profile.php?u=ojukyjab[/url]
[url=http://mc-pe.net/index.php?subaction=userinfo&user=uzarome/]mc-pe.net/index.php?subaction=userinfo&user=uzarome[/url]
[url=http://pax.nichost.ru/forum/view_profile.php?UID=133467/]pax.nichost.ru/forum/view_profile.php?UID=133467[/url]
Dnrtuva
August 6, 2024 @ 12:22 am
[b]Заказать документ[/b] о получении высшего образования вы можете у нас в Москве.
[url=http://managerhotels.com/business-sales/diplom-281350wlmat/]managerhotels.com/business-sales/diplom-281350wlmat[/url]
[url=http://bookmarkpath.com/story17609436/diplom/]bookmarkpath.com/story17609436/diplom[/url]
[url=http://socialbaskets.com/story3090087/diplom/]socialbaskets.com/story3090087/diplom[/url]
[url=http://selfhackathon.com/diplom-kupit-359385ex/]selfhackathon.com/diplom-kupit-359385ex[/url]
[url=http://iowa-bookmarks.com/story13281929/diplom/]iowa-bookmarks.com/story13281929/diplom[/url]
Lazrbpx
August 6, 2024 @ 1:37 am
[u][b] Привет, друзья![/b][/u]
[b]Мы изготавливаем дипломы[/b] любой профессии по доступным тарифам.
[url=http://rushkas-diploms.ru/kupit-diplom-v-kazani/]rushkas-diploms.ru/kupit-diplom-v-kazani[/url]
Sazrbfj
August 6, 2024 @ 2:16 am
[u][b] Добрый день![/b][/u]
Диплом магистра
[url=http://mos.flybb.ru/posting.php?mode=post&f=2/]mos.flybb.ru/posting.php?mode=post&f=2[/url]
Uazrbau
August 6, 2024 @ 2:22 am
[u][b] Добрый день![/b][/u]
[b]Заказать диплом любого университета.[/b]
[url=http://true.pahom.su/2024/07/07/podderzhka-i-faq-zakazyvaem-dokumenty-v-internete/]true.pahom.su/2024/07/07/podderzhka-i-faq-zakazyvaem-dokumenty-v-internete[/url]
Carlosses
August 6, 2024 @ 2:22 am
what is tamoxifen used for: tamoxifen and ovarian cancer – who should take tamoxifen
Carlosses
August 6, 2024 @ 2:46 am
low dose tamoxifen: tamoxifen therapy – tamoxifen side effects forum
Yrefxbs
August 6, 2024 @ 3:07 am
[u][b] Привет, друзья![/b][/u]
[b]Купить диплом о высшем образовании.[/b]
[url=http://lamerpension.co.kr/www/bbs/board.php?bo_table=bod703&wr_id=152618/]lamerpension.co.kr/www/bbs/board.php?bo_table=bod703&wr_id=152618[/url]
[url=http://madesports.net/купить-диплом-в-бульгуме-анонимно-и-по/]madesports.net/купить-диплом-в-бульгуме-анонимно-и-по[/url]
[url=http://yobasket.es/es/perfil-usuario/pluginclass/cbblogs?action=blogs&func=show&id=126/]yobasket.es/es/perfil-usuario/pluginclass/cbblogs?action=blogs&func=show&id=126[/url]
[url=http://thesocialmusic.co.uk/blogs/7254/Основные-затраты-российского-производителя-дипломов-обзор/]thesocialmusic.co.uk/blogs/7254/Основные-затраты-российского-производителя-дипломов-обзор[/url]
[url=http://blogs.rufox.ru/~worksale/48372.htm/]blogs.rufox.ru/~worksale/48372.htm[/url]
Dnrtlrt
August 6, 2024 @ 3:15 am
[b]Приобрести документ[/b] института можно у нас в столице.
[url=http://candidecoin.com/uncategorized/diplom-483151bamyp/]candidecoin.com/uncategorized/diplom-483151bamyp[/url]
[url=http://welnesbiolabs.com/diplom-kupit-697843rg/]welnesbiolabs.com/diplom-kupit-697843rg[/url]
[url=http://allyourbookmarks.com/story17664353/diplom/]allyourbookmarks.com/story17664353/diplom[/url]
[url=http://bookmarksoflife.com/story3122489/diplom/]bookmarksoflife.com/story3122489/diplom[/url]
[url=http://applysarkarinaukri.com/diplom-587060dvdpj/]applysarkarinaukri.com/diplom-587060dvdpj[/url]
StevenDak
August 6, 2024 @ 4:10 am
prednisone uk buy http://prednisonebestprice.pro/# prednisone 100 mg
prednisone 20mg prescription cost
Lewiscoata
August 6, 2024 @ 4:23 am
https://prednisonebestprice.pro/# prednisone no rx
Lazream
August 6, 2024 @ 4:43 am
[u][b] Привет![/b][/u]
[b]Мы готовы предложить дипломы[/b] психологов, юристов, экономистов и прочих профессий по доступным тарифам.
[url=http://diplomrushkan.ru/kupit-diplom-novosibirsk/]diplomrushkan.ru/kupit-diplom-novosibirsk[/url]
Trefpww
August 6, 2024 @ 5:03 am
[u][b] Привет![/b][/u]
Быстрое обучение и получение диплома магистра – возможно ли это?
[url=http://mans-diplom.ru/kupit-diplom-ekaterinburg/]mans-diplom.ru/kupit-diplom-ekaterinburg[/url]
Всегда вам поможем!.
Matthewadact
August 6, 2024 @ 5:27 am
zithromax online usa: zithromax 500mg over the counter – zithromax z-pak
Lewiscoata
August 6, 2024 @ 5:50 am
http://zithromaxbestprice.pro/# zithromax capsules price
Matthewadact
August 6, 2024 @ 5:59 am
buy cytotec over the counter: Misoprostol 200 mg buy online – buy misoprostol over the counter
Mazrrao
August 6, 2024 @ 6:07 am
[b]Приобрести документ о получении высшего образования.[/b]
[url=http://hariharparagovernmentiti.com/2024/07/26/kupit-diplom-906266ulgkp/]hariharparagovernmentiti.com/2024/07/26/kupit-diplom-906266ulgkp[/url]
[url=http://bookmarkfly.com/story17696288/kupit-diplom/]bookmarkfly.com/story17696288/kupit-diplom[/url]
[url=http://freebiznetwork.com/kupit-diplom-40330hpnsr/]freebiznetwork.com/kupit-diplom-40330hpnsr[/url]
[url=http://pr6bookmark.com/story17805454/kupit-diplom/]pr6bookmark.com/story17805454/kupit-diplom[/url]
[url=http://thecatalystapproach.com/kupit-diplom-158584bbcpr/]thecatalystapproach.com/kupit-diplom-158584bbcpr[/url]
Trefqig
August 6, 2024 @ 7:46 am
[u][b] Привет, друзья![/b][/u]
Пошаговая инструкция по официальной покупке диплома о высшем образовании
[url=http://mandiplomikx.ru/kupit-diplom-ekaterinburg/]mandiplomikx.ru/kupit-diplom-ekaterinburg[/url]
Окажем помощь!.
Oariorksr
August 6, 2024 @ 7:53 am
[u][b] Привет![/b][/u]
Купить диплом университета
[b]Наша компания предлагает[/b] выгодно и быстро заказать диплом, который выполняется на оригинальном бланке и заверен печатями, водяными знаками, подписями должностных лиц. Наш документ пройдет лубую проверку, даже при помощи профессиональных приборов. Решайте свои задачи быстро и просто с нашим сервисом.
[b]Где заказать диплом по актуальной специальности?[/b]
[url=http://silich.ru/forum/member.php?u=4810/]silich.ru/forum/member.php?u=4810[/url]
[url=http://www.nhps1914.com/wiki/Официальные_Дипломы:_Качество_и_Легальность/]www.nhps1914.com/wiki/Официальные_Дипломы:_Качество_и_Легальность[/url]
[url=http://9unity.com/read-blog/2412/]9unity.com/read-blog/2412[/url]
[url=http://obozrevatelevents.ru/diplomyi-ot-nadezhnogo-postavshhika//]obozrevatelevents.ru/diplomyi-ot-nadezhnogo-postavshhika/[/url]
[url=http://smd.mybb.ru/viewtopic.php?id=4039#p23031/]smd.mybb.ru/viewtopic.php?id=4039#p23031[/url]
Manrisw
August 6, 2024 @ 8:16 am
[u][b] Привет, друзья![/b][/u]
Приобрести документ университета
[url=http://swflpastors.org/kupit-diplom-896000bkhjl]swflpastors.org/kupit-diplom-896000bkhjl[/url]
[url=http://nervivporyadke.ru/Voprosi/kupit-diplom-728746obglc]nervivporyadke.ru/Voprosi/kupit-diplom-728746obglc[/url]
[url=http://bookmarkerz.com/story17576434/kupit-diplom]bookmarkerz.com/story17576434/kupit-diplom[/url]
[url=http://onlybookmarkings.com/story17605809/kupit-diplom]onlybookmarkings.com/story17605809/kupit-diplom[/url]
[url=http://tayarut.bkerem.org.il/kupit-diplom-902571zxcjg]tayarut.bkerem.org.il/kupit-diplom-902571zxcjg[/url]
Iariorapu
August 6, 2024 @ 10:42 am
[u][b] Здравствуйте![/b][/u]
Заказать документ института вы можете в нашей компании в столице. Мы оказываем услуги по продаже документов об окончании любых университетов РФ. Вы получите диплом по любым специальностям, включая документы старого образца. Гарантируем, что при проверке документа работодателем, подозрений не возникнет.
[url=http://wmrp.listbb.ru/index.php/]wmrp.listbb.ru/index.php[/url]
[url=http://exoltech.net/blogs/157386/На-что-конкретно-обращать-внимание-при-заказе-документов-в-сети/]exoltech.net/blogs/157386/На-что-конкретно-обращать-внимание-при-заказе-документов-в-сети[/url]
[url=http://liobio.com/blogs/1612/Главные-расходы-российского-изготовителя-дипломов-обзор/]liobio.com/blogs/1612/Главные-расходы-российского-изготовителя-дипломов-обзор[/url]
[url=http://redebrasil.app/read-blog/335/]redebrasil.app/read-blog/335[/url]
[url=http://dachaweek.ru/vash-shans-na-vyisshee-obrazovanie-kupit-diplom/]dachaweek.ru/vash-shans-na-vyisshee-obrazovanie-kupit-diplom[/url]
Uazraas
August 6, 2024 @ 11:27 am
[b]Где купить диплом специалиста?[/b]
[url=http://medexmd.com/community/profile/jakeest72729801/]medexmd.com/community/profile/jakeest72729801[/url]
[url=http://getsocialnetwork.com/story3039183/diplom/]getsocialnetwork.com/story3039183/diplom[/url]
[url=http://gaigoimoinhat.xyz/diplom-17529nfcsi/]gaigoimoinhat.xyz/diplom-17529nfcsi[/url]
[url=http://bookmarkingfeed.com/story17603732/diplom/]bookmarkingfeed.com/story17603732/diplom[/url]
[url=http://turnkeymodular.ca/diplom-665228yjked/]turnkeymodular.ca/diplom-665228yjked[/url]
Sazrrrf
August 6, 2024 @ 11:41 am
[u][b] Привет![/b][/u]
Мы можем предложить дипломы психологов, юристов, экономистов и прочих профессий.
[url=http://bookmark-rss.com/story17530112/kupit-diplom/]bookmark-rss.com/story17530112/kupit-diplom[/url]
[url=http://trottiloc.com/author/debbramclei/]trottiloc.com/author/debbramclei[/url]
[url=http://lgukapangan.gov.ph/2024/07/29/kupit-diplom-536242glrih/]lgukapangan.gov.ph/2024/07/29/kupit-diplom-536242glrih[/url]
[url=http://socialicus.com/story2974717/kupit-diplom/]socialicus.com/story2974717/kupit-diplom[/url]
[url=http://sociallytraffic.com/story2474175/kupit-diplom/]sociallytraffic.com/story2474175/kupit-diplom[/url]
Matthewadact
August 6, 2024 @ 12:34 pm
order prednisone online no prescription: prednisone 2 mg – 10 mg prednisone
Cazrvhk
August 6, 2024 @ 12:57 pm
[u][b] Добрый день![/b][/u]
Приобрести диплом о высшем образовании
[url=http://epicit.ru/oformlenie-diplomov-za-schitannyie-dni/]epicit.ru/oformlenie-diplomov-za-schitannyie-dni[/url]
[url=http://afterpad.com/forums/post.php?fid=7/]afterpad.com/forums/post.php?fid=7[/url]
[url=http://collie.fatbb.ru/ucp.php?mode=login&sid=4688d534da970ddc62e0a4d6cf8ba75b/]collie.fatbb.ru/ucp.php?mode=login&sid=4688d534da970ddc62e0a4d6cf8ba75b[/url]
[url=http://mojgorod.getbb.ru/viewtopic.php?f=12&t=1510/]mojgorod.getbb.ru/viewtopic.php?f=12&t=1510[/url]
[url=http://talkrealty.ru/vash-diplom-uzhe-segodnya/]talkrealty.ru/vash-diplom-uzhe-segodnya[/url]
Sazrbfz
August 6, 2024 @ 1:04 pm
[u][b] Привет, друзья![/b][/u]
Диплом любой специальности
[url=http://fbadult.com/blogs/3292/оформите-диплом-без-лишних-хлопот/]fbadult.com/blogs/3292/оформите-диплом-без-лишних-хлопот[/url]
Matthewadact
August 6, 2024 @ 1:09 pm
tamoxifen breast cancer prevention: tamoxifen dose – tamoxifen bone density
Mazrnmi
August 6, 2024 @ 1:38 pm
[u][b] Добрый день![/b][/u]
[b]Стоимость дипломов высшего и среднего образования и процесс их получения[/b]
[url=http://rushkadiplomik.ru/]rushkadiplomik.ru[/url]
Davidpoolo
August 6, 2024 @ 2:35 pm
Самое свежее и актуальное http://barbie-games.ru/ychenye-nashli-sposob-poimat-i-yderjivat-v-lovyshke-edinstvennye-molekyly
Jariorvxt
August 6, 2024 @ 4:01 pm
[u][b] Здравствуйте![/b][/u]
Приобрести диплом ВУЗа
[url=http://lighttur.ru/kupite-diplom-i-zabudte-o-studencheskih-zabotah/]lighttur.ru/kupite-diplom-i-zabudte-o-studencheskih-zabotah[/url]
[url=http://demo.1c-college.ru/club/user/7/blog/bolshaya-populyarnost-internetmagazinov-chto-prodayut-diplomy/]demo.1c-college.ru/club/user/7/blog/bolshaya-populyarnost-internetmagazinov-chto-prodayut-diplomy[/url]
[url=http://mojgorod.getbb.ru/viewtopic.php?f=12&t=1440/]mojgorod.getbb.ru/viewtopic.php?f=12&t=1440[/url]
[url=http://gadjetforyou.ru/diplom-za-dengi-byistro-nadezhno-bezopasno/]gadjetforyou.ru/diplom-za-dengi-byistro-nadezhno-bezopasno[/url]
[url=http://komfort.rusff.me/viewtopic.php?id=13930#p41024/]komfort.rusff.me/viewtopic.php?id=13930#p41024[/url]
Mazrwop
August 6, 2024 @ 4:34 pm
[u][b] Привет![/b][/u]
[b]Как получить диплом техникума с упрощенным обучением в Москве официально[/b]
[url=http://rushkadiplomik.ru/]rushkadiplomik.ru[/url]
Xazrpae
August 6, 2024 @ 4:47 pm
[u][b] Привет![/b][/u]
Купить документ ВУЗа.
[url=http://toplentanews.ru/kupite-diplom-i-zabudte-o-studencheskih-hlopotah/]toplentanews.ru/kupite-diplom-i-zabudte-o-studencheskih-hlopotah[/url]
[url=http://newmedtime.ru/kupite-diplom-i-dostignite-svoih-tseley/]newmedtime.ru/kupite-diplom-i-dostignite-svoih-tseley[/url]
[url=http://team-clo.mn.co/posts/62310512/]team-clo.mn.co/posts/62310512[/url]
[url=http://heyrodisscusion.listbb.ru/viewtopic.php?f=10&t=451/]heyrodisscusion.listbb.ru/viewtopic.php?f=10&t=451[/url]
[url=http://vira-ss.flybb.ru/viewtopic.php?f=9&t=439/]vira-ss.flybb.ru/viewtopic.php?f=9&t=439[/url]
Dnrtkri
August 6, 2024 @ 5:17 pm
[b]Купить документ[/b] института можно у нас в столице.
[url=http://kbkrealtors.com/diplom-kupit-357756sd/]kbkrealtors.com/diplom-kupit-357756sd[/url]
[url=http://higherranker.com/diplom-272198rtggu/]higherranker.com/diplom-272198rtggu[/url]
[url=http://pencis.com/diplom-590415dwhkz/]pencis.com/diplom-590415dwhkz[/url]
[url=http://itdongnam.com/diplom-kupit-817456hg/]itdongnam.com/diplom-kupit-817456hg[/url]
[url=http://sttimothysignal.org/groups/diplom-95082ohsup/]sttimothysignal.org/groups/diplom-95082ohsup[/url]
Danrlaa
August 6, 2024 @ 5:29 pm
[u][b] Привет, друзья![/b][/u]
Мы можем предложить дипломы психологов, юристов, экономистов и других профессий.
[url=http://worldavtonew.ru/ofitsialnyie-diplomyi-podlinnost-i-nadezhnost]worldavtonew.ru/ofitsialnyie-diplomyi-podlinnost-i-nadezhnost[/url]
Рады помочь!.
Oarioryvo
August 6, 2024 @ 6:25 pm
[u][b] Здравствуйте![/b][/u]
Купить диплом о высшем образовании
[b]Наши специалисты предлагают[/b] выгодно купить диплом, который выполнен на бланке ГОЗНАКа и заверен мокрыми печатями, водяными знаками, подписями официальных лиц. Наш документ пройдет лубую проверку, даже с использованием специального оборудования. Достигайте своих целей быстро и просто с нашими дипломами.
[b]Где купить диплом по актуальной специальности?[/b]
[url=http://linkthere.club/read-blog/8168/]linkthere.club/read-blog/8168[/url]
[url=http://www.xusyou.com/blogs/290/Советы-что-позволят-найти-проверенный-магазин-с-документами/]www.xusyou.com/blogs/290/Советы-что-позволят-найти-проверенный-магазин-с-документами[/url]
[url=http://vip.rolevaya.info/viewtopic.php?id=4961#p8497/]vip.rolevaya.info/viewtopic.php?id=4961#p8497[/url]
[url=http://wiki.nikiforov.ru/index.php/Дипломы_на_заказ:_гарантия_качества/]wiki.nikiforov.ru/index.php/Дипломы_на_заказ:_гарантия_качества[/url]
[url=http://bvf.ru/forum/showthread.php?p=25249084#post25249084/]bvf.ru/forum/showthread.php?p=25249084#post25249084[/url]
Yrefuvy
August 6, 2024 @ 7:27 pm
[u][b] Привет, друзья![/b][/u]
[b]Купить диплом университета.[/b]
[url=http://financeokey.ru/page/2/]financeokey.ru/page/2[/url]
[url=http://passneurosurgery.net/learn/blog/index.php?entryid=321177/]passneurosurgery.net/learn/blog/index.php?entryid=321177[/url]
[url=http://ebonyfaces.com/blogs/35/Что-понадобится-из-оборудования-для-создания-качественного-диплома?lang=ar_sa/]ebonyfaces.com/blogs/35/Что-понадобится-из-оборудования-для-создания-качественного-диплома?lang=ar_sa[/url]
[url=http://roqezhishaehae.almoheet-travel.com/glavnye-zatraty-proizvoditela-diplomov-specialnyj-obzor/]roqezhishaehae.almoheet-travel.com/glavnye-zatraty-proizvoditela-diplomov-specialnyj-obzor[/url]
[url=http://djcuna.social/blogs/221/Что-понадобится-для-создания-качественного-диплома-университета?lang=es_mx/]djcuna.social/blogs/221/Что-понадобится-для-создания-качественного-диплома-университета?lang=es_mx[/url]
Lazrvgk
August 6, 2024 @ 7:31 pm
[u][b] Привет![/b][/u]
[b]Мы изготавливаем дипломы[/b] любой профессии по приятным ценам.
[url=http://rushkas-diploms.ru/attestat-9-klassov/]rushkas-diploms.ru/attestat-9-klassov[/url]
Matthewadact
August 6, 2024 @ 7:33 pm
buying cheap propecia without a prescription: cost of generic propecia without a prescription – order propecia without a prescription
Manrwiz
August 6, 2024 @ 7:39 pm
[u][b] Привет, друзья![/b][/u]
Заказать документ о получении высшего образования
[url=http://ilovebookmarking.com/story17657347/kupit-diplom]ilovebookmarking.com/story17657347/kupit-diplom[/url]
[url=http://thecatalystapproach.com/kupit-diplom-248920esqan]thecatalystapproach.com/kupit-diplom-248920esqan[/url]
[url=http://singnalsocial.com/story2940161/kupit-diplom]singnalsocial.com/story2940161/kupit-diplom[/url]
[url=http://managerhotels.com/business-small-business/kupit-diplom-344795lcapi]managerhotels.com/business-small-business/kupit-diplom-344795lcapi[/url]
[url=http://a4everyone.org/kupit-diplom-707883wcpqw]a4everyone.org/kupit-diplom-707883wcpqw[/url]
Matthewadact
August 6, 2024 @ 8:06 pm
zithromax online pharmacy canada: purchase zithromax online – buy zithromax 1000 mg online
Dnrtowe
August 6, 2024 @ 8:12 pm
[b]Заказать документ[/b] о получении высшего образования можно у нас.
[url=http://webookmarks.com/story3052872/diplom/]webookmarks.com/story3052872/diplom[/url]
[url=http://socialbaskets.com/story3091852/diplom/]socialbaskets.com/story3091852/diplom[/url]
[url=http://bookmarkplaces.com/story17612461/diplom/]bookmarkplaces.com/story17612461/diplom[/url]
[url=http://shinhwaspodium.com/bbs/board.php?bo_table=free&wr_id=2180232/]shinhwaspodium.com/bbs/board.php?bo_table=free&wr_id=2180232[/url]
[url=http://youlangue.lu/blog/index.php?entryid=87863/]youlangue.lu/blog/index.php?entryid=87863[/url]
Trefgrs
August 6, 2024 @ 8:26 pm
[u][b] Привет, друзья![/b][/u]
Покупка диплома о среднем полном образовании: как избежать мошенничества?
[url=http://mandiplomikx.ru/diplom-tehnikuma/]mandiplomikx.ru/diplom-tehnikuma[/url]
Рады помочь!.
Diplomi_chSi
August 6, 2024 @ 8:30 pm
Привет!
Купить документ о получении высшего образования вы имеете возможность у нас в столице.
[url=http://rushkadiplomikx.ru/]rushkadiplomikx.ru[/url]
Eanrgez
August 6, 2024 @ 9:00 pm
[u][b] Добрый день![/b][/u]
[b]Мы изготавливаем дипломы[/b] любой профессии по доступным ценам.
[url=http://x70795vj.beget.tech/2024/07/07/faq-i-teh-podderzhka-priobretaem-dokumenty-v-internete/]x70795vj.beget.tech/2024/07/07/faq-i-teh-podderzhka-priobretaem-dokumenty-v-internete[/url]
[url=http://thesn.eu/blogs/2845/Хотите-приобрести-аттестат-у-надежного-исполнителя-Ждем-Вас/]thesn.eu/blogs/2845/Хотите-приобрести-аттестат-у-надежного-исполнителя-Ждем-Вас[/url]
[url=http://myliveroom.com/blogs/1052/Как-купить-сегодня-качественный-диплом-в-сети-интернет/]myliveroom.com/blogs/1052/Как-купить-сегодня-качественный-диплом-в-сети-интернет[/url]
[url=http://80.87.194.225/blogs/1/bystryy-vybor-internetmagazina-prodayushchego-diplomy-i-attestaty.php/]80.87.194.225/blogs/1/bystryy-vybor-internetmagazina-prodayushchego-diplomy-i-attestaty.php[/url]
[url=http://sonnick84.vblogetin.com/33766841/Ищем-проверенный-и-надежный-интернет-магазин-для-покупки-диплома/]sonnick84.vblogetin.com/33766841/Ищем-проверенный-и-надежный-интернет-магазин-для-покупки-диплома[/url]
Iariorjhy
August 6, 2024 @ 9:20 pm
[u][b] Привет![/b][/u]
Купить документ ВУЗа можно у нас в Москве. Мы предлагаем документы об окончании любых ВУЗов РФ. Вы сможете получить необходимый диплом по любым специальностям, включая документы Советского Союза. Гарантируем, что в случае проверки документов работодателем, никаких подозрений не возникнет.
[url=http://good-serial.ru/2024/07/garantirovannoe-kachestvo-diplomov-legalno-i-byistro/]good-serial.ru/2024/07/garantirovannoe-kachestvo-diplomov-legalno-i-byistro[/url]
[url=http://nc750.ru/index.php/]nc750.ru/index.php[/url]
[url=http://korrespondentweek.ru/kupit-diplom-bez-posrednikov-legko-i-byistro/]korrespondentweek.ru/kupit-diplom-bez-posrednikov-legko-i-byistro[/url]
[url=http://sky-metaverse.com/read-blog/10198/]sky-metaverse.com/read-blog/10198[/url]
[url=http://nsibirsk.ru/forum/obyavleniya/newtopic/#google_vignette/]nsibirsk.ru/forum/obyavleniya/newtopic/#google_vignette[/url]
Lewiscoata
August 6, 2024 @ 9:39 pm
http://propeciabestprice.pro/# get propecia online
Sazrsel
August 6, 2024 @ 9:42 pm
[u][b] Здравствуйте![/b][/u]
Мы готовы предложить дипломы любой профессии.
[url=http://fridayad.in/user/profile/2636958/]fridayad.in/user/profile/2636958[/url]
[url=http://kbookmarking.com/story17636739/kupit-diplom/]kbookmarking.com/story17636739/kupit-diplom[/url]
[url=http://sun-clinic.co.il/he/question/kupit-diplom-337241tlifg/]sun-clinic.co.il/he/question/kupit-diplom-337241tlifg[/url]
[url=http://bookmarksurl.com/story3006625/kupit-diplom/]bookmarksurl.com/story3006625/kupit-diplom[/url]
[url=http://bookmarklinking.com/story3102364/kupit-diplom/]bookmarklinking.com/story3102364/kupit-diplom[/url]
Lazrmzw
August 6, 2024 @ 9:49 pm
[b]Заказать диплом университета.[/b]
[url=http://odnopolchane.net/forum/member.php?u=546804/]odnopolchane.net/forum/member.php?u=546804[/url]
[url=http://mymink.5bb.ru/viewtopic.php?id=8884#p454993/]mymink.5bb.ru/viewtopic.php?id=8884#p454993[/url]
[url=http://itbitgroup.ru/kupit-diplom-s-garantiey-podlinnosti/]itbitgroup.ru/kupit-diplom-s-garantiey-podlinnosti[/url]
[url=http://avva.getbb.ru/viewtopic.php?f=3&t=571/]avva.getbb.ru/viewtopic.php?f=3&t=571[/url]
[url=http://hip-hop.ru/forum/id298234-worksale/]hip-hop.ru/forum/id298234-worksale[/url]
Mazrimh
August 6, 2024 @ 9:55 pm
[u][b] Привет, друзья![/b][/u]
[b]Парадокс, но купить диплом кандидата наук оказалось не так и сложно[/b]
[url=http://rushkas-diplomy.ru/kupit-diplom-omsk/]rushkas-diplomy.ru/kupit-diplom-omsk[/url]
Yrefknr
August 6, 2024 @ 10:17 pm
[u][b] Добрый день![/b][/u]
[b]Приобрести диплом университета.[/b]
[url=http://justbevictorious.com/где-купить-диплом/]justbevictorious.com/где-купить-диплом[/url]
[url=http://versatilecommunication.com/купить-диплом-3/]versatilecommunication.com/купить-диплом-3[/url]
[url=http://blogs.rufox.ru/~worksale/48372.htm/]blogs.rufox.ru/~worksale/48372.htm[/url]
[url=http://promed-sd.com/blog/index.php?entryid=77430/]promed-sd.com/blog/index.php?entryid=77430[/url]
[url=http://vocal.com.ua/node?page=12&destination=node/58085/]vocal.com.ua/node?page=12&destination=node/58085[/url]
Lazrtlc
August 6, 2024 @ 10:32 pm
[u][b] Привет, друзья![/b][/u]
[b]Мы предлагаем дипломы[/b] любой профессии по разумным тарифам.
[url=http://diplomrushka.ru/attestat-9-klassov/]diplomrushka.ru/attestat-9-klassov[/url]
Williamraics
August 6, 2024 @ 10:37 pm
[u][b] Привет, друзья![/b][/u]
Мы изготавливаем дипломы.
[url=http://uktuliza.ru/forum/?PAGE_NAME=profile_view&UID=17763/]uktuliza.ru/forum/?PAGE_NAME=profile_view&UID=17763[/url]
[url=http://academy4elements.mybb.ru/viewtopic.php?id=946#p2080/]academy4elements.mybb.ru/viewtopic.php?id=946#p2080[/url]
[url=http://biketrials.ru/live/blog.php?b=9122/]biketrials.ru/live/blog.php?b=9122[/url]
[url=http://shuvalduet.getbb.ru/viewtopic.php?f=3&t=573/]shuvalduet.getbb.ru/viewtopic.php?f=3&t=573[/url]
[url=http://lada-xray.net/member.php?u=2434/]lada-xray.net/member.php?u=2434[/url]
Uazrgpq
August 6, 2024 @ 10:40 pm
[b]Где приобрести диплом по актуальной специальности?[/b]
[url=http://thesocialroi.com/story7353487/diplom/]thesocialroi.com/story7353487/diplom[/url]
[url=http://pirooztak.ir/?option=com_k2&view=itemlist&task=user&id=1356889/]pirooztak.ir/?option=com_k2&view=itemlist&task=user&id=1356889[/url]
[url=http://abileneguntrader.com/author/bettesoundy/]abileneguntrader.com/author/bettesoundy[/url]
[url=http://ecrinparis.com/diplom-825154bhzdi/]ecrinparis.com/diplom-825154bhzdi[/url]
[url=http://wiishlist.com/story18207532/diplom/]wiishlist.com/story18207532/diplom[/url]
Lazrgtg
August 6, 2024 @ 10:59 pm
[u][b] Привет![/b][/u]
[b]Приобрести диплом любого ВУЗа.[/b]
[url=http://mans-diplomyxx.ru/kupit-svidetelstva/]mans-diplomyxx.ru/kupit-svidetelstva[/url]
Trefplh
August 6, 2024 @ 11:05 pm
[u][b] Добрый день![/b][/u]
Как избежать рисков при покупке диплома колледжа или ПТУ в России
[url=http://mans-diplomxx.ru/kupit-svidetelstva/]mans-diplomxx.ru/kupit-svidetelstva[/url]
Окажем помощь!.
Diplomi_utSi
August 6, 2024 @ 11:24 pm
Привет, друзья!
Приобрести документ университета вы сможете у нас.
[url=http://diplomrushkan.ru/]diplomrushkan.ru[/url]
Vazrois
August 7, 2024 @ 3:25 am
[u][b] Привет, друзья![/b][/u]
Приобрести документ ВУЗа можно в нашем сервисе. Мы предлагаем документы об окончании любых ВУЗов Российской Федерации.
[url=http://biyografiforum.10tl.net/newthread.php?fid=124/]biyografiforum.10tl.net/newthread.php?fid=124[/url]
[url=http://polimer42.ru/communication/forum/messages/forum5/topic49/message349159/?result=reply#message349159/]polimer42.ru/communication/forum/messages/forum5/topic49/message349159/?result=reply#message349159[/url]
[url=http://forumklassika.ru/showthread.php?t=129419&p=1843644#post1843644/]forumklassika.ru/showthread.php?t=129419&p=1843644#post1843644[/url]
[url=http://allods.mlx.su/forums/topic/321-купить-диплом-цена-p426g/]allods.mlx.su/forums/topic/321-купить-диплом-цена-p426g[/url]
[url=http://berzarina28a.ru/index.php?/topic/24838-не-могу-создать-новую-тему-в-berzarina28aru/]berzarina28a.ru/index.php?/topic/24838-не-могу-создать-новую-тему-в-berzarina28aru[/url]
Xazrgrs
August 7, 2024 @ 3:40 am
[u][b] Привет![/b][/u]
Приобрести документ о получении высшего образования.
[url=http://demo.advised360.com/read-blog/162165/]demo.advised360.com/read-blog/162165[/url]
[url=http://nnars.ru/forum/suggestion-box/1219-ваш-диплом-за-короткое-время-просто-и-удобно#74605/]nnars.ru/forum/suggestion-box/1219-ваш-диплом-за-короткое-время-просто-и-удобно#74605[/url]
[url=http://nsk.ukrbb.net/viewtopic.php?f=41&t=27434&sid=d210ad3b2868ecf1525d5f8cea79b987/]nsk.ukrbb.net/viewtopic.php?f=41&t=27434&sid=d210ad3b2868ecf1525d5f8cea79b987[/url]
[url=http://vintfint.com/blogs/22839/Диплом-в-кратчайшие-сроки-качественно-и-безопасно/]vintfint.com/blogs/22839/Диплом-в-кратчайшие-сроки-качественно-и-безопасно[/url]
[url=http://the-trifecta-network.mn.co/posts/62175783/]the-trifecta-network.mn.co/posts/62175783[/url]
Lariorkrx
August 7, 2024 @ 4:06 am
Приобретение школьного аттестата с официальным упрощенным обучением в Москве
[url=http://foousrp.listbb.ru/viewtopic.php?f=4&t=515/]foousrp.listbb.ru/viewtopic.php?f=4&t=515[/url]
[url=http://bestworld.getbb.ru/viewtopic.php?f=8&t=1260/]bestworld.getbb.ru/viewtopic.php?f=8&t=1260[/url]
[url=http://respectmyopinion.com/blogs/32/Советы-что-позволят-найти-проверенный-магазин-с-дипломами/]respectmyopinion.com/blogs/32/Советы-что-позволят-найти-проверенный-магазин-с-дипломами[/url]
[url=http://club.sabaylok.com/blogs/5110/Ваш-диплом-без-лишних-забот-быстро-и-безопасно/]club.sabaylok.com/blogs/5110/Ваш-диплом-без-лишних-забот-быстро-и-безопасно[/url]
[url=http://3b1bkqfhg.theideasblog.com/28725622/Советы-что-помогут-подобрать-проверенный-онлайн-магазин-с-дипломами/]3b1bkqfhg.theideasblog.com/28725622/Советы-что-помогут-подобрать-проверенный-онлайн-магазин-с-дипломами[/url]
binance signup
August 7, 2024 @ 4:11 am
Your point of view caught my eye and was very interesting. Thanks. I have a question for you.
Danruxr
August 7, 2024 @ 4:35 am
[u][b] Здравствуйте![/b][/u]
Мы предлагаем дипломы любой профессии.
[url=http://formfinance.ru/byistroe-oformlenie-diplomov-nadezhno-i-legalno]formfinance.ru/byistroe-oformlenie-diplomov-nadezhno-i-legalno[/url]
Рады оказаться полезными!.
Cazrfvg
August 7, 2024 @ 5:48 am
[u][b] Привет![/b][/u]
Мы предлагаем документы техникумов
[url=http://ya.bestbb.ru/viewtopic.php?id=3035#p6708/]ya.bestbb.ru/viewtopic.php?id=3035#p6708[/url]
[url=http://cocapal.com/read-blog/1231/]cocapal.com/read-blog/1231[/url]
[url=http://newnormalnetwork.me/create-blog/]newnormalnetwork.me/create-blog[/url]
[url=http://sky-metaverse.com/read-blog/11387/]sky-metaverse.com/read-blog/11387[/url]
[url=http://collie.fatbb.ru/viewtopic.php?f=18&t=4397/]collie.fatbb.ru/viewtopic.php?f=18&t=4397[/url]
Mazrdtq
August 7, 2024 @ 7:22 am
[u][b] Здравствуйте![/b][/u]
[b]Официальная покупка диплома вуза с сокращенной программой обучения в Москве[/b]
[url=http://rushkadiplomik.ru/]rushkadiplomik.ru[/url]
Mazrpiu
August 7, 2024 @ 8:42 am
[u][b] Привет![/b][/u]
[b]Официальная покупка школьного аттестата с упрощенным обучением в Москве[/b]
[url=http://rushkadiplomiks.ru/diplom-tekhnikuma-kolledzha/]rushkadiplomiks.ru/diplom-tekhnikuma-kolledzha[/url]
Lazrolx
August 7, 2024 @ 8:56 am
[b]Купить диплом о высшем образовании.[/b]
[url=http://peaceofficial.5nx.ru/viewtopic.php?f=111&t=805/]peaceofficial.5nx.ru/viewtopic.php?f=111&t=805[/url]
[url=http://lada-xray.net/member.php?u=2434/]lada-xray.net/member.php?u=2434[/url]
[url=http://seaman.com.ua/forum/index.php?threads/vash-put-k-vysshemu-obrazovaniju-nadezhno-i-legalno.37210/]seaman.com.ua/forum/index.php?threads/vash-put-k-vysshemu-obrazovaniju-nadezhno-i-legalno.37210[/url]
[url=http://miku-crdb.ru/forum/messages/forum1/topic333/message361/?result=new#message361/]miku-crdb.ru/forum/messages/forum1/topic333/message361/?result=new#message361[/url]
[url=http://myturtime.ru/kak-zakazat-diplom-onlayn/]myturtime.ru/kak-zakazat-diplom-onlayn[/url]
Williamraics
August 7, 2024 @ 9:49 am
[u][b] Привет, друзья![/b][/u]
Мы готовы предложить дипломы.
[url=http://wecanchat.mn.co/posts/61639341/]wecanchat.mn.co/posts/61639341[/url]
[url=http://wmrp.listbb.ru/viewtopic.php?f=51&t=666/]wmrp.listbb.ru/viewtopic.php?f=51&t=666[/url]
[url=http://elektrofahrrad-tests.de/forums/newthread.php?fid=2&processed=1/]elektrofahrrad-tests.de/forums/newthread.php?fid=2&processed=1[/url]
[url=http://dailygram.com/blog/1304353/купить-официальный-диплом-быстро-и-легально/]dailygram.com/blog/1304353/купить-официальный-диплом-быстро-и-легально[/url]
[url=http://mirmafii.ru/article/7545/]mirmafii.ru/article/7545[/url]
Lazrmky
August 7, 2024 @ 9:53 am
[u][b] Привет, друзья![/b][/u]
[b]Приобрести диплом любого университета.[/b]
[url=http://mans-diplomyxx.ru/kupit-diplom-sssr/]mans-diplomyxx.ru/kupit-diplom-sssr[/url]
Dnrtepj
August 7, 2024 @ 10:00 am
[b]Купить документ[/b] о получении высшего образования вы сможете в нашей компании в Москве.
[url=http://toprankdesign.co.uk/wiki/index.php?title=Diplom_216405lhHEt/]toprankdesign.co.uk/wiki/index.php?title=Diplom_216405lhHEt[/url]
[url=http://taondinternational.rudraserver.com/blog/index.php?entryid=133849/]taondinternational.rudraserver.com/blog/index.php?entryid=133849[/url]
[url=http://kankerala.org/uncategorized/diplom-kupit-938227fq/]kankerala.org/uncategorized/diplom-kupit-938227fq[/url]
[url=http://investicos.com/uncategorized/diplom-222212kfmnk/]investicos.com/uncategorized/diplom-222212kfmnk[/url]
[url=http://toplistar.com/story19383437/diplom/]toplistar.com/story19383437/diplom[/url]
Mazrdor
August 7, 2024 @ 10:22 am
[u][b] Привет![/b][/u]
[b]Официальная покупка аттестата о среднем образовании в Москве и других городах[/b]
[url=http://rushkadiplomik.ru/]rushkadiplomik.ru[/url]
RandyDox
August 7, 2024 @ 11:31 am
acquistare farmaci senza ricetta: Farmacie che vendono Cialis senza ricetta – top farmacia online
Trefrvf
August 7, 2024 @ 11:58 am
[u][b] Здравствуйте![/b][/u]
Как приобрести диплом о среднем образовании в Москве и других городах
[url=http://mans-diplomyxx.ru/kupit-diplom-ryazan/]mans-diplomyxx.ru/kupit-diplom-ryazan[/url]
Рады оказать помощь!.
Yrefsok
August 7, 2024 @ 11:59 am
[u][b] Привет![/b][/u]
[b]Приобрести диплом о высшем образовании.[/b]
[url=http://salon-domino.ru/]salon-domino.ru[/url]
[url=http://orbita.com.py/blogs/317/Что-требуется-из-техники-для-создания-документов?lang=en_us/]orbita.com.py/blogs/317/Что-требуется-из-техники-для-создания-документов?lang=en_us[/url]
[url=http://prikol100500.ru/kak-zakazat-diplom-onlayn-prostyie-shagi/]prikol100500.ru/kak-zakazat-diplom-onlayn-prostyie-shagi[/url]
[url=http://blogs.rufox.ru/~worksale/48372.htm/]blogs.rufox.ru/~worksale/48372.htm[/url]
[url=http://webnewsrealty.ru/page/3/]webnewsrealty.ru/page/3[/url]
Jesseoweld
August 7, 2024 @ 12:01 pm
Свежие и актуальные http://barbie-games.ru новости современной техники
Dnrttmx
August 7, 2024 @ 1:24 pm
[b]Заказать документ[/b] ВУЗа можно в нашем сервисе.
[url=http://sttimothysignal.org/groups/diplom-kupit-308215bx/]sttimothysignal.org/groups/diplom-kupit-308215bx[/url]
[url=http://ecrinparis.com/diplom-904942kwczz/]ecrinparis.com/diplom-904942kwczz[/url]
[url=http://samnetworkth.com/diplom-517251fjybl/]samnetworkth.com/diplom-517251fjybl[/url]
[url=http://bruthindustan.com/diplom-765474kthsi/]bruthindustan.com/diplom-765474kthsi[/url]
[url=http://taondinternational.rudraserver.com/blog/index.php?entryid=134382/]taondinternational.rudraserver.com/blog/index.php?entryid=134382[/url]
Lazrcbv
August 7, 2024 @ 1:37 pm
[u][b] Здравствуйте![/b][/u]
[b]Мы изготавливаем дипломы[/b] любых профессий по приятным ценам.
[url=http://rushkadiplomikx.ru/diplom-sssr/]rushkadiplomikx.ru/diplom-sssr[/url]
Tomasstync
August 7, 2024 @ 1:41 pm
viagra generico prezzo piГ№ basso: viagra prezzo – viagra generico prezzo piГ№ basso
Vazryic
August 7, 2024 @ 2:12 pm
[u][b] Привет![/b][/u]
Приобрести документ института можно у нас. Мы оказываем услуги по продаже документов об окончании любых университетов России.
[url=http://golpro.jp/userinfo.php?uid=13510#/]golpro.jp/userinfo.php?uid=13510#[/url]
[url=http://narodovmnogo-omsk.ru/forum/user/4138/]narodovmnogo-omsk.ru/forum/user/4138[/url]
[url=http://nnklad.ru/viewtopic.php?f=4&t=1720/]nnklad.ru/viewtopic.php?f=4&t=1720[/url]
[url=http://economreklama.ru/index.php?subaction=userinfo&user=ezavixava/]economreklama.ru/index.php?subaction=userinfo&user=ezavixava[/url]
[url=http://pax.nichost.ru/forum/view_profile.php?UID=133465/]pax.nichost.ru/forum/view_profile.php?UID=133465[/url]
Trefgqx
August 7, 2024 @ 2:50 pm
[u][b] Привет![/b][/u]
Возможно ли купить диплом стоматолога, и как это происходит
[url=http://mandiplomikx.ru/kupit-diplom-novosibirsk/]mandiplomikx.ru/kupit-diplom-novosibirsk[/url]
Рады оказать помощь!.
Yrefdqo
August 7, 2024 @ 3:01 pm
[u][b] Привет![/b][/u]
[b]Приобрести диплом любого университета.[/b]
[url=http://passneurosurgery.net/learn/blog/index.php?entryid=321177/]passneurosurgery.net/learn/blog/index.php?entryid=321177[/url]
[url=http://turservisnews.ru/page/2/]turservisnews.ru/page/2[/url]
[url=http://facefam.com/read-blog/333_cena-diploma-avtorskij-obzor-i-sovety-ekspertov?mode=day/]facefam.com/read-blog/333_cena-diploma-avtorskij-obzor-i-sovety-ekspertov?mode=day[/url]
[url=http://malaysiasteelinstitute.com/купить-диплом/]malaysiasteelinstitute.com/купить-диплом[/url]
[url=http://ecejoin.com/link.php?url=frees-diplom.com/]ecejoin.com/link.php?url=frees-diplom.com[/url]
RandyDox
August 7, 2024 @ 4:27 pm
п»їFarmacia online migliore: kamagra oral jelly consegna 24 ore – Farmacia online piГ№ conveniente
Lazrwzw
August 7, 2024 @ 4:35 pm
[u][b] Добрый день![/b][/u]
[b]Мы изготавливаем дипломы[/b] любой профессии по приятным тарифам.
[url=http://rushkas-diplomasxx.ru/kupit-diplom-yurista-shag-v-budushchee/]rushkas-diplomasxx.ru/kupit-diplom-yurista-shag-v-budushchee[/url]
Mazramc
August 7, 2024 @ 4:54 pm
[u][b] Привет![/b][/u]
Легальная покупка диплома ПТУ с сокращенной программой обучения
[url=http://arusak-diploms-srednee.ru/kupit-diplom-kandidata-nauk/]arusak-diploms-srednee.ru/kupit-diplom-kandidata-nauk[/url]
Cazrrdd
August 7, 2024 @ 4:59 pm
[u][b] Привет![/b][/u]
Мы предлагаем документы ВУЗов
[url=http://preceptor.flybb.ru/viewtopic.php?f=13&t=9947/]preceptor.flybb.ru/viewtopic.php?f=13&t=9947[/url]
[url=http://demo.4admins.ru/search.php?search_id=egosearch/]demo.4admins.ru/search.php?search_id=egosearch[/url]
[url=http://wo.linyway.com/read-blog/4491/]wo.linyway.com/read-blog/4491[/url]
[url=http://sampnl.listbb.ru/viewtopic.php?f=3&t=500/]sampnl.listbb.ru/viewtopic.php?f=3&t=500[/url]
[url=http://btclub.fun/read-blog/412/]btclub.fun/read-blog/412[/url]
Tomasstync
August 7, 2024 @ 5:20 pm
migliori farmacie online 2024: Cialis generico controindicazioni – farmacie online affidabili
RandyDox
August 7, 2024 @ 6:40 pm
Farmacia online miglior prezzo: kamagra gold – farmacia online piГ№ conveniente
Tomasstync
August 7, 2024 @ 9:21 pm
farmacia senza ricetta recensioni: viagra naturale in farmacia senza ricetta – esiste il viagra generico in farmacia
Mazryud
August 7, 2024 @ 9:40 pm
[b]Купить документ о получении высшего образования.[/b]
[url=http://thebookpage.com/story2935121/kupit-diplom/]thebookpage.com/story2935121/kupit-diplom[/url]
[url=http://allbookmarking.com/story17732945/kupit-diplom/]allbookmarking.com/story17732945/kupit-diplom[/url]
[url=http://redhotbookmarks.com/story17621494/kupit-diplom/]redhotbookmarks.com/story17621494/kupit-diplom[/url]
[url=http://bookmarking1.com/story17638058/kupit-diplom/]bookmarking1.com/story17638058/kupit-diplom[/url]
[url=http://bookmarkilo.com/story17526952/kupit-diplom/]bookmarkilo.com/story17526952/kupit-diplom[/url]
Diplomi_tgSi
August 7, 2024 @ 9:49 pm
Привет, друзья!
Купить документ о получении высшего образования вы имеете возможность у нас в столице.
[url=http://rushkas-diplomy.ru/]rushkas-diplomy.ru[/url]
RandyDox
August 7, 2024 @ 11:19 pm
farmacie online sicure: kamagra gel prezzo – farmaci senza ricetta elenco
Sazrzdc
August 8, 2024 @ 12:28 am
[u][b] Здравствуйте![/b][/u]
Заказать документ о получении высшего образования
[url=http://bs.listbb.ru/viewtopic.php?f=3&t=652/]bs.listbb.ru/viewtopic.php?f=3&t=652[/url]
[url=http://pipewiki.org/app/index.php/образование_мечты:_дипломы_ведущих_вузов/]pipewiki.org/app/index.php/образование_мечты:_дипломы_ведущих_вузов[/url]
[url=http://sociomixr.com/blogs/91/Что-готов-сейчас-предоставить-интернет-магазин-с-дипломами/]sociomixr.com/blogs/91/Что-готов-сейчас-предоставить-интернет-магазин-с-дипломами[/url]
[url=http://pokemon-go.onl/users/6146/]pokemon-go.onl/users/6146[/url]
[url=http://infiniterealities.listbb.ru/viewtopic.php?f=7&t=430/]infiniterealities.listbb.ru/viewtopic.php?f=7&t=430[/url]
Diplomi_sjSi
August 8, 2024 @ 12:51 am
Здравствуйте!
Купить документ о получении высшего образования можно у нас.
[url=http://rushkas-diplomasxx.ru/]rushkas-diplomasxx.ru[/url]
Tomasstync
August 8, 2024 @ 1:02 am
pillole per erezioni fortissime: viagra senza prescrizione – siti sicuri per comprare viagra online
TimothyLew
August 8, 2024 @ 1:28 am
https://farmait.store/# farmacie online affidabili
Mazrprq
August 8, 2024 @ 1:43 am
[u][b] Здравствуйте![/b][/u]
[b]Всё, что нужно знать о покупке аттестата о среднем образовании[/b]
[url=http://rushkadiplomik.ru/]rushkadiplomik.ru[/url]
NathanGriva
August 8, 2024 @ 2:08 am
Link pyramid, tier 1, tier 2, tier 3
Level 1 – 500 hyperlinks with placement contained in pieces on article domains
Secondary – 3000 web address Forwarded connections
Tier 3 – 20000 connections blend, comments, writings
Utilizing a link structure is beneficial for web crawlers.
Require:
One connection to the website.
Query Terms.
True when 1 key phrase from the page subject.
Observe the complementary offering!
Essential! Tier 1 links do not conflict with Tier 2 and Tier 3-order connections
A link hierarchy is a instrument for boosting the movement and referral sources of a website or social media platform
Dnrteph
August 8, 2024 @ 3:42 am
[b]Приобрести документ[/b] университета можно у нас.
[url=http://allbookmarking.com/story17731606/diplom/]allbookmarking.com/story17731606/diplom[/url]
[url=http://bookmarkunit.com/story17539391/diplom/]bookmarkunit.com/story17539391/diplom[/url]
[url=http://sttimothysignal.org/groups/diplom-585796aryoh/]sttimothysignal.org/groups/diplom-585796aryoh[/url]
[url=http://turnkeymodular.ca/diplom-425077sqfab/]turnkeymodular.ca/diplom-425077sqfab[/url]
[url=http://localtrusted.co.uk/wiki/index.php?title=Diplom_997986uoOXE/]localtrusted.co.uk/wiki/index.php?title=Diplom_997986uoOXE[/url]
Mazrguq
August 8, 2024 @ 4:10 am
[u][b] Здравствуйте![/b][/u]
Рекомендации по безопасной покупке диплома о высшем образовании
[url=http://landik-diploms-srednee.ru/diplom-bakalavra/]landik-diploms-srednee.ru/diplom-bakalavra[/url]
Mazrpnh
August 8, 2024 @ 4:56 am
[u][b] Добрый день![/b][/u]
[b]Легальная покупка диплома ПТУ с сокращенной программой обучения[/b]
[url=http://rushkadiplomik.ru/]rushkadiplomik.ru[/url]
Cazrovr
August 8, 2024 @ 6:22 am
[u][b] Привет![/b][/u]
Заказать диплом о высшем образовании
[url=http://weekevents.ru/diplom-o-vyisshem-obrazovanii-dlya-ambitsioznyih-professionalov/]weekevents.ru/diplom-o-vyisshem-obrazovanii-dlya-ambitsioznyih-professionalov[/url]
[url=http://dnd.listbb.ru/ucp.php?mode=login/]dnd.listbb.ru/ucp.php?mode=login[/url]
[url=http://lubov.listbb.ru/viewtopic.php?f=2&t=493/]lubov.listbb.ru/viewtopic.php?f=2&t=493[/url]
[url=http://testforum.1stbb.ru/viewtopic.php?f=3&t=444/]testforum.1stbb.ru/viewtopic.php?f=3&t=444[/url]
[url=http://koxma.4adm.ru/viewtopic.php?f=298&t=5534&view=next&sid=05f6e58705ea31cb4f7032cc409091a8/]koxma.4adm.ru/viewtopic.php?f=298&t=5534&view=next&sid=05f6e58705ea31cb4f7032cc409091a8[/url]
Sazrkhm
August 8, 2024 @ 6:29 am
[u][b] Добрый день![/b][/u]
Диплом о высшем образовании
[url=http://av.flyboard.ru/viewtopic.php?f=9&t=1452/]av.flyboard.ru/viewtopic.php?f=9&t=1452[/url]
Dnrtdor
August 8, 2024 @ 6:40 am
[b]Купить документ[/b] о получении высшего образования вы сможете в нашей компании в Москве.
[url=http://redhotbookmarks.com/story17616851/diplom/]redhotbookmarks.com/story17616851/diplom[/url]
[url=http://socialbuzzmaster.com/story3120446/diplom/]socialbuzzmaster.com/story3120446/diplom[/url]
[url=http://bookmarkahref.com/story17660322/diplom/]bookmarkahref.com/story17660322/diplom[/url]
[url=http://bookmarkunit.com/story17543128/diplom/]bookmarkunit.com/story17543128/diplom[/url]
[url=http://tuzari.shop/?option=com_k2&view=itemlist&task=user&id=1529/]tuzari.shop/?option=com_k2&view=itemlist&task=user&id=1529[/url]
Mazrgfr
August 8, 2024 @ 7:24 am
[b]Заказать документ института.[/b]
[url=http://sttimothysignal.org/groups/kupit-diplom-197448hfdcy/]sttimothysignal.org/groups/kupit-diplom-197448hfdcy[/url]
[url=http://luxuriousrentz.com/kupit-diplom-562240kjrjt-2/]luxuriousrentz.com/kupit-diplom-562240kjrjt-2[/url]
[url=http://bookmarkchamp.com/story17606643/kupit-diplom/]bookmarkchamp.com/story17606643/kupit-diplom[/url]
[url=http://orangebookmarks.com/story17714076/kupit-diplom/]orangebookmarks.com/story17714076/kupit-diplom[/url]
[url=http://bookmarklogin.com/story17758107/kupit-diplom/]bookmarklogin.com/story17758107/kupit-diplom[/url]
Lazrjly
August 8, 2024 @ 7:27 am
[u][b] Привет, друзья![/b][/u]
[b]Мы можем предложить дипломы[/b] любой профессии по приятным ценам.
[url=http://rushkas-diplomas.ru/kupit-diplom-voronezh/]rushkas-diplomas.ru/kupit-diplom-voronezh[/url]
Tomasstync
August 8, 2024 @ 9:10 am
comprare farmaci online con ricetta: Farmacie online sicure – Farmacia online piГ№ conveniente
Trefuxh
August 8, 2024 @ 10:23 am
[u][b] Здравствуйте![/b][/u]
Быстрое обучение и получение диплома магистра – возможно ли это?
[url=http://diplomyx-man.ru/kupit-diplom-v-ufe/]diplomyx-man.ru/kupit-diplom-v-ufe[/url]
Рады оказаться полезными!.
Sazrwud
August 8, 2024 @ 10:28 am
[u][b] Здравствуйте![/b][/u]
Заказать документ ВУЗа
[url=http://vladmines.dn.ua/forum/index.php/topic,38572.0/]vladmines.dn.ua/forum/index.php/topic,38572.0[/url]
[url=http://telegra.ph/zayavlenie-na-postuplenie-v-vuz-kazahstan-07-24/]telegra.ph/zayavlenie-na-postuplenie-v-vuz-kazahstan-07-24[/url]
[url=http://telegra.ph/mnogodetnye-semi-lgoty-pri-postuplenii-v-vuz-07-24/]telegra.ph/mnogodetnye-semi-lgoty-pri-postuplenii-v-vuz-07-24[/url]
[url=http://hebergementweb.org/threads/diplomy-dlja-uspeshnogo-buduschego.1531937/]hebergementweb.org/threads/diplomy-dlja-uspeshnogo-buduschego.1531937[/url]
[url=http://hl2forever.ru/member.php?u=266/]hl2forever.ru/member.php?u=266[/url]
Lazrrrx
August 8, 2024 @ 10:40 am
[u][b] Привет![/b][/u]
[b]Мы готовы предложить дипломы[/b] любой профессии по доступным тарифам.
[url=http://rushkas-diplom.ru/svidetelstvo-o-rozhdenii-sssr1/]rushkas-diplom.ru/svidetelstvo-o-rozhdenii-sssr1[/url]
Sazrays
August 8, 2024 @ 11:56 am
[u][b] Добрый день![/b][/u]
Заказать документ о получении высшего образования
[url=http://fbadult.com/blogs/3380/Дипломы-для-карьерного-продвижения/]fbadult.com/blogs/3380/Дипломы-для-карьерного-продвижения[/url]
[url=http://1001vieclam.forumvi.com/t9321-topic#10006/]1001vieclam.forumvi.com/t9321-topic#10006[/url]
[url=http://foro.rune-nifelheim.com/general-ragnarok-m/diplomy-s-garantiej-kachestva/]foro.rune-nifelheim.com/general-ragnarok-m/diplomy-s-garantiej-kachestva[/url]
[url=http://volnodumie.bbmy.ru/viewtopic.php?id=13205#p27005/]volnodumie.bbmy.ru/viewtopic.php?id=13205#p27005[/url]
[url=http://demo.advised360.com/read-blog/162493/]demo.advised360.com/read-blog/162493[/url]
Sazrwlo
August 8, 2024 @ 12:24 pm
[u][b] Привет![/b][/u]
Мы можем предложить дипломы любых профессий.
[url=http://gogogobookmarks.com/story17653557/kupit-diplom/]gogogobookmarks.com/story17653557/kupit-diplom[/url]
[url=http://speedgh.com/index.php?page=user&action=pub_profile&id=1321546/]speedgh.com/index.php?page=user&action=pub_profile&id=1321546[/url]
[url=http://atozbookmarkc.com/story17857325/kupit-diplom/]atozbookmarkc.com/story17857325/kupit-diplom[/url]
[url=http://sociallytraffic.com/story2471987/kupit-diplom/]sociallytraffic.com/story2471987/kupit-diplom[/url]
[url=http://gtj.kr/board_KtRj53/370227/]gtj.kr/board_KtRj53/370227[/url]
Trefpvv
August 8, 2024 @ 1:06 pm
[u][b] Здравствуйте![/b][/u]
Удивительно, но купить диплом кандидата наук оказалось не так сложно
[url=http://mandiplomiks.ru/kupit-diplom-ekaterinburg/]mandiplomiks.ru/kupit-diplom-ekaterinburg[/url]
Рады помочь!.
Tomasstync
August 8, 2024 @ 1:33 pm
Farmacie on line spedizione gratuita: Cialis generico 20 mg 8 compresse prezzo – comprare farmaci online con ricetta
Jeffry Proffitt
August 8, 2024 @ 2:01 pm
To the missouri.edu administrator, Your posts are always well-written and easy to understand.
RandyDox
August 8, 2024 @ 2:16 pm
Farmacia online piГ№ conveniente: Farmacie online sicure – top farmacia online
Manrsoi
August 8, 2024 @ 2:32 pm
[u][b] Добрый день![/b][/u]
Заказать документ института
[url=http://bookmarkahref.com/story17658878/kupit-diplom]bookmarkahref.com/story17658878/kupit-diplom[/url]
[url=http://bookmarklinking.com/story3088797/kupit-diplom]bookmarklinking.com/story3088797/kupit-diplom[/url]
[url=http://socialrator.com/story7880952/kupit-diplom]socialrator.com/story7880952/kupit-diplom[/url]
[url=http://tayarut.bkerem.org.il/kupit-diplom-723528smmug]tayarut.bkerem.org.il/kupit-diplom-723528smmug[/url]
[url=http://managerhotels.com/business-small-business/kupit-diplom-990914yrhez]managerhotels.com/business-small-business/kupit-diplom-990914yrhez[/url]
TimothyLew
August 8, 2024 @ 2:43 pm
http://viagragenerico.site/# esiste il viagra generico in farmacia
Oariorzje
August 8, 2024 @ 2:47 pm
[u][b] Привет, друзья![/b][/u]
Купить диплом любого университета
[b]Наша компания предлагает[/b] выгодно купить диплом, который выполнен на бланке ГОЗНАКа и заверен мокрыми печатями, штампами, подписями должностных лиц. Документ пройдет лубую проверку, даже при помощи профессиональных приборов. Достигайте цели быстро и просто с нашим сервисом.
[b]Где заказать диплом по актуальной специальности?[/b]
[url=http://import-moto.com/users/88/]import-moto.com/users/88[/url]
[url=http://comedyforme.ru/diplomyi-dlya-vashego-karernogo-rosta/]comedyforme.ru/diplomyi-dlya-vashego-karernogo-rosta[/url]
[url=http://cromia.ua/forum/user/6329/]cromia.ua/forum/user/6329[/url]
[url=http://c99596qr.beget.tech/2024/07/19/dostupnye-diplomy-s-garantiey-kachestva/]c99596qr.beget.tech/2024/07/19/dostupnye-diplomy-s-garantiey-kachestva[/url]
[url=http://p91648f6.beget.tech/2024/07/04/dlya-chego-seychas-pokupayut-diplomy-i-attestaty-v-seti-internet/]p91648f6.beget.tech/2024/07/04/dlya-chego-seychas-pokupayut-diplomy-i-attestaty-v-seti-internet[/url]
Iariormej
August 8, 2024 @ 3:23 pm
[u][b] Здравствуйте![/b][/u]
Купить документ института можно в нашей компании. Мы оказываем услуги по продаже документов об окончании любых университетов РФ. Вы получите необходимый диплом по любой специальности, включая документы старого образца. Гарантируем, что при проверке документов работодателем, подозрений не появится.
[url=http://fabnews.ru/forum/showthread.php?p=80204#post80204/]fabnews.ru/forum/showthread.php?p=80204#post80204[/url]
[url=http://noctuagg.ro/forum/index.php?/gallery/image/30-быстрый-поиск-интернет-магазина-что-реализует-дипломы/]noctuagg.ro/forum/index.php?/gallery/image/30-быстрый-поиск-интернет-магазина-что-реализует-дипломы[/url]
[url=http://snaprama.com/read-blog/1322/]snaprama.com/read-blog/1322[/url]
[url=http://jivonews.ru/byistryie-i-nadezhnyie-diplomyi/]jivonews.ru/byistryie-i-nadezhnyie-diplomyi[/url]
[url=http://telegra.ph/kogda-budut-rezultaty-postupleniya-v-vuzy-07-24/]telegra.ph/kogda-budut-rezultaty-postupleniya-v-vuzy-07-24[/url]
Yreflbx
August 8, 2024 @ 3:28 pm
[u][b] Привет, друзья![/b][/u]
[b]Заказать диплом университета.[/b]
[url=http://qyshikisoxa.lowescouponn.com/cto-potrebuetsa-iz-oborudovania-dla-izgotovlenia-diploma/]qyshikisoxa.lowescouponn.com/cto-potrebuetsa-iz-oborudovania-dla-izgotovlenia-diploma[/url]
[url=http://mdkl.ru/blogs/9204/Быстрый-поиск-онлайн-магазина-продающего-дипломы-и-аттестаты?lang=ru_ru/]mdkl.ru/blogs/9204/Быстрый-поиск-онлайн-магазина-продающего-дипломы-и-аттестаты?lang=ru_ru[/url]
[url=http://wiuwi.com/blogs/126950/Оформление-диплома-под-ключ-просто-и-безопасно/]wiuwi.com/blogs/126950/Оформление-диплома-под-ключ-просто-и-безопасно[/url]
[url=http://blogs.rufox.ru/~worksale/48372.htm/]blogs.rufox.ru/~worksale/48372.htm[/url]
[url=http://buldingnews.ru/page/3/]buldingnews.ru/page/3[/url]
Cazrqnh
August 8, 2024 @ 4:13 pm
[u][b] Добрый день![/b][/u]
Получить диплом о высшем образовании
[url=http://mf.getbb.ru/viewtopic.php?f=28&t=664/]mf.getbb.ru/viewtopic.php?f=28&t=664[/url]
[url=http://daddycow.com/blogs/view/25775/]daddycow.com/blogs/view/25775[/url]
[url=http://sontopic.com/blogs/new/]sontopic.com/blogs/new[/url]
[url=http://gromscream.80lvl.ru/viewtopic.php?f=3&t=2102/]gromscream.80lvl.ru/viewtopic.php?f=3&t=2102[/url]
[url=http://freedomrp.getbb.ru/viewtopic.php?f=110&t=814/]freedomrp.getbb.ru/viewtopic.php?f=110&t=814[/url]
Sazrxty
August 8, 2024 @ 4:25 pm
[u][b] Привет, друзья![/b][/u]
Диплом специалиста
[url=http://ya.10bb.ru/viewtopic.php?id=4091#p7301/]ya.10bb.ru/viewtopic.php?id=4091#p7301[/url]
RandyDox
August 8, 2024 @ 4:41 pm
miglior sito dove acquistare viagra: viagra senza prescrizione – miglior sito dove acquistare viagra
Uazrgoy
August 8, 2024 @ 5:04 pm
[b]Где купить диплом по актуальной специальности?[/b]
[url=http://youwantech.com/xe/board/460996/]youwantech.com/xe/board/460996[/url]
[url=http://hotbookmarkings.com/story17685684/diplom/]hotbookmarkings.com/story17685684/diplom[/url]
[url=http://kankerala.org/uncategorized/diplom-507922nikbw/]kankerala.org/uncategorized/diplom-507922nikbw[/url]
[url=http://laviehub.com/blog/diplom-856482uxwxb/]laviehub.com/blog/diplom-856482uxwxb[/url]
[url=http://tigaedu.com/blog/index.php?entryid=330539/]tigaedu.com/blog/index.php?entryid=330539[/url]
Tomasstync
August 8, 2024 @ 5:25 pm
farmaci senza ricetta elenco: Farmacie online sicure – farmacie online autorizzate elenco
Mazroaz
August 8, 2024 @ 7:48 pm
[u][b] Добрый день![/b][/u]
[b]Быстрое обучение и получение диплома магистра – возможно ли это?[/b]
[url=http://rushkadiplomik.ru/]rushkadiplomik.ru[/url]
Xazrilq
August 8, 2024 @ 8:37 pm
[u][b] Привет![/b][/u]
Заказать документ о получении высшего образования.
[url=http://human.forumieren.de/t305-topic#373/]human.forumieren.de/t305-topic#373[/url]
[url=http://demo.4admins.ru/ucp.php?mode=resend_act&sid=56d5ced6597f36cf644550e216340435/]demo.4admins.ru/ucp.php?mode=resend_act&sid=56d5ced6597f36cf644550e216340435[/url]
[url=http://1776reloaded.us/blogs/77/Получите-диплом-без-лишних-забот-и-усилий/]1776reloaded.us/blogs/77/Получите-диплом-без-лишних-забот-и-усилий[/url]
[url=http://gromscream.80lvl.ru/viewtopic.php?f=3&t=2081/]gromscream.80lvl.ru/viewtopic.php?f=3&t=2081[/url]
[url=http://ironway.ru/forum/viewtopic.php?f=5&t=23801/]ironway.ru/forum/viewtopic.php?f=5&t=23801[/url]
Dnrtljn
August 8, 2024 @ 9:01 pm
[b]Заказать документ[/b] о получении высшего образования можно в нашей компании в столице.
[url=http://njkkot.org/?document_srl=1142984/]njkkot.org/?document_srl=1142984[/url]
[url=http://bruthindustan.com/diplom-kupit-990130ds/]bruthindustan.com/diplom-kupit-990130ds[/url]
[url=http://cyberbookmarking.com/story17581168/diplom/]cyberbookmarking.com/story17581168/diplom[/url]
[url=http://zanybookmarks.com/story17737239/diplom/]zanybookmarks.com/story17737239/diplom[/url]
[url=http://yourbuyersguide.co.uk/diplom-kupit-818901od/]yourbuyersguide.co.uk/diplom-kupit-818901od[/url]
Sazruft
August 8, 2024 @ 9:37 pm
[u][b] Привет![/b][/u]
Мы можем предложить дипломы любой профессии.
[url=http://optimusbookmarks.com/story17628763/kupit-diplom/]optimusbookmarks.com/story17628763/kupit-diplom[/url]
[url=http://sb-bookmarking.com/story17710988/kupit-diplom/]sb-bookmarking.com/story17710988/kupit-diplom[/url]
[url=http://toprankdesign.co.uk/wiki/index.php?title=Kupit_Diplom_826199UpbDk/]toprankdesign.co.uk/wiki/index.php?title=Kupit_Diplom_826199UpbDk[/url]
[url=http://total-bookmark.com/story17548442/kupit-diplom/]total-bookmark.com/story17548442/kupit-diplom[/url]
[url=http://gogogobookmarks.com/story17653536/kupit-diplom/]gogogobookmarks.com/story17653536/kupit-diplom[/url]
RandyDox
August 8, 2024 @ 9:40 pm
Farmacie online sicure: Farmacie online sicure – Farmacia online piГ№ conveniente
Mazrxeo
August 8, 2024 @ 11:00 pm
[u][b] Добрый день![/b][/u]
[b]Как оказалось, купить диплом кандидата наук не так уж и сложно[/b]
[url=http://rushkadiplomik.ru/]rushkadiplomik.ru[/url]
Diplomi_kbSi
August 8, 2024 @ 11:17 pm
Добрый день!
Заказать документ о получении высшего образования можно у нас в столице.
[url=http://rushkadiplomiks.ru/]rushkadiplomiks.ru[/url]
Uazrvom
August 8, 2024 @ 11:18 pm
[u][b]Добрыйдень![/b][/u]
[b]Приобрести диплом любого ВУЗа.[/b]
[url=http://pageoftoday.com/story2981038/diplom]pageoftoday.com/story2981038/diplom[/url]
[url=http://bookmarkshome.com/story3146599/diplom]bookmarkshome.com/story3146599/diplom[/url]
[url=http://vacayla.com/2024/07/29/diplom-kupit-972383og]vacayla.com/2024/07/29/diplom-kupit-972383og[/url]
[url=http://directlynews.com/diplom-555086rlewe]directlynews.com/diplom-555086rlewe[/url]
[url=http://welnesbiolabs.com/diplom-kupit-359403bg]welnesbiolabs.com/diplom-kupit-359403bg[/url]
[url=http://stemacumen.net/blog/index.php?entryid=9275]stemacumen.net/blog/index.php?entryid=9275[/url]
[url=http://repurtech.com/diplom-785115aifcq]repurtech.com/diplom-785115aifcq[/url]
[url=http://stemacumen.net/blog/index.php?entryid=9354]stemacumen.net/blog/index.php?entryid=9354[/url]
[url=http://kankerala.org/uncategorized/diplom-kupit-128756ur]kankerala.org/uncategorized/diplom-kupit-128756ur[/url]
[url=http://taondinternational.rudraserver.com/blog/index.php?entryid=130140]taondinternational.rudraserver.com/blog/index.php?entryid=130140[/url]
[url=http://ypchina.org/diplom-kupit-26878va]ypchina.org/diplom-kupit-26878va[/url]
[url=http://ranatourandtravels.com/diplom-861271nsywx]ranatourandtravels.com/diplom-861271nsywx[/url]
[url=http://taondinternational.rudraserver.com/blog/index.php?entryid=133275]taondinternational.rudraserver.com/blog/index.php?entryid=133275[/url]
[url=http://welnesbiolabs.com/diplom-596453fsgbn]welnesbiolabs.com/diplom-596453fsgbn[/url]
[url=http://stemacumen.net/blog/index.php?entryid=7597]stemacumen.net/blog/index.php?entryid=7597[/url]
[url=http://laviehub.com/blog/diplom-892573dlsxg]laviehub.com/blog/diplom-892573dlsxg[/url]
[url=http://gamereleasetoday.com/diplom-474939zxgiq]gamereleasetoday.com/diplom-474939zxgiq[/url]
[url=http://cruxbookmarks.com/story17687036/diplom]cruxbookmarks.com/story17687036/diplom[/url]
[url=http://bookmarkmoz.com/story17699610/diplom]bookmarkmoz.com/story17699610/diplom[/url]
[url=http://thehumanbehaviour.com/diplom-955826vgwtt]thehumanbehaviour.com/diplom-955826vgwtt[/url]
Danrxuo
August 8, 2024 @ 11:18 pm
[u][b] Привет, друзья![/b][/u]
Мы предлагаем дипломы любых профессий.
[url=http://youlangue.lu/blog/index.php?entryid=84581]youlangue.lu/blog/index.php?entryid=84581[/url]
[url=http://orangebookmarks.com/story17707117/diplom]orangebookmarks.com/story17707117/diplom[/url]
[url=http://investicos.com/uncategorized/diplom-172950ocspx]investicos.com/uncategorized/diplom-172950ocspx[/url]
[url=http://syg-us.com/entrenamiento-virtual/blog/index.php?entryid=46170]syg-us.com/entrenamiento-virtual/blog/index.php?entryid=46170[/url]
[url=http://laviehub.com/blog/diplom-182708xvqkv]laviehub.com/blog/diplom-182708xvqkv[/url]
[url=http://indexedbookmarks.com/story17602783/diplom]indexedbookmarks.com/story17602783/diplom[/url]
[url=http://kankerala.org/uncategorized/diplom-370675lmkvd]kankerala.org/uncategorized/diplom-370675lmkvd[/url]
[url=http://ashwinihydropneumatics.com/diplom-362878wfmor]ashwinihydropneumatics.com/diplom-362878wfmor[/url]
[url=http://bookmarkquotes.com/story17732922/diplom]bookmarkquotes.com/story17732922/diplom[/url]
[url=http://socialistener.com/story3017338/diplom]socialistener.com/story3017338/diplom[/url]
[url=http://bookmarktiger.com/story17625189/diplom]bookmarktiger.com/story17625189/diplom[/url]
[url=http://apollobookmarks.com/story17604671/diplom]apollobookmarks.com/story17604671/diplom[/url]
[url=http://investicos.com/uncategorized/diplom-526525flbmx]investicos.com/uncategorized/diplom-526525flbmx[/url]
[url=http://expressbookmark.com/story17654178/diplom]expressbookmark.com/story17654178/diplom[/url]
[url=http://stemacumen.net/blog/index.php?entryid=7136]stemacumen.net/blog/index.php?entryid=7136[/url]
[url=http://rankuppages.com/story2998082/diplom]rankuppages.com/story2998082/diplom[/url]
[url=http://hubwebsites.com/story18916434/diplom]hubwebsites.com/story18916434/diplom[/url]
[url=http://a4everyone.org/diplom-kupit-332274bi]a4everyone.org/diplom-kupit-332274bi[/url]
[url=http://taondinternational.rudraserver.com/blog/index.php?entryid=135094]taondinternational.rudraserver.com/blog/index.php?entryid=135094[/url]
[url=http://sttimothysignal.org/groups/diplom-581012siksy]sttimothysignal.org/groups/diplom-581012siksy[/url]
Jariormhq
August 8, 2024 @ 11:30 pm
[u][b] Здравствуйте![/b][/u]
Купить диплом любого ВУЗа
[url=http://alkhazana.net/2024/07/28/kupit-diplom-684171gjdxu]alkhazana.net/2024/07/28/kupit-diplom-684171gjdxu[/url]
[url=http://williamwesley.com/kupit-diplom-986552thhlh]williamwesley.com/kupit-diplom-986552thhlh[/url]
[url=http://localtrusted.co.uk/wiki/index.php?title=Kupit_Diplom_863230LbZlW]localtrusted.co.uk/wiki/index.php?title=Kupit_Diplom_863230LbZlW[/url]
[url=http://drraphaelfonseca.com.br/kupit-diplom-875177athcm]drraphaelfonseca.com.br/kupit-diplom-875177athcm[/url]
[url=http://digitalmagazine.xyz/kupit-diplom-442832qidyf]digitalmagazine.xyz/kupit-diplom-442832qidyf[/url]
[url=http://socialinplace.com/story2962673/kupit-diplom]socialinplace.com/story2962673/kupit-diplom[/url]
[url=http://toooptarinha.ir/kupit-diplom-487743abxti]toooptarinha.ir/kupit-diplom-487743abxti[/url]
[url=http://guidemysocial.com/story2945965/kupit-diplom]guidemysocial.com/story2945965/kupit-diplom[/url]
[url=http://sociallweb.com/story3017521/kupit-diplom]sociallweb.com/story3017521/kupit-diplom[/url]
[url=http://sttimothysignal.org/groups/kupit-diplom-385829uxfpw]sttimothysignal.org/groups/kupit-diplom-385829uxfpw[/url]
Larioranw
August 8, 2024 @ 11:52 pm
[u][b]Привет![/b][/u]
[b]Заказать диплом ВУЗа.[/b]
[url=http://whitebookmarks.com/story17709501/kupit-diplom]whitebookmarks.com/story17709501/kupit-diplom[/url]
[url=http://bookmarkleader.com/story17669035/kupit-diplom]bookmarkleader.com/story17669035/kupit-diplom[/url]
[url=http://sttimothysignal.org/groups/kupit-diplom-178155iaxni]sttimothysignal.org/groups/kupit-diplom-178155iaxni[/url]
[url=http://koolegardi.ir/2024/07/27/kupit-diplom-985858dcvpk]koolegardi.ir/2024/07/27/kupit-diplom-985858dcvpk[/url]
[url=http://fam2fam.com/groups/kupit-diplom-3409fayhf]fam2fam.com/groups/kupit-diplom-3409fayhf[/url]
[url=http://olivebookmarks.com/story17758777/kupit-diplom]olivebookmarks.com/story17758777/kupit-diplom[/url]
[url=http://newtheories.info/community/profile/michellondon714]newtheories.info/community/profile/michellondon714[/url]
[url=http://laviehub.com/blog/kupit-diplom-872523xnrvg]laviehub.com/blog/kupit-diplom-872523xnrvg[/url]
[url=http://howis.info/2024/07/28/kupit-diplom-464644qkjzw]howis.info/2024/07/28/kupit-diplom-464644qkjzw[/url]
[url=http://lampcanvas.com/kupit-diplom-631201zpefq]lampcanvas.com/kupit-diplom-631201zpefq[/url]
RandyDox
August 8, 2024 @ 11:54 pm
cialis farmacia senza ricetta: acquisto viagra – viagra generico recensioni
Dnrtztk
August 9, 2024 @ 12:03 am
[b]Заказать документ[/b] о получении высшего образования вы имеете возможность в нашей компании в столице.
[url=http://comfortrent.ru/2024/07/27/diplom-537576anobb/]comfortrent.ru/2024/07/27/diplom-537576anobb[/url]
[url=http://topsocialplan.com/story3044387/diplom/]topsocialplan.com/story3044387/diplom[/url]
[url=http://pwi2.dragonicgames.com/diplom-189759kytyz/]pwi2.dragonicgames.com/diplom-189759kytyz[/url]
[url=http://stemacumen.net/blog/index.php?entryid=8508/]stemacumen.net/blog/index.php?entryid=8508[/url]
[url=http://tourxperts.com/diplom-kupit-877275kk/]tourxperts.com/diplom-kupit-877275kk[/url]
Manrjvd
August 9, 2024 @ 1:12 am
[u][b] Привет![/b][/u]
Заказать документ института
[url=http://fridayad.in/user/profile/2634311]fridayad.in/user/profile/2634311[/url]
[url=http://bookmarksfocus.com/story3106153/kupit-diplom]bookmarksfocus.com/story3106153/kupit-diplom[/url]
[url=http://kankerala.org/uncategorized/kupit-diplom-550083tlhtz]kankerala.org/uncategorized/kupit-diplom-550083tlhtz[/url]
[url=http://fridayad.in/user/profile/2634313]fridayad.in/user/profile/2634313[/url]
[url=http://laviehub.com/blog/kupit-diplom-900855njanf]laviehub.com/blog/kupit-diplom-900855njanf[/url]
Lazrhyx
August 9, 2024 @ 1:14 am
[u][b] Добрый день![/b][/u]
[b]Мы предлагаем дипломы[/b] любых профессий по приятным ценам.
[url=http://diplomrushkan.ru/kupit-diplom-stroitelya/]diplomrushkan.ru/kupit-diplom-stroitelya[/url]
Iariorcqn
August 9, 2024 @ 1:30 am
[u][b] Добрый день![/b][/u]
Заказать документ о получении высшего образования вы можете в нашей компании. Мы предлагаем документы об окончании любых ВУЗов РФ. Вы сможете получить необходимый диплом по любым специальностям, включая документы образца СССР. Даем 100% гарантию, что в случае проверки документа работодателями, никаких подозрений не возникнет.
[url=http://doomelang.com/create-blog/]doomelang.com/create-blog[/url]
[url=http://zooclub.com.ua/o-proekte/]zooclub.com.ua/o-proekte[/url]
[url=http://vetstate.ru/forum/?PAGE_NAME=profile_view&UID=126055/]vetstate.ru/forum/?PAGE_NAME=profile_view&UID=126055[/url]
[url=http://income.forum2x2.ru/t2985-topic#5305/]income.forum2x2.ru/t2985-topic#5305[/url]
[url=http://japapmessenger.com/read-blog/551/]japapmessenger.com/read-blog/551[/url]
Tomasstync
August 9, 2024 @ 1:37 am
Farmacie on line spedizione gratuita: Farmacia online miglior prezzo – Farmacia online piГ№ conveniente
Sazrlxa
August 9, 2024 @ 1:56 am
[u][b] Здравствуйте![/b][/u]
Купить документ о получении высшего образования
[url=http://sonnick84.blogzag.com/72702803/Цена-диплома-обзор-специалистов/]sonnick84.blogzag.com/72702803/Цена-диплома-обзор-специалистов[/url]
[url=http://dekortab.com/index.php?/gallery/image/158-документы-по-выгодным-ценам-в-популярном-онлайн-магазине/]dekortab.com/index.php?/gallery/image/158-документы-по-выгодным-ценам-в-популярном-онлайн-магазине[/url]
[url=http://a0oy43ata.blog-kids.com/28446509/С-легкостью-покупаем-диплом-в-магазине-russian-diplom/]a0oy43ata.blog-kids.com/28446509/С-легкостью-покупаем-диплом-в-магазине-russian-diplom[/url]
[url=http://lucylink.com/read-blog/55/]lucylink.com/read-blog/55[/url]
[url=http://autoplus74.com/club/user/170/blog/384/]autoplus74.com/club/user/170/blog/384[/url]
Trefzrv
August 9, 2024 @ 2:16 am
[u][b] Добрый день![/b][/u]
Как получить диплом техникума официально и без лишних проблем
[url=http://mans-diplomyxx.ru/kupit-diplom-ekaterinburg/]mans-diplomyxx.ru/kupit-diplom-ekaterinburg[/url]
Рады оказать помощь!.
Diplomi_fnSi
August 9, 2024 @ 2:24 am
Привет!
Купить документ ВУЗа можно у нас в Москве.
[url=http://diplomyx-rushka.ru/]diplomyx-rushka.ru[/url]
Lazrrff
August 9, 2024 @ 3:05 am
[u][b] Здравствуйте![/b][/u]
[b]Заказать диплом ВУЗа.[/b]
[url=http://mandiplomik.ru/kupit-diplom-voronezh/]mandiplomik.ru/kupit-diplom-voronezh[/url]
Lazrxhz
August 9, 2024 @ 3:08 am
[b]Приобрести диплом о высшем образовании.[/b]
[url=http://gamesfortop.ru/nadezhnyie-diplomyi-dlya-lyubyih-tseley/]gamesfortop.ru/nadezhnyie-diplomyi-dlya-lyubyih-tseley[/url]
[url=http://premium.listbb.ru/posting.php?mode=post&f=2&sid=63fe33cf50c3bda7c48067d8ff6e7bd5/]premium.listbb.ru/posting.php?mode=post&f=2&sid=63fe33cf50c3bda7c48067d8ff6e7bd5[/url]
[url=http://wayworld.listbb.ru/posting.php?mode=post&f=2/]wayworld.listbb.ru/posting.php?mode=post&f=2[/url]
[url=http://4atex.ru/blogs/2342/Быстрое-Оформление-Дипломов-Надежно-и-Легально/]4atex.ru/blogs/2342/Быстрое-Оформление-Дипломов-Надежно-и-Легально[/url]
[url=http://asktourist.ru/viewtopic.php?id=5523#p20078/]asktourist.ru/viewtopic.php?id=5523#p20078[/url]
Uazrtki
August 9, 2024 @ 4:03 am
[b]Где заказать диплом по нужной специальности?[/b]
[url=http://taondinternational.rudraserver.com/blog/index.php?entryid=135936/]taondinternational.rudraserver.com/blog/index.php?entryid=135936[/url]
[url=http://tuzari.shop/?option=com_k2&view=itemlist&task=user&id=1528/]tuzari.shop/?option=com_k2&view=itemlist&task=user&id=1528[/url]
[url=http://bigbizstuff.com/2024/07/28/diplom-kupit-526075by/]bigbizstuff.com/2024/07/28/diplom-kupit-526075by[/url]
[url=http://repurtech.com/diplom-82567xpbwr/]repurtech.com/diplom-82567xpbwr[/url]
[url=http://saveorgrieve.com/diplom-410952cuztk/]saveorgrieve.com/diplom-410952cuztk[/url]
Williamraics
August 9, 2024 @ 4:17 am
[u][b] Привет, друзья![/b][/u]
Мы можем предложить дипломы.
[url=http://kinopuk.ru/viewtopic.php?id=9068#p56633/]kinopuk.ru/viewtopic.php?id=9068#p56633[/url]
[url=http://peaceofficial.5nx.ru/posting.php?mode=post&f=111&sid=4a046ec746e47eeb8f1f9ebaf4c033cf/]peaceofficial.5nx.ru/posting.php?mode=post&f=111&sid=4a046ec746e47eeb8f1f9ebaf4c033cf[/url]
[url=http://myworldavto.ru/vash-diplom-byistro-i-legko-legalno-i-nadezhno/]myworldavto.ru/vash-diplom-byistro-i-legko-legalno-i-nadezhno[/url]
[url=http://altasugar.it/new/index.php?option=com_kunena&view=topic&catid=3&id=115742&Itemid=151/]altasugar.it/new/index.php?option=com_kunena&view=topic&catid=3&id=115742&Itemid=151[/url]
[url=http://shockmusik.ru/nastoyashhie-diplomyi-bez-ozhidaniya/]shockmusik.ru/nastoyashhie-diplomyi-bez-ozhidaniya[/url]
Lazrazl
August 9, 2024 @ 4:20 am
[u][b] Здравствуйте![/b][/u]
[b]Мы изготавливаем дипломы[/b] психологов, юристов, экономистов и прочих профессий по приятным тарифам.
[url=http://rushkas-diploms.ru/kupit-diplom-vracha/]rushkas-diploms.ru/kupit-diplom-vracha[/url]
Mazrddh
August 9, 2024 @ 4:33 am
[u][b] Здравствуйте![/b][/u]
[b]Сколько стоит диплом высшего и среднего образования и как его получить?[/b]
[url=http://rushkadiplomiks.ru/kupit-diplom-medsestry/]rushkadiplomiks.ru/kupit-diplom-medsestry[/url]
Treffaa
August 9, 2024 @ 5:04 am
[u][b] Привет![/b][/u]
Процесс получения диплома стоматолога: реально ли это сделать быстро?
[url=http://mans-diplomasxx.ru/kupit-diplom-ekaterinburg/]mans-diplomasxx.ru/kupit-diplom-ekaterinburg[/url]
Будем рады вам помочь!.
Tomasstync
August 9, 2024 @ 5:27 am
farmacia online piГ№ conveniente: Farmacie online sicure – п»їFarmacia online migliore
LouieScoow
August 9, 2024 @ 6:35 am
торт на рождение торт на заказ с доставкой
RandyDox
August 9, 2024 @ 7:23 am
Farmacia online miglior prezzo: super kamagra – п»їFarmacia online migliore
Xazryzo
August 9, 2024 @ 7:34 am
[u][b] Привет![/b][/u]
Приобрести документ университета.
[url=http://mylot.su/addblogs/]mylot.su/addblogs[/url]
[url=http://kalininabad.flyboard.ru/viewtopic.php?f=11&t=2829/]kalininabad.flyboard.ru/viewtopic.php?f=11&t=2829[/url]
[url=http://generalarminius.com/viewtopic.php?t=155979/]generalarminius.com/viewtopic.php?t=155979[/url]
[url=http://crystalroleplay.listbb.ru/viewtopic.php?f=14&t=2499/]crystalroleplay.listbb.ru/viewtopic.php?f=14&t=2499[/url]
[url=http://socialnetwork.cloudyzx.com/read-blog/14317/]socialnetwork.cloudyzx.com/read-blog/14317[/url]
Uazradg
August 9, 2024 @ 10:36 am
[u][b]Здравствуйте![/b][/u]
[b]Приобрести диплом университета .[/b]
[url=http://lyfesaverscpr.com/diplom-417639vgmer]lyfesaverscpr.com/diplom-417639vgmer[/url]
[url=http://7prbookmarks.com/story17661702/diplom]7prbookmarks.com/story17661702/diplom[/url]
[url=http://kbkrealtors.com/diplom-720808pqipa]kbkrealtors.com/diplom-720808pqipa[/url]
[url=http://bruthindustan.com/diplom-703667iqylo]bruthindustan.com/diplom-703667iqylo[/url]
[url=http://laviehub.com/blog/diplom-407646vaszt]laviehub.com/blog/diplom-407646vaszt[/url]
[url=http://bookmarkahref.com/story17659696/diplom]bookmarkahref.com/story17659696/diplom[/url]
[url=http://krovbar.ru/strashnye-istorii/diplom-908490obrrx]krovbar.ru/strashnye-istorii/diplom-908490obrrx[/url]
[url=http://itdongnam.com/diplom-781290sbadb]itdongnam.com/diplom-781290sbadb[/url]
[url=http://taondinternational.rudraserver.com/blog/index.php?entryid=135628]taondinternational.rudraserver.com/blog/index.php?entryid=135628[/url]
[url=http://taondinternational.rudraserver.com/blog/index.php?entryid=133126]taondinternational.rudraserver.com/blog/index.php?entryid=133126[/url]
[url=http://elearning.maniatech-academy.co.uk/blog/index.php?entryid=280278]elearning.maniatech-academy.co.uk/blog/index.php?entryid=280278[/url]
[url=http://yoursocialpeople.com/story2932188/diplom]yoursocialpeople.com/story2932188/diplom[/url]
[url=http://yourbookmarklist.com/story17799831/diplom]yourbookmarklist.com/story17799831/diplom[/url]
[url=http://tigaedu.com/blog/index.php?entryid=330929]tigaedu.com/blog/index.php?entryid=330929[/url]
[url=http://explorebookmarks.com/story17599395/diplom]explorebookmarks.com/story17599395/diplom[/url]
[url=http://taondinternational.rudraserver.com/blog/index.php?entryid=135393]taondinternational.rudraserver.com/blog/index.php?entryid=135393[/url]
[url=http://hariharparagovernmentiti.com/2024/07/27/diplom-667313frztz]hariharparagovernmentiti.com/2024/07/27/diplom-667313frztz[/url]
[url=http://tourxperts.com/diplom-kupit-960636gl]tourxperts.com/diplom-kupit-960636gl[/url]
[url=http://heealthy.com/question/diplom-370809cijwx]heealthy.com/question/diplom-370809cijwx[/url]
[url=http://pencis.com/diplom-108236dczee]pencis.com/diplom-108236dczee[/url]
Jariorgxs
August 9, 2024 @ 10:41 am
[u][b] Привет, друзья![/b][/u]
Купить диплом любого университета
[url=http://bookmarkcork.com/story18198329/kupit-diplom]bookmarkcork.com/story18198329/kupit-diplom[/url]
[url=http://yourbookmarklist.com/story17795483/kupit-diplom]yourbookmarklist.com/story17795483/kupit-diplom[/url]
[url=http://laviehub.com/blog/kupit-diplom-281146qhnex]laviehub.com/blog/kupit-diplom-281146qhnex[/url]
[url=http://bookmark-master.com/story17675876/kupit-diplom]bookmark-master.com/story17675876/kupit-diplom[/url]
[url=http://offmarketbusinessforsale.com/kupit-diplom-164083xrtbw]offmarketbusinessforsale.com/kupit-diplom-164083xrtbw[/url]
[url=http://bookmarkquotes.com/story17733421/kupit-diplom]bookmarkquotes.com/story17733421/kupit-diplom[/url]
[url=http://isocialfans.com/story3020090/kupit-diplom]isocialfans.com/story3020090/kupit-diplom[/url]
[url=http://trademarketclassifieds.com/user/profile/832533]trademarketclassifieds.com/user/profile/832533[/url]
[url=http://starryjeju.com/qna/5645812]starryjeju.com/qna/5645812[/url]
[url=http://onelifesocial.com/story2891194/kupit-diplom]onelifesocial.com/story2891194/kupit-diplom[/url]
Danrxko
August 9, 2024 @ 10:42 am
[u][b] Добрый день![/b][/u]
Мы изготавливаем дипломы любой профессии.
[url=http://elearning.maniatech-academy.co.uk/blog/index.php?entryid=282712]elearning.maniatech-academy.co.uk/blog/index.php?entryid=282712[/url]
[url=http://gtj.kr/board_KtRj53/369897]gtj.kr/board_KtRj53/369897[/url]
[url=http://nodlik.com/diplom-962583wbbki]nodlik.com/diplom-962583wbbki[/url]
[url=http://astradigital.co.uk/wiki/index.php?title=Diplom_207790ZVcCi]astradigital.co.uk/wiki/index.php?title=Diplom_207790ZVcCi[/url]
[url=http://bookmarkblast.com/story17684939/diplom]bookmarkblast.com/story17684939/diplom[/url]
[url=http://gen8ai.com/groups/diplom-kupit-754291jp/members/all-members]gen8ai.com/groups/diplom-kupit-754291jp/members/all-members[/url]
[url=http://xatrivietnam.vn/question/diplom-570944hbwvx]xatrivietnam.vn/question/diplom-570944hbwvx[/url]
[url=http://youlangue.lu/blog/index.php?entryid=84908]youlangue.lu/blog/index.php?entryid=84908[/url]
[url=http://leftbookmarks.com/story17717379/diplom]leftbookmarks.com/story17717379/diplom[/url]
[url=http://royalbookmarking.com/story17666763/diplom]royalbookmarking.com/story17666763/diplom[/url]
[url=http://astradigital.co.uk/wiki/index.php?title=Diplom_207790ZVcCi]astradigital.co.uk/wiki/index.php?title=Diplom_207790ZVcCi[/url]
[url=http://stemacumen.net/blog/index.php?entryid=9036]stemacumen.net/blog/index.php?entryid=9036[/url]
[url=http://redhotbookmarks.com/story17616708/diplom]redhotbookmarks.com/story17616708/diplom[/url]
[url=http://arandaasesoria.com/diplom-352337sbdqd]arandaasesoria.com/diplom-352337sbdqd[/url]
[url=http://gen8ai.com/groups/diplom-kupit-67091hu/members/all-members]gen8ai.com/groups/diplom-kupit-67091hu/members/all-members[/url]
[url=http://sttimothysignal.org/groups/diplom-kupit-314046jx]sttimothysignal.org/groups/diplom-kupit-314046jx[/url]
[url=http://bookmarkangaroo.com/story17756183/diplom]bookmarkangaroo.com/story17756183/diplom[/url]
[url=http://techhansha.com/diplom-936150uurmx]techhansha.com/diplom-936150uurmx[/url]
[url=http://manette153.com/diplom-975697blnex]manette153.com/diplom-975697blnex[/url]
[url=http://ssokhongchiam.in.th/main/index.php?name=webboard&file=read&id=695206]ssokhongchiam.in.th/main/index.php?name=webboard&file=read&id=695206[/url]
Lariorioz
August 9, 2024 @ 11:08 am
[u][b]Добрыйдень![/b][/u]
[b]Заказать диплом любогоВУЗа.[/b]
[url=http://modernbookmarks.com/story17472136/kupit-diplom]modernbookmarks.com/story17472136/kupit-diplom[/url]
[url=http://isocialfans.com/story3022285/kupit-diplom]isocialfans.com/story3022285/kupit-diplom[/url]
[url=http://bookmarkcitizen.com/story17687026/kupit-diplom]bookmarkcitizen.com/story17687026/kupit-diplom[/url]
[url=http://astradigital.co.uk/wiki/index.php?title=Kupit_Diplom_538204gCWXP]astradigital.co.uk/wiki/index.php?title=Kupit_Diplom_538204gCWXP[/url]
[url=http://taondinternational.rudraserver.com/blog/index.php?entryid=132866]taondinternational.rudraserver.com/blog/index.php?entryid=132866[/url]
[url=http://alkhazana.net/2024/07/29/kupit-diplom-214358xyryz]alkhazana.net/2024/07/29/kupit-diplom-214358xyryz[/url]
[url=http://mysterybookmarks.com/story17651197/kupit-diplom]mysterybookmarks.com/story17651197/kupit-diplom[/url]
[url=http://ecrinparis.com/kupit-diplom-667330mwntj]ecrinparis.com/kupit-diplom-667330mwntj[/url]
[url=http://webookmarks.com/story3051932/kupit-diplom]webookmarks.com/story3051932/kupit-diplom[/url]
[url=http://bookmark-vip.com/story17708679/kupit-diplom]bookmark-vip.com/story17708679/kupit-diplom[/url]
GeorgeZed
August 9, 2024 @ 11:15 am
cialis generic online uk: Generic Cialis without a doctor prescription – cialis before and after pictures
SamuelGop
August 9, 2024 @ 12:20 pm
http://tadalafil.auction/# cheap cialis online overnight shipping
RufusLoach
August 9, 2024 @ 1:21 pm
Новости высоких технологий http://barbie-games.ru/karta-glybinnyh-temperatyr новинки компьютерной техники и мобильных телефонов
CoreyCow
August 9, 2024 @ 1:21 pm
Свежие новости http://barbie-games.ru/bez-rylia технологий новинки компьютерной техники
SamuelGop
August 9, 2024 @ 1:30 pm
https://tadalafil.auction/# canada toronto cheap fase cialis
Robertpoise
August 9, 2024 @ 1:33 pm
Новости высоких технологий: новинки http://barbie-games.ru/sintez-elementov-i-iadernye-filtry компьютерной техники и мобильных телефонов
RoccoTrimi
August 9, 2024 @ 1:37 pm
Актуальные новости техники http://barbie-games.ru/ychenym-ydalos-yvidet-dvijenie-eksitonov и гаджетов.
CurtisGoW
August 9, 2024 @ 1:42 pm
Новости высоких технологий http://barbie-games.ru/ochistnoi-kombain-ykd400-dobycha-chernogo-zolota-stala-ekonomichnee и гаджетов.
Mazrcig
August 9, 2024 @ 2:03 pm
[u][b] Привет, друзья![/b][/u]
[b]Быстрое обучение и получение диплома магистра – возможно ли это?[/b]
[url=http://rushkadiplomik.ru/]rushkadiplomik.ru[/url]
Lazrwjf
August 9, 2024 @ 2:06 pm
[u][b] Здравствуйте![/b][/u]
[b]Купить диплом любого ВУЗа.[/b]
[url=http://diplomman.ru/kupit-diplom-voronezh/]diplomman.ru/kupit-diplom-voronezh[/url]
Lazrhgz
August 9, 2024 @ 2:31 pm
[b]Приобрести диплом университета.[/b]
[url=http://forum.analysisclub.ru/index.php/topic,200459.new#new/]forum.analysisclub.ru/index.php/topic,200459.new#new[/url]
[url=http://ic-info.ru/forum/user/166179/]ic-info.ru/forum/user/166179[/url]
[url=http://helix-forum.maxbb.ru/posting.php?mode=post&f=3/]helix-forum.maxbb.ru/posting.php?mode=post&f=3[/url]
[url=http://kronverskiy.ru/posting.php?mode=post&f=28/]kronverskiy.ru/posting.php?mode=post&f=28[/url]
[url=http://tatuheart.ukrbb.net/viewtopic.php?f=41&t=13530/]tatuheart.ukrbb.net/viewtopic.php?f=41&t=13530[/url]
Dnrtldk
August 9, 2024 @ 2:50 pm
[b]Купить документ[/b] университета вы можете у нас в Москве.
[url=http://njspmaca.in/2024/07/29/diplom-515059qkuth/]njspmaca.in/2024/07/29/diplom-515059qkuth[/url]
[url=http://njspmaca.in/2024/07/29/diplom-376143ptahz/]njspmaca.in/2024/07/29/diplom-376143ptahz[/url]
[url=http://socialbaskets.com/story3090025/diplom/]socialbaskets.com/story3090025/diplom[/url]
[url=http://bookmarkworm.com/story17622877/diplom/]bookmarkworm.com/story17622877/diplom[/url]
[url=http://selfhackathon.com/diplom-kupit-777852lo/]selfhackathon.com/diplom-kupit-777852lo[/url]
Mazrlyo
August 9, 2024 @ 3:26 pm
[u][b] Здравствуйте![/b][/u]
[b]Как получить диплом техникума с упрощенным обучением в Москве официально[/b]
[url=http://rushkas-diplomasxx.ru/kupit-diplom-novosibirsk/]rushkas-diplomasxx.ru/kupit-diplom-novosibirsk[/url]
GeorgeZed
August 9, 2024 @ 4:49 pm
buy cialis in cro: cialis soft tabs, united states origin – cheap cialis from australia
Mazrpmk
August 9, 2024 @ 4:59 pm
[u][b] Привет, друзья![/b][/u]
[b]Стоимость дипломов высшего и среднего образования и процесс их получения[/b]
[url=http://rushkadiplomik.ru/]rushkadiplomik.ru[/url]
Dnrtfoq
August 9, 2024 @ 5:49 pm
[b]Купить документ[/b] института можно у нас.
[url=http://kasaanamedia.com/diplom-957849acnor/]kasaanamedia.com/diplom-957849acnor[/url]
[url=http://seobookmarkpro.com/story17677523/diplom/]seobookmarkpro.com/story17677523/diplom[/url]
[url=http://healthywellness.site/diplom-171316dahxz/]healthywellness.site/diplom-171316dahxz[/url]
[url=http://bookmarketmaven.com/story18084913/diplom/]bookmarketmaven.com/story18084913/diplom[/url]
[url=http://healthywellness.site/diplom-691621wfqxp/]healthywellness.site/diplom-691621wfqxp[/url]
Cazrwez
August 9, 2024 @ 6:19 pm
[u][b] Добрый день![/b][/u]
Мы предлагаем документы техникумов
[url=http://ffxiv.one/blogs/193/Оформите-диплом-за-пару-дней/]ffxiv.one/blogs/193/Оформите-диплом-за-пару-дней[/url]
[url=http://library.kemu.ac.ke/kemuwiki/index.php/образование_мечты:_дипломы_ведущих_вузов/]library.kemu.ac.ke/kemuwiki/index.php/образование_мечты:_дипломы_ведущих_вузов[/url]
[url=http://odinzovo.rusff.me/viewtopic.php?id=4923#p8319/]odinzovo.rusff.me/viewtopic.php?id=4923#p8319[/url]
[url=http://njt.ru/forum/user/197785/]njt.ru/forum/user/197785[/url]
[url=http://agarwalconnect.com/blogs/227/Быстрое-получение-диплома-онлайн/]agarwalconnect.com/blogs/227/Быстрое-получение-диплома-онлайн[/url]
Treftkd
August 9, 2024 @ 6:37 pm
[u][b] Привет![/b][/u]
Официальная покупка диплома ПТУ с упрощенной программой обучения
[url=http://mans-diplomasxx.ru/svidetelstvo-o-rozhdenii-sssr1/]mans-diplomasxx.ru/svidetelstvo-o-rozhdenii-sssr1[/url]
Рады помочь!.
Lazrdnx
August 9, 2024 @ 7:19 pm
[u][b] Привет, друзья![/b][/u]
[b]Мы предлагаем дипломы[/b] любой профессии по приятным ценам.
[url=http://rushkas-diploms.ru/kupit-diplom-stroitelya/]rushkas-diploms.ru/kupit-diplom-stroitelya[/url]
GeorgeZed
August 9, 2024 @ 7:21 pm
viagra online: Buy Viagra online cheap – generic viagra overnight
Trefxgb
August 9, 2024 @ 9:16 pm
[u][b] Добрый день![/b][/u]
Всё, что нужно знать о покупке аттестата о среднем образовании
[url=http://mans-diplomyxx.ru/kupit-diplom-novosibirsk/]mans-diplomyxx.ru/kupit-diplom-novosibirsk[/url]
Рады оказать помощь!.
Yrefwkr
August 9, 2024 @ 9:33 pm
[u][b] Привет, друзья![/b][/u]
[b]Заказать диплом о высшем образовании.[/b]
[url=http://newmedtime.ru/page/4/]newmedtime.ru/page/4[/url]
[url=http://bbs.heyshell.com/forum.php?mod=viewthread&tid=28327/]bbs.heyshell.com/forum.php?mod=viewthread&tid=28327[/url]
[url=http://youlangue.lu/blog/index.php?entryid=60027&nonjscomment=1&comment_itemid=60027&comment_context=71954&comment_component=blog&comment_area=format_blog/]youlangue.lu/blog/index.php?entryid=60027&nonjscomment=1&comment_itemid=60027&comment_context=71954&comment_component=blog&comment_area=format_blog[/url]
[url=http://scapeacademy.com/blog/index.php?entryid=21955/]scapeacademy.com/blog/index.php?entryid=21955[/url]
[url=http://yasqueencommunity.com/blogs/347/Что-именно-необходимо-из-техники-для-производства-диплома/]yasqueencommunity.com/blogs/347/Что-именно-необходимо-из-техники-для-производства-диплома[/url]
Lazrfzl
August 9, 2024 @ 10:25 pm
[u][b] Здравствуйте![/b][/u]
[b]Мы можем предложить дипломы[/b] любых профессий по приятным ценам.
[url=http://rushkas-diploms.ru/diplom-tekhnikuma-kolledzha/]rushkas-diploms.ru/diplom-tekhnikuma-kolledzha[/url]
Mazrlbp
August 9, 2024 @ 11:00 pm
[u][b] Добрый день![/b][/u]
Официальная покупка школьного аттестата с упрощенным обучением в Москве
[url=http://landik-diploms-srednee.ru/svidetelstvo-o-razvode/]landik-diploms-srednee.ru/svidetelstvo-o-razvode[/url]
Yrefaif
August 10, 2024 @ 12:13 am
[u][b] Добрый день![/b][/u]
[b]Купить диплом университета.[/b]
[url=http://duster-clubs.ru/forum/album.php?albumid=863&pictureid=9103&commentid=4916/]duster-clubs.ru/forum/album.php?albumid=863&pictureid=9103&commentid=4916[/url]
[url=http://livestockkenya.com/index.php/cb-profile/pluginclass/cbblogs?action=blogs&func=show&id=250/]livestockkenya.com/index.php/cb-profile/pluginclass/cbblogs?action=blogs&func=show&id=250[/url]
[url=http://ktrcycleworld.com/купить-диплом/]ktrcycleworld.com/купить-диплом[/url]
[url=http://mpett.com/blogs/704/Что-нужно-для-изготовления-неотличимого-от-подлинника-диплома/]mpett.com/blogs/704/Что-нужно-для-изготовления-неотличимого-от-подлинника-диплома[/url]
[url=http://isig.ac.cd/alumni/blogs/37709/Основные-затраты-российского-изготовителя-дипломов-обзор/]isig.ac.cd/alumni/blogs/37709/Основные-затраты-российского-изготовителя-дипломов-обзор[/url]
Diplomi_xjSi
August 10, 2024 @ 12:47 am
Привет, друзья!
Купить документ о получении высшего образования можно у нас в Москве.
[url=http://rushkadiplomiks.ru/]rushkadiplomiks.ru[/url]
SamuelGop
August 10, 2024 @ 1:04 am
http://sildenafil.llc/# viagra vs cialis
GeorgeZed
August 10, 2024 @ 1:18 am
why does cialis cost so much: cheapest tadalafil – buy generic cialis
StephenBoype
August 10, 2024 @ 1:24 am
https://sildenafil.llc/# viagra professional
viagra levitra cialis which works better? [url=http://tadalafil.auction/#]Buy Cialis online[/url] how long does cialis last
SamuelGop
August 10, 2024 @ 2:30 am
https://tadalafil.auction/# cialis online mastercard
Sazrjex
August 10, 2024 @ 2:47 am
[u][b] Здравствуйте![/b][/u]
Приобрести документ о получении высшего образования
[url=http://easyinsurancehub.co.uk/forum/thread-57319/]easyinsurancehub.co.uk/forum/thread-57319[/url]
[url=http://vipka.mybb.ru/viewtopic.php?id=2930#p5212/]vipka.mybb.ru/viewtopic.php?id=2930#p5212[/url]
[url=http://telegra.ph/spravka-ot-psihiatra-dlya-postupleniya-v-vuz-07-24/]telegra.ph/spravka-ot-psihiatra-dlya-postupleniya-v-vuz-07-24[/url]
[url=http://telegra.ph/obuchenie-v-vuzah-germanii-07-24/]telegra.ph/obuchenie-v-vuzah-germanii-07-24[/url]
[url=http://umorfishki.ru/poluchite-diplom-i-stante-luchshim/]umorfishki.ru/poluchite-diplom-i-stante-luchshim[/url]
Diplomi_kxSi
August 10, 2024 @ 3:54 am
Добрый день!
Купить документ о получении высшего образования можно у нас в Москве.
[url=http://diploms-rushkas.ru/]diploms-rushkas.ru[/url]
StephenBoype
August 10, 2024 @ 4:32 am
http://tadalafil.auction/# purchase cialis in canada
natural viagra [url=http://sildenafil.llc/#]Buy Viagra online cheap[/url] viagra coupon
Cazrkug
August 10, 2024 @ 5:21 am
[u][b] Привет![/b][/u]
Мы предлагаем документы ВУЗов
[url=http://superforum.spybb.ru/viewtopic.php?id=2771#p50818/]superforum.spybb.ru/viewtopic.php?id=2771#p50818[/url]
[url=http://forexsnews.ru/byistroe-oformlenie-diplomov/]forexsnews.ru/byistroe-oformlenie-diplomov[/url]
[url=http://vintfint.com/blogs/23344/Быстрое-получение-диплома-онлайн/]vintfint.com/blogs/23344/Быстрое-получение-диплома-онлайн[/url]
[url=http://veneraroleplay.listbb.ru/viewtopic.php?f=4&t=328/]veneraroleplay.listbb.ru/viewtopic.php?f=4&t=328[/url]
[url=http://beijingpal.com/read-blog/1230/]beijingpal.com/read-blog/1230[/url]
Sazrgqd
August 10, 2024 @ 5:35 am
[u][b] Добрый день![/b][/u]
Заказать документ ВУЗа
[url=http://itstagram.ru/read-blog/937/]itstagram.ru/read-blog/937[/url]
[url=http://telegra.ph/harakteristika-na-uchenika-11-klassa-dlya-postupleniya-v-vuz-primer-07-24/]telegra.ph/harakteristika-na-uchenika-11-klassa-dlya-postupleniya-v-vuz-primer-07-24[/url]
[url=http://mystroycenter.ru/byistroe-i-legkoe-poluchenie-diploma/]mystroycenter.ru/byistroe-i-legkoe-poluchenie-diploma[/url]
[url=http://telegra.ph/kalkulyator-ballov-dlya-postupleniya-v-vuz-07-24/]telegra.ph/kalkulyator-ballov-dlya-postupleniya-v-vuz-07-24[/url]
[url=http://krugozorov.ru/forum/user/9258/]krugozorov.ru/forum/user/9258[/url]
Sazrjcz
August 10, 2024 @ 7:41 am
[u][b] Привет![/b][/u]
Мы изготавливаем дипломы психологов, юристов, экономистов и любых других профессий.
[url=http://localtrusted.co.uk/wiki/index.php?title=Kupit_Diplom_949396aciux/]localtrusted.co.uk/wiki/index.php?title=Kupit_Diplom_949396aciux[/url]
[url=http://edux.izvrsna.hr/blog/index.php?entryid=84/]edux.izvrsna.hr/blog/index.php?entryid=84[/url]
[url=http://socialmediaentry.com/story2980944/kupit-diplom/]socialmediaentry.com/story2980944/kupit-diplom[/url]
[url=http://growthbookmarks.com/story17602922/kupit-diplom/]growthbookmarks.com/story17602922/kupit-diplom[/url]
[url=http://socialwebnotes.com/story3101273/kupit-diplom/]socialwebnotes.com/story3101273/kupit-diplom[/url]
Mazrewo
August 10, 2024 @ 7:42 am
[b]Заказать документ о получении высшего образования.[/b]
[url=http://geniusbookmarks.com/story17653368/kupit-diplom/]geniusbookmarks.com/story17653368/kupit-diplom[/url]
[url=http://offmarketbusinessforsale.com/kupit-diplom-800607rwljc/]offmarketbusinessforsale.com/kupit-diplom-800607rwljc[/url]
[url=http://socialimarketing.com/story3089896/kupit-diplom/]socialimarketing.com/story3089896/kupit-diplom[/url]
[url=http://fa.earnvisits.com/index.php?page=user&action=pub_profile&id=114702/]fa.earnvisits.com/index.php?page=user&action=pub_profile&id=114702[/url]
[url=http://fam2fam.com/groups/kupit-diplom-95110ldyqe/]fam2fam.com/groups/kupit-diplom-95110ldyqe[/url]
Mazrbhu
August 10, 2024 @ 7:43 am
[u][b] Здравствуйте![/b][/u]
[b]Как официально купить аттестат 11 класса с упрощенным обучением в Москве[/b]
[url=http://rushkadiplomik.ru/]rushkadiplomik.ru[/url]
Dnrtjuc
August 10, 2024 @ 7:59 am
[b]Приобрести документ[/b] о получении высшего образования можно в нашей компании в Москве.
[url=http://rotatesites.com/story18770137/diplom/]rotatesites.com/story18770137/diplom[/url]
[url=http://totalbookmarking.com/story17682496/diplom/]totalbookmarking.com/story17682496/diplom[/url]
[url=http://thesocialintro.com/story3092960/diplom/]thesocialintro.com/story3092960/diplom[/url]
[url=http://iamwomanacademy.com/diplom-828254dfmvm/]iamwomanacademy.com/diplom-828254dfmvm[/url]
[url=http://freebookmarkpost.com/story17551481/diplom/]freebookmarkpost.com/story17551481/diplom[/url]
Sazrkvk
August 10, 2024 @ 9:48 am
[u][b] Здравствуйте![/b][/u]
Диплом любой специальности
[url=http://iaescortsmap.ixbb.ru/viewtopic.php?id=118#p118/]iaescortsmap.ixbb.ru/viewtopic.php?id=118#p118[/url]
Cazrmsm
August 10, 2024 @ 9:49 am
[u][b] Добрый день![/b][/u]
Приобрести диплом о высшем образовании
[url=http://polegasm.net/index.php/forum/welcome-mat/151625/]polegasm.net/index.php/forum/welcome-mat/151625[/url]
[url=http://local-urban-eats.mn.co/posts/62529753/]local-urban-eats.mn.co/posts/62529753[/url]
[url=http://mosday.getbb.ru/viewtopic.php?f=12&t=1343/]mosday.getbb.ru/viewtopic.php?f=12&t=1343[/url]
[url=http://baxoday.maxbb.ru/viewtopic.php?f=25&t=525/]baxoday.maxbb.ru/viewtopic.php?f=25&t=525[/url]
[url=http://tradersopen.com/login/login/]tradersopen.com/login/login[/url]
Trefjif
August 10, 2024 @ 10:24 am
[u][b] Привет, друзья![/b][/u]
Приобретение диплома ПТУ с сокращенной программой обучения в Москве
[url=http://diploms-man.ru/kupit-diplom-v-ufe/]diploms-man.ru/kupit-diplom-v-ufe[/url]
Поможем вам всегда!.
Mazrqqh
August 10, 2024 @ 10:33 am
[u][b] Добрый день![/b][/u]
Всё, что нужно знать о покупке аттестата о среднем образовании
[url=http://landik-diploms-srednee.ru/diplom-bakalavra/]landik-diploms-srednee.ru/diplom-bakalavra[/url]
GeorgeZed
August 10, 2024 @ 10:46 am
cialis next day delivery: Generic Tadalafil 20mg price – viagra cialis levitra trial pack
Dnrtxby
August 10, 2024 @ 11:04 am
[b]Заказать документ[/b] о получении высшего образования вы имеете возможность в нашем сервисе.
[url=http://starryjeju.com/qna/5649131/]starryjeju.com/qna/5649131[/url]
[url=http://socialinplace.com/story2958807/diplom/]socialinplace.com/story2958807/diplom[/url]
[url=http://pristinefleetsolution.com/diplom-kupit-885460ot/]pristinefleetsolution.com/diplom-kupit-885460ot[/url]
[url=http://welnesbiolabs.com/diplom-624438texnc/]welnesbiolabs.com/diplom-624438texnc[/url]
[url=http://maitemach.com/diplom-35140datai/]maitemach.com/diplom-35140datai[/url]
Mazratg
August 10, 2024 @ 11:04 am
[u][b] Привет![/b][/u]
[b]Покупка диплома о среднем полном образовании: как избежать мошенничества?[/b]
[url=http://rushkadiplomik.ru/]rushkadiplomik.ru[/url]
Trefhww
August 10, 2024 @ 1:05 pm
[u][b] Привет, друзья![/b][/u]
Как получить диплом стоматолога быстро и официально
[url=http://mans-diplom.ru/diplom-tehnikuma/]mans-diplom.ru/diplom-tehnikuma[/url]
Будем рады вам помочь!.
Sazrkme
August 10, 2024 @ 1:41 pm
[u][b] Добрый день![/b][/u]
Купить документ о получении высшего образования
[url=http://avtomobil1980.ixbb.ru/viewtopic.php?id=125#p125/]avtomobil1980.ixbb.ru/viewtopic.php?id=125#p125[/url]
[url=http://androidinweb.ru/kupite-diplom-po-vyigodnoy-tsene/]androidinweb.ru/kupite-diplom-po-vyigodnoy-tsene[/url]
[url=http://mirnuy.forumieren.de/t8248-topic#13523/]mirnuy.forumieren.de/t8248-topic#13523[/url]
[url=http://russianecuador.com/forum/member.php?u=13207/]russianecuador.com/forum/member.php?u=13207[/url]
[url=http://backshowtime.ru/kachestvennyie-diplomyi-na-zakaz/]backshowtime.ru/kachestvennyie-diplomyi-na-zakaz[/url]
GeorgeZed
August 10, 2024 @ 1:48 pm
blue pill viagra: Viagra online price – viagra without doctor prescription
Lazrkrw
August 10, 2024 @ 1:53 pm
[u][b] Добрый день![/b][/u]
[b]Мы предлагаем дипломы[/b] любой профессии по выгодным тарифам.
[url=http://rushkadiplomik.ru/kupit-diplom-voronezh/]rushkadiplomik.ru/kupit-diplom-voronezh[/url]
Yrefjxd
August 10, 2024 @ 1:57 pm
[u][b] Привет, друзья![/b][/u]
[b]Приобрести диплом о высшем образовании.[/b]
[url=http://proshnottor.com/купить-диплом-парикмахера/]proshnottor.com/купить-диплом-парикмахера[/url]
[url=http://tonythang.dev/blogs/94/Что-нужно-для-изготовления-неотличимого-от-оригинала-диплома?lang=fr_fr/]tonythang.dev/blogs/94/Что-нужно-для-изготовления-неотличимого-от-оригинала-диплома?lang=fr_fr[/url]
[url=http://imf1fan.com/community/profile/leopoldobeh2135/]imf1fan.com/community/profile/leopoldobeh2135[/url]
[url=http://prikol100500.ru/kak-zakazat-diplom-onlayn-prostyie-shagi/]prikol100500.ru/kak-zakazat-diplom-onlayn-prostyie-shagi[/url]
[url=http://jbnucri.com/bbs/board.php?bo_table=companylist&wr_id=6824/]jbnucri.com/bbs/board.php?bo_table=companylist&wr_id=6824[/url]
SamuelGop
August 10, 2024 @ 4:03 pm
https://tadalafil.auction/# cialis generic cipla
Sazrmqj
August 10, 2024 @ 4:46 pm
[u][b] Привет, друзья![/b][/u]
Приобрести документ института
[url=http://cittaviva.net/read-blog/86/]cittaviva.net/read-blog/86[/url]
[url=http://grp.7olimp.ru/viewforum.php?f=1/]grp.7olimp.ru/viewforum.php?f=1[/url]
[url=http://vipka.0bb.ru/viewtopic.php?id=5925#p8923/]vipka.0bb.ru/viewtopic.php?id=5925#p8923[/url]
[url=http://ukrevents.ru/diplomyi-dlya-karernyih-ambitsiy/]ukrevents.ru/diplomyi-dlya-karernyih-ambitsiy[/url]
[url=http://artrp.5nx.ru/viewtopic.php?f=3&t=9578/]artrp.5nx.ru/viewtopic.php?f=3&t=9578[/url]
Lazrlgt
August 10, 2024 @ 5:11 pm
[u][b] Здравствуйте![/b][/u]
[b]Мы предлагаем дипломы[/b] любой профессии по приятным ценам.
[url=http://diploms-rushka.ru/diplom-sssr/]diploms-rushka.ru/diplom-sssr[/url]
Oariorqhv
August 10, 2024 @ 5:25 pm
[u][b] Здравствуйте![/b][/u]
Купить диплом ВУЗа
[b]Наши специалисты предлагают[/b] максимально быстро купить диплом, который выполняется на оригинальном бланке и заверен печатями, штампами, подписями. Диплом пройдет лубую проверку, даже при помощи профессионального оборудования. Достигайте цели быстро и просто с нашим сервисом.
[b]Где заказать диплом по актуальной специальности?[/b]
[url=http://lezt.ixbb.ru/viewtopic.php?id=122#p122/]lezt.ixbb.ru/viewtopic.php?id=122#p122[/url]
[url=http://telegra.ph/kak-sozdat-lichnyj-kabinet-dlya-postupleniya-v-vuz-07-24/]telegra.ph/kak-sozdat-lichnyj-kabinet-dlya-postupleniya-v-vuz-07-24[/url]
[url=http://riftynet.com/blogs/26248/Надежное-оформление-дипломов-онлайн/]riftynet.com/blogs/26248/Надежное-оформление-дипломов-онлайн[/url]
[url=http://telegra.ph/stoimost-obucheniya-v-vuzah-tveri-07-24/]telegra.ph/stoimost-obucheniya-v-vuzah-tveri-07-24[/url]
[url=http://vip.rolevaya.info/viewtopic.php?id=5111#p8768/]vip.rolevaya.info/viewtopic.php?id=5111#p8768[/url]
SamuelGop
August 10, 2024 @ 5:27 pm
http://tadalafil.auction/# generic cialis priligy australia
Mazryan
August 10, 2024 @ 6:04 pm
[b]Приобрести документ о получении высшего образования.[/b]
[url=http://managerhotels.com/business-small-business/kupit-diplom-269065rpyxc/]managerhotels.com/business-small-business/kupit-diplom-269065rpyxc[/url]
[url=http://bookmarkize.com/story17670303/kupit-diplom/]bookmarkize.com/story17670303/kupit-diplom[/url]
[url=http://taondinternational.rudraserver.com/blog/index.php?entryid=132254/]taondinternational.rudraserver.com/blog/index.php?entryid=132254[/url]
[url=http://utahsyardsale.com/author/macredmond2/]utahsyardsale.com/author/macredmond2[/url]
[url=http://artybookmarks.com/story17578477/kupit-diplom/]artybookmarks.com/story17578477/kupit-diplom[/url]
Sazryqc
August 10, 2024 @ 6:08 pm
[u][b] Привет, друзья![/b][/u]
Мы готовы предложить дипломы психологов, юристов, экономистов и других профессий.
[url=http://moodjhomedia.com/story1916602/kupit-diplom/]moodjhomedia.com/story1916602/kupit-diplom[/url]
[url=http://gtj.kr/board_KtRj53/371070/]gtj.kr/board_KtRj53/371070[/url]
[url=http://kankerala.org/uncategorized/kupit-diplom-999897beotr/]kankerala.org/uncategorized/kupit-diplom-999897beotr[/url]
[url=http://kasaanamedia.com/kupit-diplom-533947sxzgi/]kasaanamedia.com/kupit-diplom-533947sxzgi[/url]
[url=http://thebookmarkfree.com/story17799632/kupit-diplom/]thebookmarkfree.com/story17799632/kupit-diplom[/url]
Iarioriid
August 10, 2024 @ 7:29 pm
[u][b] Добрый день![/b][/u]
Купить документ о получении высшего образования вы можете в нашей компании. Мы оказываем услуги по изготовлению и продаже документов об окончании любых университетов РФ. Вы получите диплом по любой специальности, любого года выпуска, в том числе документы СССР. Гарантируем, что при проверке документов работодателями, подозрений не возникнет.
[url=http://nachosking.com/read-blog/852/]nachosking.com/read-blog/852[/url]
[url=http://network.janenk.com/read-blog/761/]network.janenk.com/read-blog/761[/url]
[url=http://zforum.kabb.ru/viewtopic.php?f=13&t=1348/]zforum.kabb.ru/viewtopic.php?f=13&t=1348[/url]
[url=http://umorforme.ru/diplomyi-dlya-professionalnogo-rosta/]umorforme.ru/diplomyi-dlya-professionalnogo-rosta[/url]
[url=http://bintarotrojan.com/blogs/11343/оформите-диплом-для-новой-карьеры/]bintarotrojan.com/blogs/11343/оформите-диплом-для-новой-карьеры[/url]
Sazrnzu
August 10, 2024 @ 8:39 pm
[u][b] Привет![/b][/u]
Диплом о высшем образовании
[url=http://avtoweek2016.ru/dostupnyie-diplomyi-s-garantiey-kachestva/]avtoweek2016.ru/dostupnyie-diplomyi-s-garantiey-kachestva[/url]
Manrsgj
August 10, 2024 @ 9:19 pm
[u][b] Добрый день![/b][/u]
Приобрести документ института
[url=http://astradigital.co.uk/wiki/index.php?title=Kupit_Diplom_561117WfPhU]astradigital.co.uk/wiki/index.php?title=Kupit_Diplom_561117WfPhU[/url]
[url=http://alkhazana.net/2024/07/28/kupit-diplom-30254jprsx]alkhazana.net/2024/07/28/kupit-diplom-30254jprsx[/url]
[url=http://moodjhomedia.com/story1915982/kupit-diplom]moodjhomedia.com/story1915982/kupit-diplom[/url]
[url=http://seolistlinks.com/story18958123/kupit-diplom]seolistlinks.com/story18958123/kupit-diplom[/url]
[url=http://bookmarkuse.com/story17494194/kupit-diplom]bookmarkuse.com/story17494194/kupit-diplom[/url]
Douglasidomb
August 10, 2024 @ 11:41 pm
online shopping pharmacy india: indian pharmacy – best online pharmacy india
Uazrtia
August 11, 2024 @ 12:09 am
[b]Где заказать диплом специалиста?[/b]
[url=http://tigaedu.com/blog/index.php?entryid=333707/]tigaedu.com/blog/index.php?entryid=333707[/url]
[url=http://gaigoimoinhat.xyz/diplom-222832zyeww/]gaigoimoinhat.xyz/diplom-222832zyeww[/url]
[url=http://heealthy.com/question/diplom-121481aitpn/]heealthy.com/question/diplom-121481aitpn[/url]
[url=http://directlynews.com/diplom-810018cqmtx/]directlynews.com/diplom-810018cqmtx[/url]
[url=http://thebookmarkplaza.com/story17602699/diplom/]thebookmarkplaza.com/story17602699/diplom[/url]
Dnrtrgc
August 11, 2024 @ 12:39 am
[b]Купить документ[/b] о получении высшего образования можно у нас в столице.
[url=http://bookmarktune.com/story17574691/diplom/]bookmarktune.com/story17574691/diplom[/url]
[url=http://pirooztak.ir/?option=com_k2&view=itemlist&task=user&id=1357605/]pirooztak.ir/?option=com_k2&view=itemlist&task=user&id=1357605[/url]
[url=http://candidecoin.com/uncategorized/diplom-707788jvsno/]candidecoin.com/uncategorized/diplom-707788jvsno[/url]
[url=http://parathajoint.com/2024/07/29/diplom-kupit-91173qu/]parathajoint.com/2024/07/29/diplom-kupit-91173qu[/url]
[url=http://bookmarklayer.com/story17674917/diplom/]bookmarklayer.com/story17674917/diplom[/url]
Xazrewi
August 11, 2024 @ 1:11 am
[u][b] Привет![/b][/u]
Купить документ о получении высшего образования.
[url=http://rentry.co/ipe5ww38/]rentry.co/ipe5ww38[/url]
[url=http://rogerfilms.com/index.php?title=Купите_диплом_и_улучшите_свое_будущее/]rogerfilms.com/index.php?title=Купите_диплом_и_улучшите_свое_будущее[/url]
[url=http://beijingpal.com/read-blog/1026/]beijingpal.com/read-blog/1026[/url]
[url=http://boxer-forum.ru/viewtopic.php?f=17&t=4022/]boxer-forum.ru/viewtopic.php?f=17&t=4022[/url]
[url=http://collieforum.ru/posting.php?mode=post&f=17/]collieforum.ru/posting.php?mode=post&f=17[/url]
Trefwbm
August 11, 2024 @ 1:18 am
[u][b] Привет, друзья![/b][/u]
Пошаговая инструкция по безопасной покупке диплома о высшем образовании
[url=http://mans-diplomyxx.ru/kupit-diplom-ekaterinburg/]mans-diplomyxx.ru/kupit-diplom-ekaterinburg[/url]
Будем рады вам помочь!.
Diplomi_llSi
August 11, 2024 @ 1:52 am
Здравствуйте!
Заказать документ о получении высшего образования вы можете в нашей компании.
[url=http://diploms-rushkas.ru/]diploms-rushkas.ru[/url]
Leonardginge
August 11, 2024 @ 2:10 am
https://indiapharmacy.shop/# pharmacy website india
Charlescaulk
August 11, 2024 @ 2:31 am
http://mexicopharmacy.win/# buying prescription drugs in mexico
cheap ed treatment
Trefsog
August 11, 2024 @ 3:47 am
[u][b] Привет, друзья![/b][/u]
Приобретение школьного аттестата с официальным упрощенным обучением в Москве
[url=http://mans-diplomyxx.ru/kupit-diplom-voronezh/]mans-diplomyxx.ru/kupit-diplom-voronezh[/url]
Будем рады вам помочь!.
Dnrttwy
August 11, 2024 @ 4:19 am
[b]Заказать документ[/b] о получении высшего образования вы можете в нашей компании.
[url=http://kankerala.org/uncategorized/diplom-251646lrgbp/]kankerala.org/uncategorized/diplom-251646lrgbp[/url]
[url=http://trackbookmark.com/story19019233/diplom/]trackbookmark.com/story19019233/diplom[/url]
[url=http://krovbar.ru/strashnye-istorii/diplom-322235bomnd/]krovbar.ru/strashnye-istorii/diplom-322235bomnd[/url]
[url=http://investicos.com/uncategorized/diplom-kupit-546082zq/]investicos.com/uncategorized/diplom-kupit-546082zq[/url]
[url=http://campusvirtual.newlink.es/blog/index.php?entryid=16587/]campusvirtual.newlink.es/blog/index.php?entryid=16587[/url]
Mazrvok
August 11, 2024 @ 5:07 am
[u][b] Привет, друзья![/b][/u]
[b]Официальная покупка диплома вуза с сокращенной программой в Москве[/b]
[url=http://diplomrushkan.ru/]diplomrushkan.ru[/url]
Jariorwgc
August 11, 2024 @ 5:26 am
[u][b] Добрый день![/b][/u]
Купить диплом о высшем образовании
[url=http://shikhadabas.com/2024/07/27/kupit-diplom-109933tnruy]shikhadabas.com/2024/07/27/kupit-diplom-109933tnruy[/url]
[url=http://mysocialfeeder.com/story3000704/kupit-diplom]mysocialfeeder.com/story3000704/kupit-diplom[/url]
[url=http://mysocialguides.com/story2972557/kupit-diplom]mysocialguides.com/story2972557/kupit-diplom[/url]
[url=http://greatbookmarking.com/story17705936/kupit-diplom]greatbookmarking.com/story17705936/kupit-diplom[/url]
[url=http://ysa.sa/kupit-diplom-663724kmkib]ysa.sa/kupit-diplom-663724kmkib[/url]
[url=http://listingbookmarks.com/story17712596/kupit-diplom]listingbookmarks.com/story17712596/kupit-diplom[/url]
[url=http://nanobookmarking.com/story17586723/kupit-diplom]nanobookmarking.com/story17586723/kupit-diplom[/url]
[url=http://apollobookmarks.com/story17606423/kupit-diplom]apollobookmarks.com/story17606423/kupit-diplom[/url]
[url=http://nextbiz.blog/2024/07/28/kupit-diplom-832173tbmlz]nextbiz.blog/2024/07/28/kupit-diplom-832173tbmlz[/url]
[url=http://guidemysocial.com/story2945459/kupit-diplom]guidemysocial.com/story2945459/kupit-diplom[/url]
Charlescaulk
August 11, 2024 @ 5:30 am
https://indiapharmacy.shop/# buy prescription drugs from india
ed medicine online
Diplomi_alSi
August 11, 2024 @ 5:30 am
Привет, друзья!
Заказать документ о получении высшего образования можно в нашем сервисе.
[url=http://rushkas-diplomas.ru/]rushkas-diplomas.ru[/url]
Douglasidomb
August 11, 2024 @ 5:59 am
best online ed treatment: ed pills online – ed med online
Iariorvlo
August 11, 2024 @ 6:03 am
[u][b] Привет, друзья![/b][/u]
Купить документ о получении высшего образования вы сможете в нашей компании. Мы оказываем услуги по изготовлению и продаже документов об окончании любых ВУЗов России. Вы сможете получить диплом по любым специальностям, любого года выпуска, включая документы образца СССР. Гарантируем, что в случае проверки документов работодателем, никаких подозрений не возникнет.
[url=http://kinopuk.ru/viewtopic.php?id=9121#p56745/]kinopuk.ru/viewtopic.php?id=9121#p56745[/url]
[url=http://art-gymnastics.ru/users/54/]art-gymnastics.ru/users/54[/url]
[url=http://animalprotect.org/forum/index.php?topic=240258.0/]animalprotect.org/forum/index.php?topic=240258.0[/url]
[url=http://telegra.ph/postuplenie-v-vuzy-ameriki-07-24/]telegra.ph/postuplenie-v-vuzy-ameriki-07-24[/url]
[url=http://telegra.ph/kak-projti-medosmotr-dlya-postupleniya-v-vuz-07-24/]telegra.ph/kak-projti-medosmotr-dlya-postupleniya-v-vuz-07-24[/url]
Lariorzyw
August 11, 2024 @ 6:28 am
[u][b]Привет,друзья![/b][/u]
[b]Заказать диплом ВУЗа.[/b]
[url=http://alonegocio.net.br/author/dwaynemilne]alonegocio.net.br/author/dwaynemilne[/url]
[url=http://mediasocially.com/story2907890/kupit-diplom]mediasocially.com/story2907890/kupit-diplom[/url]
[url=http://thecatalystapproach.com/kupit-diplom-466076nvrii]thecatalystapproach.com/kupit-diplom-466076nvrii[/url]
[url=http://socialdummies.com/story2432145/kupit-diplom]socialdummies.com/story2432145/kupit-diplom[/url]
[url=http://freebookmarkpost.com/story17550549/kupit-diplom]freebookmarkpost.com/story17550549/kupit-diplom[/url]
[url=http://swflpastors.org/kupit-diplom-906977jcqza]swflpastors.org/kupit-diplom-906977jcqza[/url]
[url=http://seobookmarkpro.com/story17692451/kupit-diplom]seobookmarkpro.com/story17692451/kupit-diplom[/url]
[url=http://mnobookmarks.com/story17594436/kupit-diplom]mnobookmarks.com/story17594436/kupit-diplom[/url]
[url=http://kankerala.org/uncategorized/kupit-diplom-775180tfxbb]kankerala.org/uncategorized/kupit-diplom-775180tfxbb[/url]
[url=http://active-bookmarks.com/story17565495/kupit-diplom]active-bookmarks.com/story17565495/kupit-diplom[/url]
Danrqzt
August 11, 2024 @ 6:29 am
[u][b] Привет![/b][/u]
Мы можем предложить дипломы любой профессии.
[url=http://socialimarketing.com/story3074190/diplom]socialimarketing.com/story3074190/diplom[/url]
[url=http://socialwebconsult.com/story2971130/diplom]socialwebconsult.com/story2971130/diplom[/url]
[url=http://ecrinparis.com/diplom-713591crrez]ecrinparis.com/diplom-713591crrez[/url]
[url=http://bookmarkeasier.com/story17523546/diplom]bookmarkeasier.com/story17523546/diplom[/url]
[url=http://setbookmarks.com/story17707970/diplom]setbookmarks.com/story17707970/diplom[/url]
[url=http://us.alertbreakingnews.com/diplom-815820aiilq]us.alertbreakingnews.com/diplom-815820aiilq[/url]
[url=http://managerhotels.com/business-customer-service/diplom-920726ezyht]managerhotels.com/business-customer-service/diplom-920726ezyht[/url]
[url=http://medexmd.com/community/profile/wadestephensen]medexmd.com/community/profile/wadestephensen[/url]
[url=http://itdongnam.com/diplom-kupit-515763ba]itdongnam.com/diplom-kupit-515763ba[/url]
[url=http://brettswebsite.com/diplom-925162pekzd]brettswebsite.com/diplom-925162pekzd[/url]
[url=http://bookmark-group.com/story3103675/diplom]bookmark-group.com/story3103675/diplom[/url]
[url=http://escortexxx.ca/author/darwinm217]escortexxx.ca/author/darwinm217[/url]
[url=http://hariharparagovernmentiti.com/2024/07/26/diplom-901323souno]hariharparagovernmentiti.com/2024/07/26/diplom-901323souno[/url]
[url=http://socialbaskets.com/story3091727/diplom]socialbaskets.com/story3091727/diplom[/url]
[url=http://socialmphl.com/story19467905/diplom]socialmphl.com/story19467905/diplom[/url]
[url=http://socialbaskets.com/story3095980/diplom]socialbaskets.com/story3095980/diplom[/url]
[url=http://honda-sib.ru/diplom-574913ytmtg]honda-sib.ru/diplom-574913ytmtg[/url]
[url=http://thesocialintro.com/story3097097/diplom]thesocialintro.com/story3097097/diplom[/url]
[url=http://xatrivietnam.vn/question/diplom-kupit-378510su]xatrivietnam.vn/question/diplom-kupit-378510su[/url]
[url=http://iowa-bookmarks.com/story13284622/diplom]iowa-bookmarks.com/story13284622/diplom[/url]
Uazrjzw
August 11, 2024 @ 6:31 am
[u][b]Привет,друзья![/b][/u]
[b]Заказать диплом любого университета.[/b]
[url=http://reech.life/index.php/blog/4905/diplom-kupit-880584cw]reech.life/index.php/blog/4905/diplom-kupit-880584cw[/url]
[url=http://latestbusinessnew.com/diplom-kupit-216240im]latestbusinessnew.com/diplom-kupit-216240im[/url]
[url=http://offmarketbusinessforsale.com/diplom-kupit-41479ig]offmarketbusinessforsale.com/diplom-kupit-41479ig[/url]
[url=http://cometothecook.com/2024/07/29/diplom-kupit-396074io]cometothecook.com/2024/07/29/diplom-kupit-396074io[/url]
[url=http://sttimothysignal.org/groups/diplom-313360lgeug]sttimothysignal.org/groups/diplom-313360lgeug[/url]
[url=http://royalbookmarking.com/story17665051/diplom]royalbookmarking.com/story17665051/diplom[/url]
[url=http://iwanttobookmark.com/story17755971/diplom]iwanttobookmark.com/story17755971/diplom[/url]
[url=http://beautyisclassybrand.com/diplom-154189nwwmz]beautyisclassybrand.com/diplom-154189nwwmz[/url]
[url=http://yoursocialpeople.com/story2927544/diplom]yoursocialpeople.com/story2927544/diplom[/url]
[url=http://bookmarkleader.com/story17659268/diplom]bookmarkleader.com/story17659268/diplom[/url]
[url=http://investicos.com/uncategorized/diplom-470226bbjbr]investicos.com/uncategorized/diplom-470226bbjbr[/url]
[url=http://laviehub.com/blog/diplom-862372frswc]laviehub.com/blog/diplom-862372frswc[/url]
[url=http://bookmarkboom.com/story17654348/diplom]bookmarkboom.com/story17654348/diplom[/url]
[url=http://kankerala.org/uncategorized/diplom-210734duxig]kankerala.org/uncategorized/diplom-210734duxig[/url]
[url=http://bookmarkworm.com/story17626103/diplom]bookmarkworm.com/story17626103/diplom[/url]
[url=http://mirrorbookmarks.com/story17604695/diplom]mirrorbookmarks.com/story17604695/diplom[/url]
[url=http://digitalmagazine.xyz/diplom-456741tkfpu]digitalmagazine.xyz/diplom-456741tkfpu[/url]
[url=http://livebackpage.com/story2947669/diplom]livebackpage.com/story2947669/diplom[/url]
[url=http://free-bookmarking.com/story17724898/diplom]free-bookmarking.com/story17724898/diplom[/url]
[url=http://geilebookmarks.com/story17601674/diplom]geilebookmarks.com/story17601674/diplom[/url]
Lazrsqb
August 11, 2024 @ 7:34 am
[u][b] Здравствуйте![/b][/u]
[b]Мы изготавливаем дипломы[/b] любой профессии по разумным ценам.
[url=http://diploms-rushkas.ru/kupit-diplom-novosibirsk/]diploms-rushkas.ru/kupit-diplom-novosibirsk[/url]
Lazrugh
August 11, 2024 @ 8:04 am
[u][b] Привет![/b][/u]
[b]Купить диплом о высшем образовании.[/b]
[url=http://mandiplomik.ru/kupit-diplom-voronezh/]mandiplomik.ru/kupit-diplom-voronezh[/url]
Mazrcda
August 11, 2024 @ 8:07 am
[u][b] Привет![/b][/u]
[b]Как приобрести диплом о среднем образовании в Москве и других городах[/b]
[url=http://diplomrushkan.ru/]diplomrushkan.ru[/url]
Manrtyj
August 11, 2024 @ 8:34 am
[u][b] Здравствуйте![/b][/u]
Заказать документ о получении высшего образования
[url=http://socialstrategie.com/story3163176/kupit-diplom]socialstrategie.com/story3163176/kupit-diplom[/url]
[url=http://greatbookmarking.com/story17690900/kupit-diplom]greatbookmarking.com/story17690900/kupit-diplom[/url]
[url=http://thecatalystapproach.com/kupit-diplom-791039ibhfh]thecatalystapproach.com/kupit-diplom-791039ibhfh[/url]
[url=http://tayarut.bkerem.org.il/kupit-diplom-517860kfcqz]tayarut.bkerem.org.il/kupit-diplom-517860kfcqz[/url]
[url=http://annunciogratis.net/author/alysahocken]annunciogratis.net/author/alysahocken[/url]
Douglasidomb
August 11, 2024 @ 8:35 am
cheapest online pharmacy india: Indian pharmacy online – top 10 pharmacies in india
Charlescaulk
August 11, 2024 @ 8:57 am
http://edpillpharmacy.store/# п»їed pills online
ed pills for sale
Lazrzio
August 11, 2024 @ 10:14 am
[b]Заказать диплом университета.[/b]
[url=http://californiarpn2.listbb.ru/viewtopic.php?f=1&t=503/]californiarpn2.listbb.ru/viewtopic.php?f=1&t=503[/url]
[url=http://vaca-ps.org/blogs/171138/ упить-?иплом-без-ѕосредников-и-—уеты/]vaca-ps.org/blogs/171138/ упить-?иплом-без-ѕосредников-и-—уеты[/url]
[url=http://cn-stage-1.wmcloud.org/wiki/Настоящие_Дипломы:_Легально_и_Удобно/]cn-stage-1.wmcloud.org/wiki/Настоящие_Дипломы:_Легально_и_Удобно[/url]
[url=http://yourealtynews.ru/vash-nastoyashhiy-diplom-legalno-i-byistro/]yourealtynews.ru/vash-nastoyashhiy-diplom-legalno-i-byistro[/url]
[url=http://mkbox.ru/communication/forum/user/73890/]mkbox.ru/communication/forum/user/73890[/url]
Vazrrfw
August 11, 2024 @ 10:26 am
[u][b] Привет, друзья![/b][/u]
Купить документ университета можно в нашей компании в Москве. Мы предлагаем документы об окончании любых университетов РФ.
[url=http://onlineboxing.net/jforum/user/profile/295635.page/]onlineboxing.net/jforum/user/profile/295635.page[/url]
[url=http://chip-ecu.ru/threads/fados-vs-uce-ct220l-локатор-неисправностей-и-изогнутый-трассировщик-прибор-для-ремонта-эбу.422/#post-6890/]chip-ecu.ru/threads/fados-vs-uce-ct220l-локатор-неисправностей-и-изогнутый-трассировщик-прибор-для-ремонта-эбу.422/#post-6890[/url]
[url=http://ssd-astra.ru/forum/viewtopic.php?f=14&t=7014/]ssd-astra.ru/forum/viewtopic.php?f=14&t=7014[/url]
[url=http://forum.farmer.pl/profile/150711-ofaqokul/]forum.farmer.pl/profile/150711-ofaqokul[/url]
[url=http://mouur6.ru/forum/profile.php?action=show&member=3292/]mouur6.ru/forum/profile.php?action=show&member=3292[/url]
Lazrtrz
August 11, 2024 @ 11:21 am
[u][b] Привет, друзья![/b][/u]
[b]Мы изготавливаем дипломы[/b] любой профессии по приятным тарифам.
[url=http://diplomrushka.ru/kupit-diplom-instituta/]diplomrushka.ru/kupit-diplom-instituta[/url]
Williamraics
August 11, 2024 @ 11:29 am
[u][b] Добрый день![/b][/u]
Мы предлагаем дипломы.
[url=http://mirfinrealty.ru/kupit-diplom-bez-problem-legko-i-byistro/]mirfinrealty.ru/kupit-diplom-bez-problem-legko-i-byistro[/url]
[url=http://forum.resmihat.kz/viewtopic.php?f=10&t=2257463/]forum.resmihat.kz/viewtopic.php?f=10&t=2257463[/url]
[url=http://newspromworld.ru/nastoyashhie-diplomyi-dostupno-i-bezopasno/]newspromworld.ru/nastoyashhie-diplomyi-dostupno-i-bezopasno[/url]
[url=http://politikforum.ru/member.php?u=13385920/]politikforum.ru/member.php?u=13385920[/url]
[url=http://forums.worldsamba.org/showthread.php?tid=1046246/]forums.worldsamba.org/showthread.php?tid=1046246[/url]
Uazrupg
August 11, 2024 @ 11:36 am
[b]Где приобрести диплом специалиста?[/b]
[url=http://nodlik.com/diplom-798447yyxkd/]nodlik.com/diplom-798447yyxkd[/url]
[url=http://travelingvacation.com/2024/07/28/diplom-534740zuzzx/]travelingvacation.com/2024/07/28/diplom-534740zuzzx[/url]
[url=http://travelingvacation.com/2024/07/29/diplom-306821yuaxo/]travelingvacation.com/2024/07/29/diplom-306821yuaxo[/url]
[url=http://throbsocial.com/story19395787/diplom/]throbsocial.com/story19395787/diplom[/url]
[url=http://ztndz.com/story19866078/diplom/]ztndz.com/story19866078/diplom[/url]
Charlescaulk
August 11, 2024 @ 12:02 pm
http://mexicopharmacy.win/# п»їbest mexican online pharmacies
online ed meds
Xazrtqe
August 11, 2024 @ 12:28 pm
[u][b] Привет, друзья![/b][/u]
Приобрести документ университета.
[url=http://blogsfere.com/viewtopic.php?t=181808/]blogsfere.com/viewtopic.php?t=181808[/url]
[url=http://bence.net/read-blog/81/]bence.net/read-blog/81[/url]
[url=http://nosnitches.com/create-blog/]nosnitches.com/create-blog[/url]
[url=http://grp.7olimp.ru/memberlist.php?mode=viewprofile&u=68&sid=a47dd82411e288ee69f770ba462ec800/]grp.7olimp.ru/memberlist.php?mode=viewprofile&u=68&sid=a47dd82411e288ee69f770ba462ec800[/url]
[url=http://rakisochi.ru/users/20/]rakisochi.ru/users/20[/url]
Mazrldz
August 11, 2024 @ 1:44 pm
[u][b] Привет, друзья![/b][/u]
[b]Как купить аттестат 11 класса с официальным упрощенным обучением в Москве[/b]
[url=http://diplomrushkan.ru/kupit-diplom-voronezh/]diplomrushkan.ru/kupit-diplom-voronezh[/url]
Douglasidomb
August 11, 2024 @ 2:03 pm
mexican border pharmacies shipping to usa: Medicines Mexico – mexican border pharmacies shipping to usa
Yrefwva
August 11, 2024 @ 2:59 pm
[u][b] Здравствуйте![/b][/u]
[b]Купить диплом любого университета.[/b]
[url=http://wow-tour.ru/page/4/]wow-tour.ru/page/4[/url]
[url=http://neurostarcheats.com/index.php?/gallery/image/34-быстрый-выбор-онлайн-магазина-что-продает-дипломы/]neurostarcheats.com/index.php?/gallery/image/34-быстрый-выбор-онлайн-магазина-что-продает-дипломы[/url]
[url=http://balacixynu.trexgame.net/glavnye-zatraty-proizvoditela-diplomov-avtorskij-obzor/]balacixynu.trexgame.net/glavnye-zatraty-proizvoditela-diplomov-avtorskij-obzor[/url]
[url=http://taondinternational.rudraserver.com/blog/index.php?entryid=21229&nonjscomment=1&comment_itemid=21229&comment_context=23104&comment_component=blog&comment_area=format_blog/]taondinternational.rudraserver.com/blog/index.php?entryid=21229&nonjscomment=1&comment_itemid=21229&comment_context=23104&comment_component=blog&comment_area=format_blog[/url]
[url=http://sciens.ru/page/blwdr/]sciens.ru/page/blwdr[/url]
Charlescaulk
August 11, 2024 @ 3:35 pm
https://indiapharmacy.shop/# indian pharmacy
erectile dysfunction online
Douglasidomb
August 11, 2024 @ 4:35 pm
get ed meds today: Best ED pills non prescription – order ed pills
Jariorimn
August 11, 2024 @ 4:48 pm
[u][b] Привет![/b][/u]
Заказать диплом любого университета
[url=http://bookmarksparkle.com/story17772303/kupit-diplom]bookmarksparkle.com/story17772303/kupit-diplom[/url]
[url=http://axisgraf.com/2024/07/28/kupit-diplom-288021onkdh]axisgraf.com/2024/07/28/kupit-diplom-288021onkdh[/url]
[url=http://njkkot.org/?document_srl=1128174]njkkot.org/?document_srl=1128174[/url]
[url=http://thecatalystapproach.com/kupit-diplom-885478bdncb]thecatalystapproach.com/kupit-diplom-885478bdncb[/url]
[url=http://myeasybookmarks.com/story3040327/kupit-diplom]myeasybookmarks.com/story3040327/kupit-diplom[/url]
[url=http://networkbookmarks.com/story17658792/kupit-diplom]networkbookmarks.com/story17658792/kupit-diplom[/url]
[url=http://yxzbookmarks.com/story17631723/kupit-diplom]yxzbookmarks.com/story17631723/kupit-diplom[/url]
[url=http://agendabookmarks.com/story17578113/kupit-diplom]agendabookmarks.com/story17578113/kupit-diplom[/url]
[url=http://mylittlebookmark.com/story3143905/kupit-diplom]mylittlebookmark.com/story3143905/kupit-diplom[/url]
[url=http://zbookmarkhub.com/story17770693/kupit-diplom]zbookmarkhub.com/story17770693/kupit-diplom[/url]
Trefgjo
August 11, 2024 @ 5:15 pm
[u][b] Привет![/b][/u]
Можно ли купить аттестат о среднем образовании? Основные рекомендации
[url=http://mandiplomik.ru/kupit-diplom-voronezh/]mandiplomik.ru/kupit-diplom-voronezh[/url]
Поможем вам всегда!.
Sazrlya
August 11, 2024 @ 5:18 pm
[u][b] Привет![/b][/u]
Купить документ о получении высшего образования
[url=http://balacixynu.trexgame.net/glavnye-zatraty-proizvoditela-diplomov-avtorskij-obzor/]balacixynu.trexgame.net/glavnye-zatraty-proizvoditela-diplomov-avtorskij-obzor[/url]
[url=http://heimur.ru/index.php?/gallery/image/123-приобретаем-документы-в-известном-онлайн-магазине-по-выгодной-цене/]heimur.ru/index.php?/gallery/image/123-приобретаем-документы-в-известном-онлайн-магазине-по-выгодной-цене[/url]
[url=http://followgrown.com/read-blog/2645/]followgrown.com/read-blog/2645[/url]
[url=http://mighty-networks-foundation.mn.co/posts/61679287/]mighty-networks-foundation.mn.co/posts/61679287[/url]
[url=http://sonnick84.pointblog.net/Что-понадобится-для-создания-качественного-диплома-ВУЗа-69447016/]sonnick84.pointblog.net/Что-понадобится-для-создания-качественного-диплома-ВУЗа-69447016[/url]
MarcusOvara
August 11, 2024 @ 5:24 pm
бесплатное гадание бесплатные гадания онлайн
Yrefbws
August 11, 2024 @ 5:48 pm
[u][b] Здравствуйте![/b][/u]
[b]Приобрести диплом о высшем образовании.[/b]
[url=http://forum.drustvogil-galad.si/index.php?topic=14421.0/]forum.drustvogil-galad.si/index.php?topic=14421.0[/url]
[url=http://familylevel.com/blogs/167/Что-понадобится-из-техники-для-производства-качественного-диплома/]familylevel.com/blogs/167/Что-понадобится-из-техники-для-производства-качественного-диплома[/url]
[url=http://voiceof.com/купить-диплом-в-москве/]voiceof.com/купить-диплом-в-москве[/url]
[url=http://exoltech.us/blogs/207865/Диплом-любой-профессии-качественно-и-надежно/]exoltech.us/blogs/207865/Диплом-любой-профессии-качественно-и-надежно[/url]
[url=http://overfun.ru/index.php?/gallery/image/333-аттестаты-дипломы-и-свидетельства-в-лучшем-онлайн-магазине/]overfun.ru/index.php?/gallery/image/333-аттестаты-дипломы-и-свидетельства-в-лучшем-онлайн-магазине[/url]
Lariorhya
August 11, 2024 @ 5:59 pm
[u][b]Привет,друзья![/b][/u]
[b]Заказать диплом о высшем образовании.[/b]
[url=http://top10bookmark.com/story17554643/kupit-diplom]top10bookmark.com/story17554643/kupit-diplom[/url]
[url=http://bookmarkchamp.com/story17607322/kupit-diplom]bookmarkchamp.com/story17607322/kupit-diplom[/url]
[url=http://thekiwisocial.com/story3019872/kupit-diplom]thekiwisocial.com/story3019872/kupit-diplom[/url]
[url=http://managerhotels.com/business-small-business/kupit-diplom-276438adsru]managerhotels.com/business-small-business/kupit-diplom-276438adsru[/url]
[url=http://linkingbookmark.com/story17559224/kupit-diplom]linkingbookmark.com/story17559224/kupit-diplom[/url]
[url=http://hotbookmarkings.com/story17688847/kupit-diplom]hotbookmarkings.com/story17688847/kupit-diplom[/url]
[url=http://penteadistadesucesso.com/kupit-diplom-444101npxcp]penteadistadesucesso.com/kupit-diplom-444101npxcp[/url]
[url=http://bookmarkalexa.com/story3040061/kupit-diplom]bookmarkalexa.com/story3040061/kupit-diplom[/url]
[url=http://astradigital.co.uk/wiki/index.php?title=Kupit_Diplom_896957dgzYp]astradigital.co.uk/wiki/index.php?title=Kupit_Diplom_896957dgzYp[/url]
[url=http://siambookmark.com/story17688250/kupit-diplom]siambookmark.com/story17688250/kupit-diplom[/url]
Uazrsdn
August 11, 2024 @ 6:04 pm
[u][b]Здравствуйте![/b][/u]
[b]Приобрести диплом любого университета.[/b]
[url=http://bookmarkforce.com/story17740338/diplom]bookmarkforce.com/story17740338/diplom[/url]
[url=http://nowbookmarks.com/story17672601/diplom]nowbookmarks.com/story17672601/diplom[/url]
[url=http://techonpage.com/story2945960/diplom]techonpage.com/story2945960/diplom[/url]
[url=http://bookmarkstumble.com/story19204006/diplom]bookmarkstumble.com/story19204006/diplom[/url]
[url=http://ingbrick.com/diplom-808373xfqaz]ingbrick.com/diplom-808373xfqaz[/url]
[url=http://madbookmarks.com/story17641327/diplom]madbookmarks.com/story17641327/diplom[/url]
[url=http://wise-social.com/story3026085/diplom]wise-social.com/story3026085/diplom[/url]
[url=http://sttimothysignal.org/groups/diplom-kupit-364792gf]sttimothysignal.org/groups/diplom-kupit-364792gf[/url]
[url=http://welnesbiolabs.com/diplom-672096uuezn]welnesbiolabs.com/diplom-672096uuezn[/url]
[url=http://hariharparagovernmentiti.com/2024/07/27/diplom-139255xmzfy]hariharparagovernmentiti.com/2024/07/27/diplom-139255xmzfy[/url]
[url=http://travelingvacation.com/2024/07/29/diplom-729886wqdfq]travelingvacation.com/2024/07/29/diplom-729886wqdfq[/url]
[url=http://topsocialplan.com/story3042327/diplom]topsocialplan.com/story3042327/diplom[/url]
[url=http://elearning.maniatech-academy.co.uk/blog/index.php?entryid=279348]elearning.maniatech-academy.co.uk/blog/index.php?entryid=279348[/url]
[url=http://itdongnam.com/diplom-199784kvyrl]itdongnam.com/diplom-199784kvyrl[/url]
[url=http://tealbookmarks.com/story17651798/diplom]tealbookmarks.com/story17651798/diplom[/url]
[url=http://pageoftoday.com/story2977138/diplom]pageoftoday.com/story2977138/diplom[/url]
[url=http://laviehub.com/blog/diplom-46221alxou]laviehub.com/blog/diplom-46221alxou[/url]
[url=http://lampcanvas.com/diplom-126582gzqji]lampcanvas.com/diplom-126582gzqji[/url]
[url=http://lyfesaverscpr.com/diplom-52200ulpkw]lyfesaverscpr.com/diplom-52200ulpkw[/url]
[url=http://investicos.com/uncategorized/diplom-377456ycmsg]investicos.com/uncategorized/diplom-377456ycmsg[/url]
Danrgab
August 11, 2024 @ 6:11 pm
[u][b] Привет, друзья![/b][/u]
Мы изготавливаем дипломы психологов, юристов, экономистов и прочих профессий.
[url=http://bookmarkingace.com/story17641149/diplom]bookmarkingace.com/story17641149/diplom[/url]
[url=http://vibesvietnam.com/diplom-kupit-503393uj]vibesvietnam.com/diplom-kupit-503393uj[/url]
[url=http://digitalmagazine.xyz/diplom-476962eswnn]digitalmagazine.xyz/diplom-476962eswnn[/url]
[url=http://ntep2008.com/index.php?name=webboard&file=read&id=18546]ntep2008.com/index.php?name=webboard&file=read&id=18546[/url]
[url=http://cometothecook.com/2024/07/28/diplom-97044ygryc]cometothecook.com/2024/07/28/diplom-97044ygryc[/url]
[url=http://pencis.com/diplom-915113lqtnu]pencis.com/diplom-915113lqtnu[/url]
[url=http://ecrinparis.com/diplom-178199vfabu]ecrinparis.com/diplom-178199vfabu[/url]
[url=http://smiletraveling.com/2024/07/28/diplom-496755abfcr]smiletraveling.com/2024/07/28/diplom-496755abfcr[/url]
[url=http://travelingvacation.com/2024/07/28/diplom-70133fadgj]travelingvacation.com/2024/07/28/diplom-70133fadgj[/url]
[url=http://wisesocialsmedia.com/story2952469/diplom]wisesocialsmedia.com/story2952469/diplom[/url]
[url=http://xatrivietnam.vn/question/diplom-943025vapga]xatrivietnam.vn/question/diplom-943025vapga[/url]
[url=http://offmarketbusinessforsale.com/diplom-865787hydpu]offmarketbusinessforsale.com/diplom-865787hydpu[/url]
[url=http://gen8ai.com/groups/diplom-kupit-388451gl/members/all-members]gen8ai.com/groups/diplom-kupit-388451gl/members/all-members[/url]
[url=http://battat-advisors.com/?p=19937]battat-advisors.com/?p=19937[/url]
[url=http://socialwebleads.com/story2993535/diplom]socialwebleads.com/story2993535/diplom[/url]
[url=http://parathajoint.com/2024/07/29/diplom-kupit-20083lv]parathajoint.com/2024/07/29/diplom-kupit-20083lv[/url]
[url=http://cometothecook.com/2024/07/28/diplom-kupit-682747xi]cometothecook.com/2024/07/28/diplom-kupit-682747xi[/url]
[url=http://youlangue.lu/blog/index.php?entryid=89198]youlangue.lu/blog/index.php?entryid=89198[/url]
[url=http://tuzari.shop/?option=com_k2&view=itemlist&task=user&id=1420]tuzari.shop/?option=com_k2&view=itemlist&task=user&id=1420[/url]
[url=http://alonegocio.net.br/author/cezdarwin61]alonegocio.net.br/author/cezdarwin61[/url]
Charlescaulk
August 11, 2024 @ 6:37 pm
http://mexicopharmacy.win/# mexican rx online
order ed pills
Lazrcrn
August 11, 2024 @ 7:19 pm
[u][b] Здравствуйте![/b][/u]
[b]Купить диплом университета.[/b]
[url=http://mans-diplomy.ru/diplom-tehnikuma/]mans-diplomy.ru/diplom-tehnikuma[/url]
Trefzte
August 11, 2024 @ 7:55 pm
[u][b] Здравствуйте![/b][/u]
Всё, что нужно знать о покупке аттестата о среднем образовании
[url=http://mandiplomikx.ru/kupit-svidetelstva/]mandiplomikx.ru/kupit-svidetelstva[/url]
Всегда вам поможем!.
Dnrtrzt
August 11, 2024 @ 9:03 pm
[b]Приобрести документ[/b] университета вы сможете у нас в Москве.
[url=http://lampcanvas.com/diplom-438624wzhwt/]lampcanvas.com/diplom-438624wzhwt[/url]
[url=http://bookmarkpagerank.com/story17666532/diplom/]bookmarkpagerank.com/story17666532/diplom[/url]
[url=http://selfhackathon.com/diplom-261592fatls/]selfhackathon.com/diplom-261592fatls[/url]
[url=http://njkkot.org/?document_srl=1143307/]njkkot.org/?document_srl=1143307[/url]
[url=http://ypchina.org/diplom-kupit-549100un/]ypchina.org/diplom-kupit-549100un[/url]
Vazrqfi
August 11, 2024 @ 9:17 pm
[u][b] Привет![/b][/u]
Купить документ института вы можете в нашей компании в столице. Мы предлагаем документы об окончании любых университетов РФ.
[url=http://bumblebeeco.ru/index.php?subaction=userinfo&user=ynazuwytu/]bumblebeeco.ru/index.php?subaction=userinfo&user=ynazuwytu[/url]
[url=http://koreamuseum.ru/index.php?subaction=userinfo&user=omiwiqaw/]koreamuseum.ru/index.php?subaction=userinfo&user=omiwiqaw[/url]
[url=http://ka4nem.ru/index.php?subaction=userinfo&user=ovocy/]ka4nem.ru/index.php?subaction=userinfo&user=ovocy[/url]
[url=http://mkala-koncert.ru/users/ytizih/]mkala-koncert.ru/users/ytizih[/url]
[url=http://receptar.ru/forum/profile.php?action=show&member=1529/]receptar.ru/forum/profile.php?action=show&member=1529[/url]
Lazriti
August 11, 2024 @ 9:38 pm
[b]Заказать диплом любого университета.[/b]
[url=http://bestcoolfun.ru/vyisshee-obrazovanie-dlya-vas-legalno-i-byistro/]bestcoolfun.ru/vyisshee-obrazovanie-dlya-vas-legalno-i-byistro[/url]
[url=http://elfae.ruhelp.com/viewtopic.php?id=17033#p39245/]elfae.ruhelp.com/viewtopic.php?id=17033#p39245[/url]
[url=http://grot.getbb.ru/viewtopic.php?f=6&t=1051/]grot.getbb.ru/viewtopic.php?f=6&t=1051[/url]
[url=http://lighttur.ru/ofitsialnyie-diplomyi-podlinnost-i-nadezhnost/]lighttur.ru/ofitsialnyie-diplomyi-podlinnost-i-nadezhnost[/url]
[url=http://forex.g-talk.ru/viewtopic.php?f=43&t=7087/]forex.g-talk.ru/viewtopic.php?f=43&t=7087[/url]
Douglasidomb
August 11, 2024 @ 9:59 pm
buy ed pills online: ed pills online – ed meds by mail
Williamraics
August 11, 2024 @ 10:53 pm
[u][b] Привет, друзья![/b][/u]
Мы можем предложить дипломы.
[url=http://derivsocial.org/read-blog/2540/]derivsocial.org/read-blog/2540[/url]
[url=http://mdr7.ru/viewtopic.php?f=6&t=8880/]mdr7.ru/viewtopic.php?f=6&t=8880[/url]
[url=http://connectme.live/blogs/957/Официальные-Дипломы-Быстро-и-Удобно/]connectme.live/blogs/957/Официальные-Дипломы-Быстро-и-Удобно[/url]
[url=http://dogovor.top/wiki/Купить_Диплом_без_Посредников_и_Суеты/]dogovor.top/wiki/Купить_Диплом_без_Посредников_и_Суеты[/url]
[url=http://rkiyosaki.ru/discussion/9689/vysshee-obrazovanie-za-den-pokupka-diplomov/]rkiyosaki.ru/discussion/9689/vysshee-obrazovanie-za-den-pokupka-diplomov[/url]
Leonardginge
August 12, 2024 @ 12:10 am
http://mexicopharmacy.win/# medication from mexico pharmacy
Dnrtnsb
August 12, 2024 @ 12:35 am
[b]Купить документ[/b] ВУЗа можно у нас в Москве.
[url=http://managerhotels.com/business-customer-service/diplom-612216pjbqs/]managerhotels.com/business-customer-service/diplom-612216pjbqs[/url]
[url=http://vibesvietnam.com/diplom-kupit-57590zu/]vibesvietnam.com/diplom-kupit-57590zu[/url]
[url=http://bookmarkwuzz.com/story17646033/diplom/]bookmarkwuzz.com/story17646033/diplom[/url]
[url=http://bruthindustan.com/diplom-188190cbcqc/]bruthindustan.com/diplom-188190cbcqc[/url]
[url=http://pageoftoday.com/story2976976/diplom/]pageoftoday.com/story2976976/diplom[/url]
Douglasidomb
August 12, 2024 @ 12:35 am
cheapest online pharmacy india: Indian pharmacy online – online pharmacy india
Mazrwlm
August 12, 2024 @ 12:42 am
[u][b] Добрый день![/b][/u]
[b]Полезные советы по покупке диплома о высшем образовании без риска[/b]
[url=http://diploms-rushkas.ru/kupit-diplom-novosibirsk/]diploms-rushkas.ru/kupit-diplom-novosibirsk[/url]
Cazrcxa
August 12, 2024 @ 1:12 am
[u][b] Привет![/b][/u]
Мы можем предложить документы ВУЗов
[url=http://av.flyboard.ru/viewtopic.php?f=9&t=1452/]av.flyboard.ru/viewtopic.php?f=9&t=1452[/url]
[url=http://bdfeu.ukraine7.com/t20080-topic#36542/]bdfeu.ukraine7.com/t20080-topic#36542[/url]
[url=http://forum.kam.su/member.php?u=5734/]forum.kam.su/member.php?u=5734[/url]
[url=http://socialnetwork.cloudyzx.com/read-blog/14817/]socialnetwork.cloudyzx.com/read-blog/14817[/url]
[url=http://kome.maxbb.ru/viewtopic.php?f=22&t=2274/]kome.maxbb.ru/viewtopic.php?f=22&t=2274[/url]
Lazrxcm
August 12, 2024 @ 1:51 am
[u][b] Привет![/b][/u]
[b]Мы готовы предложить дипломы[/b] любой профессии по доступным тарифам.
[url=http://rushkas-diplomas.ru/kupit-diplom-s-reestrom/]rushkas-diplomas.ru/kupit-diplom-s-reestrom[/url]
Lazrqai
August 12, 2024 @ 5:37 am
[u][b] Добрый день![/b][/u]
[b]Мы предлагаем дипломы[/b] любой профессии по приятным тарифам.
[url=http://rushkas-diplomasxx.ru/kupit-diplom-vracha/]rushkas-diplomasxx.ru/kupit-diplom-vracha[/url]
Diplomi_tcSi
August 12, 2024 @ 6:08 am
Здравствуйте!
Купить документ о получении высшего образования вы можете в нашей компании.
[url=http://rushkas-diplomxx.ru/]rushkas-diplomxx.ru[/url]
Douglasidomb
August 12, 2024 @ 6:17 am
erectile dysfunction online prescription: cheap ed pills online – ed drugs online
Mazrqiv
August 12, 2024 @ 6:45 am
[u][b] Привет![/b][/u]
Всё, что нужно знать о покупке аттестата о среднем образовании без рисков
[url=http://arusak-diploms-srednee.ru/kupit-diplom-sssr/]arusak-diploms-srednee.ru/kupit-diplom-sssr[/url]
Sazruuu
August 12, 2024 @ 7:17 am
[u][b] Здравствуйте![/b][/u]
Заказать документ о получении высшего образования
[url=http://chayka.ixbb.ru/viewtopic.php?id=155#p165/]chayka.ixbb.ru/viewtopic.php?id=155#p165[/url]
[url=http://bs.listbb.ru/viewtopic.php?f=3&t=652/]bs.listbb.ru/viewtopic.php?f=3&t=652[/url]
[url=http://sport-faq.ru/zakazhite-diplom-v-kratchayshie-sroki/]sport-faq.ru/zakazhite-diplom-v-kratchayshie-sroki[/url]
[url=http://4atex.ru/blogs/2533/Легальное-получение-диплома-в-короткие-сроки/]4atex.ru/blogs/2533/Легальное-получение-диплома-в-короткие-сроки[/url]
[url=http://telegra.ph/polozhenie-o-postuplenii-v-vuz-07-24/]telegra.ph/polozhenie-o-postuplenii-v-vuz-07-24[/url]
Yrefszg
August 12, 2024 @ 7:26 am
[u][b] Привет, друзья![/b][/u]
[b]Заказать диплом любого ВУЗа.[/b]
[url=http://?????.??/rooftile/gnuboard/bbs/board.php?bo_table=bbs03&wr_id=19453/]?????.??/rooftile/gnuboard/bbs/board.php?bo_table=bbs03&wr_id=19453[/url]
[url=http://voiceof.com/купить-диплом-в-москве/]voiceof.com/купить-диплом-в-москве[/url]
[url=http://worldhealthstock.com/купить-диплом-в-волжском/]worldhealthstock.com/купить-диплом-в-волжском[/url]
[url=http://hellsms.com/cb-profile/pluginclass/cbblogs?action=blogs&func=show&id=6637/]hellsms.com/cb-profile/pluginclass/cbblogs?action=blogs&func=show&id=6637[/url]
[url=http://mevajafeke.huicopper.com/cto-neobhodimo-iz-oborudovania-dla-sozdania-diploma/]mevajafeke.huicopper.com/cto-neobhodimo-iz-oborudovania-dla-sozdania-diploma[/url]
Trefvjq
August 12, 2024 @ 8:29 am
[u][b] Привет, друзья![/b][/u]
Официальное получение диплома техникума с упрощенным обучением в Москве
[url=http://mandiplomik.ru/kupit-diplom-v-ufe/]mandiplomik.ru/kupit-diplom-v-ufe[/url]
Рады оказать помощь!.
Douglasidomb
August 12, 2024 @ 9:02 am
ed meds by mail: Best ED meds online – erectile dysfunction drugs online
Yrefsag
August 12, 2024 @ 10:26 am
[u][b] Здравствуйте![/b][/u]
[b]Заказать диплом о высшем образовании.[/b]
[url=http://elearning.ims-schulungen.de/blog/index.php?entryid=64943/]elearning.ims-schulungen.de/blog/index.php?entryid=64943[/url]
[url=http://dchatmogul.com/blogs/1414/Ключевые-затраты-любого-производителя-дипломов-обзор/]dchatmogul.com/blogs/1414/Ключевые-затраты-любого-производителя-дипломов-обзор[/url]
[url=http://remotebillpay.com/купить-диплом/]remotebillpay.com/купить-диплом[/url]
[url=http://justforkidsclinic.com/ricar/index.php?page=2122/]justforkidsclinic.com/ricar/index.php?page=2122[/url]
[url=http://operazionispeciali.it/cb-profile/pluginclass/cbblogs?action=blogs&func=show&id=257/]operazionispeciali.it/cb-profile/pluginclass/cbblogs?action=blogs&func=show&id=257[/url]
Sazrptf
August 12, 2024 @ 10:55 am
[u][b] Добрый день![/b][/u]
Мы готовы предложить дипломы любой профессии.
[url=http://thenolugroup.co.za/groups/kupit-diplom-489380yeoyt/]thenolugroup.co.za/groups/kupit-diplom-489380yeoyt[/url]
[url=http://zbookmarkhub.com/story17770058/kupit-diplom/]zbookmarkhub.com/story17770058/kupit-diplom[/url]
[url=http://thegreatbookmark.com/story17715150/kupit-diplom/]thegreatbookmark.com/story17715150/kupit-diplom[/url]
[url=http://us.alertbreakingnews.com/kupit-diplom-39590mqklb/]us.alertbreakingnews.com/kupit-diplom-39590mqklb[/url]
[url=http://socialbookmarkgs.com/story17708271/kupit-diplom/]socialbookmarkgs.com/story17708271/kupit-diplom[/url]
Mazrxge
August 12, 2024 @ 10:56 am
[b]Купить документ института.[/b]
[url=http://bookmarklogin.com/story17755008/kupit-diplom/]bookmarklogin.com/story17755008/kupit-diplom[/url]
[url=http://listingbookmarks.com/story17712245/kupit-diplom/]listingbookmarks.com/story17712245/kupit-diplom[/url]
[url=http://maitemach.com/kupit-diplom-944445tmtxu/]maitemach.com/kupit-diplom-944445tmtxu[/url]
[url=http://sttimothysignal.org/groups/kupit-diplom-88169buflm/]sttimothysignal.org/groups/kupit-diplom-88169buflm[/url]
[url=http://apollobookmarks.com/story17598376/kupit-diplom/]apollobookmarks.com/story17598376/kupit-diplom[/url]
Treflok
August 12, 2024 @ 11:28 am
[u][b] Здравствуйте![/b][/u]
Как быстро получить диплом магистра? Легальные способы
[url=http://mans-diplomxx.ru/kupit-diplom-novosibirsk/]mans-diplomxx.ru/kupit-diplom-novosibirsk[/url]
Будем рады вам помочь!.
Sazrwvf
August 12, 2024 @ 11:39 am
[u][b] Добрый день![/b][/u]
Приобрести документ ВУЗа
[url=http://itstagram.ru/read-blog/937/]itstagram.ru/read-blog/937[/url]
[url=http://magnat-matras.ru/forum/user/50884/]magnat-matras.ru/forum/user/50884[/url]
[url=http://coup.forum2x2.ru/t3821-topic#10077/]coup.forum2x2.ru/t3821-topic#10077[/url]
[url=http://silich.ru/forum/member.php?u=4810/]silich.ru/forum/member.php?u=4810[/url]
[url=http://insta.tel/read-blog/49859/]insta.tel/read-blog/49859[/url]
Leonardginge
August 12, 2024 @ 2:05 pm
https://indiapharmacy.shop/# п»їlegitimate online pharmacies india
Douglasidomb
August 12, 2024 @ 2:35 pm
pharmacies in mexico that ship to usa: Purple pharmacy online ordering – buying prescription drugs in mexico online
Douglasidomb
August 12, 2024 @ 5:02 pm
mexican drugstore online: Best pharmacy in Mexico – п»їbest mexican online pharmacies
LewisImise
August 12, 2024 @ 5:54 pm
Звуковое оборудование https://zvukovoe-oborudovanie12.ru с доставкой. Большой ассортимент звукового оборудования, доступные цены, доставка по РФ.
Sazrebf
August 12, 2024 @ 6:09 pm
[u][b] Привет![/b][/u]
Купить документ университета
[url=http://o91707v5.beget.tech/2024/07/18/vash-diplom-vash-uspeh/]o91707v5.beget.tech/2024/07/18/vash-diplom-vash-uspeh[/url]
[url=http://fsmi.wiki/index.php?title=Дипломы_для_карьерного_роста_без_лишних_усилий/]fsmi.wiki/index.php?title=Дипломы_для_карьерного_роста_без_лишних_усилий[/url]
[url=http://astroroleplay.getbb.ru/viewtopic.php?f=6&t=508/]astroroleplay.getbb.ru/viewtopic.php?f=6&t=508[/url]
[url=http://telegra.ph/individualnye-dostizheniya-pri-postuplenii-v-vuz-07-24/]telegra.ph/individualnye-dostizheniya-pri-postuplenii-v-vuz-07-24[/url]
[url=http://agrofly.ch/Быстрые_и_надежные_дипломы/]agrofly.ch/Быстрые_и_надежные_дипломы[/url]
Lazrcin
August 12, 2024 @ 8:24 pm
[u][b] Привет, друзья![/b][/u]
[b]Мы предлагаем дипломы[/b] любых профессий по приятным тарифам.
[url=http://telegra.ph/kak-prohodit-obuchenie-v-medicinskom-vuze-08-02/]telegra.ph/kak-prohodit-obuchenie-v-medicinskom-vuze-08-02[/url]
Sazrjgb
August 12, 2024 @ 9:11 pm
[u][b] Здравствуйте![/b][/u]
Мы готовы предложить дипломы психологов, юристов, экономистов и других профессий.
[url=http://tetrabookmarks.com/story17686078/kupit-diplom/]tetrabookmarks.com/story17686078/kupit-diplom[/url]
[url=http://mx.anuncioseroticos.co/author/coycollazo/]mx.anuncioseroticos.co/author/coycollazo[/url]
[url=http://hubwebsites.com/story18919061/kupit-diplom/]hubwebsites.com/story18919061/kupit-diplom[/url]
[url=http://yxzbookmarks.com/story17636091/kupit-diplom/]yxzbookmarks.com/story17636091/kupit-diplom[/url]
[url=http://bookmark-media.com/story17729813/kupit-diplom/]bookmark-media.com/story17729813/kupit-diplom[/url]
Mazrbfl
August 12, 2024 @ 9:13 pm
[b]Приобрести документ ВУЗа.[/b]
[url=http://bookmarkfly.com/story17692117/kupit-diplom/]bookmarkfly.com/story17692117/kupit-diplom[/url]
[url=http://taondinternational.rudraserver.com/blog/index.php?entryid=133047/]taondinternational.rudraserver.com/blog/index.php?entryid=133047[/url]
[url=http://laviehub.com/blog/kupit-diplom-59630xhxdn/]laviehub.com/blog/kupit-diplom-59630xhxdn[/url]
[url=http://health-lists.com/story18241075/kupit-diplom/]health-lists.com/story18241075/kupit-diplom[/url]
[url=http://kbkrealtors.com/kupit-diplom-876737hbwun/]kbkrealtors.com/kupit-diplom-876737hbwun[/url]
Diplomi_zmSi
August 12, 2024 @ 9:39 pm
Привет!
Приобрести документ института вы сможете в нашей компании.
[url=http://allrealtor.ru/threads/3407/]allrealtor.ru/threads/3407[/url]
Douglasidomb
August 12, 2024 @ 10:00 pm
mexican border pharmacies shipping to usa: Best online Mexican pharmacy – buying from online mexican pharmacy
Sazrhtx
August 12, 2024 @ 10:49 pm
[u][b] Привет![/b][/u]
Заказать документ ВУЗа
[url=http://heyhey.icu/blogs/18094/Получите-диплом-за-пару-дней/]heyhey.icu/blogs/18094/Получите-диплом-за-пару-дней[/url]
[url=http://u90517ol.beget.tech/2024/06/18/karera-bez-dolgogo-obucheniya/]u90517ol.beget.tech/2024/06/18/karera-bez-dolgogo-obucheniya[/url]
[url=http://coastalplainplants.org/wiki/index.php/Быстрый_и_надежный_способ_получить_диплом/]coastalplainplants.org/wiki/index.php/Быстрый_и_надежный_способ_получить_диплом[/url]
[url=http://nnars.ru/forum/suggestion-box/1230-ваш-диплом-Ч-ваш-успех#74616/]nnars.ru/forum/suggestion-box/1230-ваш-диплом-Ч-ваш-успех#74616[/url]
[url=http://satopttorg.ru/forum/user/3683/]satopttorg.ru/forum/user/3683[/url]
Lazrrxx
August 12, 2024 @ 11:31 pm
[u][b] Добрый день![/b][/u]
[b]Мы изготавливаем дипломы[/b] любых профессий по приятным тарифам.
[url=http://telegra.ph/sokrashchennaya-forma-obucheniya-v-vuze-posle-kolledzha-08-02/]telegra.ph/sokrashchennaya-forma-obucheniya-v-vuze-posle-kolledzha-08-02[/url]
Trefjmv
August 12, 2024 @ 11:47 pm
[u][b] Привет, друзья![/b][/u]
Как купить аттестат 11 класса с официальным упрощенным обучением в Москве
[url=http://svetlana5ege.blogspot.com/2014/12/7_16/]svetlana5ege.blogspot.com/2014/12/7_16[/url]
Всегда вам поможем!.
Iariorqzx
August 13, 2024 @ 12:05 am
[u][b] Добрый день![/b][/u]
Заказать документ ВУЗа вы можете в нашем сервисе. Мы предлагаем документы об окончании любых университетов России. Вы сможете получить необходимый диплом по любой специальности, включая документы Советского Союза. Гарантируем, что при проверке документов работодателем, никаких подозрений не возникнет.
[url=http://pdlspd.listbb.ru/viewtopic.php?f=3&t=492/]pdlspd.listbb.ru/viewtopic.php?f=3&t=492[/url]
[url=http://u2c.tv/member.php?u=30984/]u2c.tv/member.php?u=30984[/url]
[url=http://mymink.5bb.ru/viewtopic.php?id=8977#p455130/]mymink.5bb.ru/viewtopic.php?id=8977#p455130[/url]
[url=http://1776reloaded.us/blogs/173/Легальное-получение-диплома-в-короткие-сроки/]1776reloaded.us/blogs/173/Легальное-получение-диплома-в-короткие-сроки[/url]
[url=http://forexrassia.ru/diplomyi-na-zakaz-bez-lishnih-hlopot/]forexrassia.ru/diplomyi-na-zakaz-bez-lishnih-hlopot[/url]
Douglasidomb
August 13, 2024 @ 12:22 am
mexican drugstore online: Mexico pharmacy online – mexico drug stores pharmacies
Yrefrsb
August 13, 2024 @ 12:45 am
[u][b] Здравствуйте![/b][/u]
[b]Приобрести диплом любого университета.[/b]
[url=http://medlennayazvezda.blogspot.com/2014/05/wonder-cream-purity-herbs/]medlennayazvezda.blogspot.com/2014/05/wonder-cream-purity-herbs[/url]
Diplomi_jxSi
August 13, 2024 @ 1:24 am
Привет!
Приобрести документ университета вы сможете у нас в столице.
[url=http://geolan-ksl.ru/forum/user/87403/]geolan-ksl.ru/forum/user/87403[/url]
Trefyjw
August 13, 2024 @ 2:37 am
[u][b] Привет, друзья![/b][/u]
Как безопасно купить диплом колледжа или ПТУ в России, что важно знать
[url=http://allrealtor.ru/threads/3407/]allrealtor.ru/threads/3407[/url]
Окажем помощь!.
Yrefeax
August 13, 2024 @ 3:59 am
[u][b] Привет![/b][/u]
[b]Приобрести диплом университета.[/b]
[url=http://arskland.ru/people/user/16063/]arskland.ru/people/user/16063[/url]
Manrrve
August 13, 2024 @ 4:55 am
[u][b] Добрый день![/b][/u]
Купить документ института
[url=http://astradigital.co.uk/wiki/index.php?title=Kupit_Diplom_283025qrzIK]astradigital.co.uk/wiki/index.php?title=Kupit_Diplom_283025qrzIK[/url]
[url=http://bookmark-rss.com/story17540540/kupit-diplom]bookmark-rss.com/story17540540/kupit-diplom[/url]
[url=http://offmarketbusinessforsale.com/kupit-diplom-614679caqwb]offmarketbusinessforsale.com/kupit-diplom-614679caqwb[/url]
[url=http://wise-social.com/story3030930/kupit-diplom]wise-social.com/story3030930/kupit-diplom[/url]
[url=http://muhammadcenter.com/kupit-diplom-267959neaze]muhammadcenter.com/kupit-diplom-267959neaze[/url]
Douglasidomb
August 13, 2024 @ 5:23 am
buy prescription drugs from india: Online pharmacy – Online medicine home delivery
Dnrtgcg
August 13, 2024 @ 7:08 am
[b]Купить документ[/b] института вы имеете возможность в нашей компании в Москве.
[url=http://kingbookmark.com/story17729679/diplom/]kingbookmark.com/story17729679/diplom[/url]
[url=http://alkhazana.net/2024/07/28/diplom-248631iesen/]alkhazana.net/2024/07/28/diplom-248631iesen[/url]
[url=http://thesocialintro.com/story3091638/diplom/]thesocialintro.com/story3091638/diplom[/url]
[url=http://managerhotels.com/business-customer-service/diplom-808112ckzrc/]managerhotels.com/business-customer-service/diplom-808112ckzrc[/url]
[url=http://shikhadabas.com/2024/07/28/diplom-kupit-242692el/]shikhadabas.com/2024/07/28/diplom-kupit-242692el[/url]
Oariorent
August 13, 2024 @ 7:17 am
[u][b] Привет![/b][/u]
Заказать диплом университета
[b]Мы предлагаем[/b] быстро купить диплом, который выполнен на оригинальном бланке и заверен мокрыми печатями, водяными знаками, подписями. Диплом способен пройти любые проверки, даже с использованием специфических приборов. Достигайте цели быстро и просто с нашей компанией.
[b]Где заказать диплом специалиста?[/b]
[url=http://malispa.ru/users/122/]malispa.ru/users/122[/url]
[url=http://mirnuy.forumieren.de/t8244-topic#13519/]mirnuy.forumieren.de/t8244-topic#13519[/url]
[url=http://bence.net/read-blog/99/]bence.net/read-blog/99[/url]
[url=http://kondrateff.5bb.ru/viewtopic.php?id=6770#p12688/]kondrateff.5bb.ru/viewtopic.php?id=6770#p12688[/url]
[url=http://antennastar.com/blogs/483/Что-необходимо-из-техники-для-создания-документов/]antennastar.com/blogs/483/Что-необходимо-из-техники-для-создания-документов[/url]
Douglasidomb
August 13, 2024 @ 7:59 am
mail order pharmacy india: Online medicine home delivery – india online pharmacy
Iariortiq
August 13, 2024 @ 10:35 am
[u][b] Добрый день![/b][/u]
Заказать документ университета вы имеете возможность в нашей компании в Москве. Мы предлагаем документы об окончании любых университетов РФ. Вы сможете получить диплом по любой специальности, любого года выпуска, включая документы старого образца. Даем 100% гарантию, что в случае проверки документа работодателями, каких-либо подозрений не возникнет.
[url=http://politikforum.ru/member.php?u=13385920/]politikforum.ru/member.php?u=13385920[/url]
[url=http://ic-info.ru/forum/user/166684/]ic-info.ru/forum/user/166684[/url]
[url=http://ya.forum.cool/viewtopic.php?id=5893#p17555/]ya.forum.cool/viewtopic.php?id=5893#p17555[/url]
[url=http://telegra.ph/lgoty-mnogodetnym-pri-postuplenii-v-vuz-07-24/]telegra.ph/lgoty-mnogodetnym-pri-postuplenii-v-vuz-07-24[/url]
[url=http://net4women.ru/blogs/4032/ваш-ключ-к-успеху-диплом-о-высшем-образовании/]net4women.ru/blogs/4032/ваш-ключ-к-успеху-диплом-о-высшем-образовании[/url]
Sazrluf
August 13, 2024 @ 10:59 am
[u][b] Здравствуйте![/b][/u]
Приобрести документ института
[url=http://twanty2.com/read-blog/181/]twanty2.com/read-blog/181[/url]
[url=http://sonnick84.blogdun.com/28750502/Дипломы-справки-а-так-же-свидетельства-низкие-цены/]sonnick84.blogdun.com/28750502/Дипломы-справки-а-так-же-свидетельства-низкие-цены[/url]
[url=http://mizunosoccershoesfans.com/read-blog/3427/]mizunosoccershoesfans.com/read-blog/3427[/url]
[url=http://belsprosshina.by/company/personal/user/127/forum/message/1566/5812/]belsprosshina.by/company/personal/user/127/forum/message/1566/5812[/url]
[url=http://sonnick84.qowap.com/87895139/Советы-специалиста-что-помогут-легко-купить-диплом/]sonnick84.qowap.com/87895139/Советы-специалиста-что-помогут-легко-купить-диплом[/url]
Dnrtpom
August 13, 2024 @ 11:09 am
[b]Заказать документ[/b] института вы можете у нас.
[url=http://welnesbiolabs.com/diplom-kupit-600863iv/]welnesbiolabs.com/diplom-kupit-600863iv[/url]
[url=http://freebookmarkpost.com/story17554248/diplom/]freebookmarkpost.com/story17554248/diplom[/url]
[url=http://bookmarkblast.com/story17684083/diplom/]bookmarkblast.com/story17684083/diplom[/url]
[url=http://youcanautism.com/diplom-355652ruhiw/]youcanautism.com/diplom-355652ruhiw[/url]
[url=http://axisgraf.com/2024/07/27/diplom-147019mfzaw/]axisgraf.com/2024/07/27/diplom-147019mfzaw[/url]
pech_mtOa
August 13, 2024 @ 11:56 am
Выберите идеальную печь-камин для вашего дома, Купите печь-камин и создайте уют в доме, Выберите печь-камин по своим желаниям и потребностям, Создайте атмосферу уюта с помощью печи-камина, профессиональные рекомендации по выбору, Сделайте правильный выбор печи-камина для вашего дома, подборка моделей с отзывами, Выберите идеальную печь-камин для своего дома и наслаждайтесь теплом, где найти лучшую модель
Печь-камин цена [url=https://dom-35.ru/]https://dom-35.ru/[/url] .
Lariortpf
August 13, 2024 @ 1:20 pm
[u][b]Здравствуйте![/b][/u]
[b]Заказать диплом о высшем образовании.[/b]
[url=http://newtheories.info/community/profile/shielastearns92]newtheories.info/community/profile/shielastearns92[/url]
[url=http://ingbrick.com/kupit-diplom-3973jkhuq]ingbrick.com/kupit-diplom-3973jkhuq[/url]
[url=http://my-social-box.com/story2948500/kupit-diplom]my-social-box.com/story2948500/kupit-diplom[/url]
[url=http://thecatalystapproach.com/kupit-diplom-355909opqcs]thecatalystapproach.com/kupit-diplom-355909opqcs[/url]
[url=http://naturalbookmarks.com/story17676607/kupit-diplom]naturalbookmarks.com/story17676607/kupit-diplom[/url]
[url=http://tvsocialnews.com/story3030007/kupit-diplom]tvsocialnews.com/story3030007/kupit-diplom[/url]
[url=http://bookmarksparkle.com/story17767760/kupit-diplom]bookmarksparkle.com/story17767760/kupit-diplom[/url]
[url=http://mysterybookmarks.com/story17636448/kupit-diplom]mysterybookmarks.com/story17636448/kupit-diplom[/url]
[url=http://socialfactories.com/story3014507/kupit-diplom]socialfactories.com/story3014507/kupit-diplom[/url]
[url=http://penteadistadesucesso.com/kupit-diplom-372212ikran]penteadistadesucesso.com/kupit-diplom-372212ikran[/url]
Douglasidomb
August 13, 2024 @ 1:37 pm
mexico pharmacies prescription drugs: Certified Mexican pharmacy – mexico drug stores pharmacies
Uazrvxi
August 13, 2024 @ 2:08 pm
[u][b]Добрыйдень![/b][/u]
[b]Заказать диплом любого ВУЗа.[/b]
[url=http://lyfesaverscpr.com/diplom-kupit-166619uc]lyfesaverscpr.com/diplom-kupit-166619uc[/url]
[url=http://gtj.kr/board_KtRj53/372632]gtj.kr/board_KtRj53/372632[/url]
[url=http://taondinternational.rudraserver.com/blog/index.php?entryid=137386]taondinternational.rudraserver.com/blog/index.php?entryid=137386[/url]
[url=http://lyfesaverscpr.com/diplom-173865eyble]lyfesaverscpr.com/diplom-173865eyble[/url]
[url=http://njkkot.org/?document_srl=1144066]njkkot.org/?document_srl=1144066[/url]
[url=http://bookmark-dofollow.com/story19739539/diplom]bookmark-dofollow.com/story19739539/diplom[/url]
[url=http://digitalmagazine.xyz/diplom-998685yfqrh]digitalmagazine.xyz/diplom-998685yfqrh[/url]
[url=http://bookmarkindexing.com/story17554620/diplom]bookmarkindexing.com/story17554620/diplom[/url]
[url=http://thebookmarkking.com/story17637673/diplom]thebookmarkking.com/story17637673/diplom[/url]
[url=http://social4geek.com/story3099234/diplom]social4geek.com/story3099234/diplom[/url]
[url=http://socialmphl.com/story19473936/diplom]socialmphl.com/story19473936/diplom[/url]
[url=http://itdongnam.com/diplom-kupit-670990ac]itdongnam.com/diplom-kupit-670990ac[/url]
[url=http://ecrinparis.com/diplom-977239jpmvz]ecrinparis.com/diplom-977239jpmvz[/url]
[url=http://ztndz.com/story19868249/diplom]ztndz.com/story19868249/diplom[/url]
[url=http://wr1te.com/diplom-858341ozkgu]wr1te.com/diplom-858341ozkgu[/url]
[url=http://bookmarkplaces.com/story17612788/diplom]bookmarkplaces.com/story17612788/diplom[/url]
[url=http://ntep2008.com/index.php?name=webboard&file=read&id=18527]ntep2008.com/index.php?name=webboard&file=read&id=18527[/url]
[url=http://210list.com/story18182087/diplom]210list.com/story18182087/diplom[/url]
[url=http://localtrusted.co.uk/wiki/index.php?title=Diplom_618947DMuAg]localtrusted.co.uk/wiki/index.php?title=Diplom_618947DMuAg[/url]
[url=http://sobrouremedio.com.br/author/zacherydesm]sobrouremedio.com.br/author/zacherydesm[/url]
Vazrvzc
August 13, 2024 @ 2:18 pm
[u][b] Привет, друзья![/b][/u]
Приобрести документ о получении высшего образования вы можете у нас в Москве. Мы предлагаем документы об окончании любых университетов России.
[url=http://vip-forum.com/threads/kupit-diplom-o-vysshem-obrazovanii-k35o.1803/]vip-forum.com/threads/kupit-diplom-o-vysshem-obrazovanii-k35o.1803[/url]
[url=http://tamklad.ru/viewtopic.php?f=8&t=8772/]tamklad.ru/viewtopic.php?f=8&t=8772[/url]
[url=http://kiopro.ru/support/forum/view_profile.php?UID=73823/]kiopro.ru/support/forum/view_profile.php?UID=73823[/url]
[url=http://cstrike.g-service.ru/index.php?/topic/46-купить-диплом-цена-m944q/]cstrike.g-service.ru/index.php?/topic/46-купить-диплом-цена-m944q[/url]
[url=http://videopro.ru/xenforo/index.php?threads/Тестовая-тема-закреплённая.1/page-113#post-4152/]videopro.ru/xenforo/index.php?threads/Тестовая-тема-закреплённая.1/page-113#post-4152[/url]
Lazrdqh
August 13, 2024 @ 2:20 pm
[u][b] Привет![/b][/u]
[b]Мы предлагаем дипломы[/b] психологов, юристов, экономистов и других профессий по доступным ценам.
[url=http://telegra.ph/distancionnoe-obuchenie-v-inostrannyh-vuzah-08-02/]telegra.ph/distancionnoe-obuchenie-v-inostrannyh-vuzah-08-02[/url]
Danrkny
August 13, 2024 @ 2:24 pm
[u][b] Привет, друзья![/b][/u]
Мы готовы предложить дипломы любой профессии.
[url=http://magicjewels.net/diplom-kupit-427165pg]magicjewels.net/diplom-kupit-427165pg[/url]
[url=http://kankerala.org/uncategorized/diplom-888631oxlzv]kankerala.org/uncategorized/diplom-888631oxlzv[/url]
[url=http://cherryandgriffiths.co.uk/diplom-kupit-588118zj]cherryandgriffiths.co.uk/diplom-kupit-588118zj[/url]
[url=http://bookmarkpressure.com/story17612816/diplom]bookmarkpressure.com/story17612816/diplom[/url]
[url=http://enfogentraining.com/blog/index.php?entryid=82050]enfogentraining.com/blog/index.php?entryid=82050[/url]
[url=http://campusvirtual.newlink.es/blog/index.php?entryid=14898]campusvirtual.newlink.es/blog/index.php?entryid=14898[/url]
[url=http://reech.life/index.php/blog/4744/diplom-kupit-401268zq]reech.life/index.php/blog/4744/diplom-kupit-401268zq[/url]
[url=http://welnesbiolabs.com/diplom-kupit-90317dq]welnesbiolabs.com/diplom-kupit-90317dq[/url]
[url=http://allkindsofsocial.com/story2922548/diplom]allkindsofsocial.com/story2922548/diplom[/url]
[url=http://localtrusted.co.uk/wiki/index.php?title=Diplom_481186SxKuo]localtrusted.co.uk/wiki/index.php?title=Diplom_481186SxKuo[/url]
[url=http://hariharparagovernmentiti.com/2024/07/28/diplom-629290krhau]hariharparagovernmentiti.com/2024/07/28/diplom-629290krhau[/url]
[url=http://ada.waaron.org/blog/index.php?entryid=6962]ada.waaron.org/blog/index.php?entryid=6962[/url]
[url=http://kankerala.org/uncategorized/diplom-497631duvjm]kankerala.org/uncategorized/diplom-497631duvjm[/url]
[url=http://youlangue.lu/blog/index.php?entryid=83134]youlangue.lu/blog/index.php?entryid=83134[/url]
[url=http://mysocialname.com/story3017122/diplom]mysocialname.com/story3017122/diplom[/url]
[url=http://webnowmedia.com/story2941510/diplom]webnowmedia.com/story2941510/diplom[/url]
[url=http://socialislife.com/story2994058/diplom]socialislife.com/story2994058/diplom[/url]
[url=http://ingbrick.com/diplom-75884ebygz]ingbrick.com/diplom-75884ebygz[/url]
[url=http://taondinternational.rudraserver.com/blog/index.php?entryid=131773]taondinternational.rudraserver.com/blog/index.php?entryid=131773[/url]
[url=http://digitalmagazine.xyz/diplom-922861btnoc]digitalmagazine.xyz/diplom-922861btnoc[/url]
Uazrnua
August 13, 2024 @ 2:43 pm
[b]Где купить диплом по нужной специальности?[/b]
[url=http://comfortrent.ru/2024/07/29/diplom-kupit-746181ol/]comfortrent.ru/2024/07/29/diplom-kupit-746181ol[/url]
[url=http://kbookmarking.com/story17632964/diplom/]kbookmarking.com/story17632964/diplom[/url]
[url=http://tigaedu.com/blog/index.php?entryid=335740/]tigaedu.com/blog/index.php?entryid=335740[/url]
[url=http://worldlistpro.com/story19310554/diplom/]worldlistpro.com/story19310554/diplom[/url]
[url=http://mykindadoctor.com/diplom-kupit-563269qg/]mykindadoctor.com/diplom-kupit-563269qg[/url]
Eanrzko
August 13, 2024 @ 3:21 pm
[u][b] Привет, друзья![/b][/u]
[b]Мы предлагаем дипломы[/b] любых профессий по доступным ценам.
[url=http://bisshogram.com/read-blog/281/]bisshogram.com/read-blog/281[/url]
[url=http://vocal.com.ua/node/62299/]vocal.com.ua/node/62299[/url]
[url=http://ozoms.com/read-blog/469/]ozoms.com/read-blog/469[/url]
[url=http://sonnick84.blogdeazar.com/28344883/На-что-конкретно-обратить-внимание-при-заказе-документов-в-сети/]sonnick84.blogdeazar.com/28344883/На-что-конкретно-обратить-внимание-при-заказе-документов-в-сети[/url]
[url=http://guyspages.com/read-blog/783/]guyspages.com/read-blog/783[/url]
Lazrzfz
August 13, 2024 @ 3:23 pm
[b]Заказать диплом о высшем образовании.[/b]
[url=http://ros.listbb.ru/posting.php?mode=post&f=2/]ros.listbb.ru/posting.php?mode=post&f=2[/url]
[url=http://kingcityrp.moibb.ru/viewtopic.php?f=34&t=458/]kingcityrp.moibb.ru/viewtopic.php?f=34&t=458[/url]
[url=http://ladymystery.ru/forum/topic.asp?TOPIC_ID=9497/]ladymystery.ru/forum/topic.asp?TOPIC_ID=9497[/url]
[url=http://owen.ru/forum/member.php?u=106526/]owen.ru/forum/member.php?u=106526[/url]
[url=http://chaodisiaque.com/forum/viewtopic.php?pid=22027#p22027/]chaodisiaque.com/forum/viewtopic.php?pid=22027#p22027[/url]
Douglasidomb
August 13, 2024 @ 4:15 pm
buying prescription drugs in mexico online: Best online Mexican pharmacy – buying prescription drugs in mexico online
Williamraics
August 13, 2024 @ 4:27 pm
[u][b] Привет![/b][/u]
Мы готовы предложить дипломы.
[url=http://kalininabad.flyboard.ru/viewtopic.php?f=11&t=2804/]kalininabad.flyboard.ru/viewtopic.php?f=11&t=2804[/url]
[url=http://lapd.getbb.ru/posting.php?mode=post&f=5/]lapd.getbb.ru/posting.php?mode=post&f=5[/url]
[url=http://visacart.80lvl.ru/viewtopic.php?f=1&t=772/]visacart.80lvl.ru/viewtopic.php?f=1&t=772[/url]
[url=http://hip-hop.ru/forum/id298234-worksale/]hip-hop.ru/forum/id298234-worksale[/url]
[url=http://vared.flyboard.ru/viewtopic.php?f=1&t=534/]vared.flyboard.ru/viewtopic.php?f=1&t=534[/url]
Manrwfe
August 13, 2024 @ 4:31 pm
[u][b] Здравствуйте![/b][/u]
Заказать документ института
[url=http://topsocialplan.com/story3048022/kupit-diplom]topsocialplan.com/story3048022/kupit-diplom[/url]
[url=http://tealbookmarks.com/story17666273/kupit-diplom]tealbookmarks.com/story17666273/kupit-diplom[/url]
[url=http://kingbookmark.com/story17729598/kupit-diplom]kingbookmark.com/story17729598/kupit-diplom[/url]
[url=http://whitebookmarks.com/story17710870/kupit-diplom]whitebookmarks.com/story17710870/kupit-diplom[/url]
[url=http://bookmarkingbay.com/story17666799/kupit-diplom]bookmarkingbay.com/story17666799/kupit-diplom[/url]
Treftns
August 13, 2024 @ 4:31 pm
[u][b] Привет, друзья![/b][/u]
Пошаговая инструкция по безопасной покупке диплома о высшем образовании
[url=http://lada-4×4.net/member.php?u=23303/]lada-4×4.net/member.php?u=23303[/url]
Окажем помощь!.
Lazrhtg
August 13, 2024 @ 5:12 pm
[u][b] Добрый день![/b][/u]
[b]Мы изготавливаем дипломы[/b] любой профессии по приятным ценам.
[url=http://telegra.ph/distancionnoe-obuchenie-v-muzykalnyh-vuzah-08-02/]telegra.ph/distancionnoe-obuchenie-v-muzykalnyh-vuzah-08-02[/url]
Jariordjz
August 13, 2024 @ 5:13 pm
[u][b] Добрый день![/b][/u]
Приобрести диплом ВУЗа
[url=http://wr1te.com/kupit-diplom-539588cufuc]wr1te.com/kupit-diplom-539588cufuc[/url]
[url=http://koolegardi.ir/2024/07/28/kupit-diplom-825983hujtq]koolegardi.ir/2024/07/28/kupit-diplom-825983hujtq[/url]
[url=http://exactlybookmarks.com/story17616675/kupit-diplom]exactlybookmarks.com/story17616675/kupit-diplom[/url]
[url=http://thebookmarkage.com/story17636613/kupit-diplom]thebookmarkage.com/story17636613/kupit-diplom[/url]
[url=http://userbookmark.com/story17644152/kupit-diplom]userbookmark.com/story17644152/kupit-diplom[/url]
[url=http://followbookmarks.com/story17733378/kupit-diplom]followbookmarks.com/story17733378/kupit-diplom[/url]
[url=http://setbookmarks.com/story17708498/kupit-diplom]setbookmarks.com/story17708498/kupit-diplom[/url]
[url=http://worldsocialindex.com/story3029714/kupit-diplom]worldsocialindex.com/story3029714/kupit-diplom[/url]
[url=http://ztndz.com/story19887760/kupit-diplom]ztndz.com/story19887760/kupit-diplom[/url]
[url=http://isocialfans.com/story3019263/kupit-diplom]isocialfans.com/story3019263/kupit-diplom[/url]
Mazrhhv
August 13, 2024 @ 5:20 pm
[u][b] Привет![/b][/u]
[b]Купить диплом старого образца, можно ли это сделать по быстрой схеме?[/b]
[url=http://forum.dalmatin-club.ru/profile/24344-christimoope/]forum.dalmatin-club.ru/profile/24344-christimoope[/url]
svadebnyy_wqpi
August 13, 2024 @ 5:27 pm
Продление жизни свадебного букета: полезные советы
букет на свадьбу для невесты [url=https://0gorodnik.ru/]https://0gorodnik.ru/[/url] .
Leonardginge
August 13, 2024 @ 5:50 pm
https://mexicopharmacy.win/# buying from online mexican pharmacy
Cazrdew
August 13, 2024 @ 6:08 pm
[u][b] Добрый день![/b][/u]
Мы можем предложить документы техникумов
[url=http://brenderupmediterrane.free.fr/index.php?file=Members&op=detail&autor=ocasonux/]brenderupmediterrane.free.fr/index.php?file=Members&op=detail&autor=ocasonux[/url]
Xazrxor
August 13, 2024 @ 6:09 pm
[u][b] Здравствуйте![/b][/u]
Купить документ о получении высшего образования.
[url=http://boxer-forum.ru/viewtopic.php?f=17&t=4022/]boxer-forum.ru/viewtopic.php?f=17&t=4022[/url]
[url=http://cocapal.com/read-blog/1025/]cocapal.com/read-blog/1025[/url]
[url=http://dopalnenie.listbb.ru/viewtopic.php?f=3&t=712/]dopalnenie.listbb.ru/viewtopic.php?f=3&t=712[/url]
[url=http://gazeta.ekafe.ru/viewforum.php?f=48/]gazeta.ekafe.ru/viewforum.php?f=48[/url]
[url=http://farm86.com/blogs/375/Получите-диплом-за-считанные-дни/]farm86.com/blogs/375/Получите-диплом-за-считанные-дни[/url]
kompoziciy_ylpa
August 13, 2024 @ 6:32 pm
Топ-20 вариантов цветочных композиций для украшения вашего дома, как выбрать идеальную комбинацию.
5 прекрасных идей для садовых композиций из цветов, и заставят соседей восхищаться.
Идеи оригинальных композиций для вашего букета, которые покорят сердце любого цветочника.
Как выбрать идеальный букет для невесты, и заставят всех гостей восхищаться.
Идеи сезонных композиций для вашего праздника, которые заставят всех гостей восхищаться.
Как сделать необычный декор для вашего дома, и создадут атмосферу гармонии и уюта.
Как украсить рабочее место цветами, и повысят продуктивность и настроение сотрудников.
Как украсить участок свежими цветами, которые преобразят ваш дачный уголок и наполнят его ароматом.
композиция цветов [url=https://101-po3a.ru/]композиция цветов[/url] .
Lazrrln
August 13, 2024 @ 6:50 pm
[u][b] Добрый день![/b][/u]
[b]Заказать диплом о высшем образовании.[/b]
[url=http://telegra.ph/kogda-nachinaetsya-obuchenie-v-vuzah-08-02/]telegra.ph/kogda-nachinaetsya-obuchenie-v-vuzah-08-02[/url]
Yrefrex
August 13, 2024 @ 7:01 pm
[u][b] Привет![/b][/u]
[b]Приобрести диплом любого ВУЗа.[/b]
[url=http://multisupra.ru/2014/08/konkurs-egoist/]multisupra.ru/2014/08/konkurs-egoist[/url]
Trefytl
August 13, 2024 @ 7:16 pm
[u][b] Здравствуйте![/b][/u]
Аттестат школы купить официально с упрощенным обучением в Москве
[url=http://heimur.ru/index.php?/topic/6326-куплю-дипломы-повара-k139p/]heimur.ru/index.php?/topic/6326-куплю-дипломы-повара-k139p[/url]
Рады оказать помощь!.
Mazrdeh
August 13, 2024 @ 7:20 pm
[u][b] Привет, друзья![/b][/u]
Всё о покупке аттестата о среднем образовании: полезные советы
[url=http://chaek.ru/matryoshki/matreshka-zima-art-79/reviews/]chaek.ru/matryoshki/matreshka-zima-art-79/reviews[/url]
dymohody_oppi
August 13, 2024 @ 7:44 pm
Выбор качественных дымоходов для бани в Нижнем Новгороде, быстро и надежно.
Как выбрать исполнителя на установку дымоходов для бани в Нижнем Новгороде, сравнение цен и услуг.
Какие материалы лучше всего подходят для дымоходов в Нижнем Новгороде, рекомендации по выбору.
На что обратить внимание при выборе дымоходов для бани в Нижнем Новгороде, основные критерии.
Секреты долговечности дымоходов для бани в Нижнем Новгороде, рекомендации по уходу.
Выбор оптимального типа дымоходов для бани в Нижнем Новгороде, сравнение характеристик.
дымоход для банной печи купить [url=https://forum-bani.ru/]https://forum-bani.ru/[/url] .
chto_gvEr
August 13, 2024 @ 8:54 pm
История и значение розы в культуре и символике, знаменитый цветок с многовековой историей.
Какие бывают виды роз, правила посадки и ухода за розами.
Как роза влияет на человека и его эмоции, тайны и загадки розы.
Роза как идеальный подарок для любого случая, почему роза считается королевой цветов.
Розы в архитектуре и дизайне интерьера, секреты сбора и хранения розовых лепестков.
розы это [url=https://roslina.ru/]https://roslina.ru/[/url] .
Douglasidomb
August 13, 2024 @ 9:42 pm
cheap ed medicine: online ed prescription same-day – best ed meds online
Yrefeme
August 13, 2024 @ 10:03 pm
[u][b] Привет, друзья![/b][/u]
[b]Приобрести диплом любого ВУЗа.[/b]
[url=http://vestiinform.ru/forums/topic/kuplyu-diplom-o-vysshem-s173j/]vestiinform.ru/forums/topic/kuplyu-diplom-o-vysshem-s173j[/url]
kotel_odea
August 13, 2024 @ 10:08 pm
Лучшие котлы для отопления частного дома | Советы по выбору котла для отопления | Лучшие цены на котлы для отопления | Сравнение котлов для отопления дома | Секреты установки котла для отопления | Выбор лучшего котла для отопления частного дома | Выбор магазина для покупки котла для отопления | Какие котлы для отопления частного дома лучше | Советы по экономии на отоплении | Где купить недорогой котел для отопления
купить котел для отопления частного дома [url=https://sauna-manzana.ru/]https://sauna-manzana.ru/[/url] .
Cazroqh
August 13, 2024 @ 10:47 pm
[u][b] Здравствуйте![/b][/u]
Приобрести диплом института
[url=http://telegra.ph/podgotovitelnye-kursy-dlya-postupleniya-v-vuz-08-02/]telegra.ph/podgotovitelnye-kursy-dlya-postupleniya-v-vuz-08-02[/url]
Diplomi_qqSi
August 14, 2024 @ 12:17 am
Добрый день!
Приобрести документ о получении высшего образования вы имеете возможность у нас.
[url=http://fromrus.su/index.php?/topic/15712-куплю-дипломы-магистра-j654l/]fromrus.su/index.php?/topic/15712-куплю-дипломы-магистра-j654l[/url]
Lariorhsw
August 14, 2024 @ 12:30 am
[u][b]Добрыйдень![/b][/u]
[b]Заказать диплом любогоуниверситета.[/b]
[url=http://yoursocialpeople.com/story2941745/kupit-diplom]yoursocialpeople.com/story2941745/kupit-diplom[/url]
[url=http://socialeweb.com/story2928846/kupit-diplom]socialeweb.com/story2928846/kupit-diplom[/url]
[url=http://thecatalystapproach.com/kupit-diplom-268986dvrzm]thecatalystapproach.com/kupit-diplom-268986dvrzm[/url]
[url=http://socialevity.com/story19356446/kupit-diplom]socialevity.com/story19356446/kupit-diplom[/url]
[url=http://sociallweb.com/story3022871/kupit-diplom]sociallweb.com/story3022871/kupit-diplom[/url]
[url=http://push2bookmark.com/story17789622/kupit-diplom]push2bookmark.com/story17789622/kupit-diplom[/url]
[url=http://hariharparagovernmentiti.com/2024/07/27/kupit-diplom-561306miugm]hariharparagovernmentiti.com/2024/07/27/kupit-diplom-561306miugm[/url]
[url=http://agency-social.com/story2984941/kupit-diplom]agency-social.com/story2984941/kupit-diplom[/url]
[url=http://bookmarkstime.com/story17969503/kupit-diplom]bookmarkstime.com/story17969503/kupit-diplom[/url]
[url=http://bookmarkindexing.com/story17570366/kupit-diplom]bookmarkindexing.com/story17570366/kupit-diplom[/url]
Vazruwi
August 14, 2024 @ 1:14 am
[u][b] Привет![/b][/u]
Купить документ о получении высшего образования можно в нашем сервисе. Мы оказываем услуги по изготовлению и продаже документов об окончании любых университетов РФ.
[url=http://master39.net/forum/messages/forum1/topic42/message28722/?result=reply#message28722/]master39.net/forum/messages/forum1/topic42/message28722/?result=reply#message28722[/url]
[url=http://j013.koreawebcenter.com/bbs/board.php?bo_table=free&wr_id=668409/]j013.koreawebcenter.com/bbs/board.php?bo_table=free&wr_id=668409[/url]
[url=http://rytof.ru/node/16116/]rytof.ru/node/16116[/url]
[url=http://knigigo.ru/forums/topic/kupit-diplom-czena-z749d/]knigigo.ru/forums/topic/kupit-diplom-czena-z749d[/url]
[url=http://avestalaw.ru/index.php?subaction=userinfo&user=isupypi/]avestalaw.ru/index.php?subaction=userinfo&user=isupypi[/url]
Uazrnte
August 14, 2024 @ 1:25 am
[u][b]Привет,друзья![/b][/u]
[b]Купить диплом любого ВУЗа.[/b]
[url=http://directlynews.com/diplom-kupit-789991pe]directlynews.com/diplom-kupit-789991pe[/url]
[url=http://ada.waaron.org/blog/index.php?entryid=7254]ada.waaron.org/blog/index.php?entryid=7254[/url]
[url=http://geilebookmarks.com/story17598603/diplom]geilebookmarks.com/story17598603/diplom[/url]
[url=http://tandme.co.uk/author/rosalinacon]tandme.co.uk/author/rosalinacon[/url]
[url=http://tigaedu.com/blog/index.php?entryid=334308]tigaedu.com/blog/index.php?entryid=334308[/url]
[url=http://cometothecook.com/2024/07/28/diplom-260691zpyyz]cometothecook.com/2024/07/28/diplom-260691zpyyz[/url]
[url=http://bookmarkbooth.com/story17672175/diplom]bookmarkbooth.com/story17672175/diplom[/url]
[url=http://arandaasesoria.com/diplom-154597ombdf]arandaasesoria.com/diplom-154597ombdf[/url]
[url=http://enfogentraining.com/blog/index.php?entryid=81535]enfogentraining.com/blog/index.php?entryid=81535[/url]
[url=http://digibookmarks.com/story17630098/diplom]digibookmarks.com/story17630098/diplom[/url]
[url=http://rotatesites.com/story18776117/diplom]rotatesites.com/story18776117/diplom[/url]
[url=http://bookmarkusers.com/story17496596/diplom]bookmarkusers.com/story17496596/diplom[/url]
[url=http://elearning.maniatech-academy.co.uk/blog/index.php?entryid=281393]elearning.maniatech-academy.co.uk/blog/index.php?entryid=281393[/url]
[url=http://campusvirtual.newlink.es/blog/index.php?entryid=16753]campusvirtual.newlink.es/blog/index.php?entryid=16753[/url]
[url=http://taondinternational.rudraserver.com/blog/index.php?entryid=136437]taondinternational.rudraserver.com/blog/index.php?entryid=136437[/url]
[url=http://mysocialname.com/story3018367/diplom]mysocialname.com/story3018367/diplom[/url]
[url=http://youlangue.lu/blog/index.php?entryid=87490]youlangue.lu/blog/index.php?entryid=87490[/url]
[url=http://alkhazana.net/2024/07/28/diplom-892189tvrwp]alkhazana.net/2024/07/28/diplom-892189tvrwp[/url]
[url=http://turnkeymodular.ca/diplom-535347xgtxc]turnkeymodular.ca/diplom-535347xgtxc[/url]
[url=http://taondinternational.rudraserver.com/blog/index.php?entryid=133892]taondinternational.rudraserver.com/blog/index.php?entryid=133892[/url]
Danrmol
August 14, 2024 @ 1:33 am
[u][b] Привет![/b][/u]
Мы изготавливаем дипломы любых профессий.
[url=http://funbookmarking.com/story17639606/diplom]funbookmarking.com/story17639606/diplom[/url]
[url=http://bookmarksusa.com/story17673398/diplom]bookmarksusa.com/story17673398/diplom[/url]
[url=http://bookmarkzap.com/story17555246/diplom]bookmarkzap.com/story17555246/diplom[/url]
[url=http://letusbookmark.com/story19169493/diplom]letusbookmark.com/story19169493/diplom[/url]
[url=http://candidecoin.com/uncategorized/diplom-kupit-75291ik]candidecoin.com/uncategorized/diplom-kupit-75291ik[/url]
[url=http://pencis.com/diplom-287899lsudc]pencis.com/diplom-287899lsudc[/url]
[url=http://enfogentraining.com/blog/index.php?entryid=81906]enfogentraining.com/blog/index.php?entryid=81906[/url]
[url=http://ada.waaron.org/blog/index.php?entryid=7003]ada.waaron.org/blog/index.php?entryid=7003[/url]
[url=http://welnesbiolabs.com/diplom-124026frwiv]welnesbiolabs.com/diplom-124026frwiv[/url]
[url=http://captainbookmark.com/story17598951/diplom]captainbookmark.com/story17598951/diplom[/url]
[url=http://youwantech.com/xe/board/461520]youwantech.com/xe/board/461520[/url]
[url=http://vacayla.com/2024/07/28/diplom-kupit-230695vs]vacayla.com/2024/07/28/diplom-kupit-230695vs[/url]
[url=http://freeflashgamesnow.com/profile/2771908/Makayla33X3]freeflashgamesnow.com/profile/2771908/Makayla33X3[/url]
[url=http://aplamaharashtra.com/archives/1664]aplamaharashtra.com/archives/1664[/url]
[url=http://dirstop.com/story19846355/diplom]dirstop.com/story19846355/diplom[/url]
[url=http://kankerala.org/uncategorized/diplom-742948ucyxe]kankerala.org/uncategorized/diplom-742948ucyxe[/url]
[url=http://luxuriousrentz.com/diplom-347387ttopb]luxuriousrentz.com/diplom-347387ttopb[/url]
[url=http://webookmarks.com/story3051754/diplom]webookmarks.com/story3051754/diplom[/url]
[url=http://syg-us.com/entrenamiento-virtual/blog/index.php?entryid=46356]syg-us.com/entrenamiento-virtual/blog/index.php?entryid=46356[/url]
[url=http://brainit.co.in/diplom-697390ntcca]brainit.co.in/diplom-697390ntcca[/url]
Dnrtpqs
August 14, 2024 @ 1:55 am
[b]Приобрести документ[/b] института вы можете в нашем сервисе.
[url=http://bookmarkangaroo.com/story17753361/diplom/]bookmarkangaroo.com/story17753361/diplom[/url]
[url=http://heealthy.com/question/diplom-540342wkbjl/]heealthy.com/question/diplom-540342wkbjl[/url]
[url=http://bookmarkquotes.com/story17732203/diplom/]bookmarkquotes.com/story17732203/diplom[/url]
[url=http://ada.waaron.org/blog/index.php?entryid=6591/]ada.waaron.org/blog/index.php?entryid=6591[/url]
[url=http://bookmarkpagerank.com/story17664856/diplom/]bookmarkpagerank.com/story17664856/diplom[/url]
Uazrold
August 14, 2024 @ 1:59 am
[b]Где приобрести диплом по необходимой специальности?[/b]
[url=http://tvsocialnews.com/story3026616/diplom/]tvsocialnews.com/story3026616/diplom[/url]
[url=http://mediasocially.com/story2907894/diplom/]mediasocially.com/story2907894/diplom[/url]
[url=http://social-lyft.com/story7433538/diplom/]social-lyft.com/story7433538/diplom[/url]
[url=http://moodle.spice-training.com/blog/index.php?entryid=306338/]moodle.spice-training.com/blog/index.php?entryid=306338[/url]
[url=http://social-lyft.com/story7433437/diplom/]social-lyft.com/story7433437/diplom[/url]
Lazrggi
August 14, 2024 @ 2:15 am
[b]Приобрести диплом любого университета.[/b]
[url=http://russiajoy.ru/diplomyi-na-zakaz-legko-i-bezopasno/]russiajoy.ru/diplomyi-na-zakaz-legko-i-bezopasno[/url]
[url=http://kalininabad.flyboard.ru/viewtopic.php?f=28&t=2813&sid=029fdb150d994c78b02b8f8582ded9c0/]kalininabad.flyboard.ru/viewtopic.php?f=28&t=2813&sid=029fdb150d994c78b02b8f8582ded9c0[/url]
[url=http://obshenie.flybb.ru/viewtopic.php?f=2&t=676/]obshenie.flybb.ru/viewtopic.php?f=2&t=676[/url]
[url=http://twikkers.nl/blogs/238134/Настоящие-дипломы-без-лишних-вопросов/]twikkers.nl/blogs/238134/Настоящие-дипломы-без-лишних-вопросов[/url]
[url=http://ratingforex.ru/forum-forex/viewtopic.php?f=18&t=40990&sid=e64ff0e11acab3f7a177f69322469c8c/]ratingforex.ru/forum-forex/viewtopic.php?f=18&t=40990&sid=e64ff0e11acab3f7a177f69322469c8c[/url]
Eanroku
August 14, 2024 @ 2:18 am
[u][b] Добрый день![/b][/u]
[b]Мы можем предложить дипломы[/b] любых профессий по приятным ценам.
[url=http://bizinhome.mn.co/posts/61639396/]bizinhome.mn.co/posts/61639396[/url]
[url=http://yur-dom.ru/communication/forum/messages/forum3/message254/297-nizkie-tseny-i-vysokoe-kachestvo-_-priobretay-diplomy-onlayn?result=new/]yur-dom.ru/communication/forum/messages/forum3/message254/297-nizkie-tseny-i-vysokoe-kachestvo-_-priobretay-diplomy-onlayn?result=new[/url]
[url=http://jointsnmotion-hub.mn.co/posts/61748294/]jointsnmotion-hub.mn.co/posts/61748294[/url]
[url=http://sonnick84.ltfblog.com/27683502/Планируете-купить-диплом-Выбирайте-наш-магазин/]sonnick84.ltfblog.com/27683502/Планируете-купить-диплом-Выбирайте-наш-магазин[/url]
[url=http://nongkhai.online/blogs/106/Расширенное-описание-покупки-диплома-в-онлайн-магазине/]nongkhai.online/blogs/106/Расширенное-описание-покупки-диплома-в-онлайн-магазине[/url]
Williamraics
August 14, 2024 @ 3:25 am
[u][b] Привет, друзья![/b][/u]
Мы изготавливаем дипломы.
[url=http://wallazz.com/blogs/205241/Настоящие-Дипломы-Легко-и-Быстро/]wallazz.com/blogs/205241/Настоящие-Дипломы-Легко-и-Быстро[/url]
[url=http://x70795vj.beget.tech/2024/07/07/bystroe-oformlenie-diplomov-dostupno-i-nadezhno/]x70795vj.beget.tech/2024/07/07/bystroe-oformlenie-diplomov-dostupno-i-nadezhno[/url]
[url=http://selkovo.rolka.me/viewtopic.php?id=3407#p10093/]selkovo.rolka.me/viewtopic.php?id=3407#p10093[/url]
[url=http://silkhunter.com/index.php?title=Купить_Официальный_Диплом_Быстро_и_Легально/]silkhunter.com/index.php?title=Купить_Официальный_Диплом_Быстро_и_Легально[/url]
[url=http://medbereg.ru/club/user/82/blog/4146/]medbereg.ru/club/user/82/blog/4146[/url]
Diplomi_ifSi
August 14, 2024 @ 3:34 am
Привет!
Приобрести документ ВУЗа можно у нас в Москве.
[url=http://prazdnikbaby.ru/index.php/separator?view=item&item_id=10/]prazdnikbaby.ru/index.php/separator?view=item&item_id=10[/url]
Jariorchc
August 14, 2024 @ 4:08 am
[u][b] Привет![/b][/u]
Купить диплом любого университета
[url=http://expressbookmark.com/story17654916/kupit-diplom]expressbookmark.com/story17654916/kupit-diplom[/url]
[url=http://fridayad.in/user/profile/2637128]fridayad.in/user/profile/2637128[/url]
[url=http://whitebookmarks.com/story17712370/kupit-diplom]whitebookmarks.com/story17712370/kupit-diplom[/url]
[url=http://bookmarksparkle.com/story17773707/kupit-diplom]bookmarksparkle.com/story17773707/kupit-diplom[/url]
[url=http://exactlybookmarks.com/story17616639/kupit-diplom]exactlybookmarks.com/story17616639/kupit-diplom[/url]
[url=http://dentifolio.com/index.php?page=user&action=pub_profile&id=4393]dentifolio.com/index.php?page=user&action=pub_profile&id=4393[/url]
[url=http://topsocialplan.com/story3043359/kupit-diplom]topsocialplan.com/story3043359/kupit-diplom[/url]
[url=http://apollobookmarks.com/story17598483/kupit-diplom]apollobookmarks.com/story17598483/kupit-diplom[/url]
[url=http://whitebookmarks.com/story17711619/kupit-diplom]whitebookmarks.com/story17711619/kupit-diplom[/url]
[url=http://socialwebconsult.com/story2973970/kupit-diplom]socialwebconsult.com/story2973970/kupit-diplom[/url]
Mazrqzr
August 14, 2024 @ 4:08 am
[u][b] Добрый день![/b][/u]
[b]Как быстро и легально купить аттестат 11 класса в Москве[/b]
[url=http://d.mwoff.org/index.php?/topic/10451-по-ссылке-o704e/]d.mwoff.org/index.php?/topic/10451-по-ссылке-o704e[/url]
arenda_wbOt
August 14, 2024 @ 4:57 am
Аренда экскаватора погрузчика: удобно и выгодно, воспользуйтесь услугой уже сегодня.
Аренда экскаватора погрузчика: безопасность и удобство на вашем объекте, воспользуйтесь услугой уже сегодня.
Экскаватор погрузчик на прокат: удобство и эффективность на стройплощадке, арендуйте прямо сейчас.
Аренда экскаватора погрузчика: лучший выбор для строительных работ, заказывайте прямо сейчас.
Аренда экскаватора погрузчика: быстро и качественно, арендуйте прямо сейчас.
Экскаватор погрузчик на прокат: оптимальное решение для строительных работ, воспользуйтесь услугой уже сегодня.
аренда экскаватора погрузчика в москве [url=https://arenda-jekskavatora-pogruzchika-197.ru/]https://arenda-jekskavatora-pogruzchika-197.ru/[/url] .
Cazrvly
August 14, 2024 @ 5:03 am
[u][b] Здравствуйте![/b][/u]
Мы можем предложить документы ВУЗов
[url=http://london-is-passion.blogspot.com/2014/08/52/]london-is-passion.blogspot.com/2014/08/52[/url]
Douglasidomb
August 14, 2024 @ 5:18 am
best ed medication online: Best ED meds online – ed meds online
Dnrtmky
August 14, 2024 @ 5:20 am
[b]Купить документ[/b] о получении высшего образования можно в нашей компании.
[url=http://howis.info/2024/07/27/diplom-803897udmaj/]howis.info/2024/07/27/diplom-803897udmaj[/url]
[url=http://luxuriousrentz.com/diplom-kupit-795213ih/]luxuriousrentz.com/diplom-kupit-795213ih[/url]
[url=http://esocialmall.com/story2958331/diplom/]esocialmall.com/story2958331/diplom[/url]
[url=http://socialinplace.com/story2961642/diplom/]socialinplace.com/story2961642/diplom[/url]
[url=http://techonpage.com/story2946918/diplom/]techonpage.com/story2946918/diplom[/url]
Leonardginge
August 14, 2024 @ 5:29 am
http://mexicopharmacy.win/# buying prescription drugs in mexico online
Lazriss
August 14, 2024 @ 6:27 am
[u][b] Добрый день![/b][/u]
[b]Купить диплом любого ВУЗа.[/b]
[url=http://telegra.ph/postuplenie-v-tvorcheskie-vuzy-chehii-08-02/]telegra.ph/postuplenie-v-tvorcheskie-vuzy-chehii-08-02[/url]
Leonardginge
August 14, 2024 @ 6:34 am
https://edpillpharmacy.store/# low cost ed meds
Douglasidomb
August 14, 2024 @ 7:45 am
online ed medications: Best ED meds online – cheap ed
Lazrore
August 14, 2024 @ 7:55 am
[u][b] Привет, друзья![/b][/u]
[b]Мы изготавливаем дипломы[/b] психологов, юристов, экономистов и любых других профессий по приятным тарифам.
[url=http://telegra.ph/obuchenie-v-vuzah-avstrii-08-02/]telegra.ph/obuchenie-v-vuzah-avstrii-08-02[/url]
Trefysx
August 14, 2024 @ 8:39 am
[u][b] Добрый день![/b][/u]
Приобретение диплома ПТУ с сокращенной программой обучения в Москве
[url=http://matripedia.de/doku.php?id=diplomandoks/]matripedia.de/doku.php?id=diplomandoks[/url]
Рады оказаться полезными!.
Cazrchq
August 14, 2024 @ 9:06 am
[u][b] Привет, друзья![/b][/u]
Получить диплом ВУЗа
[url=http://telegra.ph/distancionnoe-obuchenie-v-vuzah-kazahstana-08-02/]telegra.ph/distancionnoe-obuchenie-v-vuzah-kazahstana-08-02[/url]
Sazrcsn
August 14, 2024 @ 9:16 am
[u][b] Добрый день![/b][/u]
Диплом экономиста
[url=http://telegra.ph/vechernyaya-forma-obucheniya-v-medicinskom-vuze-08-02/]telegra.ph/vechernyaya-forma-obucheniya-v-medicinskom-vuze-08-02[/url]
Lazrwpp
August 14, 2024 @ 11:15 am
[u][b] Здравствуйте![/b][/u]
[b]Мы изготавливаем дипломы[/b] психологов, юристов, экономистов и других профессий по выгодным тарифам.
[url=http://telegra.ph/distancionnoe-obuchenie-v-medicinskih-vuzah-08-02/]telegra.ph/distancionnoe-obuchenie-v-medicinskih-vuzah-08-02[/url]
Trefebk
August 14, 2024 @ 12:06 pm
[u][b] Привет![/b][/u]
Как получить диплом техникума официально и без лишних проблем
[url=http://knigigo.ru/forums/topic/kuplyu-diplomy-inzhenera-d74u/]knigigo.ru/forums/topic/kuplyu-diplomy-inzhenera-d74u[/url]
Рады оказать помощь!.
peretyazhka-mebeli-minsk mqSa
August 14, 2024 @ 12:50 pm
Какие выгоды дает перетяжка мягкой мебели, о которых стоит знать, для успешного обновления мебели, Как экономно обновить мягкую мебель без перетяжки, без лишних затрат, что учитывать при выборе техника для работы, для создания уютного уголка в доме
перетяжка мягкой мебели в минске [url=https://obivka-divana.ru/]перетяжка мягкой мебели в минске[/url] .
Yrefiie
August 14, 2024 @ 1:00 pm
[u][b] Здравствуйте![/b][/u]
[b]Приобрести диплом о высшем образовании.[/b]
[url=http://balaklavskiy-16.ru/user/10465/]balaklavskiy-16.ru/user/10465[/url]
Sazruoo
August 14, 2024 @ 1:09 pm
[u][b] Добрый день![/b][/u]
Мы изготавливаем дипломы любой профессии.
[url=http://olivebookmarks.com/story17758830/kupit-diplom/]olivebookmarks.com/story17758830/kupit-diplom[/url]
[url=http://koolegardi.ir/2024/07/27/kupit-diplom-384050fxtxz/]koolegardi.ir/2024/07/27/kupit-diplom-384050fxtxz[/url]
[url=http://dentifolio.com/index.php?page=user&action=pub_profile&id=4393/]dentifolio.com/index.php?page=user&action=pub_profile&id=4393[/url]
[url=http://xyzbookmarks.com/story17521068/kupit-diplom/]xyzbookmarks.com/story17521068/kupit-diplom[/url]
[url=http://socialwebleads.com/story2992939/kupit-diplom/]socialwebleads.com/story2992939/kupit-diplom[/url]
Tommyburse
August 14, 2024 @ 3:34 pm
Federation of Scout Associations of Spain https://scout.es/scout-academy-2017/ is a non-profit entity whose registered office is located in Lake
peretyazhka-mebeli-v-minske qbOi
August 14, 2024 @ 3:35 pm
Восстановление красоты и комфорта в вашем доме, попробуйте.
Новая жизнь для вашей любимой мебели, сделаем вашу мебель снова привлекательной.
Профессиональное оформление вашей мебели, оригинальные решения дизайна.
Эксклюзивные решения для вашего интерьера, сделаем вашу мебель уникальной.
Мастера перетяжки мебели в деле, сделаем вашу мебель по-новому.
перетяжка мягкой мебели в минске [url=https://peretyazhkann.ru/]https://peretyazhkann.ru/[/url] .
Yrefrfb
August 14, 2024 @ 3:53 pm
[u][b] Добрый день![/b][/u]
[b]Заказать диплом любого ВУЗа.[/b]
[url=http://atoll-filters.ru/product/umjagchitel-atoll-ecolife-s-20/reviews/]atoll-filters.ru/product/umjagchitel-atoll-ecolife-s-20/reviews[/url]
Sazroli
August 14, 2024 @ 4:45 pm
[u][b] Привет![/b][/u]
Приобрести документ института
[url=http://mirmafii.ru/article/7599/]mirmafii.ru/article/7599[/url]
[url=http://kino.10bb.ru/viewtopic.php?id=163757#p276793/]kino.10bb.ru/viewtopic.php?id=163757#p276793[/url]
[url=http://telegra.ph/konkurs-postupleniya-v-vuzy-2019-07-24/]telegra.ph/konkurs-postupleniya-v-vuzy-2019-07-24[/url]
[url=http://ulmo.ukrbb.net/index.php/]ulmo.ukrbb.net/index.php[/url]
[url=http://wiki.edu54.ru/Быстрое_решение_для_карьерного_роста/]wiki.edu54.ru/Быстрое_решение_для_карьерного_роста[/url]
Mazrjya
August 14, 2024 @ 5:55 pm
[b]Заказать документ о получении высшего образования.[/b]
[url=http://onlybookmarkings.com/story17618091/kupit-diplom/]onlybookmarkings.com/story17618091/kupit-diplom[/url]
[url=http://freeflashgamesnow.com/profile/2763118/EmmaLamaro8/]freeflashgamesnow.com/profile/2763118/EmmaLamaro8[/url]
[url=http://molettes.online/kupit-diplom-975728xfsix/]molettes.online/kupit-diplom-975728xfsix[/url]
[url=http://koolegardi.ir/2024/07/28/kupit-diplom-747033dnvxj/]koolegardi.ir/2024/07/28/kupit-diplom-747033dnvxj[/url]
[url=http://crossbookmark.com/story17572824/kupit-diplom/]crossbookmark.com/story17572824/kupit-diplom[/url]
Leonardginge
August 14, 2024 @ 5:59 pm
https://mexicopharmacy.win/# mexican online pharmacies prescription drugs
Dnrtdwi
August 14, 2024 @ 8:15 pm
[b]Приобрести документ[/b] ВУЗа вы имеете возможность у нас.
[url=http://tayarut.bkerem.org.il/diplom-442005xlfdc/]tayarut.bkerem.org.il/diplom-442005xlfdc[/url]
[url=http://canterburytaxis.co.uk/2024/07/26/diplom-457563unlaz/]canterburytaxis.co.uk/2024/07/26/diplom-457563unlaz[/url]
[url=http://tigaedu.com/blog/index.php?entryid=333870/]tigaedu.com/blog/index.php?entryid=333870[/url]
[url=http://tourxperts.com/diplom-522934ftmrx/]tourxperts.com/diplom-522934ftmrx[/url]
[url=http://agendabookmarks.com/story17576342/diplom/]agendabookmarks.com/story17576342/diplom[/url]
Douglasidomb
August 14, 2024 @ 8:37 pm
ed treatments online: ED meds online with insurance – ed meds cheap
Sazrxhu
August 14, 2024 @ 10:54 pm
[u][b] Добрый день![/b][/u]
Заказать документ о получении высшего образования
[url=http://forexrassia.ru/byistryiy-put-k-vyisshemu-obrazovaniyu-i-novyim-vozmozhnostyam/]forexrassia.ru/byistryiy-put-k-vyisshemu-obrazovaniyu-i-novyim-vozmozhnostyam[/url]
[url=http://newsofmebel.ru/diplomyi-dlya-karernyih-vyisot/]newsofmebel.ru/diplomyi-dlya-karernyih-vyisot[/url]
[url=http://kozhuhovo.forum2x2.ru/t6440-topic#13367/]kozhuhovo.forum2x2.ru/t6440-topic#13367[/url]
[url=http://vip.forums.party/viewtopic.php?id=3465#p71150/]vip.forums.party/viewtopic.php?id=3465#p71150[/url]
[url=http://innovator24.com/create-blog/]innovator24.com/create-blog[/url]
Douglasidomb
August 14, 2024 @ 11:03 pm
buying prescription drugs in mexico: Medicines Mexico – purple pharmacy mexico price list
Sazrwsg
August 14, 2024 @ 11:08 pm
[u][b] Добрый день![/b][/u]
Мы изготавливаем дипломы психологов, юристов, экономистов и прочих профессий.
[url=http://fridayad.in/user/profile/2637083/]fridayad.in/user/profile/2637083[/url]
[url=http://v98773zt.bget.ru/community/profile/roseannesmoot58/]v98773zt.bget.ru/community/profile/roseannesmoot58[/url]
[url=http://paltosuzca.com/kupit-diplom-129281kdnip/]paltosuzca.com/kupit-diplom-129281kdnip[/url]
[url=http://tvsocialnews.com/story3026842/kupit-diplom/]tvsocialnews.com/story3026842/kupit-diplom[/url]
[url=http://captainbookmark.com/story17603586/kupit-diplom/]captainbookmark.com/story17603586/kupit-diplom[/url]
Dnrttam
August 14, 2024 @ 11:20 pm
[b]Приобрести документ[/b] о получении высшего образования можно в нашем сервисе.
[url=http://kingslists.com/story18738450/diplom/]kingslists.com/story18738450/diplom[/url]
[url=http://ntep2008.com/index.php?name=webboard&file=read&id=18523/]ntep2008.com/index.php?name=webboard&file=read&id=18523[/url]
[url=http://bookmarkfly.com/story17693851/diplom/]bookmarkfly.com/story17693851/diplom[/url]
[url=http://isocialfans.com/story3023011/diplom/]isocialfans.com/story3023011/diplom[/url]
[url=http://bookmarktiger.com/story17629336/diplom/]bookmarktiger.com/story17629336/diplom[/url]
DavidGot
August 15, 2024 @ 12:10 am
inflated balloons https://balloons-sale-dubai.com
Oariorzbu
August 15, 2024 @ 12:27 am
[u][b] Привет, друзья![/b][/u]
Заказать диплом университета
[b]Мы предлагаем[/b] выгодно приобрести диплом, который выполнен на оригинальном бланке и заверен мокрыми печатями, водяными знаками, подписями официальных лиц. Документ пройдет любые проверки, даже с применением специфических приборов. Решайте свои задачи быстро и просто с нашим сервисом.
[b]Где купить диплом специалиста?[/b]
[url=http://fabnews.ru/forum/showthread.php?p=80362#post80362/]fabnews.ru/forum/showthread.php?p=80362#post80362[/url]
[url=http://kinopuk.ru/viewtopic.php?id=9107#p56712/]kinopuk.ru/viewtopic.php?id=9107#p56712[/url]
[url=http://admtobolsk-new.ru/forum/messages/forum2/topic586/message622/?result=new#message622/]admtobolsk-new.ru/forum/messages/forum2/topic586/message622/?result=new#message622[/url]
[url=http://danceway74.ru/users/36/]danceway74.ru/users/36[/url]
[url=http://ruconf.ru/forum/index.php?PAGE_NAME=profile_view&UID=104781/]ruconf.ru/forum/index.php?PAGE_NAME=profile_view&UID=104781[/url]
PatrickbiG
August 15, 2024 @ 12:35 am
трубные решетки теплообменников https://trubnye-doski.ru
Trefovy
August 15, 2024 @ 12:45 am
[u][b] Привет, друзья![/b][/u]
Всё, что нужно знать о покупке аттестата о среднем образовании без рисков
[url=http://lider-school174.com/forum/user/3104/]lider-school174.com/forum/user/3104[/url]
Поможем вам всегда!.
assist id
August 15, 2024 @ 1:44 am
Get the best medical care anytime, anywhere, with Assist id
Kampus Digital Marketing
August 15, 2024 @ 1:56 am
This article is very helpful and what is written in it has been taken care of, if you want to find similar websites you can visit to Argiaacademy
Lazrpyl
August 15, 2024 @ 2:14 am
[u][b] Здравствуйте![/b][/u]
[b]Мы можем предложить дипломы[/b] любой профессии по выгодным тарифам.
[url=http://telegra.ph/izlozhenie-dlya-postupleniya-v-vuz-08-02/]telegra.ph/izlozhenie-dlya-postupleniya-v-vuz-08-02[/url]
Diplomi_uqSi
August 15, 2024 @ 2:15 am
Привет, друзья!
Купить документ института вы можете у нас в Москве.
[url=http://radio-wave.ru/forum/member.php?3622-lucymorZEnce/]radio-wave.ru/forum/member.php?3622-lucymorZEnce[/url]
Trefrqm
August 15, 2024 @ 3:33 am
[u][b] Привет, друзья![/b][/u]
Реально ли приобрести диплом стоматолога? Основные этапы
[url=http://bestwecando.ourproject.org/dokuwiki-2013-05-10a/doku.php?id=luxdiplom/]bestwecando.ourproject.org/dokuwiki-2013-05-10a/doku.php?id=luxdiplom[/url]
Рады помочь!.
Sazrloz
August 15, 2024 @ 3:34 am
[u][b] Добрый день![/b][/u]
Купить документ о получении высшего образования
[url=http://telegra.ph/kak-sdelat-ehlektronnuyu-podpis-dlya-postupleniya-v-vuz-07-24/]telegra.ph/kak-sdelat-ehlektronnuyu-podpis-dlya-postupleniya-v-vuz-07-24[/url]
[url=http://mymink.5bb.ru/viewtopic.php?id=9007#p455164/]mymink.5bb.ru/viewtopic.php?id=9007#p455164[/url]
[url=http://wiki.3cdr.ru/wiki/index.php/Быстрое_оформление_дипломов_для_всех/]wiki.3cdr.ru/wiki/index.php/Быстрое_оформление_дипломов_для_всех[/url]
[url=http://russianecuador.com/forum/member.php?u=13207/]russianecuador.com/forum/member.php?u=13207[/url]
[url=http://telegra.ph/rejting-luchshih-shkol-dlya-postupleniya-v-vedushchie-vuzy-rossii-07-24/]telegra.ph/rejting-luchshih-shkol-dlya-postupleniya-v-vedushchie-vuzy-rossii-07-24[/url]
Mazrssv
August 15, 2024 @ 3:49 am
[b]Заказать документ о получении высшего образования.[/b]
[url=http://mysocialquiz.com/story3031381/kupit-diplom/]mysocialquiz.com/story3031381/kupit-diplom[/url]
[url=http://digitalmagazine.xyz/kupit-diplom-706250jqgib/]digitalmagazine.xyz/kupit-diplom-706250jqgib[/url]
[url=http://molettes.online/kupit-diplom-277180dxwbt/]molettes.online/kupit-diplom-277180dxwbt[/url]
[url=http://xyzbookmarks.com/story17521076/kupit-diplom/]xyzbookmarks.com/story17521076/kupit-diplom[/url]
[url=http://singnalsocial.com/story2946748/kupit-diplom/]singnalsocial.com/story2946748/kupit-diplom[/url]
Douglasidomb
August 15, 2024 @ 4:22 am
buying ed pills online: buy ed pills online – get ed meds online
Iariorqti
August 15, 2024 @ 4:33 am
[u][b] Привет![/b][/u]
Приобрести документ о получении высшего образования вы имеете возможность в нашей компании. Мы оказываем услуги по изготовлению и продаже документов об окончании любых университетов Российской Федерации. Вы сможете получить диплом по любой специальности, любого года выпуска, включая документы Советского Союза. Гарантируем, что в случае проверки документов работодателями, подозрений не возникнет.
[url=http://twoplustwoequal.com/read-blog/38476/]twoplustwoequal.com/read-blog/38476[/url]
[url=http://golf.od.ua/forum/viewtopic.php?p=77951#77951/]golf.od.ua/forum/viewtopic.php?p=77951#77951[/url]
[url=http://ukrevent.ru/diplomyi-dlya-uverennyih-v-sebe-lyudey/]ukrevent.ru/diplomyi-dlya-uverennyih-v-sebe-lyudey[/url]
[url=http://betalk.in.th/read-blog/2981/]betalk.in.th/read-blog/2981[/url]
[url=http://sklad-slabov.ru/forum/user/8462/]sklad-slabov.ru/forum/user/8462[/url]
Lazrfxi
August 15, 2024 @ 5:37 am
[u][b] Здравствуйте![/b][/u]
[b]Мы готовы предложить дипломы[/b] любой профессии по разумным тарифам.
[url=http://telegra.ph/vechernyaya-forma-obucheniya-v-medicinskom-vuze-08-02/]telegra.ph/vechernyaya-forma-obucheniya-v-medicinskom-vuze-08-02[/url]
Douglasidomb
August 15, 2024 @ 6:50 am
mexican border pharmacies shipping to usa: Best pharmacy in Mexico – buying prescription drugs in mexico
Douglasidomb
August 15, 2024 @ 11:58 am
low cost ed meds: ED meds online with insurance – get ed meds online
Douglasidomb
August 15, 2024 @ 2:23 pm
buy ed medication online: Best ED pills non prescription – ed medicines
Dnrtftt
August 15, 2024 @ 2:37 pm
[b]Купить документ[/b] ВУЗа вы сможете в нашей компании в Москве.
[url=http://iwanttobookmark.com/story17761228/diplom/]iwanttobookmark.com/story17761228/diplom[/url]
[url=http://hylistings.com/story18655694/diplom/]hylistings.com/story18655694/diplom[/url]
[url=http://throbsocial.com/story19400544/diplom/]throbsocial.com/story19400544/diplom[/url]
[url=http://bookmarkingalpha.com/story17659886/diplom/]bookmarkingalpha.com/story17659886/diplom[/url]
[url=http://minibookmarking.com/story17767653/diplom/]minibookmarking.com/story17767653/diplom[/url]
Leonardginge
August 15, 2024 @ 3:01 pm
http://edpillpharmacy.store/# buying ed pills online
Danrava
August 15, 2024 @ 3:27 pm
[u][b] Привет![/b][/u]
Мы предлагаем дипломы любой профессии.
[url=http://utahsyardsale.com/author/peterktu997]utahsyardsale.com/author/peterktu997[/url]
[url=http://hariharparagovernmentiti.com/2024/08/14/diplom-kupit-772573je]hariharparagovernmentiti.com/2024/08/14/diplom-kupit-772573je[/url]
[url=http://investicos.com/uncategorized/diplom-512757dzpyp]investicos.com/uncategorized/diplom-512757dzpyp[/url]
[url=http://laviehub.com/blog/diplom-354863ltwtl]laviehub.com/blog/diplom-354863ltwtl[/url]
[url=http://parathajoint.com/2024/07/29/diplom-kupit-914795kp]parathajoint.com/2024/07/29/diplom-kupit-914795kp[/url]
Iariornck
August 15, 2024 @ 4:00 pm
[u][b] Привет, друзья![/b][/u]
Приобрести документ университета можно у нас в столице. Мы оказываем услуги по изготовлению и продаже документов об окончании любых ВУЗов РФ. Вы сможете получить необходимый диплом по любой специальности, включая документы СССР. Даем 100% гарантию, что в случае проверки документа работодателем, подозрений не возникнет.
[url=http://geoamor.com/read-blog/30212/]geoamor.com/read-blog/30212[/url]
[url=http://malispa.ru/users/122/]malispa.ru/users/122[/url]
[url=http://beijingpal.com/read-blog/1147/]beijingpal.com/read-blog/1147[/url]
[url=http://telegra.ph/vozvrat-ndfl-za-obuchenie-v-vuze-07-24/]telegra.ph/vozvrat-ndfl-za-obuchenie-v-vuze-07-24[/url]
[url=http://domovou.3nx.ru/viewtopic.php?p=7083#7083/]domovou.3nx.ru/viewtopic.php?p=7083#7083[/url]
Trefnnt
August 15, 2024 @ 5:02 pm
[u][b] Привет![/b][/u]
Как официально купить диплом вуза с упрощенным обучением в Москве
[url=http://politikforum.ru/member.php?u=13387572/]politikforum.ru/member.php?u=13387572[/url]
Будем рады вам помочь!.
Dnrtctb
August 15, 2024 @ 5:40 pm
[b]Приобрести документ[/b] о получении высшего образования можно у нас в столице.
[url=http://paltosuzca.com/diplom-kupit-683979ha/]paltosuzca.com/diplom-kupit-683979ha[/url]
[url=http://toplistar.com/story19382702/diplom/]toplistar.com/story19382702/diplom[/url]
[url=http://localtrusted.co.uk/wiki/index.php?title=Diplom_577202RhRlA/]localtrusted.co.uk/wiki/index.php?title=Diplom_577202RhRlA[/url]
[url=http://natural-bookmark.com/story17635681/diplom/]natural-bookmark.com/story17635681/diplom[/url]
[url=http://taondinternational.rudraserver.com/blog/index.php?entryid=132322/]taondinternational.rudraserver.com/blog/index.php?entryid=132322[/url]
Douglasidomb
August 15, 2024 @ 6:54 pm
world pharmacy india: indian pharmacy – Online medicine home delivery
Cazrius
August 15, 2024 @ 7:28 pm
[u][b] Здравствуйте![/b][/u]
Мы предлагаем документы ВУЗов
[url=http://telegra.ph/gde-mozhno-kupit-diplom-08-13-2 ]telegra.ph/gde-mozhno-kupit-diplom-08-13-2[/url]
Uazritd
August 15, 2024 @ 7:37 pm
[u][b]Привет![/b][/u]
[b]Приобрести диплом ВУЗа .[/b]
[url=http://telegra.ph/kupit-diplom-vuza-vo-vladivostoke-08-13-7]telegra.ph/kupit-diplom-vuza-vo-vladivostoke-08-13-7[/url]
Trefszk
August 15, 2024 @ 7:47 pm
[u][b] Привет, друзья![/b][/u]
Аттестат школы купить официально с упрощенным обучением в Москве
[url=http://politikforum.ru/member.php?u=13387572/]politikforum.ru/member.php?u=13387572[/url]
Окажем помощь!.
Lariorvkh
August 15, 2024 @ 7:49 pm
[u][b]Привет,друзья![/b][/u]
[b]Купить диплом о высшем образовании.[/b]
[url=http://telegra.ph/kupit-diplom-v-arhangelske-08-13-9]telegra.ph/kupit-diplom-v-arhangelske-08-13-9[/url]
Lazrkdg
August 15, 2024 @ 8:45 pm
[b]Приобрести диплом любого университета.[/b]
[url=http://av.flyboard.ru/viewtopic.php?f=9&t=1416&sid=504f164ecf84d2c2cbcdbde6a86e3b75/]av.flyboard.ru/viewtopic.php?f=9&t=1416&sid=504f164ecf84d2c2cbcdbde6a86e3b75[/url]
[url=http://forum.kam.su/member.php?u=5734/]forum.kam.su/member.php?u=5734[/url]
[url=http://talkrealty.ru/vyisshee-obrazovanie-dlya-vseh-byistro-i-legko/]talkrealty.ru/vyisshee-obrazovanie-dlya-vseh-byistro-i-legko[/url]
[url=http://netcallvoip.com/wiki/index.php/Настоящие_дипломы_без_лишних_вопросов/]netcallvoip.com/wiki/index.php/Настоящие_дипломы_без_лишних_вопросов[/url]
[url=http://d91652pj.beget.tech/2024/07/07/nastoyaschie-diplomy-konfidencialno-i-nadezhno/]d91652pj.beget.tech/2024/07/07/nastoyaschie-diplomy-konfidencialno-i-nadezhno[/url]
Yrefqet
August 15, 2024 @ 8:46 pm
[u][b] Добрый день![/b][/u]
[b]Заказать диплом любого университета.[/b]
[url=http://zamok.druzya.org/index.php?/profile/605888-markedmalisy/]zamok.druzya.org/index.php?/profile/605888-markedmalisy[/url]
Eanrjcb
August 15, 2024 @ 8:46 pm
[u][b] Привет![/b][/u]
[b]Мы изготавливаем дипломы[/b] любой профессии по доступным ценам.
[url=http://biocenter.pro/club/blogs/sonnick84-blog-bc/faq-i-tekhpodderzhka-pokupaem-diplom-v-seti/]biocenter.pro/club/blogs/sonnick84-blog-bc/faq-i-tekhpodderzhka-pokupaem-diplom-v-seti[/url]
[url=http://forum.ro-ox.com/index.php?/gallery/image/187-почему-сегодня-гораздо-дешевле-будет-приобрести-диплом-университета/]forum.ro-ox.com/index.php?/gallery/image/187-почему-сегодня-гораздо-дешевле-будет-приобрести-диплом-университета[/url]
[url=http://asepbook.com/read-blog/3797/]asepbook.com/read-blog/3797[/url]
[url=http://forum.redzmax.ro/index.php?/gallery/image/69-тех-поддержка-и-faq-покупаем-документы-в-интернете/]forum.redzmax.ro/index.php?/gallery/image/69-тех-поддержка-и-faq-покупаем-документы-в-интернете[/url]
[url=http://create-lifestyle.mn.co/posts/61748481/]create-lifestyle.mn.co/posts/61748481[/url]
Uazrxgc
August 15, 2024 @ 8:47 pm
[b]Где заказать диплом специалиста?[/b]
[url=http://artybookmarks.com/story17561541/diplom/]artybookmarks.com/story17561541/diplom[/url]
[url=http://letsbookmarkit.com/story17612863/diplom/]letsbookmarkit.com/story17612863/diplom[/url]
[url=http://pencis.com/diplom-108236dczee/]pencis.com/diplom-108236dczee[/url]
[url=http://brightbookmarks.com/story17820684/diplom/]brightbookmarks.com/story17820684/diplom[/url]
[url=http://socialmediainuk.com/story18161268/diplom/]socialmediainuk.com/story18161268/diplom[/url]
Douglasidomb
August 15, 2024 @ 9:00 pm
top online pharmacy india: Best Indian pharmacy – online shopping pharmacy india
Lazrfzc
August 15, 2024 @ 9:08 pm
[u][b] Здравствуйте![/b][/u]
[b]Мы изготавливаем дипломы[/b] любой профессии по разумным ценам.
[url=http://telegra.ph/v-kakih-voennyh-vuzah-est-zaochnoe-obuchenie-08-02/]telegra.ph/v-kakih-voennyh-vuzah-est-zaochnoe-obuchenie-08-02[/url]
Jamesbip
August 15, 2024 @ 10:19 pm
Federation of Scout Associations of Spain https://scout.es/scout-academy-2017 is a non-profit entity whose registered office is located in Lake
WilliamBluse
August 15, 2024 @ 10:21 pm
A collection of 18-25-year olds from around the UK https://www.scouts.org.uk/volunteers/running-things-locally/recruiting-and-managing-volunteers/role-descriptions/uk-rep-pool/ who are trained to represent the UK Scouts at events in the UK and abroad.
Jamesbem
August 15, 2024 @ 10:37 pm
грузовой подъемник https://gruzovye-podemniki-zakazat.ru
Williamraics
August 15, 2024 @ 10:42 pm
[u][b] Привет, друзья![/b][/u]
Мы готовы предложить дипломы.
[url=http://thelspr.listbb.ru/posting.php?mode=post&f=13/]thelspr.listbb.ru/posting.php?mode=post&f=13[/url]
[url=http://mos.flybb.ru/posting.php?mode=post&f=2&sid=c33bb4d8f97cadeef1a70e8444bd540a/]mos.flybb.ru/posting.php?mode=post&f=2&sid=c33bb4d8f97cadeef1a70e8444bd540a[/url]
[url=http://flystarlady.listbb.ru/ucp.php?mode=login&sid=373068622930fbff35d2e3ed8c7c0e17/]flystarlady.listbb.ru/ucp.php?mode=login&sid=373068622930fbff35d2e3ed8c7c0e17[/url]
[url=http://o91746bp.beget.tech/2024/07/09/kak-kupit-diplom-bez-lishnih-hlopot/]o91746bp.beget.tech/2024/07/09/kak-kupit-diplom-bez-lishnih-hlopot[/url]
[url=http://msfo-soft.ru/msfo/forum/messages/forum24/topic10279/message285871/?result=new#message285871/]msfo-soft.ru/msfo/forum/messages/forum24/topic10279/message285871/?result=new#message285871[/url]
Danromr
August 15, 2024 @ 10:52 pm
[u][b] Привет, друзья![/b][/u]
Мы изготавливаем дипломы любой профессии.
[url=http://ntep2008.com/index.php?name=webboard&file=read&id=18801]ntep2008.com/index.php?name=webboard&file=read&id=18801[/url]
[url=http://garagesale.es/author/paigeulrich]garagesale.es/author/paigeulrich[/url]
[url=http://cherryandgriffiths.co.uk/diplom-kupit-553126ob]cherryandgriffiths.co.uk/diplom-kupit-553126ob[/url]
[url=http://smiletraveling.com/2024/08/10/diplom-kupit-117727hl]smiletraveling.com/2024/08/10/diplom-kupit-117727hl[/url]
[url=http://arandaasesoria.com/diplom-88059yfshh]arandaasesoria.com/diplom-88059yfshh[/url]
Yreftpr
August 15, 2024 @ 11:50 pm
[u][b] Добрый день![/b][/u]
[b]Приобрести диплом любого университета.[/b]
[url=http://freeschool.ru/index.php?option=com_easybookreloaded/]freeschool.ru/index.php?option=com_easybookreloaded[/url]
Lazrgbb
August 16, 2024 @ 12:08 am
[u][b] Здравствуйте![/b][/u]
[b]Мы готовы предложить дипломы[/b] любых профессий по приятным ценам.
[url=http://telegra.ph/dlya-chego-nuzhna-spravka-ob-obuchenii-v-vuze-08-02/]telegra.ph/dlya-chego-nuzhna-spravka-ob-obuchenii-v-vuze-08-02[/url]
Mazrjgh
August 16, 2024 @ 12:46 am
[u][b] Здравствуйте![/b][/u]
Как оказалось, купить диплом кандидата наук не так уж и сложно
[url=http://okprint.kz/uslugi/13-korporativnye-podarki]okprint.kz/uslugi/13-korporativnye-podarki[/url]
Manrvzg
August 16, 2024 @ 1:00 am
[u][b] Привет, друзья![/b][/u]
Купить документ о получении высшего образования
[url=http://gtj.kr/board_KtRj53/552337]gtj.kr/board_KtRj53/552337[/url]
[url=http://welnesbiolabs.com/diplom-16325kxbaw]welnesbiolabs.com/diplom-16325kxbaw[/url]
[url=http://garagesale.es/author/lolitaspeed]garagesale.es/author/lolitaspeed[/url]
[url=http://offmarketbusinessforsale.com/diplom-kupit-835290ss]offmarketbusinessforsale.com/diplom-kupit-835290ss[/url]
[url=http://laviehub.com/blog/diplom-562229wbrkl]laviehub.com/blog/diplom-562229wbrkl[/url]
Cazrdqv
August 16, 2024 @ 1:45 am
[u][b] Привет, друзья![/b][/u]
Мы предлагаем документы ВУЗов
[url=http://suitsandsuitsblog.com/writer/2019/02/28/20190315-3/#comment-387983]suitsandsuitsblog.com/writer/2019/02/28/20190315-3/#comment-387983[/url]
Lazriyk
August 16, 2024 @ 2:11 am
[u][b] Здравствуйте![/b][/u]
[b]Приобрести диплом любого университета.[/b]
[url=http://telegra.ph/obuchenie-v-vuzah-polshi-na-russkom-yazyke-08-02]telegra.ph/obuchenie-v-vuzah-polshi-na-russkom-yazyke-08-02[/url]
Sazrmxp
August 16, 2024 @ 4:20 am
[u][b] Привет![/b][/u]
Диплом бакалавра
[url=http://telegra.ph/onlajn-testy-po-matematike-dlya-postupleniya-v-vuz-08-02/]telegra.ph/onlajn-testy-po-matematike-dlya-postupleniya-v-vuz-08-02[/url]
Cazrija
August 16, 2024 @ 4:23 am
[u][b] Добрый день![/b][/u]
Купить диплом ВУЗа
[url=http://telegra.ph/postuplenie-v-vuz-italii-08-02/]telegra.ph/postuplenie-v-vuz-italii-08-02[/url]
DavidNup
August 16, 2024 @ 4:58 am
вызвать такси в новочеркасске https://vyzvat-taxi-novocherkassk.ru
Travisnof
August 16, 2024 @ 5:01 am
вызвать такси новочеркасск номер служба такси
Robertmeela
August 16, 2024 @ 5:13 am
дешевое такси шахтах такси шахты цены
Samuelfluse
August 16, 2024 @ 5:16 am
номер такси город вызвать такси в шахтах
Xazrtuc
August 16, 2024 @ 5:48 am
[u][b] Привет![/b][/u]
Купить документ о получении высшего образования.
Мы предлагаем [b]дипломы[/b] любой профессии по доступным ценам.
[url=http://zzatem.com/read-blog/4134]zzatem.com/read-blog/4134[/url]
[b]Будем рады вам помочь![/b].
Diplomi_mcSi
August 16, 2024 @ 6:04 am
Привет!
Заказать документ института вы сможете в нашей компании.
[url=http://zakusim.blogspot.com/2014/04/blog-post/]zakusim.blogspot.com/2014/04/blog-post[/url]
Uazrmkb
August 16, 2024 @ 6:39 am
[u][b]Привет![/b][/u]
[b]Заказать диплом любого университета.[/b]
[url=http://telegra.ph/diplom-mgu-kupit-08-13-9]telegra.ph/diplom-mgu-kupit-08-13-9[/url]
Mazrsrv
August 16, 2024 @ 8:02 am
[u][b] Добрый день![/b][/u]
[b]Легальная покупка диплома о среднем образовании в Москве и регионах[/b]
[url=http://mathematics-time.blogspot.com/2013/02/blog-post_9352]mathematics-time.blogspot.com/2013/02/blog-post_9352[/url]
Douglasidomb
August 16, 2024 @ 8:12 am
indian pharmacy online: Online India pharmacy – pharmacy website india
Cazrkhp
August 16, 2024 @ 8:42 am
[u][b] Привет, друзья![/b][/u]
Мы можем предложить документы техникумов
[url=http://telegra.ph/kupit-diplom-nizhnij-tagil-08-13-3 ]telegra.ph/kupit-diplom-nizhnij-tagil-08-13-3[/url]
Lariorgzq
August 16, 2024 @ 8:55 am
[u][b]Привет,друзья![/b][/u]
[b]Приобрести диплом университета.[/b]
[url=http://telegra.ph/kupit-diplom-o-vysshem-obrazovanii-ukraina-08-13-6]telegra.ph/kupit-diplom-o-vysshem-obrazovanii-ukraina-08-13-6[/url]
Diplomi_gySi
August 16, 2024 @ 9:14 am
Здравствуйте!
Приобрести документ о получении высшего образования можно у нас в столице.
[url=http://octaniumsw.site/2018/01/ssd-m2-acer-nitro-5-an515-41-f9wl/]octaniumsw.site/2018/01/ssd-m2-acer-nitro-5-an515-41-f9wl[/url]
Lazrpwt
August 16, 2024 @ 9:54 am
[b]Заказать диплом о высшем образовании.[/b]
[url=http://igenplan.ru/forum/user/94242/]igenplan.ru/forum/user/94242[/url]
[url=http://drovokol.ru/forum/user/12134/]drovokol.ru/forum/user/12134[/url]
[url=http://raiter.flyboard.ru/viewtopic.php?f=3&t=2368/]raiter.flyboard.ru/viewtopic.php?f=3&t=2368[/url]
[url=http://borderforum.ru/viewtopic.php?f=7&t=10280/]borderforum.ru/viewtopic.php?f=7&t=10280[/url]
[url=http://pnevmokzn.80lvl.ru/ucp.php?mode=register&sid=7e45131f73087ea43c607504fe2aff1c/]pnevmokzn.80lvl.ru/ucp.php?mode=register&sid=7e45131f73087ea43c607504fe2aff1c[/url]
Dnrtpvz
August 16, 2024 @ 10:07 am
[b]Приобрести документ[/b] о получении высшего образования можно в нашей компании.
[url=http://kbookmarking.com/story17632075/diplom/]kbookmarking.com/story17632075/diplom[/url]
[url=http://vacayla.com/2024/07/29/diplom-kupit-327506cs/]vacayla.com/2024/07/29/diplom-kupit-327506cs[/url]
[url=http://kitcart.ae/2024/07/29/diplom-kupit-108085vv/]kitcart.ae/2024/07/29/diplom-kupit-108085vv[/url]
[url=http://voiceof.com/diplom-464540tvtku/]voiceof.com/diplom-464540tvtku[/url]
[url=http://orangebookmarks.com/story17705872/diplom/]orangebookmarks.com/story17705872/diplom[/url]
Douglasidomb
August 16, 2024 @ 10:22 am
Online medicine order: Top online pharmacy in India – indian pharmacies safe
Trefabu
August 16, 2024 @ 10:48 am
[u][b] Здравствуйте![/b][/u]
Всё, что нужно знать о покупке аттестата о среднем образовании без рисков
[url=http://ninokuni.ru/showthread.php?tid=4132/]ninokuni.ru/showthread.php?tid=4132[/url]
Будем рады вам помочь!.
Williamraics
August 16, 2024 @ 11:45 am
[u][b] Привет![/b][/u]
Мы можем предложить дипломы.
[url=http://worksale.jofo.me/2279076?_ga=2.224405722.150041222.1720601601-78685845.1720601601/]worksale.jofo.me/2279076?_ga=2.224405722.150041222.1720601601-78685845.1720601601[/url]
[url=http://4yo.us/blogs/141452/Купить-диплом-все-что-вам-нужно-знать/]4yo.us/blogs/141452/Купить-диплом-все-что-вам-нужно-знать[/url]
[url=http://mosday.getbb.ru/viewtopic.php?f=12&t=1245/]mosday.getbb.ru/viewtopic.php?f=12&t=1245[/url]
[url=http://planetasp.ru/forum/post.php?fid=8/]planetasp.ru/forum/post.php?fid=8[/url]
[url=http://bn.tobase.ru/viewtopic.php?f=30&t=3346/]bn.tobase.ru/viewtopic.php?f=30&t=3346[/url]
Dnrttks
August 16, 2024 @ 1:04 pm
[b]Заказать документ[/b] института можно в нашей компании в столице.
[url=http://tigaedu.com/blog/index.php?entryid=327717/]tigaedu.com/blog/index.php?entryid=327717[/url]
[url=http://digitalmagazine.xyz/diplom-274481nrcfk/]digitalmagazine.xyz/diplom-274481nrcfk[/url]
[url=http://social40.com/story2998084/diplom/]social40.com/story2998084/diplom[/url]
[url=http://xatrivietnam.vn/question/diplom-481105aidrr/]xatrivietnam.vn/question/diplom-481105aidrr[/url]
[url=http://a4everyone.org/diplom-kupit-135270lt/]a4everyone.org/diplom-kupit-135270lt[/url]
Mazrhmc
August 16, 2024 @ 1:32 pm
[u][b] Здравствуйте![/b][/u]
Как правильно приобрести диплом колледжа или ПТУ в России, важные моменты
[url=http://civilpatrol.info/?q=petition/81346/premialniediploms]civilpatrol.info/?q=petition/81346/premialniediploms[/url]
Trefvuh
August 16, 2024 @ 1:36 pm
[u][b] Привет![/b][/u]
Полезные советы по безопасной покупке диплома о высшем образовании
[url=http://miupsik.ru/forums/showthread.php?tid=49745/]miupsik.ru/forums/showthread.php?tid=49745[/url]
Рады оказать помощь!.
Manrhtb
August 16, 2024 @ 1:54 pm
[u][b] Добрый день![/b][/u]
Купить документ о получении высшего образования
[url=http://njkkot.org/video/1246238]njkkot.org/video/1246238[/url]
[url=http://starryjeju.com/qna/5958379]starryjeju.com/qna/5958379[/url]
[url=http://welnesbiolabs.com/diplom-734905dmpba]welnesbiolabs.com/diplom-734905dmpba[/url]
[url=http://investicos.com/uncategorized/diplom-295850tapxt]investicos.com/uncategorized/diplom-295850tapxt[/url]
[url=http://yourbuyersguide.co.uk/diplom-54369xjlpx]yourbuyersguide.co.uk/diplom-54369xjlpx[/url]
Cazrrhk
August 16, 2024 @ 2:26 pm
[u][b] Привет![/b][/u]
Мы предлагаем документы техникумов
[url=http://rive-import.ru/privet-mir/#comment-15691]rive-import.ru/privet-mir/#comment-15691[/url]
Cazrwro
August 16, 2024 @ 2:51 pm
[u][b] Привет, друзья![/b][/u]
Получить диплом института
[url=http://telegra.ph/minimalnye-bally-dlya-postupleniya-v-vuz-2017-08-02/]telegra.ph/minimalnye-bally-dlya-postupleniya-v-vuz-2017-08-02[/url]
Sazrruj
August 16, 2024 @ 2:53 pm
[u][b] Добрый день![/b][/u]
Диплом специалиста
[url=http://telegra.ph/preimushchestva-zolotoj-medali-pri-postuplenii-v-vuz-08-02/]telegra.ph/preimushchestva-zolotoj-medali-pri-postuplenii-v-vuz-08-02[/url]
Lazrkjx
August 16, 2024 @ 3:33 pm
[u][b] Привет, друзья![/b][/u]
[b]Приобрести диплом любого университета.[/b]
[url=http://telegra.ph/vhodit-li-obuchenie-v-vuze-v-obshchij-trudovoj-stazh-08-02]telegra.ph/vhodit-li-obuchenie-v-vuze-v-obshchij-trudovoj-stazh-08-02[/url]
Yrefior
August 16, 2024 @ 4:07 pm
[u][b] Здравствуйте![/b][/u]
[b]Купить диплом любого ВУЗа.[/b]
[url=http://smeshlivaja.blogspot.com/2014/04/blog-post/]smeshlivaja.blogspot.com/2014/04/blog-post[/url]
Xazrrat
August 16, 2024 @ 4:56 pm
[u][b] Добрый день![/b][/u]
Заказать документ о получении высшего образования.
Мы изготавливаем [b]дипломы[/b] любой профессии по приятным ценам.
[url=http://emoneyspace.com/paulsmith7]emoneyspace.com/paulsmith7[/url]
[b]Поможем вам всегда![/b].
Lazropy
August 16, 2024 @ 4:59 pm
[u][b] Здравствуйте![/b][/u]
[b]Мы изготавливаем дипломы[/b] любой профессии по выгодным ценам.
[url=http://telegra.ph/dogovor-ob-obuchenii-v-vuze-08-02/]telegra.ph/dogovor-ob-obuchenii-v-vuze-08-02[/url]
Mazrzsj
August 16, 2024 @ 7:01 pm
[u][b] Привет, друзья![/b][/u]
[b]Как официально приобрести аттестат 11 класса с минимальными затратами времени[/b]
[url=http://j95370pe.bget.ru/main/263-vot-kudy-otvechala-pomeschica-moe-takoe]j95370pe.bget.ru/main/263-vot-kudy-otvechala-pomeschica-moe-takoe[/url]
Yreffoc
August 16, 2024 @ 7:08 pm
[u][b] Привет, друзья![/b][/u]
[b]Заказать диплом университета.[/b]
[url=http://pagenotfound94.blogspot.com/2017/03/construct-2_97/]pagenotfound94.blogspot.com/2017/03/construct-2_97[/url]
Sazrgwp
August 16, 2024 @ 7:41 pm
[u][b] Добрый день![/b][/u]
Приобрести документ института
[url=http://telegra.ph/besplatnoe-obuchenie-v-vuzah-germanii-07-24/]telegra.ph/besplatnoe-obuchenie-v-vuzah-germanii-07-24[/url]
[url=http://telegra.ph/kak-uznat-pro-postuplenie-v-vuz-07-24/]telegra.ph/kak-uznat-pro-postuplenie-v-vuz-07-24[/url]
[url=http://kharkov-balka.com/showthread.php?p=155503#post155503/]kharkov-balka.com/showthread.php?p=155503#post155503[/url]
[url=http://4mark.net/story/12411296/купить-диплом-о-высшем-образовании-гознак-любого-вуза-россии/]4mark.net/story/12411296/купить-диплом-о-высшем-образовании-гознак-любого-вуза-россии[/url]
[url=http://forum.mybb.ru/viewtopic.php?id=38567#p939931/]forum.mybb.ru/viewtopic.php?id=38567#p939931[/url]
Sazrncw
August 16, 2024 @ 7:54 pm
[u][b] Привет![/b][/u]
Мы готовы предложить [b]дипломы[/b] любой профессии.
Заказ [b]диплома[/b], который подтверждает обучение в ВУЗе, – это выгодное решение.
[url=http://mykinotime.ru/page/5]mykinotime.ru/page/5[/url]
[b]Рады оказать помощь![/b].
Lazrtha
August 16, 2024 @ 7:56 pm
[u][b] Привет![/b][/u]
[b]Мы можем предложить дипломы[/b] любых профессий по доступным ценам.
[url=http://telegra.ph/kto-imeet-lgoty-pri-postuplenii-v-vuz-08-02/]telegra.ph/kto-imeet-lgoty-pri-postuplenii-v-vuz-08-02[/url]
Douglasidomb
August 16, 2024 @ 9:36 pm
mexican online pharmacies prescription drugs: Mexico pharmacy online – mexico drug stores pharmacies
Cazrawr
August 17, 2024 @ 12:09 am
[u][b] Привет![/b][/u]
Мы готовы предложить документы ВУЗов
[url=http://divephotoguide.com/user/danielpark]divephotoguide.com/user/danielpark[/url]
Trefvxh
August 17, 2024 @ 2:20 am
[u][b] Привет, друзья![/b][/u]
Стоимость дипломов высшего и среднего образования и как избежать подделок
[url=http://rf-4fun.ru/index.php?/profile/1774-michellthymn/]rf-4fun.ru/index.php?/profile/1774-michellthymn[/url]
Всегда вам поможем!.
Dnrtoin
August 17, 2024 @ 3:50 am
[u][b]Добрый день![/b][/u]
[b]Приобрести документ[/b] о получении высшего образования вы сможете в нашей компании в Москве.
[url=http://velopiter.spb.ru/profile/108271-hnfmrfdtg/?tab=field_core_pfield_1]velopiter.spb.ru/profile/108271-hnfmrfdtg/?tab=field_core_pfield_1[/url]
Trefzuf
August 17, 2024 @ 5:09 am
[u][b] Добрый день![/b][/u]
Вопросы и ответы: можно ли быстро купить диплом старого образца?
[url=http://radio-wave.ru/forum/member.php?3622-lucymorZEnce/]radio-wave.ru/forum/member.php?3622-lucymorZEnce[/url]
Поможем вам всегда!.
Danrskh
August 17, 2024 @ 6:04 am
[u][b] Привет![/b][/u]
Мы изготавливаем дипломы любой профессии.
[url=http://youwantech.com/xe/board/700488]youwantech.com/xe/board/700488[/url]
[url=http://laviehub.com/blog/diplom-kupit-328426ic]laviehub.com/blog/diplom-kupit-328426ic[/url]
[url=http://welnesbiolabs.com/diplom-837413obggk]welnesbiolabs.com/diplom-837413obggk[/url]
[url=http://managerhotels.com/business-small-business/diplom-kupit-148426jm]managerhotels.com/business-small-business/diplom-kupit-148426jm[/url]
[url=http://gen8ai.com/groups/diplom-kupit-153607ez/members/all-members]gen8ai.com/groups/diplom-kupit-153607ez/members/all-members[/url]
Sazrifk
August 17, 2024 @ 6:04 am
[u][b] Привет, друзья![/b][/u]
Мы можем предложить [b]дипломы[/b] психологов, юристов, экономистов и других профессий.
Приобретение [b]диплома[/b], который подтверждает обучение в ВУЗе, – это грамотное решение.
[url=http://aurorahcs.com/forum/viewtopic.php?t=682524]aurorahcs.com/forum/viewtopic.php?t=682524[/url]
[b]Поможем вам всегда![/b].
Diplomi_weSi
August 17, 2024 @ 6:10 am
Здравствуйте!
Приобрести документ ВУЗа вы можете у нас в столице.
[url=http://ashraegoldcoast.com/sponsors-daikin/#comment-792677/]ashraegoldcoast.com/sponsors-daikin/#comment-792677[/url]
Sazrfzd
August 17, 2024 @ 6:30 am
[u][b] Добрый день![/b][/u]
Заказать документ университета
[url=http://bdfeu.ukraine7.com/t20091-topic#36555/]bdfeu.ukraine7.com/t20091-topic#36555[/url]
[url=http://a90275db.beget.tech/2024/07/22/poluchite-diplom-i-dostignite-vysot/]a90275db.beget.tech/2024/07/22/poluchite-diplom-i-dostignite-vysot[/url]
[url=http://rabotavinternete.forum2x2.ru/t55667-topic#138484/]rabotavinternete.forum2x2.ru/t55667-topic#138484[/url]
[url=http://telegra.ph/distancionnoe-obuchenie-v-vuzah-rossii-07-24/]telegra.ph/distancionnoe-obuchenie-v-vuzah-rossii-07-24[/url]
[url=http://deadreckoninggame.com/index.php/Ваш_путь_к_успеху:_диплом_о_высшем_образовании/]deadreckoninggame.com/index.php/Ваш_путь_к_успеху:_диплом_о_высшем_образовании[/url]
Dnrtfup
August 17, 2024 @ 7:01 am
[u][b]Привет![/b][/u]
[b]Купить документ[/b] о получении высшего образования можно в нашей компании в Москве.
[url=http://armrus.org/extranet/forum/user/155439/]armrus.org/extranet/forum/user/155439/[/url]
Mazrxci
August 17, 2024 @ 7:12 am
[u][b] Привет![/b][/u]
[b]Быстрое обучение и получение диплома магистра – возможно ли это?[/b]
[url=http://clicky.buzz/index.php?do=/public/blog/view/id_119666/title_/]clicky.buzz/index.php?do=/public/blog/view/id_119666/title_/[/url]
Oariorqpw
August 17, 2024 @ 7:28 am
[u][b] Привет![/b][/u]
Приобрести диплом университета
[b]Мы предлагаем[/b] быстро и выгодно заказать диплом, который выполняется на бланке ГОЗНАКа и заверен мокрыми печатями, водяными знаками, подписями должностных лиц. Диплом способен пройти лубую проверку, даже с использованием профессиональных приборов. Достигайте цели быстро и просто с нашей компанией.
[b]Где заказать диплом по актуальной специальности?[/b]
[url=http://biiut.com/read-blog/65173/]biiut.com/read-blog/65173[/url]
[url=http://animesocial.su/blogs/3876/Оформите-диплом-и-начните-карьеру/]animesocial.su/blogs/3876/Оформите-диплом-и-начните-карьеру[/url]
[url=http://lighttur.ru/diplomyi-dlya-vashego-uspeha/]lighttur.ru/diplomyi-dlya-vashego-uspeha[/url]
[url=http://cv53297-livestreet-1.tw1.ru/2024/07/23/diplomy-vseh-urovney-obrazovaniya/]cv53297-livestreet-1.tw1.ru/2024/07/23/diplomy-vseh-urovney-obrazovaniya[/url]
[url=http://superforum.spybb.ru/viewtopic.php?id=2771#p50818/]superforum.spybb.ru/viewtopic.php?id=2771#p50818[/url]
Yreflwj
August 17, 2024 @ 9:04 am
[u][b] Привет, друзья![/b][/u]
[b]Приобрести диплом о высшем образовании.[/b]
[url=http://sergei-cheremushkin.blogspot.com/2010/03/blog-post/]sergei-cheremushkin.blogspot.com/2010/03/blog-post[/url]
Diplomi_bySi
August 17, 2024 @ 9:09 am
Добрый день!
Купить документ института вы сможете в нашем сервисе.
[url=http://shtsh.blogspot.com/2012/01/asus-eee-pc-1201nl/]shtsh.blogspot.com/2012/01/asus-eee-pc-1201nl[/url]
Lazrzxq
August 17, 2024 @ 10:39 am
[u][b] Добрый день![/b][/u]
[b]Мы можем предложить дипломы[/b] психологов, юристов, экономистов и любых других профессий по доступным ценам.
[url=http://telegra.ph/kakie-dokumenty-dlya-postupleniya-v-vuz-08-02/]telegra.ph/kakie-dokumenty-dlya-postupleniya-v-vuz-08-02[/url]
Cazrqsn
August 17, 2024 @ 11:11 am
[u][b] Привет, друзья![/b][/u]
Мы предлагаем документы техникумов
[url=http://divephotoguide.com/user/danielpark/]divephotoguide.com/user/danielpark[/url]
Yrefoac
August 17, 2024 @ 12:11 pm
[u][b] Привет, друзья![/b][/u]
[b]Заказать диплом о высшем образовании.[/b]
[url=http://elektrofahrrad-tests.de/forums/showthread.php?tid=226033/]elektrofahrrad-tests.de/forums/showthread.php?tid=226033[/url]
Iariorwyi
August 17, 2024 @ 12:35 pm
[u][b] Привет![/b][/u]
Заказать документ университета можно у нас в столице. Мы предлагаем документы об окончании любых ВУЗов России. Вы сможете получить диплом по любым специальностям, включая документы Советского Союза. Гарантируем, что в случае проверки документа работодателем, подозрений не возникнет.
[url=http://leadrp.listbb.ru/viewtopic.php?f=3&t=415/]leadrp.listbb.ru/viewtopic.php?f=3&t=415[/url]
[url=http://minimoo.eu/index.php/en/forum/welcome-mat/667418/]minimoo.eu/index.php/en/forum/welcome-mat/667418[/url]
[url=http://u2c.tv/member.php?u=30984/]u2c.tv/member.php?u=30984[/url]
[url=http://dragonlivers.ixbb.ru/viewtopic.php?id=126#p126/]dragonlivers.ixbb.ru/viewtopic.php?id=126#p126[/url]
[url=http://a90275db.beget.tech/2024/06/18/pochemu-oficialnyy-diplom-vazhen-v-karere/]a90275db.beget.tech/2024/06/18/pochemu-oficialnyy-diplom-vazhen-v-karere[/url]
Danrqqy
August 17, 2024 @ 1:34 pm
[u][b] Добрый день![/b][/u]
Мы предлагаем дипломы любой профессии.
[url=http://eythar.org/blog/index.php?entryid=655797]eythar.org/blog/index.php?entryid=655797[/url]
[url=http://starryjeju.com/?document_srl=5899563]starryjeju.com/?document_srl=5899563[/url]
[url=http://voiceof.com/diplom-kupit-165147sw]voiceof.com/diplom-kupit-165147sw[/url]
[url=http://utahsyardsale.com/author/hughpink38]utahsyardsale.com/author/hughpink38[/url]
[url=http://offmarketbusinessforsale.com/diplom-kupit-908005nx]offmarketbusinessforsale.com/diplom-kupit-908005nx[/url]
Lazrcjf
August 17, 2024 @ 1:46 pm
[u][b] Здравствуйте![/b][/u]
[b]Мы изготавливаем дипломы[/b] любых профессий по доступным тарифам.
[url=http://telegra.ph/nalogovyj-vychet-na-obuchenie-rebenka-v-vuze-08-02/]telegra.ph/nalogovyj-vychet-na-obuchenie-rebenka-v-vuze-08-02[/url]
Mazrmow
August 17, 2024 @ 3:47 pm
[u][b] Добрый день![/b][/u]
[b]Официальная покупка школьного аттестата с упрощенным обучением в Москве[/b]
[url=http://danceway74.ru/users/36?page=3]danceway74.ru/users/36?page=3[/url]
Iariornev
August 17, 2024 @ 4:28 pm
[u][b] Здравствуйте![/b][/u]
Приобрести документ ВУЗа вы имеете возможность в нашем сервисе. Мы предлагаем документы об окончании любых университетов Российской Федерации. Вы получите необходимый диплом по любой специальности, любого года выпуска, в том числе документы СССР. Гарантируем, что в случае проверки документа работодателем, подозрений не возникнет.
[url=http://georgiantheatre.ge/index.php?subaction=userinfo&user=ibilys]georgiantheatre.ge/index.php?subaction=userinfo&user=ibilys[/url]
[url=http://vivamedia.ca/5-benefits-of-promotional-videos-for-your-company/#comment-854171]vivamedia.ca/5-benefits-of-promotional-videos-for-your-company/#comment-854171[/url]
[url=http://forumru.uniutilis.com/profile.php?action=show&member=6613]forumru.uniutilis.com/profile.php?action=show&member=6613[/url]
[url=http://metal-tracker.com/user/activate/id/927429197/secret/4b51abacb0105b29aeb2a6c3c7d61718]metal-tracker.com/user/activate/id/927429197/secret/4b51abacb0105b29aeb2a6c3c7d61718[/url]
[url=http://chaek.ru/matryoshki/matreshka-zima-art-79/reviews]chaek.ru/matryoshki/matreshka-zima-art-79/reviews[/url]
Trefhah
August 17, 2024 @ 5:56 pm
[u][b] Здравствуйте![/b][/u]
Официальная покупка диплома вуза с сокращенной программой обучения в Москве
[url=http://reisezielforum.de/viewtopic.php?t=157052/]reisezielforum.de/viewtopic.php?t=157052[/url]
Всегда вам поможем!.
Trefktz
August 17, 2024 @ 8:41 pm
[u][b] Здравствуйте![/b][/u]
Как оказалось, купить диплом кандидата наук не так уж и сложно
[url=http://speicher-photovoltaik.de/profile/?u=31803/]speicher-photovoltaik.de/profile/?u=31803[/url]
Рады оказать помощь!.
Dnrtdot
August 17, 2024 @ 9:21 pm
[u][b]Привет![/b][/u]
[b]Приобрести документ[/b] о получении высшего образования вы можете у нас в столице.
[url=http://diplom-moskva-f166630.promportal.su/about]diplom-moskva-f166630.promportal.su/about[/url]
fryd
August 17, 2024 @ 11:31 pm
Thank you for the step-by-step instructions. As a beginner, I really appreciate how easy you make it to follow along.please vifrydsit my website https://www.officialfryd.com/
Iarioresc
August 17, 2024 @ 11:37 pm
[u][b] Добрый день![/b][/u]
Заказать документ о получении высшего образования можно в нашей компании. Мы предлагаем документы об окончании любых университетов Российской Федерации. Вы сможете получить необходимый диплом по любой специальности, любого года выпуска, включая документы старого образца. Гарантируем, что в случае проверки документов работодателем, каких-либо подозрений не возникнет.
[url=http://climat-cold.ru/forum/user/3872/]climat-cold.ru/forum/user/3872[/url]
[url=http://twikkers.nl/blogs/242417/Дипломы-для-успешной-карьеры/]twikkers.nl/blogs/242417/Дипломы-для-успешной-карьеры[/url]
[url=http://vipka.0bb.ru/viewtopic.php?id=5937#p8950/]vipka.0bb.ru/viewtopic.php?id=5937#p8950[/url]
[url=http://avtolux48.ru/people/user/374/blog/8001/]avtolux48.ru/people/user/374/blog/8001[/url]
[url=http://newrealgames.ru/diplomyi-na-zakaz-bez-lishnih-hlopot/]newrealgames.ru/diplomyi-na-zakaz-bez-lishnih-hlopot[/url]
Dnrtvim
August 18, 2024 @ 12:28 am
[u][b]Здравствуйте![/b][/u]
[b]Приобрести документ[/b] института вы можете у нас в Москве.
[url=http://forumrabota.0pk.me/viewtopic.php?id=7320#p16792]forumrabota.0pk.me/viewtopic.php?id=7320#p16792[/url]
Williamraics
August 18, 2024 @ 2:09 am
[u][b] Добрый день![/b][/u]
Мы изготавливаем дипломы.
[url=http://zehov.3nx.ru/viewtopic.php?p=6799#6799]zehov.3nx.ru/viewtopic.php?p=6799#6799[/url]
indiiskii pasyans _wqSn
August 18, 2024 @ 3:20 am
индийский пасьянс гадать [url=https://indiyskiy-pasyans-online.ru/]индийский пасьянс гадать[/url] .
Iariorypt
August 18, 2024 @ 3:33 am
[u][b] Привет, друзья![/b][/u]
Купить документ ВУЗа можно у нас в столице. Мы оказываем услуги по продаже документов об окончании любых университетов России. Вы сможете получить диплом по любой специальности, включая документы СССР. Гарантируем, что в случае проверки документов работодателем, подозрений не появится.
[url=http://mc-pe.net/index.php?subaction=userinfo&user=ytozes]mc-pe.net/index.php?subaction=userinfo&user=ytozes[/url]
[url=http://forum.computest.ru/post/711574/#p711574]forum.computest.ru/post/711574/#p711574[/url]
[url=http://ffil.blogspot.com/2011/01/40]ffil.blogspot.com/2011/01/40[/url]
[url=http://e-book-ss-elen-khitrova.blogspot.com/2013/12/blog-post_7320]e-book-ss-elen-khitrova.blogspot.com/2013/12/blog-post_7320[/url]
[url=http://f1-f1helpe.1gb.ru/component]f1-f1helpe.1gb.ru/component[/url]
Yrefgjt
August 18, 2024 @ 5:30 am
[u][b] Привет, друзья![/b][/u]
[b]Заказать диплом любого ВУЗа.[/b]
[url=http://mlybovm.blogspot.com/2018/03/s-dobrym-utrom-8-marta/]mlybovm.blogspot.com/2018/03/s-dobrym-utrom-8-marta[/url]
Diplomi_zwSi
August 18, 2024 @ 5:41 am
Здравствуйте!
Заказать документ ВУЗа вы сможете в нашей компании в Москве.
[url=http://kvartirker.ru/profile/olohosub/]kvartirker.ru/profile/olohosub[/url]
Cazrgke
August 18, 2024 @ 6:38 am
[u][b] Здравствуйте![/b][/u]
Купить диплом академии
[url=http://telegra.ph/postuplenie-v-medicinskij-vuz-posle-kolledzha-08-02/]telegra.ph/postuplenie-v-medicinskij-vuz-posle-kolledzha-08-02[/url]
Sazrucs
August 18, 2024 @ 7:19 am
[u][b] Привет![/b][/u]
Диплом бакалавра
[url=http://telegra.ph/interaktivnye-metody-obucheniya-v-vuze-08-02/]telegra.ph/interaktivnye-metody-obucheniya-v-vuze-08-02[/url]
Mazrvgo
August 18, 2024 @ 7:52 am
[u][b] Здравствуйте![/b][/u]
Легальная покупка диплома ПТУ с сокращенной программой обучения
[url=http://198.199.122.222/doku.php?id=luxdiplomru/]198.199.122.222/doku.php?id=luxdiplomru[/url]
svadebnyy_ugpi
August 18, 2024 @ 7:56 am
Продление жизни свадебного букета: полезные советы
букет невесты своими руками [url=https://0gorodnik.ru/]букет невесты своими руками[/url] .
Yrefuwy
August 18, 2024 @ 8:34 am
[u][b] Добрый день![/b][/u]
[b]Приобрести диплом о высшем образовании.[/b]
[url=http://news2.ru/profile/markedmjub/]news2.ru/profile/markedmjub[/url]
Diplomi_ktSi
August 18, 2024 @ 8:38 am
Привет!
Приобрести документ о получении высшего образования вы имеете возможность у нас.
[url=http://mebelnyvkus.ru/kuhnja/kuxonnye-stulya/stul-ccpe-m/]mebelnyvkus.ru/kuhnja/kuxonnye-stulya/stul-ccpe-m[/url]
kompoziciy_oppa
August 18, 2024 @ 9:32 am
Топ-20 вариантов цветочных композиций для украшения вашего дома, советы по подбору.
Очаровательные цветочные композиции для вашего участка, которые придадут вашему саду очарование.
Идеи оригинальных композиций для вашего букета, и удивят своим необычным сочетанием.
Топ-10 самых красивых композиций из цветов для свадьбы, и заставят всех гостей восхищаться.
Уникальные идеи для оформления цветочных композиций на праздник, которые заставят всех гостей восхищаться.
Как сделать необычный декор для вашего дома, и подчеркнут ваш неповторимый стиль.
Советы по созданию комфортной атмосферы в офисе, и подчеркнут стиль вашего бизнеса.
Идеи для создания оригинальных композиций из цветов на даче, и создадут атмосферу праздника на природе.
композиция цветов [url=https://101-po3a.ru/]https://101-po3a.ru/[/url] .
Cazrdfa
August 18, 2024 @ 11:05 am
[u][b] Здравствуйте![/b][/u]
Мы можем предложить документы техникумов
[url=http://metal-tracker.com/forum/index.php/]metal-tracker.com/forum/index.php[/url]
arenda_bdOt
August 18, 2024 @ 11:06 am
Экскаватор погрузчик в аренду: лучший выбор для строительных работ, воспользуйтесь услугой уже сегодня.
Экскаватор погрузчик на прокат: надежное решение для стройки, воспользуйтесь услугой уже сегодня.
Экскаватор погрузчик на прокат: удобство и эффективность на стройплощадке, воспользуйтесь услугой уже сегодня.
Экскаватор погрузчик в аренду: выгодное предложение для строительства, заказывайте прямо сейчас.
Аренда экскаватора погрузчика: надежное решение для строительства, закажите прокат сегодня.
Аренда экскаватора погрузчика: удобство и эффективность на стройке, заказывайте прямо сейчас.
аренда экскаватора погрузчика в москве [url=https://arenda-jekskavatora-pogruzchika-197.ru/]https://arenda-jekskavatora-pogruzchika-197.ru/[/url] .
dymohody_txpi
August 18, 2024 @ 12:37 pm
Выбор качественных дымоходов для бани в Нижнем Новгороде, экономно и эффективно.
Как выбрать исполнителя на установку дымоходов для бани в Нижнем Новгороде, сравнение цен и услуг.
Плюсы и минусы различных типов дымоходов для бани в Нижнем Новгороде, подбор оптимального варианта.
Дымоходы для бани в Нижнем Новгороде: какие ошибки избегать, специализированные рекомендации.
Секреты долговечности дымоходов для бани в Нижнем Новгороде, практические рекомендации.
Преимущества и недостатки распространенных дымоходов для бани в Нижнем Новгороде, подбор идеального варианта.
дымоход для печки купить в нижнем новгороде [url=https://forum-bani.ru/]https://forum-bani.ru/[/url] .
Lazrbgz
August 18, 2024 @ 1:03 pm
[u][b] Привет![/b][/u]
Заказать [b]диплом[/b] ВУЗа.
[url=http://obmenka.forum2x2.ru/t16293-topic#37488]obmenka.forum2x2.ru/t16293-topic#37488[/url]
Williamraics
August 18, 2024 @ 1:55 pm
[u][b] Привет, друзья![/b][/u]
Мы изготавливаем дипломы.
[url=http://1abakan.ru/forum/showthread-62291/]1abakan.ru/forum/showthread-62291/[/url]
chto_wbEr
August 18, 2024 @ 2:12 pm
История и значение розы в культуре и символике, прекрасное растение, воспеваемое многими поэтами и художниками.
Какие бывают виды роз, правила посадки и ухода за розами.
Как роза влияет на человека и его эмоции, роза в религии и мифологии.
Роза как идеальный подарок для любого случая, почему роза считается королевой цветов.
Роза в мифах и легендах разных народов, изысканные букеты из роз для особого случая.
описание розы цветка [url=https://roslina.ru/]https://roslina.ru/[/url] .
Manrsfe
August 18, 2024 @ 2:16 pm
[u][b] Добрый день![/b][/u]
Купить документ ВУЗа
[url=http://gtj.kr/board_KtRj53/485586/]gtj.kr/board_KtRj53/485586[/url]
[url=http://ranatourandtravels.com/diplom-kupit-400026ou/]ranatourandtravels.com/diplom-kupit-400026ou[/url]
[url=http://battat-advisors.com/?p=22233/]battat-advisors.com/?p=22233[/url]
[url=http://hariharparagovernmentiti.com/2024/08/14/diplom-211797vbhwz/]hariharparagovernmentiti.com/2024/08/14/diplom-211797vbhwz[/url]
[url=http://starryjeju.com/qna/5788786/]starryjeju.com/qna/5788786[/url]
kotel_udea
August 18, 2024 @ 3:54 pm
Выбор котла для частного дома | Какой котел для отопления дома выбрать | Лучшие цены на котлы для отопления | Сравнение котлов для отопления дома | Как правильно подключить котел для отопления | Рейтинг котлов для отопления | Где купить котел для отопления частного дома с доставкой | Лучшие котлы для отопления: какой выбрать? | Советы по экономии на отоплении | Где купить недорогой котел для отопления
купить котел для отопления частного [url=https://sauna-manzana.ru/]https://sauna-manzana.ru/[/url] .
Dnrttum
August 18, 2024 @ 3:56 pm
[u][b]Добрый день![/b][/u]
[b]Заказать документ[/b] ВУЗа можно в нашей компании в Москве.
[url=http://x70795vj.beget.tech/2024/03/09/hotite-zakazat-originalnyy-diplom]x70795vj.beget.tech/2024/03/09/hotite-zakazat-originalnyy-diplom[/url]
Mazrsqa
August 18, 2024 @ 4:07 pm
[b]Приобрести документ о получении высшего образования.[/b]
[url=http://obzs.blogspot.com/2014/06/kak-perenesti-ranenogo/]obzs.blogspot.com/2014/06/kak-perenesti-ranenogo[/url]
[url=http://shinyoungwood.co.kr/bbs/board.php?bo_table=free&wr_id=923300/]shinyoungwood.co.kr/bbs/board.php?bo_table=free&wr_id=923300[/url]
[url=http://notebooks.ru/forum/index.php?PAGE_NAME=profile_view&UID=77988/]notebooks.ru/forum/index.php?PAGE_NAME=profile_view&UID=77988[/url]
[url=http://m.7ooo.ru/o/rebeccamog/profile/]m.7ooo.ru/o/rebeccamog/profile[/url]
[url=http://rfgrasso.com/radiologia-interventistica-muscoloscheletrica/#comment-85291/]rfgrasso.com/radiologia-interventistica-muscoloscheletrica/#comment-85291[/url]
Elektrokarniz_fmmr
August 18, 2024 @ 4:07 pm
электрокарнизы купить в москве [url=www.provorota.su/]электрокарнизы купить в москве[/url] .
Larioravu
August 18, 2024 @ 5:57 pm
Официальная покупка школьного аттестата с упрощенным обучением в Москве
[url=http://telegra.ph/diplom-o-nepolnom-vysshem-obrazovanii-kupit-08-13-3/]telegra.ph/diplom-o-nepolnom-vysshem-obrazovanii-kupit-08-13-3[/url]
Lariorfen
August 18, 2024 @ 6:10 pm
Всё, что нужно знать о покупке аттестата о среднем образовании
[url=http://telegra.ph/kupit-diplom-internatury-08-13-6/]telegra.ph/kupit-diplom-internatury-08-13-6[/url]
Sazruws
August 18, 2024 @ 6:37 pm
[u][b] Здравствуйте![/b][/u]
Приобрести документ о получении высшего образования
[url=http://stonecraft.diary.ru/p221944096_zhelaete-kupit-diplom-universiteta.htm]stonecraft.diary.ru/p221944096_zhelaete-kupit-diplom-universiteta.htm[/url]
Lariorsgn
August 18, 2024 @ 6:43 pm
Официальная покупка школьного аттестата с упрощенным обучением в Москве
[url=http://telegra.ph/kupit-diplom-s-zaneseniem-v-reestr-nizhnij-novgorod-08-13-5/]telegra.ph/kupit-diplom-s-zaneseniem-v-reestr-nizhnij-novgorod-08-13-5[/url]
Mazrmxm
August 18, 2024 @ 6:46 pm
[u][b] Здравствуйте![/b][/u]
[b]Пошаговая инструкция по официальной покупке диплома о высшем образовании[/b]
[url=http://odnopolchane.net/forum/member.php?u=621054/]odnopolchane.net/forum/member.php?u=621054[/url]
Dnrtsuv
August 18, 2024 @ 6:53 pm
[u][b]Здравствуйте![/b][/u]
[b]Заказать документ[/b] института можно в нашей компании.
[url=http://bx24.avers35.ru/company/personal/user/38/blog/341/]bx24.avers35.ru/company/personal/user/38/blog/341/[/url]
Uazruvh
August 18, 2024 @ 7:02 pm
[u][b]Добрыйдень![/b][/u]
[b]Купить диплом о высшем образовании.[/b]
[url=http://telegra.ph/kupit-diplom-medicinskogo-kolledzha-08-13-10/]telegra.ph/kupit-diplom-medicinskogo-kolledzha-08-13-10[/url]
Cazrqdh
August 18, 2024 @ 7:32 pm
[u][b] Привет![/b][/u]
Приобрести диплом о высшем образовании
[url=http://telegra.ph/chto-nuzhno-dlya-postupleniya-v-vuz-08-02/]telegra.ph/chto-nuzhno-dlya-postupleniya-v-vuz-08-02[/url]
Trevojnaya knopka rosgvardiya_ksol
August 18, 2024 @ 7:36 pm
ктс вневедомственная охрана [url=www.trknpk.ru/]www.trknpk.ru/[/url] .
Sazrgwm
August 18, 2024 @ 8:25 pm
[u][b] Здравствуйте![/b][/u]
Диплом для вас
[url=http://telegra.ph/stoimost-obucheniya-v-vuzah-moskvy-2018-08-02/]telegra.ph/stoimost-obucheniya-v-vuzah-moskvy-2018-08-02[/url]
Vivod iz zapoya rostov_xwsa
August 18, 2024 @ 9:00 pm
вывод из запоя дешево ростов [url=http://www.vyvod-iz-zapoya-rostov111.ru]http://www.vyvod-iz-zapoya-rostov111.ru[/url] .
Danrivs
August 18, 2024 @ 9:36 pm
[u][b] Привет, друзья![/b][/u]
Мы изготавливаем дипломы любой профессии.
[url=http://awsomevedeos.xyz/davisvalenci/]awsomevedeos.xyz/davisvalenci[/url]
[url=http://syg-us.com/entrenamiento-virtual/blog/index.php?entryid=46941/]syg-us.com/entrenamiento-virtual/blog/index.php?entryid=46941[/url]
[url=http://investicos.com/uncategorized/diplom-546535udcvc/]investicos.com/uncategorized/diplom-546535udcvc[/url]
[url=http://gen8ai.com/groups/diplom-644259ibguy/members/all-members/]gen8ai.com/groups/diplom-644259ibguy/members/all-members[/url]
[url=http://managerhotels.com/business-small-business/diplom-301686amjnl/]managerhotels.com/business-small-business/diplom-301686amjnl[/url]
Mazrjgp
August 18, 2024 @ 9:40 pm
[u][b] Добрый день![/b][/u]
Парадокс, но купить диплом кандидата наук оказалось не так и сложно
[url=http://w99925ro.bget.ru/main/1-post1/]w99925ro.bget.ru/main/1-post1[/url]
StephenTip
August 18, 2024 @ 11:29 pm
https://tamoxifen.bid/# tamoxifen and ovarian cancer
StephenTip
August 19, 2024 @ 12:32 am
https://furosemide.win/# lasix for sale
Yrefnym
August 19, 2024 @ 1:22 am
[u][b] Здравствуйте![/b][/u]
[b]Заказать диплом о высшем образовании.[/b]
[url=http://soccerform.ru/myachi/myach-nike-pl-strike/reviews/]soccerform.ru/myachi/myach-nike-pl-strike/reviews[/url]
Trefmul
August 19, 2024 @ 1:23 am
[u][b] Здравствуйте![/b][/u]
Покупка диплома о среднем полном образовании: как избежать мошенничества?
[url=http://dobrknigi.ru/howto/?PAGE_NAME=profile_view&UID=26918/]dobrknigi.ru/howto/?PAGE_NAME=profile_view&UID=26918[/url]
Поможем вам всегда!.
DanielTam
August 19, 2024 @ 1:33 am
buy cytotec https://cytotec.pro/# buy cytotec in usa
lasix 100 mg
DanielTam
August 19, 2024 @ 3:08 am
cytotec online https://cytotec.pro/# buy cytotec
lasix generic name
Stanleyquota
August 19, 2024 @ 3:18 am
cytotec pills buy online [url=https://cytotec.pro/#]Cytotec 200mcg price[/url] buy misoprostol over the counter
JamesTah
August 19, 2024 @ 3:45 am
buy furosemide online: cheap lasix – lasix
Manrxvs
August 19, 2024 @ 4:19 am
[u][b] Здравствуйте![/b][/u]
Приобрести документ университета
[url=http://pirooztak.ir/?option=com_k2&view=itemlist&task=user&id=1379944/]pirooztak.ir/?option=com_k2&view=itemlist&task=user&id=1379944[/url]
[url=http://starryjeju.com/qna/5833095/]starryjeju.com/qna/5833095[/url]
[url=http://hariharparagovernmentiti.com/2024/07/28/diplom-kupit-657028qk/]hariharparagovernmentiti.com/2024/07/28/diplom-kupit-657028qk[/url]
[url=http://gtj.kr/board_KtRj53/607489/]gtj.kr/board_KtRj53/607489[/url]
[url=http://hsgd.kr/board_giKf40/158537/]hsgd.kr/board_giKf40/158537[/url]
Trefjbu
August 19, 2024 @ 4:27 am
[u][b] Привет![/b][/u]
Как официально приобрести аттестат 11 класса с минимальными затратами времени
[url=http://svetlana5ege.blogspot.com/2014/12/7_16/]svetlana5ege.blogspot.com/2014/12/7_16[/url]
Рады помочь!.
Juliuscep
August 19, 2024 @ 4:47 am
CVzen https://cvzen.it e il servizio leader per la scrittura di CV e il coaching di carriera, scelto da milioni di candidati globalmente. Offriamo supporto completo: redazione di CV e lettere di presentazione, ottimizzazione di LinkedIn e coaching personalizzato per sbloccare il tuo potenziale e nuove opportunita.
Mazrzoz
August 19, 2024 @ 4:50 am
[b]Приобрести документ ВУЗа.[/b]
[url=http://sad-tver.ru/2017/05/24/my-otkrylis/#comment-51787/]sad-tver.ru/2017/05/24/my-otkrylis/#comment-51787[/url]
[url=http://unix.t7y.ru/user/rebeccaenvef/]unix.t7y.ru/user/rebeccaenvef[/url]
[url=http://zpu-journal.ru/forum/view_profile.php?UID=309894/]zpu-journal.ru/forum/view_profile.php?UID=309894[/url]
[url=http://notasothers.blogspot.com/2013/02/blog-post/]notasothers.blogspot.com/2013/02/blog-post[/url]
[url=http://megashipping.ru/user/sydnyttsoz323/]megashipping.ru/user/sydnyttsoz323[/url]
Sazrtdh
August 19, 2024 @ 4:53 am
[u][b] Привет, друзья![/b][/u]
Мы изготавливаем [b]дипломы[/b] любой профессии.
Заказ [b]диплома[/b], подтверждающего обучение в университете, – это грамотное решение.
[url=http://bodybuilding.net/members/worksale-page2/]bodybuilding.net/members/worksale-page2[/url]
[b]Рады оказать помощь![/b].
Mazrmsp
August 19, 2024 @ 5:23 am
[u][b] Добрый день![/b][/u]
[b]Как получить диплом стоматолога быстро и официально[/b]
[url=http://medicinacinesenews.it/forum/memberlist.php?mode=viewprofile&u=44717]medicinacinesenews.it/forum/memberlist.php?mode=viewprofile&u=44717[/url]
Diplomi_vfSi
August 19, 2024 @ 5:33 am
Привет!
Купить документ ВУЗа вы имеете возможность в нашем сервисе.
[url=http://zsaek.tsu.ru/user/2490/]zsaek.tsu.ru/user/2490[/url]
Danrghp
August 19, 2024 @ 6:04 am
[u][b] Здравствуйте![/b][/u]
Мы готовы предложить дипломы любой профессии.
[url=http://shikhadabas.com/2024/08/06/diplom-100353xdghk/]shikhadabas.com/2024/08/06/diplom-100353xdghk[/url]
[url=http://kankerala.org/uncategorized/diplom-kupit-627037fz/]kankerala.org/uncategorized/diplom-kupit-627037fz[/url]
[url=http://moodle.spice-training.com/blog/index.php?entryid=309874/]moodle.spice-training.com/blog/index.php?entryid=309874[/url]
[url=http://njkkot.org/video/1225759/]njkkot.org/video/1225759[/url]
[url=http://gamereleasetoday.com/diplom-720107yzwht/]gamereleasetoday.com/diplom-720107yzwht[/url]
1xbet promo code bangladesh
August 19, 2024 @ 7:09 am
Thanks for sharing your journey with us. Your words are always so encouraging and uplifting.
Lariorwqm
August 19, 2024 @ 7:13 am
Пошаговая инструкция по официальной покупке диплома о высшем образовании
[url=http://telegra.ph/kupit-diplom-ukraina-nedorogo-08-13-6/]telegra.ph/kupit-diplom-ukraina-nedorogo-08-13-6[/url]
Sazrunr
August 19, 2024 @ 7:19 am
[u][b] Здравствуйте![/b][/u]
Приобрести документ института
[url=http://a-bcd.ru/news/200281/]a-bcd.ru/news/200281/[/url]
Lariorlcj
August 19, 2024 @ 7:26 am
Парадокс, но купить диплом кандидата наук оказалось не так и сложно
[url=http://telegra.ph/kupit-diplom-ob-okonchanii-universiteta-08-13-3/]telegra.ph/kupit-diplom-ob-okonchanii-universiteta-08-13-3[/url]
Mazrnkq
August 19, 2024 @ 7:31 am
[u][b] Добрый день![/b][/u]
[b]Пошаговая инструкция по официальной покупке диплома о высшем образовании[/b]
[url=http://mebelnyvkus.ru/kuhnja/kuxonnye-stulya/stul-ccpe-m/]mebelnyvkus.ru/kuhnja/kuxonnye-stulya/stul-ccpe-m[/url]
Uazrixs
August 19, 2024 @ 7:42 am
[u][b]Здравствуйте![/b][/u]
[b]Купить диплом университета .[/b]
[url=http://telegra.ph/kupit-diplom-povara-v-moskve-08-13-5/]telegra.ph/kupit-diplom-povara-v-moskve-08-13-5[/url]
Lariordgz
August 19, 2024 @ 7:51 am
Процесс получения диплома стоматолога: реально ли это сделать быстро?
[url=http://telegra.ph/kupit-diplom-medicinskij-08-13-5/]telegra.ph/kupit-diplom-medicinskij-08-13-5[/url]
arenda_elEn
August 19, 2024 @ 8:07 am
Аренда экскаватора погрузчика в Москве, надежно и выгодно.
Экскаватор-погрузчик на любой вкус и бюджет, для вашего удобства.
Где арендовать экскаватор-погрузчик в Москве?, ждет вас.
Эффективные решения для строительства, доступно в нашем сервисе.
Оптимальные условия аренды спецтехники, с нами выгодно.
Как выбрать технику для строительства, у нас в сервисе.
Гибкие условия проката техники, воспользуйтесь нашими услугами.
Как выбрать оптимальный вариант, в столице.
Где найти надежную аренду экскаватора-погрузчика в Москве?, в Москве.
Выбор качественного проката, в столице.
Плюсы аренды экскаватора-погрузчика в Москве, в столице.
Советы по оформлению проката, в столице.
Аренда экскаватора-погрузчика в Москве: лучшие предложения, в Москве.
Плюсы и минусы проката техники, в Москве.
Экскаватор-погрузчик в аренду в Москве: оптимальное решение, в столице.
Лучшие предложения по прокату, в Москве.
Лучшие предложения по аренде, в столице.
Услуги по аренде техники в столице, в столице.
нанять экскаватор погрузчик [url=https://arenda-ekskavatora-pogruzchika197.ru/]https://arenda-ekskavatora-pogruzchika197.ru/[/url] .
Diplomi_tnSi
August 19, 2024 @ 8:45 am
Здравствуйте!
Заказать документ о получении высшего образования можно в нашей компании в столице.
[url=http://sakryukin.blogspot.com/2012/11/custom-attributes-user-profile-sap-nw/]sakryukin.blogspot.com/2012/11/custom-attributes-user-profile-sap-nw[/url]
Eanrlrz
August 19, 2024 @ 9:43 am
[u][b] Добрый день![/b][/u]
[b]Мы можем предложить дипломы[/b] любой профессии по невысоким тарифам.
[url=http://mazda-demio.ru/forums/index.php?autocom=gallery&req=si&img=4139]mazda-demio.ru/forums/index.php?autocom=gallery&req=si&img=4139[/url]
Cazrwwm
August 19, 2024 @ 9:48 am
[u][b] Добрый день![/b][/u]
Мы готовы предложить документы техникумов
[url=http://elephantjournal.com/profile/lutroduyda/]elephantjournal.com/profile/lutroduyda[/url]
Dnrtqjt
August 19, 2024 @ 9:50 am
[u][b]Добрый день![/b][/u]
[b]Купить документ[/b] ВУЗа вы имеете возможность у нас в Москве.
[url=http://nowyiforum.listbb.ru/viewtopic.php?f=45&t=9533]nowyiforum.listbb.ru/viewtopic.php?f=45&t=9533[/url]
StephenTip
August 19, 2024 @ 11:02 am
https://furosemide.win/# lasix 20 mg
svadebnyy_yspi
August 19, 2024 @ 11:03 am
Как сделать свадебный букет своими руками
букет на свадьбу для невесты [url=https://0gorodnik.ru/]https://0gorodnik.ru/[/url] .
DanielTam
August 19, 2024 @ 11:50 am
buy cytotec online fast delivery https://lisinopril.guru/# lisinopril 5 mg price
lasix pills
Stanleyquota
August 19, 2024 @ 11:51 am
cytotec online [url=https://cytotec.pro/#]Cytotec 200mcg price[/url] Abortion pills online
StephenTip
August 19, 2024 @ 12:09 pm
https://cytotec.pro/# cytotec pills buy online
DanielTam
August 19, 2024 @ 1:25 pm
buy cytotec online https://lisinopril.guru/# cheapest price for lisinopril india
lasix furosemide 40 mg
kompoziciy_nspa
August 19, 2024 @ 1:34 pm
Идеальная композиция из цветов для вашего дома, советы по подбору.
Очаровательные цветочные композиции для вашего участка, которые придадут вашему саду очарование.
Как сделать необычный подарок из цветов, и удивят своим необычным сочетанием.
Как выбрать идеальный букет для невесты, и заставят всех гостей восхищаться.
Уникальные идеи для оформления цветочных композиций на праздник, которые заставят всех гостей восхищаться.
Секреты создания стильных композиций из живых цветов, и подчеркнут ваш неповторимый стиль.
Как украсить рабочее место цветами, которые позволят сделать рабочее пространство более приятным.
Идеи для создания оригинальных композиций из цветов на даче, которые преобразят ваш дачный уголок и наполнят его ароматом.
флористика для начинающих пошагово сборка букетов [url=https://101-po3a.ru/]https://101-po3a.ru/[/url] .
Lazronk
August 19, 2024 @ 1:48 pm
[u][b] Привет![/b][/u]
[b]Мы изготавливаем дипломы[/b] любых профессий по выгодным тарифам.
[url=http://telegra.ph/stoimost-obucheniya-v-vuzah-kanady-08-02/]telegra.ph/stoimost-obucheniya-v-vuzah-kanady-08-02[/url]
Dnrtrme
August 19, 2024 @ 1:49 pm
[u][b]Привет, друзья![/b][/u]
[b]Приобрести документ[/b] института вы можете у нас.
[url=http://mlmwmzmillioner.rolevaya.com/viewtopic.php?id=14918#p26814]mlmwmzmillioner.rolevaya.com/viewtopic.php?id=14918#p26814[/url]
Mazrogu
August 19, 2024 @ 3:41 pm
[u][b] Добрый день![/b][/u]
[b]Официальная покупка диплома ПТУ с упрощенной программой обучения[/b]
[url=http://images.google.se/url?q=aurus-diploms.com]images.google.se/url?q=aurus-diploms.com[/url]
arenda_nlPi
August 19, 2024 @ 4:10 pm
Лучший выбор для аренды автобуса в СПб|Передвигайтесь по Санкт-Петербургу в удобстве и безопасности|Широкий выбор автобусов в аренду в СПб|Приятные цены на аренду автобусов в Санкт-Петербурге|Аренда автобуса для праздника в СПб – идеальное решение|Быстрая и удобная аренда автобуса в СПб|Идеальный автобус для туристических поездок по СПб|Аренда автобуса для корпоративных мероприятий в СПб|Необычный транспорт для свадьбы – арендованный автобус в СПб|Комфортный и безопасный транспорт – наши автобусы в аренде в СПб|Наши автобусы оборудованы всем необходимым для комфортной поездки по Санкт-Петербургу|Разнообразие поездок на автобусе в СПб|Не упустите выгодные акции на аренду автобусов в СПб|Индивидуальные маршруты на арендованном автобусе в СПб|24/7 поддержка для клиентов аренды автобусов в СПб|Необычайное удовольствие от поездок на арендованных автобусах в СПб|Гибкая система тарифов на аренду автобуса в СПб|Доверьте свои поездки по Санкт-Петербургу профессионалам со всеми необходимыми документами на арендованные автобусы|Интересные предложения для аренды автобуса в СПб|Быстрая и удобная аренда автобуса в СПб
аренда автобуса [url=https://arenda-avtobusa-178.ru/]https://arenda-avtobusa-178.ru/[/url] .
Sazrnmn
August 19, 2024 @ 4:18 pm
[u][b] Добрый день![/b][/u]
Приобрести документ о получении высшего образования
[url=http://agarwalconnect.com/blogs/238/ко-и-быстро/]agarwalconnect.com/blogs/238/ко-и-быстро[/url]
[url=http://daddycow.com/blogs/view/25662/]daddycow.com/blogs/view/25662[/url]
[url=http://g95334gq.beget.tech/2024/06/18/pochemu-diplom-trebuetsya-dlya-professionalnogo-rosta/]g95334gq.beget.tech/2024/06/18/pochemu-diplom-trebuetsya-dlya-professionalnogo-rosta[/url]
[url=http://telegra.ph/lgoty-medalistam-pri-postuplenii-v-vuz-07-24/]telegra.ph/lgoty-medalistam-pri-postuplenii-v-vuz-07-24[/url]
[url=http://wik.co.kr/master4/86826/]wik.co.kr/master4/86826[/url]
vivod iz zapoya rostov_suen
August 19, 2024 @ 4:33 pm
вывод из запоя ростов [url=http://vyvod-iz-zapoya-rostov11.ru]вывод из запоя ростов[/url] .
Oariordfr
August 19, 2024 @ 5:01 pm
[u][b] Здравствуйте![/b][/u]
Заказать диплом о высшем образовании
[b]Наши специалисты предлагают[/b] выгодно приобрести диплом, который выполнен на оригинальном бланке и заверен печатями, штампами, подписями должностных лиц. Данный диплом способен пройти любые проверки, даже при помощи специально предназначенного оборудования. Решите свои задачи быстро с нашим сервисом.
[b]Где приобрести диплом по актуальной специальности?[/b]
[url=http://stroimsa.forum2x2.ru/t1835-topic#4743/]stroimsa.forum2x2.ru/t1835-topic#4743[/url]
[url=http://finttech.ru/ofitsialnyie-diplomyi-dlya-vashey-kareryi/]finttech.ru/ofitsialnyie-diplomyi-dlya-vashey-kareryi[/url]
[url=http://safdfpb.ixbb.ru/viewtopic.php?id=122#p122/]safdfpb.ixbb.ru/viewtopic.php?id=122#p122[/url]
[url=http://insta.tel/read-blog/49859/]insta.tel/read-blog/49859[/url]
[url=http://talkitter.com/read-blog/204139/]talkitter.com/read-blog/204139[/url]
Lazrxyc
August 19, 2024 @ 5:30 pm
[u][b] Привет, друзья![/b][/u]
[b]Мы изготавливаем дипломы[/b] любой профессии по приятным ценам.
[url=http://telegra.ph/stoimost-obucheniya-v-vuzah-kazani-08-02/]telegra.ph/stoimost-obucheniya-v-vuzah-kazani-08-02[/url]
arenda_gben
August 19, 2024 @ 6:47 pm
Преимущества аренды экскаватора погрузчика
взять в аренду экскаватор погрузчик [url=https://arenda-ekskavatora-pogruzchika197.ru/]https://arenda-ekskavatora-pogruzchika197.ru/[/url] .
Lazrreu
August 19, 2024 @ 6:47 pm
[u][b] Добрый день![/b][/u]
[b]Купить диплом любого университета.[/b]
[url=http://telegra.ph/nalogovyj-vychet-na-obuchenie-v-vuze-08-02/]telegra.ph/nalogovyj-vychet-na-obuchenie-v-vuze-08-02[/url]
Stanleyquota
August 19, 2024 @ 6:58 pm
furosemida [url=https://furosemide.win/#]cheap lasix[/url] lasix
Xazrsgc
August 19, 2024 @ 8:43 pm
[u][b] Добрый день![/b][/u]
Купить документ о получении высшего образования.
Мы изготавливаем [b]дипломы[/b] любой профессии по выгодным ценам.
[url=http://ekonty.com/blogs/view/58699/]ekonty.com/blogs/view/58699[/url]
[b]Рады оказаться полезными![/b].
Cazrrmz
August 19, 2024 @ 8:57 pm
[u][b] Привет![/b][/u]
Мы готовы предложить документы техникумов
[url=http://rhabits.io/read-blog/2004/]rhabits.io/read-blog/2004[/url]
DanielTam
August 19, 2024 @ 8:57 pm
buy cytotec online https://lisinopril.guru/# order lisinopril 10 mg
lasix tablet
JustinIsone
August 19, 2024 @ 9:20 pm
поисковая оптимизация https://process-seo.ru
arenda_mwOt
August 19, 2024 @ 9:23 pm
Аренда экскаватора погрузчика: выгодное предложение для строительства, воспользуйтесь услугой уже сегодня.
Экскаватор погрузчик в аренду: быстро и качественно, арендуйте прямо сейчас.
Аренда экскаватора погрузчика: оптимальное решение для строительных работ, арендуйте прямо сейчас.
Экскаватор погрузчик на прокат: удобство и профессионализм, бронируйте аренду сейчас.
Аренда экскаватора погрузчика: быстро и качественно, арендуйте прямо сейчас.
Аренда экскаватора погрузчика: удобство и эффективность на стройке, бронируйте аренду сегодня.
взять в аренду экскаватор погрузчик [url=https://arenda-jekskavatora-pogruzchika-197.ru/]https://arenda-jekskavatora-pogruzchika-197.ru/[/url] .
DanielTam
August 19, 2024 @ 10:20 pm
Misoprostol 200 mg buy online https://lisinopril.guru/# lisinopril online without a prescription
lasix for sale
Iariorthj
August 19, 2024 @ 10:29 pm
[u][b] Привет![/b][/u]
Приобрести документ о получении высшего образования можно в нашем сервисе. Мы предлагаем документы об окончании любых ВУЗов РФ. Вы получите диплом по любой специальности, включая документы Советского Союза. Даем 100% гарантию, что при проверке документов работодателями, подозрений не появится.
[url=http://terorizam.listbb.ru/viewtopic.php?f=3&t=395/]terorizam.listbb.ru/viewtopic.php?f=3&t=395[/url]
[url=http://japapmessenger.com/read-blog/551/]japapmessenger.com/read-blog/551[/url]
[url=http://u90517ol.beget.tech/2024/07/20/kupit-diplom-6/]u90517ol.beget.tech/2024/07/20/kupit-diplom-6[/url]
[url=http://zdsim.ru/wiki/index.php/Купите_диплом_и_начните_новую_карьеру/]zdsim.ru/wiki/index.php/Купите_диплом_и_начните_новую_карьеру[/url]
[url=http://rpinnovative.1stbb.ru/viewtopic.php?f=8&t=373/]rpinnovative.1stbb.ru/viewtopic.php?f=8&t=373[/url]
Mazrbuj
August 19, 2024 @ 11:09 pm
[u][b] Привет![/b][/u]
[b]Рекомендации по безопасной покупке диплома о высшем образовании[/b]
[url=http://sumkin.ru/forum/member.php?u=39200/]sumkin.ru/forum/member.php?u=39200[/url]
StephenTip
August 19, 2024 @ 11:50 pm
http://tamoxifen.bid/# tamoxifen cyp2d6
arenda_msOn
August 20, 2024 @ 12:03 am
Экономьте время и деньги с арендой трактора,
Опытные водители и надежная техника на аренду,
Быстрая аренда трактора по выгодным условиям,
Профессиональные услуги по аренде тракторов для фермеров,
Скидки на аренду тракторов для постоянных клиентов,
Специализированная аренда тракторов для строительства,
Аренда трактора на длительный срок,
Безопасная и надежная аренда тракторов с водителем,
Аренда трактора под ключ
аренда трактора [url=https://arenda-traktora77.ru/]https://arenda-traktora77.ru/[/url] .
Iariorzys
August 20, 2024 @ 12:28 am
[u][b] Здравствуйте![/b][/u]
Приобрести документ ВУЗа можно у нас. Мы оказываем услуги по изготовлению и продаже документов об окончании любых университетов Российской Федерации. Вы сможете получить диплом по любым специальностям, включая документы Советского Союза. Гарантируем, что при проверке документов работодателем, подозрений не появится.
[url=http://fire-team.ru/forum/showthread.php?p=44207#post44207/]fire-team.ru/forum/showthread.php?p=44207#post44207[/url]
[url=http://lampogolovii.blogspot.com/2010/10/blog-post/]lampogolovii.blogspot.com/2010/10/blog-post[/url]
[url=http://aquapromstroy.ru/sp-70-13330-2012-nesushhie-i-ograzhdajushhie-konstrukcii/#comment-80768/]aquapromstroy.ru/sp-70-13330-2012-nesushhie-i-ograzhdajushhie-konstrukcii/#comment-80768[/url]
[url=http://krok.biz/forum/memberlist.php?mode=viewprofile&u=38923/]krok.biz/forum/memberlist.php?mode=viewprofile&u=38923[/url]
[url=http://buhtarma.ru/users/ojepug/]buhtarma.ru/users/ojepug[/url]
StephenTip
August 20, 2024 @ 1:15 am
https://lipitor.guru/# lipitor no prescription
Trefpzc
August 20, 2024 @ 2:05 am
[u][b] Добрый день![/b][/u]
Официальная покупка школьного аттестата с упрощенным обучением в Москве
[url=http://geolan-ksl.ru/forum/user/87640/]geolan-ksl.ru/forum/user/87640[/url]
Рады помочь!.
Yrefyed
August 20, 2024 @ 2:25 am
[u][b] Привет![/b][/u]
[b]Приобрести диплом университета.[/b]
[url=http://vecmir.ru/index.php/vecmirlife/58925-ybyrufe/profile/]vecmir.ru/index.php/vecmirlife/58925-ybyrufe/profile[/url]
arenda_umSn
August 20, 2024 @ 2:40 am
Что нужно знать при выборе трактора для аренды|Лучшие предложения по аренде тракторов|Оценка экономической целесообразности аренды трактора|Онлайн-платформы для аренды тракторов: как выбрать лучшую|Плюсы и минусы аренды трактора|Секреты долгосрочной аренды трактора|Что необходимо учесть, чтобы избежать ошибок при аренде трактора|Частные лица и аренда тракторов: реальность и перспективы|Уникальная возможность аренды трактора без посещения офиса|Как выбрать идеальный мини-трактор для аренды|Как выбрать надежного арендодателя тракторов|Краткосрочная аренда тракторов: что нужно знать|Аренда тракторов с водителем: безопасное и профессиональное обслуживание|Что нужно знать, чтобы сделать правильный выбор трактора|Сравнение популярных моделей тракторов для аренды|Аренда тракторов по городу: удобство и доступность|Как оценить качество услуг по аренде трактора|Идеи использования трактора в качестве декора на свадьбе|Советы по подбору идеального трактора для вашего проекта|Как арендовать бетономешалку в комплексе с трактором|Аренда спецтехники: широкий выбор тракторов под любые задачи|Советы по подбору трактора для строительных работ|Аренда тракторов для сельского хозяйства: особенности и нюансы|Как оценить профессионализм компании по аренде тракторов|Аренда тракторов с доставкой: удобство и экономия времени|Как выбрать трактор для работ на даче
аренда трактора [url=https://arenda-traktora-skovshom.ru/]https://arenda-traktora-skovshom.ru/[/url] .
Oariorayg
August 20, 2024 @ 3:31 am
[u][b] Добрый день![/b][/u]
Заказать диплом любого ВУЗа
[b]Наши специалисты предлагают[/b] выгодно и быстро купить диплом, который выполнен на оригинальном бланке и заверен мокрыми печатями, штампами, подписями. Диплом способен пройти любые проверки, даже с применением профессионального оборудования. Достигайте свои цели максимально быстро с нашими дипломами.
[b]Где заказать диплом по актуальной специальности?[/b]
[url=http://safdfpb.ixbb.ru/viewtopic.php?id=127#p127/]safdfpb.ixbb.ru/viewtopic.php?id=127#p127[/url]
[url=http://pdapal.com/read-blog/1316/]pdapal.com/read-blog/1316[/url]
[url=http://njt.ru/forum/user/198184/]njt.ru/forum/user/198184[/url]
[url=http://riftynet.com/blogs/26230/Дипломы-для-уверенного-будущего/]riftynet.com/blogs/26230/Дипломы-для-уверенного-будущего[/url]
[url=http://lubercy.ixbb.ru/viewtopic.php?id=7416#p16026/]lubercy.ixbb.ru/viewtopic.php?id=7416#p16026[/url]
evakuator_dwpl
August 20, 2024 @ 5:22 am
Эвакуаторы в Москве на высшем уровне, доступные цены|Профессиональные услуги эвакуатора в Москве, круглосуточно|Дешевый эвакуатор в Москве: не ждите, звоните сейчас|Специализированный эвакуатор в Москве|Эвакуатор в Москве для легковых автомобилей|Лучший эвакуатор Москвы ждет вашего звонка|Эвакуатор Москва: доверьте свой автомобиль профессионалам|Эвакуация мотоциклов в Москве: надежно и быстро|Экстренная эвакуация в Москве: без лишних хлопот|Эвакуатор Москва: профессиональный подход к каждому клиенту|Эвакуация автомобилей в Москве: безопасно и оперативно|Эвакуация мотоциклов в Москве: безопасно и надежно|Эвакуатор Москва: всегда на связи|Эвакуатор Москва: помощь на дорогах|Круглосуточный эвакуатор в Москве|Эвакуатор Москва: ваша безопасность на первом месте|Эвакуатор Москва: надежность и профессионализм
вызвать эвакуатор [url=https://ewacuator-moscow.ru/]вызвать эвакуатор[/url] .
Trefaub
August 20, 2024 @ 5:32 am
[u][b] Привет![/b][/u]
Как безопасно купить диплом колледжа или ПТУ в России, что важно знать
[url=http://kemavto.kuzbass.net/index.php?subaction=userinfo&user=afuvid/]kemavto.kuzbass.net/index.php?subaction=userinfo&user=afuvid[/url]
Поможем вам всегда!.
pay someone to do my online accounting class for me
August 20, 2024 @ 5:37 am
Learners can find the right equilibrium between asking for online accounting class help and fostering their growth by upholding their dedication to higher learning integrity, developing their unique abilities, and taking financial responsibility into account. It is a great way to get an education and pay someone to do my online accounting class for me and can be used to advance academic success and improve how students learn.
pay someone to do my online class for me
August 20, 2024 @ 5:38 am
The query, “Can I pay someone to do my online class for me?” is no longer just a basic one; it now represents the evolving nature of contemporary schooling. Online course help assistance programs have become important tools for pupils who want to do well in school and successfully manage their commitments. These websites allow students to get professional advice, save valuable time, and strike a greater equilibrium between work and personal life. However, learners should use extra caution when using these services and make sure they view them as educational and development tools rather than as quick fixes for achievement.
Lazrgkc
August 20, 2024 @ 6:00 am
[u][b] Здравствуйте![/b][/u]
[b]Приобрести диплом ВУЗа.[/b]
[url=http://telegra.ph/alimenty-na-obuchenie-v-vuze-08-02/]telegra.ph/alimenty-na-obuchenie-v-vuze-08-02[/url]
Diplomi_rnSi
August 20, 2024 @ 6:01 am
Здравствуйте!
Заказать документ о получении высшего образования вы сможете в нашем сервисе.
[url=http://unifyusnow.org/senate-healthcare-bill-screws-regardless-party/#comment-37850/]unifyusnow.org/senate-healthcare-bill-screws-regardless-party/#comment-37850[/url]
Yrefyfo
August 20, 2024 @ 6:01 am
[u][b] Привет, друзья![/b][/u]
[b]Купить диплом о высшем образовании.[/b]
[url=http://lider-school174.com/forum/user/3099/]lider-school174.com/forum/user/3099[/url]
Stanleyquota
August 20, 2024 @ 7:13 am
tamoxifen men [url=https://tamoxifen.bid/#]buy tamoxifen online[/url] tamoxifen and uterine thickening
Mazrdvs
August 20, 2024 @ 7:54 am
[u][b] Привет, друзья![/b][/u]
[b]Официальное получение диплома техникума с упрощенным обучением в Москве[/b]
[url=http://izmailskiypark.ru/ekonomika/sdelka-po-prodaje-magazinov-decathlon-v-rossii-eshe-ne-zavershena/]izmailskiypark.ru/ekonomika/sdelka-po-prodaje-magazinov-decathlon-v-rossii-eshe-ne-zavershena[/url]
Xazrkcz
August 20, 2024 @ 7:56 am
[u][b] Добрый день![/b][/u]
Приобрести документ ВУЗа.
Мы можем предложить [b]дипломы[/b] любых профессий по приятным ценам.
[url=http://elephantjournal.com/profile/poydicikko/]elephantjournal.com/profile/poydicikko[/url]
[b]Поможем вам всегда![/b].
Dnrtwda
August 20, 2024 @ 7:57 am
[u][b]Привет, друзья![/b][/u]
[b]Купить документ[/b] о получении высшего образования вы имеете возможность в нашей компании в столице.
[url=http://nemoskvichi.ru/forum/viewtopic.php?f=42&t=145893/]nemoskvichi.ru/forum/viewtopic.php?f=42&t=145893[/url]
dymohody_hypi
August 20, 2024 @ 7:59 am
Установка дымоходов для бани в Нижнем Новгороде, быстро и надежно.
Как выбрать исполнителя на установку дымоходов для бани в Нижнем Новгороде, гарантии качества работы.
Сравнение различных видов дымоходов для бани в Нижнем Новгороде, советы от экспертов.
На что обратить внимание при выборе дымоходов для бани в Нижнем Новгороде, основные критерии.
Простые способы поддержания работы дымоходов для бани в Нижнем Новгороде, экспертные советы.
Какие популярные виды дымоходов для бани в Нижнем Новгороде существуют, сравнение характеристик.
дымоход для банной печи купить [url=https://forum-bani.ru/]дымоход для банной печи купить[/url] .
Lazrlae
August 20, 2024 @ 8:59 am
[u][b] Добрый день![/b][/u]
[b]Мы готовы предложить дипломы[/b] любых профессий по выгодным ценам.
[url=http://telegra.ph/kak-vernut-dengi-za-obuchenie-v-vuze-pri-otchislenii-08-02/]telegra.ph/kak-vernut-dengi-za-obuchenie-v-vuze-pri-otchislenii-08-02[/url]
Iariordcv
August 20, 2024 @ 9:14 am
[u][b] Добрый день![/b][/u]
Заказать документ ВУЗа можно у нас. Мы оказываем услуги по изготовлению и продаже документов об окончании любых университетов РФ. Вы получите необходимый диплом по любой специальности, любого года выпуска, в том числе документы СССР. Гарантируем, что при проверке документа работодателями, подозрений не возникнет.
[url=http://kpilib.ru/forum.php?tema=7496/]kpilib.ru/forum.php?tema=7496[/url]
[url=http://antennastar.com/blogs/570/Экспресс-оформление-дипломов/]antennastar.com/blogs/570/Экспресс-оформление-дипломов[/url]
[url=http://montrealpal.com/read-blog/1320/]montrealpal.com/read-blog/1320[/url]
[url=http://horordark.ru/oformite-diplom-dlya-novoy-kareryi/]horordark.ru/oformite-diplom-dlya-novoy-kareryi[/url]
[url=http://telegra.ph/kak-podat-zayavku-na-postuplenie-v-vuz-cherez-internet-07-24/]telegra.ph/kak-podat-zayavku-na-postuplenie-v-vuz-cherez-internet-07-24[/url]
Diplomi_soSi
August 20, 2024 @ 9:39 am
Привет!
Купить документ о получении высшего образования вы можете у нас.
[url=http://perevozchikovaes.blogspot.com/2014/04/3-things-i-couldnt-live-without-write/]perevozchikovaes.blogspot.com/2014/04/3-things-i-couldnt-live-without-write[/url]
DanielTam
August 20, 2024 @ 9:52 am
buy cytotec over the counter http://furosemide.win/# furosemide 40 mg
buy furosemide online
kuhnya_xwel
August 20, 2024 @ 10:42 am
Какой стиль выбрать для кухни на заказ
кухни по индивидуальному заказу [url=https://kuhninazakaz177.ru/]https://kuhninazakaz177.ru/[/url] .
MatthewInalf
August 20, 2024 @ 11:14 am
ремонт айфонов в москве
Iarioruzu
August 20, 2024 @ 11:24 am
[u][b] Здравствуйте![/b][/u]
Приобрести документ о получении высшего образования можно у нас в Москве. Мы оказываем услуги по производству и продаже документов об окончании любых ВУЗов России. Вы получите необходимый диплом по любой специальности, включая документы старого образца. Гарантируем, что при проверке документов работодателями, каких-либо подозрений не появится.
[url=http://realtor-cheb.ru/index.php?subaction=userinfo&user=igalata/]realtor-cheb.ru/index.php?subaction=userinfo&user=igalata[/url]
[url=http://ntsr.info/forum/user/98427/]ntsr.info/forum/user/98427[/url]
[url=http://dikadesign.cz/gravirovani-a-rezani-laserem/?unapproved=580898&moderation-hash=4ec6a39364b6ef3b9831ecd2f0da203b#comment-580898/]dikadesign.cz/gravirovani-a-rezani-laserem/?unapproved=580898&moderation-hash=4ec6a39364b6ef3b9831ecd2f0da203b#comment-580898[/url]
[url=http://tvpolska.pl/konfederacja-jakub-kulesza-piotr-czak-zukowski-tomasz-grabarczyk-najdrozsze-boze-narodzenie-stulecia/10978/#comment-434028/]tvpolska.pl/konfederacja-jakub-kulesza-piotr-czak-zukowski-tomasz-grabarczyk-najdrozsze-boze-narodzenie-stulecia/10978/#comment-434028[/url]
[url=http://forum.mama.by/member.php?u=38003/]forum.mama.by/member.php?u=38003[/url]
Dnrtmnq
August 20, 2024 @ 11:50 am
[u][b]Привет, друзья![/b][/u]
[b]Купить документ[/b] института вы имеете возможность у нас.
[url=http://vladmines.dn.ua/forum/index.php/topic,35219.0/]vladmines.dn.ua/forum/index.php/topic,35219.0[/url]
DanielTam
August 20, 2024 @ 12:14 pm
purchase cytotec https://lisinopril.guru/# zestoretic 20 25mg
lasix medication
Lazradd
August 20, 2024 @ 12:19 pm
[u][b] Привет, друзья![/b][/u]
[b]Мы готовы предложить дипломы[/b] психологов, юристов, экономистов и прочих профессий по приятным тарифам.
[url=http://telegra.ph/obuchenie-v-vuzah-kitaya-08-02/]telegra.ph/obuchenie-v-vuzah-kitaya-08-02[/url]
JustinIsone
August 20, 2024 @ 12:21 pm
продвижение сайта заказать https://process-seo.ru
kuhnya_gkPa
August 20, 2024 @ 1:16 pm
Идеальные кухни на заказ в Москве, подберем вам лучшее решение.
У нас самые доступные цены на кухни на заказ в Москве.
Закажите кухню своей мечты прямо сейчас.
У нас вы найдете самый большой выбор кухонь на заказ в Москве.
Лучшие кухни на заказ только в Москве.
Мы предлагаем кухню на заказ для каждого клиента.
Создайте уют в своем доме с кухней на заказ.
Выбирайте кухни на заказ в Москве у профессионалов.
Уникальные решения для вашей кухни только у нас.
кухни на заказ москва [url=https://kuhny-na-zakaz77.ru/]https://kuhny-na-zakaz77.ru/[/url] .
Mazrcjs
August 20, 2024 @ 1:58 pm
[u][b] Добрый день![/b][/u]
[b]Покупка диплома о среднем полном образовании: как избежать мошенничества?[/b]
[url=http://lenta.ru/news/2008/05/21/honourable/]lenta.ru/news/2008/05/21/honourable[/url]
StephenTip
August 20, 2024 @ 2:28 pm
https://tamoxifen.bid/# tamoxifen for breast cancer prevention
Cazreqt
August 20, 2024 @ 2:46 pm
[u][b] Здравствуйте![/b][/u]
Заказать диплом института
[url=http://telegra.ph/distancionnoe-obuchenie-v-gosudarstvennyh-vuzah-moskvy-08-02/]telegra.ph/distancionnoe-obuchenie-v-gosudarstvennyh-vuzah-moskvy-08-02[/url]
Sazrfrl
August 20, 2024 @ 3:35 pm
[u][b] Привет![/b][/u]
Диплом специалиста
[url=http://telegra.ph/poryadok-postupleniya-v-voennyj-vuz-08-02/]telegra.ph/poryadok-postupleniya-v-voennyj-vuz-08-02[/url]
StephenTip
August 20, 2024 @ 3:50 pm
https://lipitor.guru/# lipitor 10mg price singapore
kuhnya_ndSn
August 20, 2024 @ 4:38 pm
Идеальная кухня на заказ для вашего дома, у профессионалов.
Уникальный дизайн кухни на заказ по вашим желаниям, превратим ваши идеи в жизнь.
Индивидуальный дизайн кухни, который подчеркнет ваш стиль, эксклюзивно для вас.
Воплотим в жизнь ваши самые смелые кулинарные фантазии, воплотите свои мечты в реальность.
Создайте уют и комфорт на своей кухне с помощью заказа, доверьтесь профессионалам.
Эксклюзивная кухня, созданная специально для вас, лучший выбор для вашего дома.
Закажите кухню своей мечты у нас, наслаждайтесь каждым моментом на своей новой кухне.
Мы создаем кухни на заказ, которые радуют глаз, с любовью к деталям.
кухни под заказ [url=https://kuhny-na-zakaz-msk.ru/]https://kuhny-na-zakaz-msk.ru/[/url] .
Mazrenq
August 20, 2024 @ 5:53 pm
[u][b] Привет![/b][/u]
Как не стать жертвой мошенников при покупке диплома о среднем полном образовании
[url=http://85sucai.com/member/index.php?uid=ukygit/]85sucai.com/member/index.php?uid=ukygit[/url]
Sazrlhm
August 20, 2024 @ 6:36 pm
[u][b] Добрый день![/b][/u]
Заказать документ ВУЗа
[url=http://forumrabota.0pk.me/viewtopic.php?id=7320#p16792/]forumrabota.0pk.me/viewtopic.php?id=7320#p16792[/url]
arenda_aypr
August 20, 2024 @ 7:19 pm
Прокат автобусов в СПб на выгодных условиях, заказать для трансфера.
Доступные цены на аренду автобуса в СПб, пользуйтесь нашими услугами.
Лучшие автобусы для аренды в СПб, езжайте с комфортом.
Организуйте праздник с нашим автобусом в СПб, весело и ярко.
Доставка из аэропорта в центр СПБ на комфортабельном автобусе, пунктуально и качественно.
Корпоративный праздник на нашем автобусе в СПб, оригинально и ярко.
Отдельный тур на автобусе в СПб, познавательно и интересно.
Организуйте школьную экскурсию с арендованным автобусом в СПб, весело и обучающе.
Свадебный автобус в СПб, сказочно и беззаботно.
Советы по выбору автобуса для проката в Санкт-Петербурге, подсказки от наших экспертов.
Способы сэкономить на аренде автобуса в Санкт-Петербурге, с максимальной выгодой.
Какие услуги включены в аренду автобуса в СПб, узнайте перед заказом.
Преимущества аренды автобуса с шофером в Санкт-Петербурге, подробное сравнение.
Анализ цен на аренду автобуса в Санкт-Петербурге: где дешевле, важные аспекты.
Прокат мини-автобусов для узкого круга пассажиров в СПб, удобно и экономно.
Аренда транспорта для фестиваля в Санкт-Петербурге, с учетом всех особенностей.
Аренда автобуса для корпоративного веселья в Санкт-Петербурге
аренда автобуса спб [url=https://arenda-avtobusa-v-spb.ru/]https://arenda-avtobusa-v-spb.ru/[/url] .
vivod iz zapoya rostov_uuMt
August 20, 2024 @ 7:33 pm
вывод из запоя круглосуточно ростов на дону [url=www.vyvod-iz-zapoya-rostov112.ru/]вывод из запоя круглосуточно ростов на дону[/url] .
vivod iz zapoya rostov_ytKn
August 20, 2024 @ 7:56 pm
вывод из запоя цены ростов на дону [url=vyvod-iz-zapoya-rostov11.ru]vyvod-iz-zapoya-rostov11.ru[/url] .
Trefcjn
August 20, 2024 @ 9:13 pm
[u][b] Привет, друзья![/b][/u]
Реально ли приобрести диплом стоматолога? Основные шаги
[url=http://legkohod.ru/viewtopic.php?f=5&t=66990&p=89002#p89002/]legkohod.ru/viewtopic.php?f=5&t=66990&p=89002#p89002[/url]
Всегда вам поможем!.
Sazrbde
August 20, 2024 @ 9:15 pm
[u][b] Привет, друзья![/b][/u]
Мы изготавливаем [b]дипломы[/b] психологов, юристов, экономистов и других профессий.
Заказ [b]диплома[/b], подтверждающего обучение в университете, – это рациональное решение.
[url=http://ultras.lv/topic1815/]ultras.lv/topic1815[/url]
[b]Окажем помощь![/b].
Mazrtse
August 20, 2024 @ 9:18 pm
[b]Заказать документ университета.[/b]
[url=http://city-hall.nvkb.ru/forum/index.php/user/104010/]city-hall.nvkb.ru/forum/index.php/user/104010[/url]
[url=http://carforum.info/topic/3238-купить-диплом-волгоград-n426v/]carforum.info/topic/3238-купить-диплом-волгоград-n426v[/url]
[url=http://laflore.ru/sale/urinastop#comment-213324/]laflore.ru/sale/urinastop#comment-213324[/url]
[url=http://opendelreyoaks.net/doku.php?id=premialniediplomix/]opendelreyoaks.net/doku.php?id=premialniediplomix[/url]
[url=http://automobile-blogs.blogspot.com/2014/09/volkswagen-golf-gti-h/]automobile-blogs.blogspot.com/2014/09/volkswagen-golf-gti-h[/url]
chto_mlEr
August 20, 2024 @ 10:06 pm
Роза – один из самых популярных цветов в мире, прекрасное растение, воспеваемое многими поэтами и художниками.
Какие бывают виды роз, как ухаживать за розами в саду.
Значение розы в разных культурах, роза в религии и мифологии.
Роза как идеальный подарок для любого случая, какие чувства вызывает роза у людей.
Розы в архитектуре и дизайне интерьера, секреты сбора и хранения розовых лепестков.
роза википедия [url=https://roslina.ru/]роза википедия[/url] .
Cazrubl
August 20, 2024 @ 10:12 pm
[u][b] Добрый день![/b][/u]
Мы предлагаем документы техникумов
[url=http://d90894u1.bget.ru/o-skripte/1-post1/]d90894u1.bget.ru/o-skripte/1-post1[/url]
Mazrokb
August 20, 2024 @ 10:40 pm
[u][b] Добрый день![/b][/u]
[b]Рекомендации по безопасной покупке диплома о высшем образовании[/b]
[url=http://tekst-pesen.ru/2018/04/04/]tekst-pesen.ru/2018/04/04[/url]
Stanleyquota
August 20, 2024 @ 10:48 pm
buy misoprostol over the counter [url=https://cytotec.pro/#]buy cytotec pills online cheap[/url] buy cytotec online fast delivery
Narkolog na dom krasnodar_tmmt
August 20, 2024 @ 11:51 pm
нарколог на дом стоимость [url=http://www.narkolog-na-dom-krasnodar11.ru]нарколог на дом стоимость[/url] .
DanielTam
August 21, 2024 @ 12:33 am
buy cytotec over the counter https://cytotec.pro/# buy cytotec online
generic lasix
Trefaog
August 21, 2024 @ 12:38 am
[u][b] Добрый день![/b][/u]
Всё, что нужно знать о покупке аттестата о среднем образовании
[url=http://msfo-soft.ru/msfo/forum/user/41085/]msfo-soft.ru/msfo/forum/user/41085[/url]
Всегда вам поможем!.
kotel_trea
August 21, 2024 @ 12:52 am
Где купить котел для отопления дома | Какой котел для отопления дома выбрать | Лучшие цены на котлы для отопления | Сравнение котлов для отопления дома | Профессиональная установка котла для отопления | Рейтинг котлов для отопления | Выбор магазина для покупки котла для отопления | Какие котлы для отопления частного дома лучше | Как правильно расходовать топливо для котла | Лучшие цены на котлы для отопления дома
купить котел для отопления [url=https://sauna-manzana.ru/]https://sauna-manzana.ru/[/url] .
Cazrdti
August 21, 2024 @ 1:15 am
[u][b] Здравствуйте![/b][/u]
Получить диплом о высшем образовании
[url=http://telegra.ph/stoimost-obucheniya-v-vuzah-ust-kamenogorska-08-02/]telegra.ph/stoimost-obucheniya-v-vuzah-ust-kamenogorska-08-02[/url]
Lariorgvi
August 21, 2024 @ 1:40 am
Как быстро и легально купить аттестат 11 класса в Москве
[url=http://telegra.ph/kupit-diplom-vracha-v-moskve-08-13-8/]telegra.ph/kupit-diplom-vracha-v-moskve-08-13-8[/url]
Information Security Analysis
August 21, 2024 @ 1:50 am
Online classes refer to courses in history that are offered through online education platforms. These courses can cover a wide range of historical topics and can be taken by students from all over the world. Online education has become increasingly popular in recent years, and online history classes are no exception. Online history class covers courses in history that are taught over the internet through online medium. These courses may include pre-recorded lectures, interactive discussion forums, virtual office hours, and online exams. carefully to catch any homophone errors.
Sazrsdz
August 21, 2024 @ 2:00 am
[u][b] Привет, друзья![/b][/u]
Диплом для вас
[url=http://telegra.ph/o-postuplenii-v-teatralnyj-vuz-08-02/]telegra.ph/o-postuplenii-v-teatralnyj-vuz-08-02[/url]
Lariorvmj
August 21, 2024 @ 2:02 am
Официальная покупка диплома вуза с сокращенной программой обучения в Москве
[url=http://telegra.ph/kupit-diplom-toefl-08-13-8/]telegra.ph/kupit-diplom-toefl-08-13-8[/url]
Uazrezj
August 21, 2024 @ 2:18 am
[u][b]Здравствуйте![/b][/u]
[b]Приобрести диплом ВУЗа .[/b]
[url=http://telegra.ph/chto-budet-esli-kupit-diplom-08-13-4/]telegra.ph/chto-budet-esli-kupit-diplom-08-13-4[/url]
Yrefnlq
August 21, 2024 @ 2:36 am
[u][b] Добрый день![/b][/u]
[b]Приобрести диплом любого ВУЗа.[/b]
[url=http://propertyclaimspain.co.uk/hello-world/#comment-62000/]propertyclaimspain.co.uk/hello-world/#comment-62000[/url]
Lariormvv
August 21, 2024 @ 2:40 am
Как правильно купить диплом колледжа и пту в России, подводные камни
[url=http://telegra.ph/diplom-kupit-ufa-08-13-2/]telegra.ph/diplom-kupit-ufa-08-13-2[/url]
DanielTam
August 21, 2024 @ 2:49 am
Cytotec 200mcg price https://lisinopril.guru/# lisinopril tabs
generic lasix
shkaf odOl
August 21, 2024 @ 3:29 am
Индивидуальный дизайн и высокое качество в вашем шкафе купе на заказ
купе на заказ [url=https://shkaf-kupe-nazakaz177.ru/]https://shkaf-kupe-nazakaz177.ru/[/url] .
Lazrwem
August 21, 2024 @ 3:45 am
[u][b] Привет![/b][/u]
[b]Мы изготавливаем дипломы[/b] психологов, юристов, экономистов и других профессий по приятным ценам.
[url=http://telegra.ph/nalogovyj-vychet-za-obuchenie-v-vuze-08-02/]telegra.ph/nalogovyj-vychet-za-obuchenie-v-vuze-08-02[/url]
Mazrstt
August 21, 2024 @ 4:47 am
[u][b] Привет![/b][/u]
Полезная информация как официально купить диплом о высшем образовании
[url=http://198.199.122.222/doku.php?id=luxdiplomru/]198.199.122.222/doku.php?id=luxdiplomru[/url]
Dnrtmmi
August 21, 2024 @ 4:50 am
[u][b]Добрый день![/b][/u]
[b]Заказать документ[/b] ВУЗа вы имеете возможность у нас в столице.
[url=http://vrn.best-city.ru/forum/thread540108312/]vrn.best-city.ru/forum/thread540108312[/url]
Lazrbbp
August 21, 2024 @ 4:53 am
[u][b] Здравствуйте![/b][/u]
Приобрести [b]диплом[/b] ВУЗа.
[url=http://x70795vj.beget.tech/2024/03/09/hotite-zakazat-originalnyy-diplom/]x70795vj.beget.tech/2024/03/09/hotite-zakazat-originalnyy-diplom[/url]
Mazrkrg
August 21, 2024 @ 5:03 am
[u][b] Здравствуйте![/b][/u]
[b]Диплом вуза купить официально с упрощенным обучением в Москве[/b]
[url=http://profbrus-optom.ru/articles/kak-postroit-banyu-iz-brusa/]profbrus-optom.ru/articles/kak-postroit-banyu-iz-brusa[/url]
StephenTip
August 21, 2024 @ 5:08 am
http://tamoxifen.bid/# alternatives to tamoxifen
Sazrzhn
August 21, 2024 @ 5:15 am
[u][b] Добрый день![/b][/u]
Приобрести документ о получении высшего образования
[url=http://sovetushka.forum2x2.ru/t28199-topic#62837/]sovetushka.forum2x2.ru/t28199-topic#62837[/url]
Sazrlng
August 21, 2024 @ 5:23 am
[u][b] Привет, друзья![/b][/u]
Приобрести документ университета
[url=http://100velikih.ru/?cat=35/]100velikih.ru/?cat=35[/url]
JamesTah
August 21, 2024 @ 5:34 am
nolvadex during cycle: Purchase Nolvadex Online – natural alternatives to tamoxifen
Mazruiu
August 21, 2024 @ 5:57 am
[u][b] Здравствуйте![/b][/u]
[b]Полезная информация как купить диплом о высшем образовании без рисков[/b]
[url=http://blagodarova.blogspot.com/2016/03/blog-post_7/]blagodarova.blogspot.com/2016/03/blog-post_7[/url]
shkafy_ujea
August 21, 2024 @ 6:03 am
Топовые шкафы купе на заказ в Москве, Индивидуальные шкафы купе на заказ в Москве
шкаф купе на заказ [url=https://shkafy-kupe-na-zakaz77.ru/]https://shkafy-kupe-na-zakaz77.ru/[/url] .
StephenTip
August 21, 2024 @ 6:28 am
http://lipitor.guru/# buy lipitor online
Lazrdzc
August 21, 2024 @ 7:01 am
[u][b] Привет, друзья![/b][/u]
[b]Мы изготавливаем дипломы[/b] психологов, юристов, экономистов и любых других профессий по невысоким тарифам.
[url=http://telegra.ph/socialnye-seti-i-ehlektronnoe-obuchenie-v-vuze-referat-08-02/]telegra.ph/socialnye-seti-i-ehlektronnoe-obuchenie-v-vuze-referat-08-02[/url]
Mazrksh
August 21, 2024 @ 7:29 am
[b]Заказать документ о получении высшего образования.[/b]
[url=http://forexforum.cz/newthread.php?do=newthread&f=21/]forexforum.cz/newthread.php?do=newthread&f=21[/url]
[url=http://old.pokvesti.ru/forum/viewtopic.php?f=42&t=140092/]old.pokvesti.ru/forum/viewtopic.php?f=42&t=140092[/url]
[url=http://dmitriysmirnov.blogspot.com/2013/02/2/]dmitriysmirnov.blogspot.com/2013/02/2[/url]
[url=http://stranasp.ru/forum/viewtopic.php?pid=181340#p181340/]stranasp.ru/forum/viewtopic.php?pid=181340#p181340[/url]
[url=http://sport.taminfo.ru/index.php?subaction=userinfo&user=ucexat/]sport.taminfo.ru/index.php?subaction=userinfo&user=ucexat[/url]
Sazrgvh
August 21, 2024 @ 7:30 am
[u][b] Здравствуйте![/b][/u]
Мы изготавливаем [b]дипломы[/b] психологов, юристов, экономистов и других профессий.
Приобретение [b]документа[/b], который подтверждает обучение в ВУЗе, – это выгодное решение.
[url=http://jeepgarage.ru/forum/topic.php?forum=24&topic=765/]jeepgarage.ru/forum/topic.php?forum=24&topic=765[/url]
[b]Рады помочь![/b].
Dnrtojg
August 21, 2024 @ 8:38 am
[u][b]Здравствуйте![/b][/u]
[b]Заказать документ[/b] университета вы имеете возможность у нас.
[url=http://cars.teamforum.ru/viewtopic.php?f=2&t=1602/]cars.teamforum.ru/viewtopic.php?f=2&t=1602[/url]
Diplomi_ubSi
August 21, 2024 @ 8:51 am
Привет, друзья!
Купить документ института вы можете в нашей компании в Москве.
zoolife55.ru/product/urinary-so-feline-kurica/reviews
Cazrmfq
August 21, 2024 @ 9:03 am
[u][b] Привет![/b][/u]
Мы предлагаем документы техникумов
[url=http://5чудес.рф/product/igra-stol-forma/reviews/]5чудес.рф/product/igra-stol-forma/reviews[/url]
EdwardTum
August 21, 2024 @ 9:56 am
купить комплектующие для вентиляции каталог вентиляционного оборудования
Diplomi_lgSi
August 21, 2024 @ 12:30 pm
Добрый день!
Купить документ о получении высшего образования вы можете у нас.
[url=http://fb7707ng.bget.ru/main/1-post1/]fb7707ng.bget.ru/main/1-post1[/url]
Lariorjtv
August 21, 2024 @ 12:32 pm
Диплом пту купить официально с упрощенным обучением в Москве
[url=http://telegra.ph/kupit-diplom-vracha-v-ukraine-08-13-2/]telegra.ph/kupit-diplom-vracha-v-ukraine-08-13-2[/url]
Lariorgre
August 21, 2024 @ 1:00 pm
Приобретение диплома ПТУ с сокращенной программой обучения в Москве
[url=http://telegra.ph/kak-kupit-diplom-vysshego-obrazovaniya-08-13-5/]telegra.ph/kak-kupit-diplom-vysshego-obrazovaniya-08-13-5[/url]
Uazrjtm
August 21, 2024 @ 1:04 pm
[u][b]Здравствуйте![/b][/u]
[b]Заказать диплом любого ВУЗа.[/b]
[url=http://telegra.ph/kupit-diplom-yurista-08-13-6/]telegra.ph/kupit-diplom-yurista-08-13-6[/url]
Lariorrws
August 21, 2024 @ 1:38 pm
Аттестат школы купить официально с упрощенным обучением в Москве
[url=http://telegra.ph/kupit-diplom-turizm-08-13-10/]telegra.ph/kupit-diplom-turizm-08-13-10[/url]
Mazrthn
August 21, 2024 @ 1:53 pm
[u][b] Здравствуйте![/b][/u]
[b]Как избежать рисков при покупке диплома колледжа или ПТУ в России[/b]
[url=http://teh-lib.ru/mss/poverka-i-kalibrovka-sredstv-izmerenija/]teh-lib.ru/mss/poverka-i-kalibrovka-sredstv-izmerenija[/url]
Trefqqt
August 21, 2024 @ 1:57 pm
[u][b] Привет, друзья![/b][/u]
Вопросы и ответы: можно ли быстро купить диплом старого образца?
[url=http://fromrus.su/index.php?/topic/15689-источник-p888t/]fromrus.su/index.php?/topic/15689-источник-p888t[/url]
Рады помочь!.
DanielTam
August 21, 2024 @ 3:01 pm
Misoprostol 200 mg buy online https://cytotec.pro/# cytotec buy online usa
lasix 100mg
Lazrlym
August 21, 2024 @ 3:48 pm
[u][b] Привет, друзья![/b][/u]
Приобрести [b]диплом[/b] любого ВУЗа.
[url=http://forum.ssa.ru/cat-novosti.-stati./topic-33008/]forum.ssa.ru/cat-novosti.-stati./topic-33008[/url]
Xazrpff
August 21, 2024 @ 4:11 pm
[u][b] Привет![/b][/u]
Купить документ о получении высшего образования.
Мы изготавливаем [b]дипломы[/b] психологов, юристов, экономистов и прочих профессий по приятным ценам.
[url=http://strata.com/forums/users/HeatherCastro//]strata.com/forums/users/HeatherCastro/[/url]
[b]Рады оказаться полезными![/b].
Sazrokw
August 21, 2024 @ 4:17 pm
[u][b] Здравствуйте![/b][/u]
Приобрести документ о получении высшего образования
[url=http://exler.me/user/Strider/]exler.me/user/Strider[/url]
Yrefkdh
August 21, 2024 @ 4:43 pm
[u][b] Здравствуйте![/b][/u]
[b]Купить диплом любого университета.[/b]
[url=http://lasergrafics.de/handel/artikel-b/#comment-92212/]lasergrafics.de/handel/artikel-b/#comment-92212[/url]
Trefimy
August 21, 2024 @ 4:47 pm
[u][b] Здравствуйте![/b][/u]
Как купить аттестат 11 класса с официальным упрощенным обучением в Москве
[url=http://ajcc-conf.net/2020/07/05/ajcc2021-call-for-papers/#comment-35400/]ajcc-conf.net/2020/07/05/ajcc2021-call-for-papers/#comment-35400[/url]
Окажем помощь!.
Mazrehb
August 21, 2024 @ 4:48 pm
[u][b] Здравствуйте![/b][/u]
[b]Всё, что нужно знать о покупке аттестата о среднем образовании[/b]
[url=http://cardano.ru/forum/index.php/]cardano.ru/forum/index.php[/url]
DanielTam
August 21, 2024 @ 5:18 pm
buy cytotec pills https://tamoxifen.bid/# tamoxifen endometriosis
lasix 100 mg tablet
StephenTip
August 21, 2024 @ 7:20 pm
https://cytotec.pro/# Cytotec 200mcg price
Williamraics
August 21, 2024 @ 7:38 pm
[u][b] Здравствуйте![/b][/u]
Мы готовы предложить дипломы.
[url=http://vrn.best-city.ru/forum/thread540106043/]vrn.best-city.ru/forum/thread540106043[/url]
Eanrdqv
August 21, 2024 @ 7:47 pm
[u][b] Добрый день![/b][/u]
[b]Мы изготавливаем дипломы[/b] любой профессии по невысоким ценам.
[url=http://petcreche.com.br/fotos-e-videos/fotos/page/4/]petcreche.com.br/fotos-e-videos/fotos/page/4[/url]
Mazrjwd
August 21, 2024 @ 7:53 pm
[u][b] Добрый день![/b][/u]
[b]Легальная покупка диплома о среднем образовании в Москве и регионах[/b]
[url=http://vsesoch.ru/a-p-chexov?start=8/]vsesoch.ru/a-p-chexov?start=8[/url]
Oariorrpb
August 21, 2024 @ 8:23 pm
[u][b] Добрый день![/b][/u]
Заказать диплом университета
[b]Наша компания предлагает[/b] выгодно и быстро заказать диплом, который выполнен на оригинальной бумаге и заверен мокрыми печатями, водяными знаками, подписями должностных лиц. Диплом способен пройти любые проверки, даже с применением специального оборудования. Решите свои задачи максимально быстро с нашим сервисом.
[b]Где приобрести диплом по необходимой специальности?[/b]
[url=http://nc750.ru/member.php?u=2812/]nc750.ru/member.php?u=2812[/url]
[url=http://telegra.ph/lgoty-dlya-krymchan-pri-postuplenii-v-vuz-07-24/]telegra.ph/lgoty-dlya-krymchan-pri-postuplenii-v-vuz-07-24[/url]
[url=http://rodme.ru/obrazovanie-dlya-karernyh-vysot-t12778/]rodme.ru/obrazovanie-dlya-karernyh-vysot-t12778[/url]
[url=http://telegra.ph/rasschitat-postuplenie-v-vuz-07-24/]telegra.ph/rasschitat-postuplenie-v-vuz-07-24[/url]
[url=http://animalprotect.org/forum/index.php?topic=239219.0/]animalprotect.org/forum/index.php?topic=239219.0[/url]
StephenTip
August 21, 2024 @ 8:37 pm
https://furosemide.win/# lasix furosemide
Sazrbqi
August 21, 2024 @ 9:04 pm
[u][b] Здравствуйте![/b][/u]
Приобрести документ о получении высшего образования
[url=http://telegra.ph/raschet-stoimosti-obucheniya-v-vuze-07-24/]telegra.ph/raschet-stoimosti-obucheniya-v-vuze-07-24[/url]
[url=http://c904581v.beget.tech/2024/07/07/teh-podderzhka-i-faq-pokupaem-dokumenty-v-seti-internet/]c904581v.beget.tech/2024/07/07/teh-podderzhka-i-faq-pokupaem-dokumenty-v-seti-internet[/url]
[url=http://telegra.ph/kak-sdat-egeh-dlya-postupleniya-v-vuz-07-24/]telegra.ph/kak-sdat-egeh-dlya-postupleniya-v-vuz-07-24[/url]
[url=http://astroroleplay.getbb.ru/viewtopic.php?f=6&t=501/]astroroleplay.getbb.ru/viewtopic.php?f=6&t=501[/url]
[url=http://vaeaem.profiforum.ru/t4090-topic#7385/]vaeaem.profiforum.ru/t4090-topic#7385[/url]
Lazrent
August 21, 2024 @ 10:38 pm
[u][b] Привет![/b][/u]
[b]Мы изготавливаем дипломы[/b] психологов, юристов, экономистов и любых других профессий по приятным тарифам.
[url=http://telegra.ph/pri-obuchenii-v-magistrature-magistrant-kak-pravilo-provodit-v-vuze-08-02/]telegra.ph/pri-obuchenii-v-magistrature-magistrant-kak-pravilo-provodit-v-vuze-08-02[/url]
Xazrhgl
August 21, 2024 @ 11:53 pm
[u][b] Добрый день![/b][/u]
Заказать документ ВУЗа.
Мы изготавливаем [b]дипломы[/b] любых профессий по приятным ценам.
[url=http://synfig.org/issues/thebuggenie/synfig/issues/7925/]synfig.org/issues/thebuggenie/synfig/issues/7925[/url]
[b]Рады помочь![/b].
Lazrrnu
August 22, 2024 @ 1:47 am
[u][b] Привет![/b][/u]
[b]Купить диплом о высшем образовании.[/b]
[url=http://telegra.ph/formy-obucheniya-v-vuzah-08-02/]telegra.ph/formy-obucheniya-v-vuzah-08-02[/url]
Plastikovie okna v sochi_elSr
August 22, 2024 @ 1:51 am
пластиковые окна в сочи цена [url=https://remstroyokna.ru]пластиковые окна в сочи цена [/url] .
Dnrtzyo
August 22, 2024 @ 1:52 am
[u][b]Привет, друзья![/b][/u]
[b]Приобрести документ[/b] о получении высшего образования вы сможете в нашей компании.
[url=http://mamadona.ru/blogs/hotite_kupit_diplom_o_vysshem_obrazovani/]mamadona.ru/blogs/hotite_kupit_diplom_o_vysshem_obrazovani[/url]
Lazrtvj
August 22, 2024 @ 1:58 am
[u][b] Добрый день![/b][/u]
[b]Мы можем предложить дипломы[/b] любой профессии по доступным тарифам.
[url=http://telegra.ph/vechernyaya-forma-obucheniya-v-vuze-08-02/]telegra.ph/vechernyaya-forma-obucheniya-v-vuze-08-02[/url]
Lazrmjt
August 22, 2024 @ 2:30 am
[u][b] Привет, друзья![/b][/u]
[b]Купить диплом любого ВУЗа.[/b]
[url=http://sdiplom.ru/kupit-attestat-o-srednem-obrazovanii-bez-lishnix-problem/]sdiplom.ru/kupit-attestat-o-srednem-obrazovanii-bez-lishnix-problem[/url]
Xazrbbr
August 22, 2024 @ 2:58 am
[u][b] Привет, друзья![/b][/u]
Купить документ о получении высшего образования.
Мы изготавливаем [b]дипломы[/b] любых профессий по приятным тарифам.
[url=http://dzone.com/users/5166875/leonardchambers/]dzone.com/users/5166875/leonardchambers[/url]
[b]Рады помочь![/b].
Iariorktc
August 22, 2024 @ 3:03 am
[u][b] Добрый день![/b][/u]
Купить документ о получении высшего образования вы имеете возможность в нашей компании. Мы предлагаем документы об окончании любых университетов России. Вы получите диплом по любой специальности, любого года выпуска, в том числе документы образца СССР. Даем 100% гарантию, что в случае проверки документов работодателями, подозрений не появится.
[url=http://newsinweek.ru/kachestvennyie-diplomyi-bez-posrednikov/]newsinweek.ru/kachestvennyie-diplomyi-bez-posrednikov[/url]
[url=http://telegra.ph/postuplenie-v-vuz-posle-1-kursa-kolledzha-07-24/]telegra.ph/postuplenie-v-vuz-posle-1-kursa-kolledzha-07-24[/url]
[url=http://telearchaeology.org/TAWiki/index.php/Дипломы_для_уверенного_будущего/]telearchaeology.org/TAWiki/index.php/Дипломы_для_уверенного_будущего[/url]
[url=http://comedyforme.ru/vash-diplom-vash-uspeh/]comedyforme.ru/vash-diplom-vash-uspeh[/url]
[url=http://heroes.app/blogs/new/]heroes.app/blogs/new[/url]
Mazrxvj
August 22, 2024 @ 5:10 am
[u][b] Привет![/b][/u]
[b]Купить диплом о среднем образовании в Москве и любом другом городе[/b]
[url=http://sport-faq.ru/put-k-uspehu-kupi-diplom-i-preuspey-v-karere/]sport-faq.ru/put-k-uspehu-kupi-diplom-i-preuspey-v-karere[/url]
AndrewSwarf
August 22, 2024 @ 5:21 am
продать скины кс 2 на карту https://prodat-skiny-ks2.ru
DanielTam
August 22, 2024 @ 5:28 am
buy cytotec pills https://lisinopril.guru/# lisinopril 10 mg
furosemide 100mg
Dnrtrut
August 22, 2024 @ 5:41 am
[u][b]Добрый день![/b][/u]
[b]Заказать документ[/b] ВУЗа вы можете в нашем сервисе.
[url=http://thesims2thebest.0pk.me/viewtopic.php?id=727#p1300/]thesims2thebest.0pk.me/viewtopic.php?id=727#p1300[/url]
Trefxpz
August 22, 2024 @ 6:04 am
[u][b] Добрый день![/b][/u]
Как официально приобрести аттестат 11 класса с минимальными затратами времени
[url=http://kamerton72.ru/users/michellshivy/]kamerton72.ru/users/michellshivy[/url]
Рады оказать помощь!.
Williamraics
August 22, 2024 @ 6:45 am
[u][b] Привет![/b][/u]
Мы готовы предложить дипломы.
[url=http://konivet.2bb.ru/viewtopic.php?id=6880#p32741/]konivet.2bb.ru/viewtopic.php?id=6880#p32741[/url]
Oarioruaz
August 22, 2024 @ 6:52 am
[u][b] Привет, друзья![/b][/u]
Заказать диплом о высшем образовании
[b]Мы предлагаем[/b] быстро и выгодно приобрести диплом, который выполняется на оригинальном бланке и заверен мокрыми печатями, штампами, подписями официальных лиц. Диплом пройдет лубую проверку, даже при использовании специального оборудования. Достигайте свои цели быстро и просто с нашим сервисом.
[b]Где заказать диплом по необходимой специальности?[/b]
[url=http://yexanin202.ixbb.ru/viewtopic.php?id=125#p125/]yexanin202.ixbb.ru/viewtopic.php?id=125#p125[/url]
[url=http://animalplanetnews.ru/poluchite-diplom-i-stante-uspeshnyim/]animalplanetnews.ru/poluchite-diplom-i-stante-uspeshnyim[/url]
[url=http://allonlinesport.ru/kupite-diplom-i-nachnite-novuyu-zhizn/]allonlinesport.ru/kupite-diplom-i-nachnite-novuyu-zhizn[/url]
[url=http://epistles.ru/blogs/1719/Купите-диплом-быстро-и-легко/]epistles.ru/blogs/1719/Купите-диплом-быстро-и-легко[/url]
[url=http://income.forum2x2.ru/t2983-topic#5303/]income.forum2x2.ru/t2983-topic#5303[/url]
Eanrrby
August 22, 2024 @ 6:53 am
[u][b] Добрый день![/b][/u]
[b]Мы можем предложить дипломы[/b] психологов, юристов, экономистов и других профессий по приятным тарифам.
[url=http://clickforart.com/artist/view/ilona-d.-veresk/?type=2/]clickforart.com/artist/view/ilona-d.-veresk/?type=2[/url]
DanielTam
August 22, 2024 @ 7:45 am
buy cytotec online https://tamoxifen.bid/# how to prevent hair loss while on tamoxifen
furosemide 40mg
Trefjss
August 22, 2024 @ 9:07 am
[u][b] Привет![/b][/u]
Сколько стоит получить диплом высшего и среднего образования легально?
[url=http://mycddiary.blogspot.com/2012/03/blog-post_4098/]mycddiary.blogspot.com/2012/03/blog-post_4098[/url]
Рады оказаться полезными!.
Diplomi_weSi
August 22, 2024 @ 11:45 am
Здравствуйте!
Заказать документ ВУЗа вы имеете возможность у нас в Москве.
[url=http://dxzw.com/member/index.php?uid=iwuhun/]dxzw.com/member/index.php?uid=iwuhun[/url]
Mazriyw
August 22, 2024 @ 12:09 pm
[u][b] Привет![/b][/u]
[b]Как оказалось, купить диплом кандидата наук не так уж и сложно[/b]
[url=http://saluttunisia.com/jomsocial/groups/viewbulletin/44-izvestnyj-internet-magazin-s-ogromnym-katalogom-diplomov?groupid=101/]saluttunisia.com/jomsocial/groups/viewbulletin/44-izvestnyj-internet-magazin-s-ogromnym-katalogom-diplomov?groupid=101[/url]
Lazrwjt
August 22, 2024 @ 1:28 pm
[u][b] Добрый день![/b][/u]
[b]Купить диплом о высшем образовании.[/b]
[url=http://telegra.ph/s-kakogo-chisla-nachinaetsya-postuplenie-v-vuzy-08-02/]telegra.ph/s-kakogo-chisla-nachinaetsya-postuplenie-v-vuzy-08-02[/url]
Lazrcum
August 22, 2024 @ 1:54 pm
[u][b] Привет![/b][/u]
[b]Приобрести диплом любого университета.[/b]
[url=http://diplom45.ru/gde-kupit-shkolnyj-attestat-legalno-i-bystro/]diplom45.ru/gde-kupit-shkolnyj-attestat-legalno-i-bystro[/url]
Iariorezy
August 22, 2024 @ 2:09 pm
[u][b] Здравствуйте![/b][/u]
Приобрести документ института вы сможете у нас. Мы оказываем услуги по продаже документов об окончании любых университетов России. Вы получите необходимый диплом по любой специальности, любого года выпуска, включая документы Советского Союза. Гарантируем, что в случае проверки документа работодателями, подозрений не появится.
[url=http://poetzinc.com/read-blog/12086/]poetzinc.com/read-blog/12086[/url]
[url=http://telegra.ph/kakoj-srednij-ball-attestata-dlya-postupleniya-v-vuz-07-24/]telegra.ph/kakoj-srednij-ball-attestata-dlya-postupleniya-v-vuz-07-24[/url]
[url=http://vseogirls.ru/byistroe-poluchenie-diploma-onlayn/]vseogirls.ru/byistroe-poluchenie-diploma-onlayn[/url]
[url=http://x91392sl.beget.tech/2024/07/18/vysokokachestvennye-diplomy-s-garantiey/]x91392sl.beget.tech/2024/07/18/vysokokachestvennye-diplomy-s-garantiey[/url]
[url=http://sankt-peterburg.forum2x2.ru/t20691-topic#77591/]sankt-peterburg.forum2x2.ru/t20691-topic#77591[/url]
Xazrrgw
August 22, 2024 @ 2:57 pm
[u][b] Привет![/b][/u]
Заказать документ института.
Мы предлагаем [b]дипломы[/b] любых профессий по приятным ценам.
[url=http://camp-fire.jp/profile/BernardCruz/]camp-fire.jp/profile/BernardCruz[/url]
[b]Будем рады вам помочь![/b].
Diplomi_nnSi
August 22, 2024 @ 3:30 pm
Добрый день!
Купить документ университета вы имеете возможность у нас в Москве.
[url=http://старый.ркэ.рф/index.php/component/k2/itemlist/user/127688/]старый.ркэ.рф/index.php/component/k2/itemlist/user/127688[/url]
Narkolog na dom krasnodar_orSa
August 22, 2024 @ 6:10 pm
нарколог краснодар [url=https://narkolog-na-dom-krasnodar12.ru/]нарколог краснодар[/url] .
Lazrwsp
August 22, 2024 @ 6:28 pm
[u][b] Здравствуйте![/b][/u]
[b]Мы готовы предложить дипломы[/b] психологов, юристов, экономистов и прочих профессий по приятным тарифам.
[url=http://telegra.ph/aktivnye-metody-obucheniya-v-vuze-primery-08-02/]telegra.ph/aktivnye-metody-obucheniya-v-vuze-primery-08-02[/url]
vivod iz zapoya_ztSt
August 22, 2024 @ 6:41 pm
кодировка гипнозом [url=https://xn——7cdhaozbh1ayqhot7ooa6e.xn--p1ai]https://xn——7cdhaozbh1ayqhot7ooa6e.xn--p1ai[/url] .
DanielTam
August 22, 2024 @ 6:52 pm
buy cytotec pills http://furosemide.win/# lasix generic
lasix online
Cazrcvz
August 22, 2024 @ 7:09 pm
[u][b] Здравствуйте![/b][/u]
Купить диплом ВУЗа
[url=http://telegra.ph/postuplenie-v-vuz-2020-08-02/]telegra.ph/postuplenie-v-vuz-2020-08-02[/url]
Sazrrgz
August 22, 2024 @ 7:17 pm
[u][b] Привет![/b][/u]
Мы готовы предложить [b]дипломы[/b] любой профессии.
Заказ [b]документа[/b], который подтверждает окончание института, – это грамотное решение.
[url=http://jamierandall.com/contact/#comment-12149/]jamierandall.com/contact/#comment-12149[/url]
[url=http://doktorpishet.ru/index.php?subaction=userinfo&user=ymaru/]doktorpishet.ru/index.php?subaction=userinfo&user=ymaru[/url]
[url=http://uralremput.ru/index.php?subaction=userinfo&user=oqoryf/]uralremput.ru/index.php?subaction=userinfo&user=oqoryf[/url]
[url=http://elkin-geo.com/index.php?option=com_akobook/]elkin-geo.com/index.php?option=com_akobook[/url]
[url=http://forum.mbprinteddroids.com/member.php?action=profile&uid=73750/]forum.mbprinteddroids.com/member.php?action=profile&uid=73750[/url]
DanielTam
August 22, 2024 @ 8:58 pm
order cytotec online http://cytotec.pro/# buy cytotec online fast delivery
furosemida 40 mg
Lazrrih
August 22, 2024 @ 10:21 pm
[u][b] Привет, друзья![/b][/u]
[b]Мы предлагаем дипломы[/b] любых профессий по приятным тарифам.
[url=http://telegra.ph/kakoj-srednij-ball-attestata-dlya-postupleniya-v-vuz-08-02/]telegra.ph/kakoj-srednij-ball-attestata-dlya-postupleniya-v-vuz-08-02[/url]
Mazrify
August 22, 2024 @ 10:37 pm
[u][b] Привет, друзья![/b][/u]
[b]Официальная покупка школьного аттестата с упрощенным обучением в Москве[/b]
[url=http://pipeclub.net/index.php?showuser=110652&tab=blog&k=880ea6a14ea49e853634fbdc5015a024&setlanguage=1&langurlbits=showuser=110652&cal_id=&langid=2/]pipeclub.net/index.php?showuser=110652&tab=blog&k=880ea6a14ea49e853634fbdc5015a024&setlanguage=1&langurlbits=showuser=110652&cal_id=&langid=2[/url]
Sazrcdb
August 22, 2024 @ 10:55 pm
[u][b] Здравствуйте![/b][/u]
Купить документ университета
[url=http://ixcjuleogr.fxmag.ru/aendr/]ixcjuleogr.fxmag.ru/aendr[/url]
Cazrmrj
August 22, 2024 @ 11:47 pm
[u][b] Добрый день![/b][/u]
Мы предлагаем документы техникумов
[url=http://slliver.getbb.ru/viewtopic.php?f=62&t=951/]slliver.getbb.ru/viewtopic.php?f=62&t=951[/url]
Mazrzsr
August 23, 2024 @ 12:29 am
[u][b] Здравствуйте![/b][/u]
Быстрое обучение и получение диплома магистра – возможно ли это?
[url=http://anomalycat.blogspot.ru/2017/03/blog-post_37/]anomalycat.blogspot.ru/2017/03/blog-post_37[/url]
Dnrtpht
August 23, 2024 @ 12:55 am
[u][b]Привет, друзья![/b][/u]
[b]Купить документ[/b] о получении высшего образования можно в нашей компании.
[url=http://bakinsky-dvorik.ru/communication/forum/user/183037/]bakinsky-dvorik.ru/communication/forum/user/183037[/url]
Trefpdi
August 23, 2024 @ 1:07 am
[u][b] Добрый день![/b][/u]
Приобретение школьного аттестата с официальным упрощенным обучением в Москве
[url=http://tastebuds.fm/topics/7635-hei-tarvitsen-apuasi/]tastebuds.fm/topics/7635-hei-tarvitsen-apuasi[/url]
Рады оказать помощь!.
pansionat dlya pojilih_xsKt
August 23, 2024 @ 2:52 am
дом пенсионеров в береговом [url=www.xn—–1-43da3arnf4adrboggk3ay6e3gtd.xn--p1ai/]дом пенсионеров в береговом[/url] .
Trefvjh
August 23, 2024 @ 4:37 am
[u][b] Здравствуйте![/b][/u]
Официальная покупка диплома вуза с упрощенной программой обучения
[url=http://1q5p6.blogspot.com/2015/11/fann/]1q5p6.blogspot.com/2015/11/fann[/url]
Будем рады вам помочь!.
Dnrtlfn
August 23, 2024 @ 4:57 am
[u][b]Привет, друзья![/b][/u]
[b]Приобрести документ[/b] ВУЗа вы сможете в нашей компании в столице.
[url=http://wiki.gptel.ru/index.php/Участник:Hnfmrfdtg/]wiki.gptel.ru/index.php/Участник:Hnfmrfdtg[/url]
Justinmew
August 23, 2024 @ 5:39 am
[url=][/url]
Sale of non ferrous metal alloys
At Cliffton Trading, we pride ourselves on being a leading international supplier of non-ferrous metals. Based in Dubai, our company specializes in providing high-quality materials such as copper powder, copper ingots, selenium powder, and nickel wire to industries around the world. Our commitment to excellence ensures that every product we offer meets the highest standards, backed by certifications from top chemical laboratories.
[url=https://cliffton-group.com/catalog/copper-powder/]Copper powder[/url]
Pure high-quality copper powder (Cu) with consistent particle size.
[url=https://cliffton-group.com/catalog/copper-ingots/]Copper ingots[/url]
These bullions have high purity and excellent conductivity, making them indispensable in electronics manufacturing, construction and mechanical engineering.
[url=https://cliffton-group.com/catalog/selenium-powder/]Selenium powder[/url]
Our products include high purity metal dust and microfine powder.
[url=https://cliffton-group.com/catalog/nickel-wire]Nickel wire[/url]
Nickel wire is ideal for use in a variety of industrial applications.
With a strong global distribution network, we serve over 15 countries, catering to diverse sectors like construction, automotive, and electronics. Our approach is centered on reliability, quality, and customer satisfaction, which allows us to build lasting relationships with our clients. We understand the importance of tailored solutions, so we work closely with our customers to meet their specific needs, ensuring that they receive the best possible value.
Our team of experienced professionals is dedicated to maintaining the integrity and efficiency of our supply chains, allowing us to deliver products consistently and on time. Competitive pricing, combined with our extensive product range, makes us a preferred partner for businesses seeking dependable suppliers of non-ferrous metals. At Cliffton Trading, we are not just about transactions; we are about building partnerships that drive success for both our company and our clients.
You can view the products and place an order on our website [url=https://cliffton-group.com/]cliffton-group.com[/url].
[url=][/url]
Mazrdfk
August 23, 2024 @ 5:41 am
[u][b] Привет![/b][/u]
[b]Приобретение диплома ПТУ с сокращенной программой обучения в Москве[/b]
[url=http://rbg-azimut.com/forum/viewtopic.php?f=86&t=2294&st=0&sk=t&sd=a&start=90/]rbg-azimut.com/forum/viewtopic.php?f=86&t=2294&st=0&sk=t&sd=a&start=90[/url]
Cazracx
August 23, 2024 @ 6:34 am
[u][b] Привет![/b][/u]
Мы можем предложить документы техникумов
[url=http://danceway74.ru/posts/47581-zarabotok-o655q/]danceway74.ru/posts/47581-zarabotok-o655q[/url]
Sazroaw
August 23, 2024 @ 6:48 am
[u][b] Здравствуйте![/b][/u]
Мы готовы предложить [b]дипломы[/b] любой профессии.
Приобретение [b]диплома[/b], подтверждающего окончание ВУЗа, – это грамотное решение.
[url=http://kulinariya123.blogspot.com/2011/01/228?m=1/]kulinariya123.blogspot.com/2011/01/228?m=1[/url]
[url=http://doktorpishet.ru/index.php?subaction=userinfo&user=iqeloc/]doktorpishet.ru/index.php?subaction=userinfo&user=iqeloc[/url]
[url=http://apbuon.blogspot.com/2013/05/blog-post_830/]apbuon.blogspot.com/2013/05/blog-post_830[/url]
[url=http://imperiasport.ru/product/utjazheliteli-2h45-kg-reguliruemye-bstaw20-/reviews/]imperiasport.ru/product/utjazheliteli-2h45-kg-reguliruemye-bstaw20-/reviews[/url]
[url=http://whitepuppywhite.blogspot.ru/2013/06/givenchy-skin-targetters-active-pure/]whitepuppywhite.blogspot.ru/2013/06/givenchy-skin-targetters-active-pure[/url]
Sazrjbh
August 23, 2024 @ 7:04 am
[u][b] Добрый день![/b][/u]
Мы можем предложить [b]дипломы[/b] любых профессий.
Заказ [b]диплома[/b], подтверждающего обучение в ВУЗе, – это грамотное решение.
[url=http://koxma.4adm.ru/viewtopic.php?f=298&t=5523/]koxma.4adm.ru/viewtopic.php?f=298&t=5523[/url]
[b]Рады оказаться полезными![/b].
Mazrmfm
August 23, 2024 @ 7:04 am
[b]Приобрести документ университета.[/b]
[url=http://vestiinform.ru/forums/topic/na-sajte-j427c/]vestiinform.ru/forums/topic/na-sajte-j427c[/url]
[url=http://6986986.com/forum.php/]6986986.com/forum.php[/url]
[url=http://avtoserviskovrov.ru/index.php?subaction=userinfo&user=ojurilan/]avtoserviskovrov.ru/index.php?subaction=userinfo&user=ojurilan[/url]
[url=http://sims4mods.ru/lots/simsy-uchastok-brodyagi-chellendzh/#comment-120/]sims4mods.ru/lots/simsy-uchastok-brodyagi-chellendzh/#comment-120[/url]
[url=http://xn—-7sbqaaodvuyaxc7e.xn--p1ai/reviews/?unapproved=622172&moderation-hash=2d706e2f904f51edab38a09c133f256a#comment-622172/]xn—-7sbqaaodvuyaxc7e.xn--p1ai/reviews/?unapproved=622172&moderation-hash=2d706e2f904f51edab38a09c133f256a#comment-622172[/url]
Lariorjpm
August 23, 2024 @ 7:40 am
Покупка диплома о среднем полном образовании: как избежать мошенничества?
[url=http://telegra.ph/skolko-stoit-kupit-diplom-08-13/]telegra.ph/skolko-stoit-kupit-diplom-08-13[/url]
DanielTam
August 23, 2024 @ 7:50 am
buy cytotec pills online cheap https://tamoxifen.bid/# tamoxifen mechanism of action
furosemide 40mg
Lariorzmx
August 23, 2024 @ 8:05 am
Официальная покупка школьного аттестата с упрощенным обучением в Москве
[url=http://telegra.ph/kupit-diplom-zakryvshegosya-vuza-08-13-8/]telegra.ph/kupit-diplom-zakryvshegosya-vuza-08-13-8[/url]
Uazrutc
August 23, 2024 @ 8:17 am
[u][b]Здравствуйте![/b][/u]
[b]Приобрести диплом любого университета.[/b]
[url=http://telegra.ph/kupit-nastoyashchij-diplom-08-13/]telegra.ph/kupit-nastoyashchij-diplom-08-13[/url]
Lariorxae
August 23, 2024 @ 8:32 am
Удивительно, но купить диплом кандидата наук оказалось не так сложно
[url=http://telegra.ph/kupit-diplom-ob-okonchanii-uchilishcha-08-13-9/]telegra.ph/kupit-diplom-ob-okonchanii-uchilishcha-08-13-9[/url]
generac 7145
August 23, 2024 @ 9:29 am
Секреты выбора лучшего генератора Generac, как выбрать генератора Generac.
В чем отличие генератора Generac от других моделей?, подробный обзор генератора Generac.
Генератор Generac для надежного источника энергии, подробный обзор.
Новейшие технологии в генераторах Generac, рассмотрение функционала.
Генератор Generac: надежность и долговечность, подробный анализ.
Эффективное решение для энергетической безопасности: генераторы Generac, советы эксперта.
Энергия без перебоев: генераторы Generac для дома, плюсы использования.
Как выбрать генератор Generac для эффективного резервного энергоснабжения, подробный обзор.
Безопасность и надежность: генераторы Generac, рекомендации.
Энергия в вашем доме: генераторы Generac, особенности.
generac 7189 [url=https://generac-generatory1.ru/]https://generac-generatory1.ru/[/url] .
DanielTam
August 23, 2024 @ 9:52 am
cytotec abortion pill https://lipitor.guru/# canadian pharmacy lipitor
lasix
Cazrrwg
August 23, 2024 @ 10:58 am
[u][b] Добрый день![/b][/u]
Мы можем предложить документы техникумов
[url=http://telegra.ph/kupitspravkuobobucheniivvuzevmoskve07242/]telegra.ph/kupitspravkuobobucheniivvuzevmoskve07242[/url]
Lazrsgq
August 23, 2024 @ 10:58 am
[u][b] Привет, друзья![/b][/u]
Купить [b]диплом[/b] университета.
[url=http://orgi.biz/org_diplom_moskva_/]orgi.biz/org_diplom_moskva_[/url]
Kak zarabotat dengi_jter
August 23, 2024 @ 11:20 am
темы для заработка [url=https://www.kak-zarabotat-dengi11.ru]https://www.kak-zarabotat-dengi11.ru[/url] .
Sazrgsy
August 23, 2024 @ 11:25 am
[u][b] Добрый день![/b][/u]
Приобрести документ о получении высшего образования
[url=http://tvserver.ru/forum/sending.php?mode=reply&t=5166/]tvserver.ru/forum/sending.php?mode=reply&t=5166[/url]
Mazrbkj
August 23, 2024 @ 11:45 am
[u][b] Привет![/b][/u]
Как не стать жертвой мошенников при покупке диплома о среднем полном образовании
[url=http://newis.biz/kak-rabotaet-sarafannoe-radio-v-vkontakte/#comment-334423/]newis.biz/kak-rabotaet-sarafannoe-radio-v-vkontakte/#comment-334423[/url]
Mazruwf
August 23, 2024 @ 2:28 pm
[u][b] Добрый день![/b][/u]
[b]Пошаговая инструкция по безопасной покупке диплома о высшем образовании[/b]
[url=http://trans-asvt.ru/raznoe/kakuyu-odezhdu-vzyat-v-konnyiy-pohod/]trans-asvt.ru/raznoe/kakuyu-odezhdu-vzyat-v-konnyiy-pohod[/url]
generac 7189 купить
August 23, 2024 @ 3:00 pm
Как не ошибиться при покупке генератора Generac, как выбрать генератора Generac.
Почему стоит выбрать генератор Generac?, подробный обзор генератора Generac.
Использование генератора Generac для обеспечения надежной работы электроприборов, советы по использованию.
Настоящее качество: генераторы Generac, рассмотрение функционала.
Почему генераторы Generac так популярны?, подробный анализ.
Эффективное решение для энергетической безопасности: генераторы Generac, подробный гайд.
Генератор Generac: лучший источник резервного питания, характеристики.
Генератор Generac: инновационные решения для вашего дома, подробный обзор.
Генератор Generac для обеспечения непрерывного электроснабжения, особенности использования.
Обеспечение надежного энергоснабжения с помощью генератора Generac, подбор модели.
7145 generac [url=https://generac-generatory1.ru/]https://generac-generatory1.ru/[/url] .
Yrefeab
August 23, 2024 @ 3:52 pm
[u][b] Привет![/b][/u]
[b]Купить диплом любого ВУЗа.[/b]
[url=http://dverkivdom.ru/lu-23-satinat/reviews/]dverkivdom.ru/lu-23-satinat/reviews[/url]
Sazrgii
August 23, 2024 @ 3:55 pm
[u][b] Добрый день![/b][/u]
Приобрести документ ВУЗа
[url=http://forum-pmr.net/member.php?u=42748/]forum-pmr.net/member.php?u=42748[/url]
[url=http://vxengine.ru/blogs/543/Легкий-поиск-интернет-магазина-который-продает-дипломы-и-аттестаты/]vxengine.ru/blogs/543/Легкий-поиск-интернет-магазина-который-продает-дипломы-и-аттестаты[/url]
[url=http://1-click.pl/excelforum/viewtopic.php?f=3&t=256376/]1-click.pl/excelforum/viewtopic.php?f=3&t=256376[/url]
[url=http://mindclub.online/wiki/index.php/Дипломы_для_карьерных_высот/]mindclub.online/wiki/index.php/Дипломы_для_карьерных_высот[/url]
[url=http://t98223u0.beget.tech/2024/07/19/legko-i-bystro-poluchite-diplom/]t98223u0.beget.tech/2024/07/19/legko-i-bystro-poluchite-diplom[/url]
Sazrgrz
August 23, 2024 @ 5:15 pm
[u][b] Добрый день![/b][/u]
Мы изготавливаем [b]дипломы[/b] любой профессии.
Приобретение [b]диплома[/b], подтверждающего окончание ВУЗа, – это выгодное решение.
[url=http://jurnalbest.com/index.php/mrbest/comment/view/162/0/93495/]jurnalbest.com/index.php/mrbest/comment/view/162/0/93495[/url]
[b]Поможем вам всегда![/b].
Elliotkix
August 23, 2024 @ 5:18 pm
Джип туры по Крыму https://м-драйв.рф/tours/krysha-kryma/ уникальные маршруты и яркие эмоции. Погрузитесь в увлекательнее приключение вместе с нами. Горные, лесные, подземные экскурсии, джиппинг в Крыму с максимальным комфортом.
Cazruin
August 23, 2024 @ 5:26 pm
[u][b] Добрый день![/b][/u]
Мы можем предложить документы ВУЗов
[url=http://newlifeincz.blogspot.com/2013/06/blog-post_4/]newlifeincz.blogspot.com/2013/06/blog-post_4[/url]
Mazrxwf
August 23, 2024 @ 5:27 pm
[b]Купить документ о получении высшего образования.[/b]
[url=http://wp1065308.server-he.de/doku.php?id=aurusdiplomscom/]wp1065308.server-he.de/doku.php?id=aurusdiplomscom[/url]
[url=http://altajska.cz/forum/user/2578-uzuwuru/]altajska.cz/forum/user/2578-uzuwuru[/url]
[url=http://dv1930.ru/konkurs/vnimanie-novyj-konkurs-ot-dvinovazhya-cvetochnye-radosti-vinogradovskogo-rajona/#comment-548243/]dv1930.ru/konkurs/vnimanie-novyj-konkurs-ot-dvinovazhya-cvetochnye-radosti-vinogradovskogo-rajona/#comment-548243[/url]
[url=http://led119.ru/forum/user/108849/]led119.ru/forum/user/108849[/url]
[url=http://clean-ace8.com/bbs/board.php?bo_table=free&wr_id=384410/]clean-ace8.com/bbs/board.php?bo_table=free&wr_id=384410[/url]
Trefflk
August 23, 2024 @ 5:31 pm
[u][b] Добрый день![/b][/u]
Узнайте стоимость диплома высшего и среднего образования и процесс получения
[url=http://hramprorokailii.ru/news/rozhdenie-prixoda#comment-6507/]hramprorokailii.ru/news/rozhdenie-prixoda#comment-6507[/url]
Рады оказать помощь!.
купить генератор generac
August 23, 2024 @ 6:00 pm
Как не ошибиться при покупке генератора Generac, советы по выбору генератора Generac.
В чем отличие генератора Generac от других моделей?, анализ генератора Generac.
Использование генератора Generac для обеспечения надежной работы электроприборов, подробный обзор.
Настоящее качество: генераторы Generac, рассмотрение функционала.
Почему генераторы Generac так популярны?, подробный анализ.
Эффективное решение для энергетической безопасности: генераторы Generac, советы эксперта.
Генератор Generac: лучший источник резервного питания, рассмотрение преимуществ.
Как выбрать генератор Generac для эффективного резервного энергоснабжения, подробный обзор.
Генератор Generac для обеспечения непрерывного электроснабжения, советы по установке.
Как выбрать генератор Generac для вашего дома?, характеристики.
дженерак генераторы [url=https://generac-generatory1.ru/]дженерак генераторы[/url] .
MatthewInalf
August 23, 2024 @ 6:23 pm
срочный ремонт телефонов москва
Yrefdrl
August 23, 2024 @ 6:52 pm
[u][b] Привет, друзья![/b][/u]
[b]Купить диплом о высшем образовании.[/b]
[url=http://davetalksbaseball.com/amazing-reasons-why-baseball-is-a-great-sport/#comment-118384/]davetalksbaseball.com/amazing-reasons-why-baseball-is-a-great-sport/#comment-118384[/url]
Dnrtnrk
August 23, 2024 @ 6:56 pm
[u][b]Привет![/b][/u]
[b]Купить документ[/b] о получении высшего образования вы сможете у нас.
[url=http://molbiol.ru/forums/index.php?showtopic=1090044/]molbiol.ru/forums/index.php?showtopic=1090044[/url]
Uazrdic
August 23, 2024 @ 7:19 pm
[u][b]Привет![/b][/u]
[b]Заказать диплом о высшем образовании.[/b]
[url=http://telegra.ph/kupit-diplom-telohranitelya-08-13-2/]telegra.ph/kupit-diplom-telohranitelya-08-13-2[/url]
Lazrvwe
August 23, 2024 @ 7:37 pm
[u][b] Здравствуйте![/b][/u]
[b]Мы изготавливаем дипломы[/b] любых профессий по невысоким тарифам.
[url=http://obozrevatelevents.ru/byistryiy-put-k-karere-kak-kupit-diplom-i-dostich-uspeha/]obozrevatelevents.ru/byistryiy-put-k-karere-kak-kupit-diplom-i-dostich-uspeha[/url]
Trefxfb
August 23, 2024 @ 8:23 pm
[u][b] Привет![/b][/u]
Приобретение диплома ПТУ с сокращенной программой обучения в Москве
[url=http://windxp.com.ru/memddr.htm/]windxp.com.ru/memddr.htm[/url]
Поможем вам всегда!.
Mazrgfj
August 23, 2024 @ 8:30 pm
[u][b] Привет, друзья![/b][/u]
[b]Всё, что нужно знать о покупке аттестата о среднем образовании без рисков[/b]
[url=http://clients1.google.com/url?sa=t&url=aurus-diploms.com/]clients1.google.com/url?sa=t&url=aurus-diploms.com[/url]
DanielTam
August 23, 2024 @ 8:55 pm
buy cytotec pills online cheap https://cytotec.pro/# buy cytotec online
lasix for sale
Lazrqeo
August 23, 2024 @ 9:56 pm
[u][b] Здравствуйте![/b][/u]
Купить [b]диплом[/b] любого университета.
[url=http://velopiter.spb.ru/profile/108271-hnfmrfdtg/?tab=field_core_pfield_1/]velopiter.spb.ru/profile/108271-hnfmrfdtg/?tab=field_core_pfield_1[/url]
Dnrtgja
August 23, 2024 @ 10:06 pm
[u][b]Привет![/b][/u]
[b]Купить документ[/b] о получении высшего образования можно в нашей компании в столице.
[url=http://t.me/jevev/7852/]t.me/jevev/7852[/url]
Sazrfeb
August 23, 2024 @ 10:10 pm
[u][b] Привет![/b][/u]
Диплом для вас
[url=http://telegra.ph/kupit-diplom-ehlektrika-08-22/]telegra.ph/kupit-diplom-ehlektrika-08-22[/url]
Sazrohe
August 23, 2024 @ 10:27 pm
[u][b] Привет, друзья![/b][/u]
Заказать документ о получении высшего образования
[url=http://3drus.ru/forum/topic_34577//]3drus.ru/forum/topic_34577/[/url]
DanielTam
August 23, 2024 @ 10:50 pm
buy cytotec over the counter https://lisinopril.guru/# lisinopril online without prescription
lasix furosemide 40 mg
Oariorszf
August 23, 2024 @ 11:52 pm
[u][b] Добрый день![/b][/u]
Приобрести диплом университета
[b]Мы предлагаем[/b] быстро заказать диплом, который выполняется на оригинальной бумаге и заверен мокрыми печатями, водяными знаками, подписями официальных лиц. Диплом способен пройти любые проверки, даже при помощи специальных приборов. Решайте свои задачи максимально быстро с нашим сервисом.
[b]Где приобрести диплом по необходимой специальности?[/b]
[url=http://telegra.ph/kak-napisat-zayavlenie-pri-postuplenii-v-vuz-07-24/]telegra.ph/kak-napisat-zayavlenie-pri-postuplenii-v-vuz-07-24[/url]
[url=http://w77515cs.beget.tech/2024/06/18/diplom-kak-pervyy-shag-k-karere/]w77515cs.beget.tech/2024/06/18/diplom-kak-pervyy-shag-k-karere[/url]
[url=http://faithstreamer.com/blogs/3328/Легко-получите-диплом-о-высшем-образовании/]faithstreamer.com/blogs/3328/Легко-получите-диплом-о-высшем-образовании[/url]
[url=http://selkovo.rolka.me/viewtopic.php?id=3519#p10325/]selkovo.rolka.me/viewtopic.php?id=3519#p10325[/url]
[url=http://test.kuli4kam.net/gallery/image/511-альтернативное-образование-для-современных-людей/]test.kuli4kam.net/gallery/image/511-альтернативное-образование-для-современных-людей[/url]
Diplomi_gwSi
August 24, 2024 @ 12:11 am
Привет, друзья!
Заказать документ о получении высшего образования можно в нашем сервисе.
[url=http://rustam-kot.blogspot.com/2013/03/unity-3d/]rustam-kot.blogspot.com/2013/03/unity-3d[/url]
vivod iz zapoya v stacionare_rcKi
August 24, 2024 @ 12:41 am
вывод из запоя стационар (для вывода из запоя в стационаре) [url=www.vyvod-iz-zapoya-v-stacionare-voronezh.ru]вывод из запоя стационар (для вывода из запоя в стационаре)[/url] .
Williamraics
August 24, 2024 @ 2:25 am
[u][b] Привет, друзья![/b][/u]
Мы готовы предложить дипломы.
[url=http://druzia.0pk.me/viewtopic.php?id=2563#p18242/]druzia.0pk.me/viewtopic.php?id=2563#p18242[/url]
Sazrkot
August 24, 2024 @ 2:52 am
[u][b] Привет, друзья![/b][/u]
Приобрести документ о получении высшего образования
[url=http://forexgroupx.ru/dostupnyie-diplomyi-byistro-i-nadezhno/]forexgroupx.ru/dostupnyie-diplomyi-byistro-i-nadezhno[/url]
[url=http://medbereg.ru/club/user/82/blog/4190/]medbereg.ru/club/user/82/blog/4190[/url]
[url=http://graph.org/Bystroe-i-nadezhnoe-poluchenie-diploma-07-22/]graph.org/Bystroe-i-nadezhnoe-poluchenie-diploma-07-22[/url]
[url=http://faithstreamer.com/blogs/3264/Быстрый-и-надежный-способ-получить-диплом/]faithstreamer.com/blogs/3264/Быстрый-и-надежный-способ-получить-диплом[/url]
[url=http://1-click.pl/excelforum/viewtopic.php?f=3&t=256147/]1-click.pl/excelforum/viewtopic.php?f=3&t=256147[/url]
Eanrpvy
August 24, 2024 @ 3:02 am
[u][b] Здравствуйте![/b][/u]
[b]Мы изготавливаем дипломы[/b] любой профессии по доступным ценам.
[url=http://ditichlichsuvanhoa.com/dttc/DEN-VA-DINH-CAO-DAI-a407/]ditichlichsuvanhoa.com/dttc/DEN-VA-DINH-CAO-DAI-a407[/url]
Diplomi_gcSi
August 24, 2024 @ 3:23 am
Привет!
Купить документ института можно в нашем сервисе.
[url=http://rsfdrive.com/forum/index.php/]rsfdrive.com/forum/index.php[/url]
Mazrgre
August 24, 2024 @ 5:50 am
[u][b] Здравствуйте![/b][/u]
[b]Можно ли купить аттестат о среднем образовании, основные моменты и вопросы[/b]
[url=http://google.com.tr/url?q=aurus-diploms.com/]google.com.tr/url?q=aurus-diploms.com[/url]
Iariormvw
August 24, 2024 @ 7:30 am
[u][b] Привет, друзья![/b][/u]
Заказать документ о получении высшего образования можно в нашей компании. Мы оказываем услуги по изготовлению и продаже документов об окончании любых ВУЗов России. Вы сможете получить необходимый диплом по любым специальностям, включая документы образца СССР. Даем 100% гарантию, что в случае проверки документов работодателями, подозрений не возникнет.
[url=http://komfort.rusff.me/viewtopic.php?id=14177#p41698/]komfort.rusff.me/viewtopic.php?id=14177#p41698[/url]
[url=http://russianecuador.com/forum/member.php?u=13207/]russianecuador.com/forum/member.php?u=13207[/url]
[url=http://myweektour.ru/poluchite-diplom-i-stante-luchshim/]myweektour.ru/poluchite-diplom-i-stante-luchshim[/url]
[url=http://telegra.ph/sonnik-postuplenie-v-vuz-07-24/]telegra.ph/sonnik-postuplenie-v-vuz-07-24[/url]
[url=http://volgogradsky.ru/forum/viewtopic.php?p=539448#539448/]volgogradsky.ru/forum/viewtopic.php?p=539448#539448[/url]
Elliotkix
August 24, 2024 @ 7:35 am
Джип туры по Крыму https://м-драйв.рф/tours/kuluarami-poluostrova/ уникальные маршруты и яркие эмоции. Погрузитесь в увлекательнее приключение вместе с нами. Горные, лесные, подземные экскурсии, джиппинг в Крыму с максимальным комфортом.
Davidjam
August 24, 2024 @ 8:00 am
Джип туры по Крыму https://м-драйв.рф/tours/yaltinskij-drajv/ уникальные маршруты и яркие эмоции. Погрузитесь в увлекательнее приключение вместе с нами. Горные, лесные, подземные экскурсии, джиппинг в Крыму с максимальным комфортом.
vivod iz zapoya v stacionare_mcEr
August 24, 2024 @ 8:59 am
вывод из запоя воронеж (вывод из запоя в воронеже) [url=http://vyvod-iz-zapoya-v-stacionare-voronezh11.ru]http://vyvod-iz-zapoya-v-stacionare-voronezh11.ru[/url] .
Sazrehb
August 24, 2024 @ 9:20 am
[u][b] Добрый день![/b][/u]
Диплом магистра
[url=http://telegra.ph/kupit-diplom-v-arhangelske-08-22-2/]telegra.ph/kupit-diplom-v-arhangelske-08-22-2[/url]
Trefbrj
August 24, 2024 @ 12:53 pm
[u][b] Привет![/b][/u]
Как получить диплом стоматолога быстро и официально
[url=http://heimur.ru/index.php?/topic/6328-купить-диплом-стоматолога-g702i/]heimur.ru/index.php?/topic/6328-купить-диплом-стоматолога-g702i[/url]
Рады помочь!.
Dnrtanx
August 24, 2024 @ 1:13 pm
[u][b]Добрый день![/b][/u]
[b]Заказать документ[/b] о получении высшего образования можно в нашем сервисе.
[url=http://nemoskvichi.ru/forum/viewtopic.php?f=42&t=145893/]nemoskvichi.ru/forum/viewtopic.php?f=42&t=145893[/url]
Mazrehf
August 24, 2024 @ 1:13 pm
[u][b] Привет, друзья![/b][/u]
[b]Как официально купить аттестат 11 класса с упрощенным обучением в Москве[/b]
[url=http://ekra.kz/user/danilowwitek/]ekra.kz/user/danilowwitek[/url]
Williamraics
August 24, 2024 @ 2:34 pm
[u][b] Привет, друзья![/b][/u]
Мы изготавливаем дипломы.
[url=http://31pokupki.rx22.ru/viewtopic.php?f=74&t=24952/]31pokupki.rx22.ru/viewtopic.php?f=74&t=24952[/url]
สล็อตเว็บตรง100
August 24, 2024 @ 2:56 pm
Howdy would you mind stating which blog platform you’re using?
I’m planning to start my own blog soon but I’m having a tough time deciding
between BlogEngine/Wordpress/B2evolution and Drupal.
The reason I ask is because your design seems
different then most blogs and I’m looking for something completely
unique. P.S Apologies for
getting off-topic but I had to ask!
Lazrkri
August 24, 2024 @ 3:08 pm
[u][b] Добрый день![/b][/u]
[b]Мы можем предложить дипломы[/b] психологов, юристов, экономистов и прочих профессий по невысоким ценам.
[url=http://cse.google.dm/url?sa=t&url=aurus-diploms.com/]cse.google.dm/url?sa=t&url=aurus-diploms.com[/url]
Eanrtyr
August 24, 2024 @ 3:17 pm
Привет!
Мы предлагаем дипломы любых профессий по доступным ценам.
[url=http://ivyfoods.co.uk/product/?category=Non+Basmati+rice&product=Sona+Masoori++Rice/]ivyfoods.co.uk/product/?category=Non+Basmati+rice&product=Sona+Masoori++Rice[/url]
MatthewInalf
August 24, 2024 @ 3:45 pm
сервисный центр ремонт телефонов
Dnrtmcc
August 24, 2024 @ 4:25 pm
[u][b]Привет![/b][/u]
[b]Купить документ[/b] о получении высшего образования можно в нашей компании в Москве.
[url=http://diplom-moskva-f166630.promportal.su/about/]diplom-moskva-f166630.promportal.su/about[/url]
Vivod iz zapoya krasnodar_lrEn
August 24, 2024 @ 5:28 pm
вывод из запоя бесплатно [url=https://vyvod-iz-zapoya-krasnodar12.ru]вывод из запоя бесплатно[/url] .
Vivod iz zapoya krasnodar_isei
August 24, 2024 @ 5:51 pm
вывод из запоя краснодар на дому анонимно [url=vyvod-iz-zapoya-krasnodar11.ru]vyvod-iz-zapoya-krasnodar11.ru[/url] .
Lazrxgx
August 24, 2024 @ 6:09 pm
[u][b] Привет, друзья![/b][/u]
[b]Мы предлагаем дипломы[/b] любой профессии по приятным тарифам.
[url=http://psyanima.ru/about/index/]psyanima.ru/about/index[/url]
Iariorbke
August 24, 2024 @ 7:00 pm
[u][b] Привет![/b][/u]
Купить документ университета вы можете у нас в столице. Мы предлагаем документы об окончании любых университетов Российской Федерации. Вы сможете получить необходимый диплом по любой специальности, включая документы Советского Союза. Гарантируем, что в случае проверки документов работодателями, подозрений не возникнет.
[url=http://ruconf.ru/forum/index.php?PAGE_NAME=profile_view&UID=104806/]ruconf.ru/forum/index.php?PAGE_NAME=profile_view&UID=104806[/url]
[url=http://1abakan.ru/forum/showthread-238916/]1abakan.ru/forum/showthread-238916[/url]
[url=http://odinzovo.rusff.me/viewtopic.php?id=4942#p8355/]odinzovo.rusff.me/viewtopic.php?id=4942#p8355[/url]
[url=http://x70795vj.beget.tech/2024/07/22/vash-klyuch-k-uspehu-diplom-o-vysshem-obrazovanii/]x70795vj.beget.tech/2024/07/22/vash-klyuch-k-uspehu-diplom-o-vysshem-obrazovanii[/url]
[url=http://fforum.ixbb.ru/viewtopic.php?id=201#p4079/]fforum.ixbb.ru/viewtopic.php?id=201#p4079[/url]
Sazrukd
August 24, 2024 @ 9:52 pm
[u][b] Привет![/b][/u]
Диплом по специальности на выбор
[url=http://telegra.ph/vozmozhno-li-kupit-diplom-08-13-9/]telegra.ph/vozmozhno-li-kupit-diplom-08-13-9[/url]
Mazrhxr
August 24, 2024 @ 9:56 pm
[u][b] Привет![/b][/u]
[b]Как правильно купить диплом колледжа и пту в России, подводные камни[/b]
[url=http://boomingwebsitetraffic.com/]boomingwebsitetraffic.com[/url]
BryantRaics
August 24, 2024 @ 11:12 pm
canadadrugpharmacy com Easy Rx Canada buying from canadian pharmacies
pet meds without vet prescription canada
BryantRaics
August 24, 2024 @ 11:16 pm
canadian drugs online read canadian online pharmacy reviews
canadianpharmacymeds com
Lazrprk
August 24, 2024 @ 11:18 pm
Привет!
Приобрести диплом любого ВУЗа
[url=http://instgsii.ru/]instgsii.ru[/url]
BryantRaics
August 24, 2024 @ 11:25 pm
trusted canadian pharmacy EasyRxCanada online pharmacy canadian family pharmacy
best rated canadian pharmacy
BryantRaics
August 24, 2024 @ 11:28 pm
real canadian pharmacy website canadian pharmacy india
canadian mail order pharmacy
BryantRaics
August 24, 2024 @ 11:37 pm
onlinecanadianpharmacy buy here canadian pharmacies compare
best online canadian pharmacy
BryantRaics
August 24, 2024 @ 11:40 pm
canadian pharmacy online reviews Easy Rx Canada canadian pharmacy reviews
canadian mail order pharmacy
Rickynairm
August 24, 2024 @ 11:41 pm
best online pharmacies in mexico here п»їbest mexican online pharmacies
п»їbest mexican online pharmacies
BryantRaics
August 24, 2024 @ 11:50 pm
my canadian pharmacy website canadian pharmacies comparison
canada pharmacy online
BryantRaics
August 24, 2024 @ 11:53 pm
cheapest pharmacy canada EasyRxCanada.com legitimate canadian online pharmacies
northwest canadian pharmacy
BryantRaics
August 25, 2024 @ 12:02 am
canadian valley pharmacy here canada drugs
ed meds online canada
Rickynairm
August 25, 2024 @ 12:04 am
best online pharmacies in mexico more medication from mexico pharmacy
mexico drug stores pharmacies
BryantRaics
August 25, 2024 @ 12:05 am
cheap canadian pharmacy online rx canada canadian drugstore online
canadian pharmacy world reviews
Jamesmalge
August 25, 2024 @ 12:07 am
top online pharmacy india more indian pharmacy paypal
indian pharmacy
Rickynairm
August 25, 2024 @ 12:11 am
best online pharmacies in mexico online pharmacy mexico pharmacies in mexico that ship to usa
mexican online pharmacies prescription drugs
Sazrckz
August 25, 2024 @ 12:13 am
[u][b] Здравствуйте![/b][/u]
Диплом специалиста
[url=http://1776reloaded.us/blogs/7/Where-to-buy-a-diploma-or-certificate-at-an-adequate/]1776reloaded.us/blogs/7/Where-to-buy-a-diploma-or-certificate-at-an-adequate[/url]
BryantRaics
August 25, 2024 @ 12:14 am
maple leaf pharmacy in canada website legal to buy prescription drugs from canada
pet meds without vet prescription canada
BryantRaics
August 25, 2024 @ 12:18 am
rate canadian pharmacies here onlinepharmaciescanada com
canada drugs online
Rickynairm
August 25, 2024 @ 12:35 am
best online pharmacies in mexico more mexican mail order pharmacies
mexican rx online
BryantRaics
August 25, 2024 @ 12:40 am
northwest canadian pharmacy website canadianpharmacyworld
canadian pharmacies comparison
BryantRaics
August 25, 2024 @ 12:43 am
canadian mail order pharmacy EasyRxCanada online pharmacy safe reliable canadian pharmacy
canadian compounding pharmacy
Rickynairm
August 25, 2024 @ 12:46 am
reputable mexican pharmacies online mexstarpharma.com pharmacies in mexico that ship to usa
mexico drug stores pharmacies
BryantRaics
August 25, 2024 @ 12:53 am
buy canadian drugs EasyRxCanada online canadian drugstore
best canadian pharmacy
BryantRaics
August 25, 2024 @ 12:56 am
drugs from canada buy here canadadrugpharmacy com
canada pharmacy
Rickynairm
August 25, 2024 @ 1:00 am
mexican pharmaceuticals online view reputable mexican pharmacies online
buying prescription drugs in mexico online
BryantRaics
August 25, 2024 @ 1:05 am
canadian family pharmacy buy here canadian pharmacy king reviews
canadian mail order pharmacy
Vivod iz zapoya ekaterinbyrg_zjpi
August 25, 2024 @ 1:06 am
вывод из запоя на дому екатеринбург [url=https://www.vyvod-iz-zapoya-ekaterinburg.ru]вывод из запоя на дому екатеринбург[/url] .
Jamesmalge
August 25, 2024 @ 1:08 am
indian pharmacy paypal view buy medicines online in india
best online pharmacy india
BryantRaics
August 25, 2024 @ 1:08 am
vipps approved canadian online pharmacy EasyRxCanada online pharmacy canadian neighbor pharmacy
buying drugs from canada
Cazrrcr
August 25, 2024 @ 1:12 am
[u][b] Привет![/b][/u]
Получить диплом о высшем образовании
[url=http://polkasocial.org/read-blog/27296?mode=day/]polkasocial.org/read-blog/27296?mode=day[/url]
Xazrlue
August 25, 2024 @ 1:13 am
Привет, друзья!
Приобрести документ о получении высшего образования.
Мы готовы предложить дипломы любых профессий по выгодным тарифам.
[url=http://c-courses.ru/]c-courses.ru[/url]
Рады оказать помощь!.
BryantRaics
August 25, 2024 @ 1:18 am
canadian pharmacy in canada here canadian pharmacy ltd
drugs from canada
Jamesmalge
August 25, 2024 @ 1:21 am
india online pharmacy view pharmacy website india
cheapest online pharmacy india
BryantRaics
August 25, 2024 @ 1:21 am
my canadian pharmacy review rx canada canadian pharmacy cheap
cheap canadian pharmacy online
Rickynairm
August 25, 2024 @ 1:24 am
mexican mail order pharmacies view mexico drug stores pharmacies
mexico drug stores pharmacies
BryantRaics
August 25, 2024 @ 1:31 am
northwest canadian pharmacy EasyRxCanada.com canadian valley pharmacy
canadian pharmacy checker
BryantRaics
August 25, 2024 @ 1:34 am
reputable canadian pharmacy buy here canadian discount pharmacy
canadapharmacyonline legit
Trefgfi
August 25, 2024 @ 1:35 am
[u][b] Добрый день![/b][/u]
Как получить диплом о среднем образовании в Москве и других городах
[url=http://zonnebloemwedstrijd.nl/2020/02/20/hello-world/#comment-21789/]zonnebloemwedstrijd.nl/2020/02/20/hello-world/#comment-21789[/url]
Поможем вам всегда!.
BryantRaics
August 25, 2024 @ 1:44 am
canadian pharmacies compare EasyRxCanada.com canadian pharmacy 1 internet online drugstore
canadian pharmacy meds
BryantRaics
August 25, 2024 @ 1:47 am
legal to buy prescription drugs from canada read canadian pharmacy tampa
canadian pharmacy online reviews
Rickynairm
August 25, 2024 @ 1:56 am
buying prescription drugs in mexico online pharmacy mexico mexican border pharmacies shipping to usa
medicine in mexico pharmacies
BryantRaics
August 25, 2024 @ 1:57 am
legit canadian pharmacy read canadianpharmacymeds
ordering drugs from canada
BryantRaics
August 25, 2024 @ 2:10 am
canadian drug here canadian pharmacy world
canadian drug stores
BryantRaics
August 25, 2024 @ 2:13 am
canadian pharmacy prices EasyRxCanada canadian online drugs
canada drugs online
Diplomi_vaSi
August 25, 2024 @ 2:15 am
Добрый день!
Заказать документ о получении высшего образования можно в нашей компании.
[url=http://zakusim.blogspot.com/2014/04/blog-post/]zakusim.blogspot.com/2014/04/blog-post[/url]
BryantRaics
August 25, 2024 @ 2:23 am
canadian pharmacy phone number buy here canadian pharmacy price checker
buy canadian drugs
BryantRaics
August 25, 2024 @ 2:26 am
canadian pharmacy 24 com buy here legit canadian pharmacy
legitimate canadian online pharmacies
Sazrwow
August 25, 2024 @ 2:39 am
[u][b] Здравствуйте![/b][/u]
Мы готовы предложить [b]дипломы[/b] психологов, юристов, экономистов и прочих профессий.
Приобретение [b]документа[/b], подтверждающего обучение в университете, – это грамотное решение.
[url=http://domknigi.blogspot.com/2015/02/blog-post_75/]domknigi.blogspot.com/2015/02/blog-post_75[/url]
[url=http://test.krestikom.net/profile/331-rebeccaduh/]test.krestikom.net/profile/331-rebeccaduh[/url]
[url=http://web.symbol.rs/forum/member.php?action=profile&uid=716255/]web.symbol.rs/forum/member.php?action=profile&uid=716255[/url]
[url=http://mineralgroup.ru/index.php?subaction=userinfo&user=obuxetu/]mineralgroup.ru/index.php?subaction=userinfo&user=obuxetu[/url]
[url=http://community.checkinpro-hotel-software.com/memberlist.php?mode=viewprofile&u=358887/]community.checkinpro-hotel-software.com/memberlist.php?mode=viewprofile&u=358887[/url]
MelvinBog
August 25, 2024 @ 2:40 am
canadianpharmacyworld com http://easyrxcanada.com/ canada pharmacy online legit
canada pharmacy online
MelvinBog
August 25, 2024 @ 2:43 am
escrow pharmacy canada https://easyrxcanada.online/ canadian pharmacy 1 internet online drugstore
legitimate canadian online pharmacies
MelvinBog
August 25, 2024 @ 2:54 am
reddit canadian pharmacy http://easyrxcanada.com/ canadapharmacyonline legit
canadian pharmacy 365
MelvinBog
August 25, 2024 @ 2:57 am
canadianpharmacymeds https://easyrxcanada.com/ onlinecanadianpharmacy
canadian pharmacy 24
MelvinNop
August 25, 2024 @ 3:00 am
mexico drug stores pharmacies https://mexstarpharma.online/ mexican pharmaceuticals online
mexican border pharmacies shipping to usa
MelvinBog
August 25, 2024 @ 3:10 am
canada drugs https://easyrxcanada.online/ canadian neighbor pharmacy
legal to buy prescription drugs from canada
MelvinNop
August 25, 2024 @ 3:18 am
mexico drug stores pharmacies https://mexstarpharma.com/ mexico drug stores pharmacies
mexico drug stores pharmacies
MelvinBog
August 25, 2024 @ 3:20 am
canada drugstore pharmacy rx https://easyrxcanada.online/ safe online pharmacies in canada
canadian drug
MelvinBog
August 25, 2024 @ 3:23 am
canada drugs http://easyrxcanada.com/ canadian online drugs
best canadian pharmacy to order from
MelvinBog
August 25, 2024 @ 3:33 am
northwest canadian pharmacy https://easyrxcanada.online/ canadian pharmacy online reviews
canadian pharmacy online
MelvinBog
August 25, 2024 @ 3:37 am
canadianpharmacyworld com http://easyrxcanada.com/ canadian drug prices
onlinecanadianpharmacy
Mazrsfj
August 25, 2024 @ 3:40 am
[u][b] Добрый день![/b][/u]
[b]Всё о покупке аттестата о среднем образовании: полезные советы[/b]
[url=http://aerochayka.ru/?p=1025/]aerochayka.ru/?p=1025[/url]
PeterMeeft
August 25, 2024 @ 3:40 am
top online pharmacy india https://easyrxindia.com/ top 10 pharmacies in india
online pharmacy india
MelvinBog
August 25, 2024 @ 3:46 am
canadian medications https://easyrxcanada.online/ canadian pharmacy 24h com
canadian valley pharmacy
MelvinBog
August 25, 2024 @ 3:49 am
online canadian pharmacy https://easyrxcanada.com/ canadapharmacyonline legit
best online canadian pharmacy
MelvinNop
August 25, 2024 @ 3:50 am
mexico drug stores pharmacies https://mexstarpharma.com/ buying from online mexican pharmacy
mexican rx online
MelvinNop
August 25, 2024 @ 3:53 am
mexico drug stores pharmacies https://mexstarpharma.online/ buying prescription drugs in mexico online
mexico drug stores pharmacies
MelvinBog
August 25, 2024 @ 3:59 am
canada rx pharmacy https://easyrxcanada.com/ canadian 24 hour pharmacy
canada pharmacy reviews
PeterMeeft
August 25, 2024 @ 4:06 am
online pharmacy india https://easyrxindia.shop/ cheapest online pharmacy india
Online medicine home delivery
MelvinBog
August 25, 2024 @ 4:13 am
canadian pharmacy ratings https://easyrxcanada.online/ legitimate canadian mail order pharmacy
canadian online drugstore
MelvinBog
August 25, 2024 @ 4:16 am
canadian pharmacy https://easyrxcanada.online/ online canadian drugstore
canadian pharmacy 24
Trefjnv
August 25, 2024 @ 4:19 am
[u][b] Привет![/b][/u]
Покупка школьного аттестата с упрощенной программой: что важно знать
[url=http://shtsh.blogspot.com/2012/01/asus-eee-pc-1201nl/]shtsh.blogspot.com/2012/01/asus-eee-pc-1201nl[/url]
Рады оказать помощь!.
MelvinNop
August 25, 2024 @ 4:20 am
reputable mexican pharmacies online https://mexstarpharma.online/ mexican pharmaceuticals online
purple pharmacy mexico price list
MelvinBog
August 25, 2024 @ 4:26 am
trusted canadian pharmacy https://easyrxcanada.online/ canadian pharmacy in canada
northwest canadian pharmacy
MelvinBog
August 25, 2024 @ 4:29 am
best online canadian pharmacy https://easyrxcanada.com/ buy canadian drugs
canada pharmacy world
PeterMeeft
August 25, 2024 @ 4:31 am
buy medicines online in india https://easyrxindia.com/ cheapest online pharmacy india
india pharmacy mail order
MelvinNop
August 25, 2024 @ 4:35 am
medicine in mexico pharmacies http://mexstarpharma.com/ mexican rx online
mexican drugstore online
MelvinBog
August 25, 2024 @ 4:38 am
canada online pharmacy https://easyrxcanada.online/ certified canadian pharmacy
legit canadian pharmacy
MelvinBog
August 25, 2024 @ 4:41 am
canadian pharmacy india http://easyrxcanada.com/ canadian pharmacy cheap
canadian pharmacy world
MelvinBog
August 25, 2024 @ 4:51 am
safe online pharmacies in canada https://easyrxcanada.com/ www canadianonlinepharmacy
is canadian pharmacy legit
MelvinBog
August 25, 2024 @ 4:54 am
canadian drug stores http://easyrxcanada.com/ canadian pharmacy in canada
canada drugs online review
MelvinNop
August 25, 2024 @ 4:55 am
mexican border pharmacies shipping to usa http://mexstarpharma.com/ mexican pharmaceuticals online
medicine in mexico pharmacies
MelvinNop
August 25, 2024 @ 4:57 am
medicine in mexico pharmacies https://mexstarpharma.online/ mexico drug stores pharmacies
reputable mexican pharmacies online
MelvinBog
August 25, 2024 @ 5:04 am
pharmacy rx world canada https://easyrxcanada.com/ online canadian pharmacy reviews
legit canadian pharmacy online
MelvinBog
August 25, 2024 @ 5:08 am
canadian pharmacy com https://easyrxcanada.online/ canadian king pharmacy
online canadian pharmacy
MelvinNop
August 25, 2024 @ 5:10 am
mexican rx online https://mexstarpharma.online/ purple pharmacy mexico price list
mexican mail order pharmacies
MelvinNop
August 25, 2024 @ 5:15 am
mexico drug stores pharmacies https://mexstarpharma.com/ mexican border pharmacies shipping to usa
buying prescription drugs in mexico
MelvinBog
August 25, 2024 @ 5:17 am
is canadian pharmacy legit https://easyrxcanada.online/ precription drugs from canada
legitimate canadian pharmacy
MelvinBog
August 25, 2024 @ 5:21 am
canadian drugstore online http://easyrxcanada.com/ canadian pharmacy online reviews
canada drugs online review
Diplomi_tmSi
August 25, 2024 @ 5:21 am
Привет!
Купить документ о получении высшего образования вы имеете возможность в нашей компании.
[url=http://prof-aksay.ru/personal/profile/index.php?PAGE_NAME=profile_view&UID=104/]prof-aksay.ru/personal/profile/index.php?PAGE_NAME=profile_view&UID=104[/url]
MelvinBog
August 25, 2024 @ 5:31 am
canadian pharmacy http://easyrxcanada.com/ safe canadian pharmacy
canadian pharmacy meds
MelvinNop
August 25, 2024 @ 5:31 am
mexico drug stores pharmacies https://mexstarpharma.com/ mexican pharmaceuticals online
mexico drug stores pharmacies
MelvinBog
August 25, 2024 @ 5:34 am
canadian 24 hour pharmacy https://easyrxcanada.com/ canadian pharmacies
canadian pharmacy ltd
MelvinNop
August 25, 2024 @ 5:36 am
mexican rx online http://mexstarpharma.com/ buying prescription drugs in mexico online
п»їbest mexican online pharmacies
MelvinBog
August 25, 2024 @ 5:44 am
canadian pharmacy in canada https://easyrxcanada.online/ canadian pharmacy online
canadian drug pharmacy
MelvinBog
August 25, 2024 @ 5:47 am
canada rx pharmacy http://easyrxcanada.com/ my canadian pharmacy
canadian valley pharmacy
Mazrmip
August 25, 2024 @ 5:51 am
[u][b] Здравствуйте![/b][/u]
[b]Официальная покупка диплома вуза с сокращенной программой в Москве[/b]
[url=http://kemavto.kuzbass.net/index.php?subaction=userinfo&user=ixupogety/]kemavto.kuzbass.net/index.php?subaction=userinfo&user=ixupogety[/url]
MelvinBog
August 25, 2024 @ 5:58 am
my canadian pharmacy http://easyrxcanada.com/ 77 canadian pharmacy
canadian drug pharmacy
MelvinBog
August 25, 2024 @ 6:01 am
canadian pharmacy 24 com http://easyrxcanada.com/ pharmacy in canada
global pharmacy canada
MelvinBog
August 25, 2024 @ 6:11 am
canadadrugpharmacy com http://easyrxcanada.com/ canadian pharmacy reviews
canadian pharmacies online
MelvinBog
August 25, 2024 @ 6:14 am
canadian pharmacy store https://easyrxcanada.online/ canadian drug
canada pharmacy online
Dnrtwmt
August 25, 2024 @ 6:22 am
[u][b]Добрый день![/b][/u]
[b]Купить документ[/b] о получении высшего образования можно в нашей компании в Москве.
[url=http://vladmines.dn.ua/forum/index.php/topic,35219.0/]vladmines.dn.ua/forum/index.php/topic,35219.0[/url]
MelvinBog
August 25, 2024 @ 6:25 am
my canadian pharmacy review https://easyrxcanada.com/ canada cloud pharmacy
canadian pharmacy no scripts
MelvinBog
August 25, 2024 @ 6:28 am
pharmacy canadian https://easyrxcanada.com/ buying from canadian pharmacies
canadian pharmacy 24
MelvinNop
August 25, 2024 @ 6:31 am
mexico drug stores pharmacies https://mexstarpharma.online/ mexican mail order pharmacies
best online pharmacies in mexico
MelvinBog
August 25, 2024 @ 6:38 am
canada pharmacy online https://easyrxcanada.online/ canadian online drugstore
canadian drug prices
MelvinNop
August 25, 2024 @ 6:41 am
buying from online mexican pharmacy https://mexstarpharma.online/ medicine in mexico pharmacies
purple pharmacy mexico price list
MelvinBog
August 25, 2024 @ 6:41 am
canadian pharmacy store https://easyrxcanada.com/ northwest canadian pharmacy
buy drugs from canada
MelvinBog
August 25, 2024 @ 6:51 am
canada drugstore pharmacy rx https://easyrxcanada.online/ canadian drug
online canadian pharmacy
MelvinBog
August 25, 2024 @ 6:54 am
safe canadian pharmacy https://easyrxcanada.com/ canada drugs online reviews
ordering drugs from canada
Sazrvse
August 25, 2024 @ 7:03 am
[u][b] Здравствуйте![/b][/u]
Диплом любой специальности
[url=http://telegra.ph/diplom-ob-okonchanii-detskogo-sada-kupit-v-moskve-08-13-6/]telegra.ph/diplom-ob-okonchanii-detskogo-sada-kupit-v-moskve-08-13-6[/url]
MelvinBog
August 25, 2024 @ 7:04 am
the canadian pharmacy https://easyrxcanada.com/ canadian pharmacy world
canadian pharmacies that deliver to the us
MelvinNop
August 25, 2024 @ 7:07 am
buying prescription drugs in mexico online https://mexstarpharma.online/ mexican rx online
buying from online mexican pharmacy
MelvinBog
August 25, 2024 @ 7:07 am
buy drugs from canada https://easyrxcanada.com/ buying from canadian pharmacies
canadian pharmacy reviews
arenda_mbpr
August 25, 2024 @ 8:54 am
Прокат автобусов в СПб на выгодных условиях, шаттл для экскурсии.
Оптимальные цены на аренду автобуса в СПб, пользуйтесь нашими услугами.
Предоставляем качественные автобусы для аренды в Санкт-Петербурге, путешествуйте с комфортом.
Аренда автобуса для торжества в Санкт-Петербурге, без лишних хлопот.
Удобный трансфер в Санкт-Петербурге на автобусе, быстро и безопасно.
Аренда автобуса для корпоративного мероприятия в СПб, оригинально и ярко.
Индивидуальный тур на арендованном автобусе по СПб, ярко и насыщенно.
Аренда автобуса для школьной поездки в Санкт-Петербурге, безопасно и познавательно.
Свадебный автобус в СПб, красиво и романтично.
Как выбрать автобус для аренды в СПб, подсказки от наших экспертов.
Недорогая аренда автобуса в СПб: как сэкономить, со всеми выгодами.
Какие услуги включены в аренду автобуса в СПб, подробно изучите перед заказом.
Аренда автобуса с водителем в СПб: плюсы и минусы, честный рейтинг.
Сравнение стоимости аренды автобуса в СПб: как выбрать выгодное предложение, важные аспекты.
Маленький автобус в аренду для поездки по Санкт-Петербургу, компактно и удобно.
Прокат автобуса для музыкального фестиваля в СПб, безопасно и комфортно.
Вечеринка на автобусе в СПб
аренда автобуса [url=https://arenda-avtobusa-v-spb.ru/]https://arenda-avtobusa-v-spb.ru/[/url] .
Dnrthgy
August 25, 2024 @ 9:24 am
[u][b]Здравствуйте![/b][/u]
[b]Купить документ[/b] о получении высшего образования можно в нашем сервисе.
[url=http://nemoskvichi.ru/forum/viewtopic.php?f=42&t=145893/]nemoskvichi.ru/forum/viewtopic.php?f=42&t=145893[/url]
Vivod iz zapoya ekaterinbyrg_znPi
August 25, 2024 @ 9:48 am
нарколог на дом екатеринбург цены [url=https://vyvod-iz-zapoya-ekaterinburg11.ru]https://vyvod-iz-zapoya-ekaterinburg11.ru[/url] .
Lazrhxa
August 25, 2024 @ 10:32 am
Привет, друзья!
Купить диплом о высшем образовании
[url=http://mobilelink.ru/]mobilelink.ru[/url]
Sazrtov
August 25, 2024 @ 11:12 am
[u][b] Здравствуйте![/b][/u]
Диплом для вас
[url=http://ediblehomegardensresorts.com/index.php?do=/public/blog/view/id_119634/title_-2024/]ediblehomegardensresorts.com/index.php?do=/public/blog/view/id_119634/title_-2024[/url]
arenda_vpEn
August 25, 2024 @ 11:22 am
Прокат техники для строительства в столице, по выгодным ценам.
Экскаватор-погрузчик на любой вкус и бюджет, только у нас.
Лучшие компании по аренде спецтехники в столице, готовы к сотрудничеству.
Аренда экскаватора-погрузчика – это просто, в столице.
Выберите лучший вариант для своего проекта, заказывайте у нас.
Как выбрать технику для строительства, в Москве.
Экономьте время и деньги, обращайтесь к нам.
Аренда экскаватора-погрузчика в Москве: важная информация, в Москве.
Лучшие предложения для строительства, в Москве.
Выбор качественного проката, у нас в сервисе.
Как сэкономить на строительстве, в нашем сервисе.
Как выбрать экскаватор-погрузчик для аренды в Москве?, в Москве.
Аренда экскаватора-погрузчика в Москве: лучшие предложения, в столице.
Какие условия аренды экскаватора-погрузчика в Москве?, в Москве.
Экскаватор-погрузчик в аренду в Москве: оптимальное решение, в столице.
Лучшие предложения по прокату, у нас в сервисе.
Где арендовать экскаватор-погрузчик в Москве с выгодой?, в столице.
Выбор экскаватора-погрузчика в Москве: где найти лучшее предложение?, у нас в сервисе.
нанять экскаватор погрузчик [url=https://arenda-ekskavatora-pogruzchika197.ru/]https://arenda-ekskavatora-pogruzchika197.ru/[/url] .
Mazrlqd
August 25, 2024 @ 11:55 am
[u][b] Привет, друзья![/b][/u]
[b]Приобретение школьного аттестата с официальным упрощенным обучением в Москве[/b]
[url=http://aspirantura.spb.ru/forum/showthread.php?t=13850&page=2/]aspirantura.spb.ru/forum/showthread.php?t=13850&page=2[/url]
AnthonyGep
August 25, 2024 @ 12:01 pm
reputable indian pharmacies [url=https://easyrxindia.shop/#]top 10 pharmacies in india[/url] online pharmacy india
Sazrdhq
August 25, 2024 @ 12:06 pm
[u][b] Добрый день![/b][/u]
Мы изготавливаем [b]дипломы[/b] любой профессии.
Покупка [b]документа[/b], который подтверждает обучение в ВУЗе, – это выгодное решение.
[url=http://sz.80lvl.ru/viewtopic.php?f=9&t=1937/]sz.80lvl.ru/viewtopic.php?f=9&t=1937[/url]
[b]Рады оказать помощь![/b].
Xazrbia
August 25, 2024 @ 12:36 pm
Привет, друзья!
Купить документ о получении высшего образования.
Мы можем предложить дипломы психологов, юристов, экономистов и любых других профессий по приятным тарифам.
[url=http://deti-journal.ru/]deti-journal.ru[/url]
Поможем вам всегда!.
AnthonyGep
August 25, 2024 @ 1:10 pm
pharmacy website india [url=http://easyrxindia.com/#]buy prescription drugs from india[/url] mail order pharmacy india
arenda_elPi
August 25, 2024 @ 1:56 pm
Лучший выбор для аренды автобуса в СПб|Аренда автобуса в СПб – залог комфортной поездки|Широкий выбор автобусов в аренду в СПб|Найдите лучшие предложения по аренде автобусов в Санкт-Петербурге|Аренда автобуса для праздника в СПб – идеальное решение|Забронируйте автобус в Санкт-Петербурге всего в несколько кликов|Насладитесь туристическими достопримечательностями Санкт-Петербурга на комфортабельном автобусе|Идеальное решение для корпоративного транспорта в СПб|Необычный транспорт для свадьбы – арендованный автобус в СПб|Доверьте свое безопасное перемещение профессионалам с опытом на арендованных автобусах в Санкт-Петербурге|Автобусы с кондиционером и Wi-Fi в аренде в СПб|Разнообразие поездок на автобусе в СПб|Не упустите выгодные акции на аренду автобусов в СПб|Адаптируйте маршрут поездки по Санкт-Петербургу под свои потребности с помощью аренды автобуса|Надежная и оперативная поддержка для клиентов аренды автобусов в СПб|Почувствуйте настоящий комфорт в поездках по Санкт-Петербургу на наших автобусах в аренде|Удобные условия аренды автобуса в СПб для каждого клиента|Доверьте свои поездки по Санкт-Петербургу профессионалам со всеми необходимыми документами на арендованные автобусы|Ознакомьтесь с нашими уникальными предложениями по аренде автобуса в Санкт-Петербурге|Моментальное оформление аренды автобуса
аренда автобуса [url=https://arenda-avtobusa-178.ru/]https://arenda-avtobusa-178.ru/[/url] .
Uazrfrz
August 25, 2024 @ 3:00 pm
[u][b]Привет![/b][/u]
[b]Купить диплом ВУЗа .[/b]
[url=http://telegra.ph/gde-kupit-diplom-v-krasnoyarske-08-13-6/]telegra.ph/gde-kupit-diplom-v-krasnoyarske-08-13-6[/url]
Stanleygot
August 25, 2024 @ 3:06 pm
If you suffer from heart problems, are a minor or have moral problems, do not enter (very exciting scenes) https://youradultplay.top/?u=p6kk602&o=377hl2y&t=n7
arenda_bmOn
August 25, 2024 @ 4:26 pm
Экономьте время и деньги с арендой трактора,
Безопасная аренда тракторов,
Аренда трактора с оперативной доставкой,
Сельскохозяйственные тракторы в аренду,
Эксклюзивные предложения по аренде трактора,
Специализированная аренда тракторов для строительства,
Индивидуальный подход к каждому клиенту в аренде трактора,
Аренда трактора с опытными водителями,
Выгодные условия аренды трактора
трактор в аренду с водителем цена москва [url=https://arenda-traktora77.ru/]https://arenda-traktora77.ru/[/url] .
Trefljk
August 25, 2024 @ 4:52 pm
[u][b] Привет![/b][/u]
Быстрая схема покупки диплома старого образца: что важно знать?
[url=http://balaklavskiy-16.ru/user/10465/]balaklavskiy-16.ru/user/10465[/url]
Рады оказаться полезными!.
Mazrysl
August 25, 2024 @ 5:43 pm
[u][b] Привет, друзья![/b][/u]
[b]Как купить аттестат 11 класса с официальным упрощенным обучением в Москве[/b]
[url=http://dzschool18.ru/obrazdeat/biblioteka/6151/]dzschool18.ru/obrazdeat/biblioteka/6151[/url]
Sazrnpk
August 25, 2024 @ 5:44 pm
[u][b] Привет, друзья![/b][/u]
Купить документ о получении высшего образования
[url=http://camnangbenh.com//]camnangbenh.com/[/url]
Lazruii
August 25, 2024 @ 6:03 pm
[u][b] Здравствуйте![/b][/u]
Заказать [b]диплом[/b] любого ВУЗа.
[url=http://ba.rolka.me/viewtopic.php?id=15483#p18803/]ba.rolka.me/viewtopic.php?id=15483#p18803[/url]
AnthonyGep
August 25, 2024 @ 7:24 pm
best rated canadian pharmacy [url=https://easyrxcanada.online/#]rate canadian pharmacies[/url] canada drugstore pharmacy rx
Trefeew
August 25, 2024 @ 7:38 pm
[u][b] Здравствуйте![/b][/u]
Сколько стоит диплом высшего и среднего образования и как это происходит?
[url=http://frichinvestigation.org/node/4731/]frichinvestigation.org/node/4731[/url]
Рады оказаться полезными!.
AnthonyGep
August 25, 2024 @ 8:31 pm
india pharmacy [url=https://easyrxindia.shop/#]buy prescription drugs from india[/url] world pharmacy india
Cazrhll
August 25, 2024 @ 10:18 pm
[u][b] Здравствуйте![/b][/u]
Получить диплом о высшем образовании
[url=http://hinduismdecoded.blogspot.ru/2014/06/maya-inca-aztec-civilizations-decoded/]купить диплом в северодвинске[/url]
[url=http://kkuzovenko.blogspot.de/2015/07/face-2-change-photo-ready-cushion-bb/]купить диплом в буденновске[/url]
[url=http://relapso.com/?p=1552&cpage=6071#comment-433199/]купить диплом в москве[/url]
[url=http://alternatio.org/index.php?option=com_k2&view=itemlist&task=user&id=160297/]купить диплом маляра[/url]
[url=http://vyvoj.barvinek.net/doku.php?id=premialniediplomix/]купить диплом в зеленодольске[/url]
Sazrghf
August 25, 2024 @ 10:32 pm
[u][b] Привет![/b][/u]
Мы изготавливаем [b]дипломы[/b] психологов, юристов, экономистов и любых других профессий.
Покупка [b]документа[/b], который подтверждает обучение в университете, – это выгодное решение.
[url=http://sitebs.ru/blogs/106630/]sitebs.ru/blogs/106630[/url]
[b]Рады помочь![/b].
Gukovo
August 25, 2024 @ 11:11 pm
[url=https://gukovo.news161.ru]Гуково[/url]
Свежие новости города Гуково Ростовской области
http://gukovo.news161.ru
Dnrtwqe
August 25, 2024 @ 11:35 pm
[u][b]Привет, друзья![/b][/u]
[b]Приобрести документ[/b] университета вы сможете в нашей компании.
[url=http://hnfmrfdtg.gallery.ru/]hnfmrfdtg.gallery.ru[/url]
Smeshnie kartinki_aipi
August 25, 2024 @ 11:47 pm
смешные фотки [url=www.kartinkitop.ru]www.kartinkitop.ru[/url] .
Sazrmjx
August 25, 2024 @ 11:52 pm
[u][b] Здравствуйте![/b][/u]
Диплом о высшем образовании
[url=http://telegra.ph/vuzydlyapostupleniyaposle11klassa0724/]telegra.ph/vuzydlyapostupleniyaposle11klassa0724[/url]
taganrog
August 26, 2024 @ 12:23 am
[url=https://taganrog.news161.ru]https://taganrog.news161.ru[/url]
Свежие новости города Таганрога Ростовской области
http://www.taganrog.news161.ru
Uazrtqh
August 26, 2024 @ 1:39 am
[u][b]Привет![/b][/u]
[b]Приобрести диплом университета .[/b]
[url=http://telegra.ph/gde-kupit-diplom-stroitelya-08-13-8/]telegra.ph/gde-kupit-diplom-stroitelya-08-13-8[/url]
Azovdargy
August 26, 2024 @ 1:40 am
[url=https://azov.news161.ru/]https://azov.news161.ru[/url]
Свежие новости города Азов Ростовской области
azov.news161.ru/
Mazrwlr
August 26, 2024 @ 2:00 am
[u][b] Привет, друзья![/b][/u]
[b]Официальная покупка школьного аттестата с упрощенным обучением в Москве[/b]
[url=http://bankiuslugi.ru/]bankiuslugi.ru[/url]
Sazrzht
August 26, 2024 @ 2:38 am
[u][b] Добрый день![/b][/u]
Диплом магистра
[url=http://telegra.ph/kupit-diplom-moryaka-08-22-3 ]telegra.ph/kupit-diplom-moryaka-08-22-3[/url]
мастерская по ремонту кофемашин
August 26, 2024 @ 2:43 am
Профессиональный ремонт кофемашин в Москве?
ремонт кофемашин в москве с выездом мастера [url=https://remont-kofemashin-las.ru/]ремонт кофемашин стоимость[/url] .
Dnrtivo
August 26, 2024 @ 2:48 am
[u][b]Здравствуйте![/b][/u]
[b]Заказать документ[/b] о получении высшего образования можно в нашей компании.
[url=http://forumrabota.0pk.me/viewtopic.php?id=7320#p16792/]forumrabota.0pk.me/viewtopic.php?id=7320#p16792[/url]
Diplomi_leSi
August 26, 2024 @ 3:33 am
Добрый день!
Приобрести документ о получении высшего образования можно в нашей компании в Москве.
[url=http://l91632su.bget.ru/forums/]l91632su.bget.ru/forums[/url]
Sazrbfe
August 26, 2024 @ 3:37 am
Как избежать рисков при покупке диплома колледжа или ПТУ в России
mahrezDiz
August 26, 2024 @ 4:11 am
[url=https://riyad-mahrez-cz.biz]www.riyad-mahrez-cz.biz[/url]
last news about Riyad Mahrez
mahrez
Sazrqze
August 26, 2024 @ 4:29 am
[u][b] Привет![/b][/u]
Заказать документ о получении высшего образования
[url=http://d-harms.ru/postanovki/sinfoniya-n-2/]d-harms.ru/postanovki/sinfoniya-n-2[/url]
AnthonyGep
August 26, 2024 @ 5:50 am
buying from canadian pharmacies [url=https://easyrxcanada.online/#]canadian pharmacy world[/url] legitimate canadian mail order pharmacy
Diplomi_oxSi
August 26, 2024 @ 6:48 am
Здравствуйте!
Приобрести документ института можно у нас в Москве.
[url=http://papa-english.blogspot.com/2016/11/18/]papa-english.blogspot.com/2016/11/18[/url]
AnthonyGep
August 26, 2024 @ 7:42 am
buying from online mexican pharmacy [url=http://mexstarpharma.com/#]buying from online mexican pharmacy[/url] best online pharmacies in mexico
Mazrzxk
August 26, 2024 @ 8:06 am
[u][b] Добрый день![/b][/u]
[b]Быстрая покупка диплома старого образца: возможные риски[/b]
[url=http://seattlereignfc.com/]seattlereignfc.com[/url]
Treftvg
August 26, 2024 @ 8:14 am
[u][b] Привет, друзья![/b][/u]
Полезные советы по покупке диплома о высшем образовании без риска
[url=http://nontota.com/wikicorporativa/doku.php?id=gosznacdiplom/]nontota.com/wikicorporativa/doku.php?id=gosznacdiplom[/url]
Рады оказать помощь!.
Cazrgis
August 26, 2024 @ 8:42 am
[u][b] Добрый день![/b][/u]
Заказать диплом о высшем образовании
[url=http://kick.gain.tw/space.php?uid=4747813/]купить диплом штукатура[/url]
[url=http://d90894u1.bget.ru/o-skripte/1-post1/]купить диплом кандидата наук[/url]
[url=http://moy-toy.ru/index.php?subaction=userinfo&user=ysulym/]купить диплом кандидата наук[/url]
[url=http://forums.worldsamba.org/showthread.php?tid=1061715/]купить диплом в магадане[/url]
[url=http://forumfajerwerki.pl/topic/16855-pyromagic-2018-11-miedzynarodowy-festiwal-sztucznych-ogni/]купить диплом в уфе[/url]
AKSAJ
August 26, 2024 @ 9:09 am
[url=https://aksaj.news161.ru]http://aksaj.news161.ru[/url]
Свежие новости города Аксай Ростовской области
https://www.aksaj.news161.ru
Williamraics
August 26, 2024 @ 9:36 am
[u][b] Привет, друзья![/b][/u]
Мы можем предложить дипломы.
[url=http://mfd.ru/blogs/posts/view/?id=216551/]mfd.ru/blogs/posts/view/?id=216551[/url]
Volgodonsk
August 26, 2024 @ 9:37 am
[url=https://volgodonsk.news161.ru]http://volgodonsk.news161.ru[/url]
Свежие новости города Волгодонска Ростовской области
Волгодонск
Millerovo
August 26, 2024 @ 9:48 am
[url=https://millerovo.news161.ru]http://www.millerovo.news161.ru[/url]
Свежие новости города Миллерово Ростовской области
http://www.millerovo.news161.ru
Eanrvst
August 26, 2024 @ 10:30 am
Здравствуйте!
Мы изготавливаем дипломы любых профессий по доступным ценам.
[url=http://hellohome.ir/en/agent/mahyar-shahbazi/]hellohome.ir/en/agent/mahyar-shahbazi[/url]
Sazrxvw
August 26, 2024 @ 10:43 am
[u][b] Здравствуйте![/b][/u]
Диплом экономиста
[url=http://orthograf.getbb.ru/viewtopic.php?f=2&t=482/]orthograf.getbb.ru/viewtopic.php?f=2&t=482[/url]
Cazrlec
August 26, 2024 @ 10:53 am
[u][b] Привет, друзья![/b][/u]
Купить диплом о высшем образовании
[url=http://b24activities.ru/club/forum/messages/forum4/topic76/message105/?result=reply#message105/]b24activities.ru/club/forum/messages/forum4/topic76/message105/?result=reply#message105[/url]
doneczk
August 26, 2024 @ 11:15 am
[url=https://doneczk.news161.ru]Донецк[/url]
Свежие новости города Донецк Ростовской области
новости донецка
Jeremytop
August 26, 2024 @ 11:20 am
https://easyrxindia.shop/# indianpharmacy com
BelayaKalitva
August 26, 2024 @ 11:21 am
[url=https://belaya-kalitva.news161.ru]новости белой калитвы[/url]
Свежие новости города Белая калитва Ростовской области
новости белой калитвы
kamensk
August 26, 2024 @ 11:35 am
[url=https://kamensk-shahtinskij.news161.ru]https://www.kamensk-shahtinskij.news161.ru[/url]
Свежие новости города Каменск шахтинский Ростовской области
https://kamensk-shahtinskij.news161.ru
Novocherkassk
August 26, 2024 @ 11:46 am
[url=https://novocherkassk.news161.ru]novocherkassk.news161.ru[/url]
свежие новости города Новочеркасска Ростовской области
http://www.novocherkassk.news161.ru
Morozovsk
August 26, 2024 @ 11:49 am
[url=https://morozovsk.news161.ru]https://www.morozovsk.news161.ru[/url]
Свежие новости города Морозовска Ростовской области
морозовск
novoshahtinsk
August 26, 2024 @ 11:56 am
[url=https://novoshahtinsk.news161.ru]novoshahtinsk.news161.ru[/url]
Свежие новости города Новошахтинска Ростовской области
новости новошахтинска
Iariorphf
August 26, 2024 @ 12:29 pm
[u][b] Здравствуйте![/b][/u]
Купить документ университета вы имеете возможность в нашей компании в столице. Мы оказываем услуги по производству и продаже документов об окончании любых ВУЗов России. Вы получите диплом по любым специальностям, любого года выпуска, в том числе документы образца СССР. Даем гарантию, что в случае проверки документа работодателями, каких-либо подозрений не возникнет.
[url=http://ya.10bb.ru/viewtopic.php?id=4144#p7377/]ya.10bb.ru/viewtopic.php?id=4144#p7377[/url]
[url=http://ai-db.science/wiki/Быстрое_получение_диплома_онлайн/]ai-db.science/wiki/Быстрое_получение_диплома_онлайн[/url]
[url=http://moneysweet.listbb.ru/viewtopic.php?f=2&t=1971/]moneysweet.listbb.ru/viewtopic.php?f=2&t=1971[/url]
[url=http://fabnews.ru/forum/showthread.php?p=80276#post80276/]fabnews.ru/forum/showthread.php?p=80276#post80276[/url]
[url=http://formfinance.ru/kupit-diplom-professionalnyiy-podhod/]formfinance.ru/kupit-diplom-professionalnyiy-podhod[/url]
batajsk
August 26, 2024 @ 1:02 pm
[url=https://batajsk.news161.ru]www.batajsk.news161.ru[/url]
Свежие новости города Батайск Ростовской области
Батайск
мастер по ремонту телевизоров москва
August 26, 2024 @ 1:07 pm
ремонт телевизоров в москве
Rostov
August 26, 2024 @ 1:07 pm
[url=https://Rostov-na-Donu.news161.ru]www.Rostov-na-Donu.news161.ru[/url]
Свежие новости города Ростов на Дону Ростовской области
новости ростова на дону
Sazrhqb
August 26, 2024 @ 1:08 pm
[u][b] Привет![/b][/u]
Диплом экономиста
[url=http://telegra.ph/kupit-diplom-kandidata-nauk-provedennyj-08-22-3/]telegra.ph/kupit-diplom-kandidata-nauk-provedennyj-08-22-3[/url]
Hermanlor
August 26, 2024 @ 2:14 pm
online pharmacy india: Online medicine order – online pharmacy india
Shahty
August 26, 2024 @ 2:16 pm
[url=https://shahty.news161.ru]https://www.shahty.news161.ru[/url]
Свежие новости города Шахты Ростовской области
shahty.news161.ru
Zverevo
August 26, 2024 @ 2:18 pm
[url=https://zverevo.news161.ru]https://zverevo.news161.ru[/url]
Свежие новости города Зверево Ростовской области
http://www.zverevo.news161.ru
czimlyansk
August 26, 2024 @ 2:22 pm
[url=https://czimlyansk.news161.ru]http://czimlyansk.news161.ru[/url]
Свежие новости города Цимлянска Ростовской области
czimlyansk.news161.ru
ремонт macbook
August 26, 2024 @ 2:22 pm
Профессиональный сервисный центр по ремонту ноутбуков, макбуков и другой компьютерной техники.
Мы предлагаем:ремонт apple macbook
Наши мастера оперативно устранят неисправности вашего устройства в сервисе или с выездом на дом!
Sazryav
August 26, 2024 @ 2:43 pm
Парадокс, но купить диплом кандидата наук оказалось не так и сложно
Salsk
August 26, 2024 @ 2:46 pm
[url=https://salsk.news161.ru]https://salsk.news161.ru[/url]
Свежие новости города Сальска Ростовской области
новости сальска
semikarakorsk
August 26, 2024 @ 3:02 pm
[url=https://semikarakorsk.news161.ru]semikarakorsk.news161.ru[/url]
Свежие новости города Семикаракорска Ростовской области
http://www.semikarakorsk.news161.ru
Oarioryiz
August 26, 2024 @ 4:10 pm
[u][b] Здравствуйте![/b][/u]
Заказать диплом любого университета
[b]Наши специалисты предлагают[/b] выгодно заказать диплом, который выполняется на оригинальной бумаге и заверен мокрыми печатями, штампами, подписями. Диплом пройдет лубую проверку, даже при использовании специально предназначенного оборудования. Достигайте цели быстро с нашей компанией.
[b]Где купить диплом по необходимой специальности?[/b]
[url=http://avtomobil1980.ixbb.ru/viewtopic.php?id=129#p129/]avtomobil1980.ixbb.ru/viewtopic.php?id=129#p129[/url]
[url=http://affiliatemetro.com/read-blog/1134/]affiliatemetro.com/read-blog/1134[/url]
[url=http://guiltyworld.getbb.ru/viewtopic.php?f=3&t=3735/]guiltyworld.getbb.ru/viewtopic.php?f=3&t=3735[/url]
[url=http://farm86.com/blogs/393/Быстрый-и-надежный-способ-получить-диплом/]farm86.com/blogs/393/Быстрый-и-надежный-способ-получить-диплом[/url]
[url=http://socialnetwork.cloudyzx.com/read-blog/15145/]socialnetwork.cloudyzx.com/read-blog/15145[/url]
Trefgev
August 26, 2024 @ 4:17 pm
[u][b] Привет![/b][/u]
Сколько стоит диплом высшего и среднего образования и как его получить?
[url=http://coool-craft.4fan.cz/profile.php?lookup=10310/]coool-craft.4fan.cz/profile.php?lookup=10310[/url]
Всегда вам поможем!.
Ремонт смартфонов
August 26, 2024 @ 4:52 pm
Профессиональный сервисный центр по ремонту сотовых телефонов, смартфонов и мобильных устройств.
Мы предлагаем: сервисный центр мобильных телефонов
Наши мастера оперативно устранят неисправности вашего устройства в сервисе или с выездом на дом!
Rickyhaw
August 26, 2024 @ 4:58 pm
Экскурсии и джип-туры по Крыму https://м-драйв.рф/tours/pervyj-krymskij/
Mazroid
August 26, 2024 @ 5:10 pm
[u][b] Добрый день![/b][/u]
[b]Официальная покупка диплома ПТУ с упрощенной программой обучения[/b]
[url=http://mybaltika.info/ru/blogs/571/7520/]mybaltika.info/ru/blogs/571/7520[/url]
Cazrlqo
August 26, 2024 @ 5:22 pm
Купить диплом старого образца, можно ли это сделать по быстрой схеме?
Dnrtdun
August 26, 2024 @ 5:34 pm
[u][b]Привет, друзья![/b][/u]
[b]Купить документ[/b] института вы можете в нашей компании.
[url=http://club.7ya.ru/hnfmrfdtg/]club.7ya.ru/hnfmrfdtg[/url]
Anonymous
August 26, 2024 @ 5:39 pm
Thanks for sharing. I read many of your blog posts, cool, your blog is very good. https://accounts.binance.info/en-IN/register-person?ref=UM6SMJM3
Ремонт смартфонов
August 26, 2024 @ 6:48 pm
Профессиональный сервисный центр по ремонту сотовых телефонов, смартфонов и мобильных устройств.
Мы предлагаем: ремонт сотовых телефонов
Наши мастера оперативно устранят неисправности вашего устройства в сервисе или с выездом на дом!
Sazrjus
August 26, 2024 @ 7:29 pm
[u][b] Привет, друзья![/b][/u]
Диплом любой специальности
[url=http://telegra.ph/kupit-diplom-mfyua-08-13-7/]telegra.ph/kupit-diplom-mfyua-08-13-7[/url]
Williamraics
August 26, 2024 @ 8:38 pm
[u][b] Привет![/b][/u]
Мы изготавливаем дипломы.
[url=http://thesims2thebest.0pk.me/viewtopic.php?id=475#p1006/]thesims2thebest.0pk.me/viewtopic.php?id=475#p1006[/url]
Dnrtnvr
August 26, 2024 @ 8:41 pm
[u][b]Добрый день![/b][/u]
[b]Заказать документ[/b] университета вы можете в нашей компании.
[url=http://lastdemo.primepix.ru/club/user/26712/blog/7066/]lastdemo.primepix.ru/club/user/26712/blog/7066[/url]
Prodvijenie saitov v Moskve_zsOa
August 26, 2024 @ 8:58 pm
раскрутка сайтов в москве [url=http://prodvizhenie-sajtov-v-moskve213.ru/]http://prodvizhenie-sajtov-v-moskve213.ru/[/url] .
Sazrstn
August 26, 2024 @ 9:28 pm
Как официально купить аттестат 11 класса с упрощенным обучением в Москве
Eanroke
August 26, 2024 @ 9:30 pm
Здравствуйте!
Мы предлагаем дипломы психологов, юристов, экономистов и любых других профессий по выгодным тарифам.
[url=http://mazda-demio.ru/forums/index.php?autocom=gallery&req=si&img=4139/]mazda-demio.ru/forums/index.php?autocom=gallery&req=si&img=4139[/url]
RostovNews
August 26, 2024 @ 9:31 pm
[url=https://rostovskaya-oblast.news161.ru]https://rostovskaya-oblast.news161.ru[/url]
Свежие новости Ростовской области
ростовская область новости
Lazrymo
August 26, 2024 @ 9:38 pm
[u][b] Здравствуйте![/b][/u]
[b]Мы изготавливаем дипломы[/b] любых профессий по выгодным ценам.
[url=http://indiarefix.in/memberlist.php?mode=viewprofile&u=14405/]indiarefix.in/memberlist.php?mode=viewprofile&u=14405[/url]
Konstantinovsk
August 26, 2024 @ 11:03 pm
[url=https://konstantinovsk.news161.ru]http://konstantinovsk.news161.ru[/url]
свежие новости города Константиновска Ростовской области
http://konstantinovsk.news161.ru
Iarioruaw
August 26, 2024 @ 11:16 pm
[u][b] Добрый день![/b][/u]
Приобрести документ о получении высшего образования вы имеете возможность у нас в Москве. Мы предлагаем документы об окончании любых ВУЗов Российской Федерации. Вы получите диплом по любым специальностям, любого года выпуска, в том числе документы СССР. Гарантируем, что при проверке документов работодателем, подозрений не появится.
[url=http://asg-amt.phorum.pl/posting.php/]asg-amt.phorum.pl/posting.php[/url]
[url=http://bahchisaray.org.ua/index.php?showtopic=33629/]bahchisaray.org.ua/index.php?showtopic=33629[/url]
[url=http://antennastar.com/blogs/568/оформите-диплом-без-лишних-хлопот/]antennastar.com/blogs/568/оформите-диплом-без-лишних-хлопот[/url]
[url=http://beep.pk/blogs/879/Which-mobile-apps-and-services-are-used-to-bet-in/]beep.pk/blogs/879/Which-mobile-apps-and-services-are-used-to-bet-in[/url]
[url=http://hip-hop.ru/forum/id298234-worksale/]hip-hop.ru/forum/id298234-worksale[/url]
Mazryvh
August 26, 2024 @ 11:20 pm
[u][b] Привет, друзья![/b][/u]
[b]Как избежать рисков при покупке диплома колледжа или ПТУ в России[/b]
[url=http://blog-doma.ru/]blog-doma.ru[/url]
Lazrxai
August 27, 2024 @ 12:54 am
[u][b] Привет![/b][/u]
[b]Мы готовы предложить дипломы[/b] психологов, юристов, экономистов и любых других профессий по невысоким тарифам.
[url=http://job.tltnews.ru/newpass.php/]job.tltnews.ru/newpass.php[/url]
Sazrsky
August 27, 2024 @ 1:24 am
Как не стать жертвой мошенников при покупке диплома о среднем полном образовании
Oariorcjj
August 27, 2024 @ 2:47 am
[u][b] Привет![/b][/u]
Заказать диплом о высшем образовании
[b]Наша компания предлагает[/b] быстро и выгодно заказать диплом, который выполнен на оригинальном бланке и заверен мокрыми печатями, штампами, подписями должностных лиц. Документ способен пройти лубую проверку, даже с применением профессионального оборудования. Решите свои задачи быстро и просто с нашим сервисом.
[b]Где купить диплом по необходимой специальности?[/b]
[url=http://kuvandyk.ru/message.php?msg=151/]kuvandyk.ru/message.php?msg=151[/url]
[url=http://vsetutonline.com/forum/member.php?u=76509/]vsetutonline.com/forum/member.php?u=76509[/url]
[url=http://biznas.com/Biz-postsm337716_Default.aspx#post337716/]biznas.com/Biz-postsm337716_Default.aspx#post337716[/url]
[url=http://abris-zip.ru/company/personal/user/190/forum/message/1299/1309/#message1309/]abris-zip.ru/company/personal/user/190/forum/message/1299/1309/#message1309[/url]
[url=http://club.sabaylok.com/blogs/5632/Быстрое-получение-диплома-онлайн/]club.sabaylok.com/blogs/5632/Быстрое-получение-диплома-онлайн[/url]
Mazrjga
August 27, 2024 @ 3:16 am
[u][b] Привет![/b][/u]
[b]Вопросы и ответы: можно ли быстро купить диплом старого образца?[/b]
[url=http://kiopro.ru/support/forum/view_profile.php?UID=74955/]kiopro.ru/support/forum/view_profile.php?UID=74955[/url]
Transport
August 27, 2024 @ 3:25 am
[url=https://avtotransport.news161.ru]http://avtotransport.news161.ru[/url]
Новости транспорта в Ростовской области
https://avtotransport.news161.ru
business
August 27, 2024 @ 3:32 am
[url=https://business.news161.ru]www.business.news161.ru[/url]
Свежие новости бизнеса в Ростовской области
https://www.business.news161.ru
Cazrdtp
August 27, 2024 @ 3:45 am
Можно ли купить аттестат о среднем образовании, основные моменты и вопросы
turizm
August 27, 2024 @ 3:55 am
[url=https://turizm.news161.ru]turizm.news161.ru[/url]
Свежие новости туризма в Ростовской области
https://www.turizm.news161.ru
Sazrnhc
August 27, 2024 @ 4:45 am
[u][b] Добрый день![/b][/u]
Диплом юриста
[url=http://telegra.ph/gotovyj-diplom-kupit-08-13-10/]telegra.ph/gotovyj-diplom-kupit-08-13-10[/url]
kultura
August 27, 2024 @ 5:06 am
[url=https://kultura.news161.ru/]https://kultura.news161.ru[/url]
Свежие новости культуры в Ростовской области
https://www.kultura.news161.ru/
Diplomi_fvSi
August 27, 2024 @ 5:32 am
Здравствуйте!
Приобрести документ университета можно в нашей компании в столице.
[url=http://forums.indexrise.com/user-357710/]forums.indexrise.com/user-357710[/url]
ремонт квадрокоптеров
August 27, 2024 @ 5:36 am
Профессиональный сервисный центр по ремонту квадрокоптеров и радиоуправляемых дронов.
Мы предлагаем:ремонт дронов
Наши мастера оперативно устранят неисправности вашего устройства в сервисе или с выездом на дом!
riyadmahrezcz
August 27, 2024 @ 5:42 am
[url=https://riyadmahrezcz.biz]riyadmahrezcz.biz[/url]
last news about riyad mahrezcz
https://riyadmahrezcz.biz
Trefigk
August 27, 2024 @ 5:44 am
[u][b] Добрый день![/b][/u]
Процесс получения диплома стоматолога: реально ли это сделать быстро?
[url=http://barlophotos.blogspot.com/2018/06/avto-marshrut-lagonaki/]barlophotos.blogspot.com/2018/06/avto-marshrut-lagonaki[/url]
Окажем помощь!.
Sazrbws
August 27, 2024 @ 5:56 am
[u][b] Привет![/b][/u]
Диплом о высшем образовании
[url=http://glenns.org/book-buzz/forum/topic/hot-and-beauty-naked-girls/?part=3851/]glenns.org/book-buzz/forum/topic/hot-and-beauty-naked-girls/?part=3851[/url]
pogoda
August 27, 2024 @ 6:01 am
[url=https://pogoda.news161.ru]https://www.pogoda.news161.ru[/url]
свежие новости погоды в Ростовской области
http://www.novocherkassk.news161.ru
Lazrnsw
August 27, 2024 @ 6:01 am
Добрый день!
Купить диплом ВУЗа
[url=http://1dip.ru/]1dip.ru[/url]
ekonomika
August 27, 2024 @ 6:22 am
[url=https://ekonomika.news161.ru]https://ekonomika.news161.ru[/url]
Свежие новости экономики Ростовской области
экономика
agriculture
August 27, 2024 @ 6:24 am
[url=https://agriculture.news161.ru]новости сельского хозяйства[/url]
Свежие новости сельского хозяйства Ростовской области
https://agriculture.news161.ru
science
August 27, 2024 @ 6:31 am
[url=https://nauka.news161.ru]новости науки[/url]
Свежие новости науки в Ростовской области
http://www.nauka.news161.ru
mahrezriyadcz
August 27, 2024 @ 6:38 am
[url=https://mahrezriyad-cz.biz]http://www.mahrezriyad-cz.biz[/url]
last news about mahrez riyad
mahrezriyad-cz.biz
mahrezriyad
August 27, 2024 @ 6:57 am
[url=https://mahrez-riyad-cz.biz]http://www.mahrez-riyad-cz.biz[/url]
last news about mahrez riyad
http://www.mahrez-riyad-cz.biz
obrazovanie
August 27, 2024 @ 7:15 am
[url=https://obrazovanie.news161.ru]новости образования[/url]
Свежие новости образования Ростовской области
obrazovanie.news161.ru
Sport
August 27, 2024 @ 7:19 am
[url=https://sports.news161.ru]https://sports.news161.ru[/url]
Свежие новости спорта Ростовской области
спорт
expibre
August 27, 2024 @ 7:21 am
The message they are sending is a clear one they are not anti gun, they just want what they call reasonable gun control priligy
mediczina
August 27, 2024 @ 7:37 am
[url=https://mediczina.news161.ru]www.mediczina.news161.ru[/url]
Свежие новости медицины в Ростовской области
http://www.mediczina.news161.ru
society
August 27, 2024 @ 8:02 am
[url=https://society.news161.ru]http://society.news161.ru[/url]
Свежие общественные новости Ростовской области
общество
Mazruod
August 27, 2024 @ 8:11 am
[u][b] Привет![/b][/u]
[b]Как избежать рисков при покупке диплома колледжа или ПТУ в России[/b]
[url=http://trans-asvt.ru/raznoe/kakuyu-odezhdu-vzyat-v-konnyiy-pohod/]trans-asvt.ru/raznoe/kakuyu-odezhdu-vzyat-v-konnyiy-pohod[/url]
Xazrvcm
August 27, 2024 @ 8:17 am
Добрый день!
Купить документ о получении высшего образования.
Мы изготавливаем дипломы психологов, юристов, экономистов и прочих профессий по приятным ценам.
[url=http://ugues.ru/]ugues.ru[/url]
Окажем помощь!.
politics
August 27, 2024 @ 8:26 am
[url=https://politics.news161.ru]https://www.politics.news161.ru[/url]
Свежие политические новости Ростовской области
https://politics.news161.ru
Trefnoy
August 27, 2024 @ 8:42 am
[u][b] Привет![/b][/u]
Стоимость дипломов высшего и среднего образования и процесс их получения
[url=http://host-ko.ru/скидка-50-на-три-месяца/#comment-3446/]host-ko.ru/скидка-50-на-три-месяца/#comment-3446[/url]
Рады оказать помощь!.
Diplomi_whSi
August 27, 2024 @ 8:44 am
Привет, друзья!
Купить документ института вы сможете в нашем сервисе.
[url=http://magadan-mebel.ru/forum/user/1005/]magadan-mebel.ru/forum/user/1005[/url]
peretyazhka-mebeli-minsk fjSa
August 27, 2024 @ 10:15 am
Преимущества перетяжки мягкой мебели, которые важно учитывать, для успешного обновления мебели, и придать дому уникальный вид, Почему стоит обратиться к профессионалам для перетяжки дивана, Как улучшить комфорт мебели при перетяжке, с помощью правильного выбора материалов
перетяжка мебели [url=https://obivka-divana.ru/]перетяжка мебели[/url] .
sex
August 27, 2024 @ 11:10 am
Thank you very much for sharing, I learned a lot from your article. Very cool. Thanks.
Dnrthbl
August 27, 2024 @ 11:11 am
[u][b]Здравствуйте![/b][/u]
[b]Приобрести документ[/b] университета вы можете у нас в Москве.
[url=http://salda.ws/meet/notes.php?id=12692/]salda.ws/meet/notes.php?id=12692[/url]
Jesusdop
August 27, 2024 @ 12:28 pm
deneme bonusu: deneme bonusu veren siteler – bonus veren siteler
peretyazhka-mebeli-minsk eiSa
August 27, 2024 @ 1:14 pm
Преимущества перетяжки мягкой мебели, Советы по выбору ткани для перетяжки мебели, которые помогут сделать правильный выбор, и придать дому уникальный вид, чтобы сэкономить бюджет, Как сделать мебель более уютной и комфортной, для создания уютного уголка в доме
перетяжка мягкой мебели [url=https://obivka-divana.ru/]перетяжка мягкой мебели[/url] .
Jesusdop
August 27, 2024 @ 1:41 pm
sweet bonanza 100 tl: sweet bonanza free spin demo – sweet bonanza oyna
Mazrstz
August 27, 2024 @ 2:02 pm
[u][b] Привет![/b][/u]
[b]Купить диплом о среднем образовании в Москве и любом другом городе[/b]
[url=http://rogerfilms.com/index.php?title=diplomandokscom/]rogerfilms.com/index.php?title=diplomandokscom[/url]
Darrelnog
August 27, 2024 @ 2:11 pm
en yeni slot siteleri: en iyi slot siteleri 2024 – en cok kazandiran slot siteleri
Dnrtclp
August 27, 2024 @ 2:21 pm
[u][b]Здравствуйте![/b][/u]
[b]Приобрести документ[/b] о получении высшего образования вы имеете возможность у нас.
[url=http://ractis.ru/forums/index.php?showtopic=4522/]ractis.ru/forums/index.php?showtopic=4522[/url]
Mazrpan
August 27, 2024 @ 2:31 pm
[u][b] Привет![/b][/u]
[b]Официальная покупка аттестата о среднем образовании в Москве и других городах[/b]
[url=http://cse.google.com.af/url?sa=t&url=aurus-diploms.com/]cse.google.com.af/url?sa=t&url=aurus-diploms.com[/url]
Sazrfus
August 27, 2024 @ 3:03 pm
[u][b] Здравствуйте![/b][/u]
Мы изготавливаем [b]дипломы[/b] любых профессий.
Заказ [b]диплома[/b], который подтверждает окончание института, – это грамотное решение.
[url=http://zemli.com/index.php?subaction=userinfo&user=inojuqu/]zemli.com/index.php?subaction=userinfo&user=inojuqu[/url]
[url=http://getamped.yxhi.com/doku.php?id=radiplomcom/]getamped.yxhi.com/doku.php?id=radiplomcom[/url]
[url=http://4eproduction.com/galleries/slub-zanety-i-krzysztofa/#comment-367884/]4eproduction.com/galleries/slub-zanety-i-krzysztofa/#comment-367884[/url]
[url=http://fromrus.su/index.php?/topic/17130-купить-диплом-о-среднем-образовании-в-волгограде-m744g/]fromrus.su/index.php?/topic/17130-купить-диплом-о-среднем-образовании-в-волгограде-m744g[/url]
[url=http://pa-trade.blogspot.com/2014/05/blog-post/]pa-trade.blogspot.com/2014/05/blog-post[/url]
Darrelnog
August 27, 2024 @ 3:36 pm
slot siteleri: bonus veren casino slot siteleri – bonus veren slot siteleri
Sazrskn
August 27, 2024 @ 3:41 pm
Как оказалось, купить диплом кандидата наук не так уж и сложно
Sazrwpd
August 27, 2024 @ 4:55 pm
[u][b] Здравствуйте![/b][/u]
Диплом о высшем образовании
[url=http://execulink.com/~mstowe/wwwboard/messages/14528/]execulink.com/~mstowe/wwwboard/messages/14528[/url]
Lazrldk
August 27, 2024 @ 5:22 pm
Привет, друзья!
Купить диплом о высшем образовании
[url=http://школа6.рф/]школа6.рф[/url]
Sazrmlh
August 27, 2024 @ 6:21 pm
[u][b] Добрый день![/b][/u]
Купить документ института
[url=http://finttech.ru/diplomyi-dlya-karernyih-vyisot/]finttech.ru/diplomyi-dlya-karernyih-vyisot[/url]
[url=http://kondrateff.5bb.ru/viewtopic.php?id=6818#p12795/]kondrateff.5bb.ru/viewtopic.php?id=6818#p12795[/url]
[url=http://cocapal.com/read-blog/1278/]cocapal.com/read-blog/1278[/url]
[url=http://kaiztech.net/IPB/index.php?/gallery/image/410-как-достичь-успеха-без-классического-образования/]kaiztech.net/IPB/index.php?/gallery/image/410-как-достичь-успеха-без-классического-образования[/url]
[url=http://fat-girls.ru/diplomyi-vseh-urovney-i-spetsialnostey/]fat-girls.ru/diplomyi-vseh-urovney-i-spetsialnostey[/url]
Sazrase
August 27, 2024 @ 6:29 pm
Как официально купить аттестат 11 класса с упрощенным обучением в Москве
Xazrtbp
August 27, 2024 @ 7:27 pm
Привет!
Приобрести документ университета.
Мы изготавливаем дипломы любых профессий по приятным ценам.
[url=http://knowledgestream.ru/]knowledgestream.ru[/url]
Окажем помощь!.
Uazriwa
August 27, 2024 @ 7:33 pm
[u][b]Привет![/b][/u]
[b]Заказать диплом любого ВУЗа.[/b]
[url=http://telegra.ph/kupit-diplom-landshaftnogo-dizajnera-08-13-4/]telegra.ph/kupit-diplom-landshaftnogo-dizajnera-08-13-4[/url]
Lazruzy
August 27, 2024 @ 8:21 pm
[u][b] Привет![/b][/u]
[b]Мы можем предложить дипломы[/b] любых профессий по приятным ценам.
[url=http://community.checkinpro-hotel-software.com/memberlist.php?mode=viewprofile&u=358251/]community.checkinpro-hotel-software.com/memberlist.php?mode=viewprofile&u=358251[/url]
Sazrzlt
August 27, 2024 @ 9:04 pm
[u][b] Привет, друзья![/b][/u]
Мы изготавливаем [b]дипломы[/b] любой профессии.
Приобретение [b]диплома[/b], который подтверждает окончание ВУЗа, – это разумное решение.
[url=http://4yo.us/blogs/143391/Дипломы-без-ожидания-и-проблем/]4yo.us/blogs/143391/Дипломы-без-ожидания-и-проблем[/url]
[b]Всегда вам поможем![/b].
Sazrnxb
August 27, 2024 @ 10:16 pm
[u][b] Привет, друзья![/b][/u]
Заказать документ ВУЗа
[url=http://google.rs/url?q=aurus-diploms.com/]google.rs/url?q=aurus-diploms.com[/url]
Treftwo
August 27, 2024 @ 10:20 pm
[u][b] Привет, друзья![/b][/u]
Можно ли купить аттестат о среднем образовании? Основные рекомендации
[url=http://biyografiforum.10tl.net/member.php?action=profile&uid=3111/]biyografiforum.10tl.net/member.php?action=profile&uid=3111[/url]
Рады оказать помощь!.
fashion
August 27, 2024 @ 10:55 pm
[url=https://fashion.news161.ru]http://fashion.news161.ru[/url]
Свежие новости моды в Ростовской области
https://fashion.news161.ru
Mazrclx
August 27, 2024 @ 11:25 pm
[u][b] Добрый день![/b][/u]
[b]Приобретение диплома ПТУ с сокращенной программой обучения в Москве[/b]
[url=http://bouffordca.com/]bouffordca.com[/url]
Lazrnkv
August 27, 2024 @ 11:36 pm
[u][b] Привет, друзья![/b][/u]
Приобрести [b]диплом[/b] о высшем образовании.
[url=http://mlmwmzmillioner.rolevaya.com/viewtopic.php?id=14918#p26814/]mlmwmzmillioner.rolevaya.com/viewtopic.php?id=14918#p26814[/url]
Lazrxbp
August 27, 2024 @ 11:37 pm
[u][b] Привет, друзья![/b][/u]
[b]Мы готовы предложить дипломы[/b] любой профессии по приятным ценам.
[url=http://sharypovo.today/news/society/page/65/]sharypovo.today/news/society/page/65[/url]
Kevinnut
August 27, 2024 @ 11:54 pm
http://slotsiteleri.bid/# slot siteleri guvenilir
Darrelnog
August 28, 2024 @ 1:00 am
yeni slot siteleri: slot casino siteleri – en iyi slot siteler
Cazrena
August 28, 2024 @ 1:05 am
[u][b] Привет![/b][/u]
Купить диплом ВУЗа
[url=http://amritaadhikary.com/monica-from-california/#comment-451231/]купить диплом в перми[/url]
[url=http://quasix.ru/user/zhaleypetrooz632/]купить диплом во владивостоке[/url]
[url=http://igram.net/index.php?name=Account&op=userinfo&user_name=ivogenod/]купить диплом продавца[/url]
[url=http://genezis-servis.ru/2018/06/28/rostov-na-donu-adresa-bolnits-mfts-i-kladbishh-goroda-rostov-na-donu/#comment-194890/]купить диплом в новочебоксарске[/url]
[url=http://xn--48-6kcd0fg.xn--p1ai/forums.php?m=posts&q=23175&n=last#bottom/]купить диплом в дзержинске[/url]
Trefvue
August 28, 2024 @ 1:05 am
[u][b] Привет![/b][/u]
Быстрое обучение и получение диплома магистра – возможно ли это?
[url=http://vpereplete.blogspot.com/2013/05/blog-post_9/]vpereplete.blogspot.com/2013/05/blog-post_9[/url]
Будем рады вам помочь!.
Sazrszv
August 28, 2024 @ 1:51 am
[u][b] Привет![/b][/u]
Мы готовы предложить [b]дипломы[/b] любой профессии.
Заказ [b]диплома[/b], который подтверждает обучение в университете, – это выгодное решение.
[url=http://inplainenglish121.blogspot.com/2017/05/exam-belarus/]inplainenglish121.blogspot.com/2017/05/exam-belarus[/url]
[url=http://wwassociation.ru/user/11711/]wwassociation.ru/user/11711[/url]
[url=http://zpu-journal.ru/forum/view_profile.php?UID=310086/]zpu-journal.ru/forum/view_profile.php?UID=310086[/url]
[url=http://nedv-revda.ru/index.php?subaction=userinfo&user=atagybo/]nedv-revda.ru/index.php?subaction=userinfo&user=atagybo[/url]
[url=http://6986986.com/forum.php/]6986986.com/forum.php[/url]
Darrelnog
August 28, 2024 @ 3:13 am
slot siteleri: guvenilir slot siteleri 2024 – slot siteleri
Jesusdop
August 28, 2024 @ 4:04 am
en cok kazandiran slot siteleri: en cok kazandiran slot siteleri – en iyi slot siteler
Cazrdag
August 28, 2024 @ 4:56 am
[u][b] Здравствуйте![/b][/u]
Купить диплом академии
[url=http://michiya-cs.com/userinfo.php?uid=30979/]michiya-cs.com/userinfo.php?uid=30979[/url]
Sazrokc
August 28, 2024 @ 5:05 am
[u][b] Здравствуйте![/b][/u]
Приобрести документ ВУЗа
[url=http://kolhos.listbb.ru/viewtopic.php?f=2&t=637/]kolhos.listbb.ru/viewtopic.php?f=2&t=637[/url]
[url=http://ya.2bb.ru/viewtopic.php?id=19726#p241293/]ya.2bb.ru/viewtopic.php?id=19726#p241293[/url]
[url=http://cv53297-livestreet-1.tw1.ru/2024/07/09/kupit-diplom-s-garantiey-podlinnosti/]cv53297-livestreet-1.tw1.ru/2024/07/09/kupit-diplom-s-garantiey-podlinnosti[/url]
[url=http://jogibaba.com/blogs/264/Легальное-получение-диплома-в-короткие-сроки/]jogibaba.com/blogs/264/Легальное-получение-диплома-в-короткие-сроки[/url]
[url=http://matusya.ukraine7.com/t4549-topic#21785/]matusya.ukraine7.com/t4549-topic#21785[/url]
Smeshnie shytki_zqpt
August 28, 2024 @ 5:24 am
прикольные шутки [url=https://korotkieshutki.ru/]korotkieshutki.ru[/url] .
Mazrzvt
August 28, 2024 @ 5:33 am
[u][b] Привет![/b][/u]
[b]Покупка диплома о среднем полном образовании: как избежать мошенничества?[/b]
[url=http://studentshop.ru/kontrolnaya/zadaniya-po-distsipline-predprinimatelskoe-pravo-3/]studentshop.ru/kontrolnaya/zadaniya-po-distsipline-predprinimatelskoe-pravo-3[/url]
Sazrtyg
August 28, 2024 @ 5:55 am
[u][b] Привет![/b][/u]
Диплом бакалавра
[url=http://telegra.ph/kupit-diplom-v-smolenske-08-22/]telegra.ph/kupit-diplom-v-smolenske-08-22[/url]
Uazrgtl
August 28, 2024 @ 6:32 am
[u][b]Здравствуйте![/b][/u]
[b]Купить диплом любого университета.[/b]
[url=http://telegra.ph/kupit-diplom-povara-4-razryada-08-13/]telegra.ph/kupit-diplom-povara-4-razryada-08-13[/url]
Diplomi_bfSi
August 28, 2024 @ 7:19 am
Здравствуйте!
Заказать документ о получении высшего образования вы сможете в нашей компании в Москве.
[url=http://mir-pozitiv.ru/forums/users/christinamatty2001-2/]mir-pozitiv.ru/forums/users/christinamatty2001-2[/url]
Sazromc
August 28, 2024 @ 7:30 am
[u][b] Привет![/b][/u]
Мы изготавливаем [b]дипломы[/b] любой профессии.
Покупка [b]диплома[/b], который подтверждает обучение в университете, – это разумное решение.
[url=http://passneurosurgery.net/learn/blog/index.php?entryid=536629/]passneurosurgery.net/learn/blog/index.php?entryid=536629[/url]
[b]Будем рады вам помочь![/b].
Sazrloo
August 28, 2024 @ 9:08 am
[u][b] Здравствуйте![/b][/u]
Купить документ университета
[url=http://alan.az//]alan.az/[/url]
Diplomi_mpSi
August 28, 2024 @ 10:15 am
Привет, друзья!
Купить документ института вы можете в нашей компании в столице.
[url=http://ntwk.ru/forum/index.php/]ntwk.ru/forum/index.php[/url]
Lazrqst
August 28, 2024 @ 10:42 am
[u][b] Привет![/b][/u]
Приобрести [b]диплом[/b] любого университета.
[url=http://postila.ru/post/78541538/]postila.ru/post/78541538[/url]
Cazrjog
August 28, 2024 @ 11:28 am
[u][b] Привет![/b][/u]
Получить диплом университета
[url=http://posysaev.blogspot.com/2012/03/11_08/]купить диплом в ессентуках[/url]
[url=http://prosperology.blogspot.com/2016/01/Filosofiya-prostoty/]купить диплом в иркутске[/url]
[url=http://xn--33-6kc4bza.xn--p1ai/users/itoliq/]купить диплом в уфе[/url]
[url=http://amatagroup.ru/forum/messages/forum1/topic59/message357308/?result=reply#message357308/]купить диплом в мытищах[/url]
[url=http://vovkainfo.ru/forum/viewtopic.php?t=19584/]купить диплом в великих луках[/url]
Sazrayr
August 28, 2024 @ 12:26 pm
Как правильно приобрести диплом колледжа или ПТУ в России, важные моменты
mahrezDiz
August 28, 2024 @ 2:20 pm
[url=https://riyadmahrez-cz.biz]riyadmahrez-cz.biz[/url]
last news about Riyad Mahrez
http://www.riyadmahrez-cz.biz
ремонт аймак в москве
August 28, 2024 @ 2:40 pm
Профессиональный сервисный центр по ремонту ноутбуков, imac и другой компьютерной техники.
Мы предлагаем:внешний жесткий диск для imac
Наши мастера оперативно устранят неисправности вашего устройства в сервисе или с выездом на дом!
Mazrbgg
August 28, 2024 @ 2:41 pm
[u][b] Привет![/b][/u]
[b]Полезная информация как купить диплом о высшем образовании без рисков[/b]
[url=http://randori.lv/index.php?q=node/1767/]randori.lv/index.php?q=node/1767[/url]
Treflml
August 28, 2024 @ 2:47 pm
[u][b] Привет![/b][/u]
Легальные способы покупки диплома о среднем полном образовании
[url=http://fire-team.ru/forum/showthread.php?p=39920#post39920/]fire-team.ru/forum/showthread.php?p=39920#post39920[/url]
Рады оказаться полезными!.
Williamraics
August 28, 2024 @ 3:19 pm
[u][b] Привет![/b][/u]
Мы можем предложить дипломы.
[url=http://newsrbc.ru/blogs/post-4020-zhelaete-kupit-diplom-sssr-/]newsrbc.ru/blogs/post-4020-zhelaete-kupit-diplom-sssr-[/url]
Lazrnyz
August 28, 2024 @ 3:22 pm
[u][b] Здравствуйте![/b][/u]
[b]Мы предлагаем дипломы[/b] любых профессий по доступным ценам.
[url=http://weekevents.ru/obrazovanie-vashey-mechtyi-legko-i-dostupno/]weekevents.ru/obrazovanie-vashey-mechtyi-legko-i-dostupno[/url]
Sazrjre
August 28, 2024 @ 3:26 pm
Приобретение диплома ПТУ с сокращенной программой обучения в Москве
Sazrbfd
August 28, 2024 @ 3:45 pm
[u][b] Добрый день![/b][/u]
Диплом бакалавра
[url=http://power.ekafe.ru/viewtopic.php?f=2&t=1766/]power.ekafe.ru/viewtopic.php?f=2&t=1766[/url]
Cazrozj
August 28, 2024 @ 3:47 pm
[u][b] Добрый день![/b][/u]
Заказать диплом о высшем образовании
[url=http://fromrus.su/index.php?/topic/15713-смотри-l430e/]fromrus.su/index.php?/topic/15713-смотри-l430e[/url]
Sazrgrg
August 28, 2024 @ 4:22 pm
[u][b] Привет![/b][/u]
Диплом любой специальности
[url=http://telegra.ph/diplom-ob-obrazovanii-kupit-08-22/]telegra.ph/diplom-ob-obrazovanii-kupit-08-22[/url]
Ремонт смартфонов
August 28, 2024 @ 5:03 pm
Профессиональный сервисный центр по ремонту сотовых телефонов, смартфонов и мобильных устройств.
Мы предлагаем: ремонт смартфонов москва
Наши мастера оперативно устранят неисправности вашего устройства в сервисе или с выездом на дом!
antoniorudiger
August 28, 2024 @ 5:25 pm
[url=https://antonio-rudiger-cz.biz]http://www.antonio-rudiger-cz.biz[/url]
last news about antonio rudiger
http://www.antonio-rudiger-cz.biz
Treflxw
August 28, 2024 @ 5:40 pm
[u][b] Привет![/b][/u]
Аттестат 11 класса купить официально с упрощенным обучением в Москве
[url=http://m.endoinfo.ru/consultations/]m.endoinfo.ru/consultations[/url]
Рады оказать помощь!.
Lazritf
August 28, 2024 @ 6:41 pm
[u][b] Здравствуйте![/b][/u]
[b]Мы можем предложить дипломы[/b] любой профессии по доступным тарифам.
[url=http://by-volt.com/e_feedback/?page=2260/]by-volt.com/e_feedback/?page=2260[/url]
Sazrbtn
August 28, 2024 @ 7:29 pm
Официальная покупка школьного аттестата с упрощенным обучением в Москве
antoniorudigerczbiz
August 28, 2024 @ 7:32 pm
[url=https://antoniorudiger-cz.biz]www.antoniorudiger-cz.biz[/url]
last news about antonio rudiger
antoniorudiger-cz.biz
Oariormmf
August 28, 2024 @ 8:00 pm
[u][b] Привет![/b][/u]
[b]Где заказать диплом по актуальной специальности?[/b]
[url=http://telegra.ph/kakie-stranicy-attestata-nuzhno-kopirovat-dlya-postupleniya-v-vuz-07-24/]telegra.ph/kakie-stranicy-attestata-nuzhno-kopirovat-dlya-postupleniya-v-vuz-07-24[/url]
[url=http://telegra.ph/kakie-foto-dlya-postupleniya-v-vuz-07-24/]telegra.ph/kakie-foto-dlya-postupleniya-v-vuz-07-24[/url]
[url=http://telegra.ph/kakie-foto-nuzhny-pri-postuplenii-v-vuz-07-24/]telegra.ph/kakie-foto-nuzhny-pri-postuplenii-v-vuz-07-24[/url]
[url=http://telegra.ph/kakih-vrachej-nuzhno-projti-dlya-postupleniya-v-vuz-v-belarusi-07-24/]telegra.ph/kakih-vrachej-nuzhno-projti-dlya-postupleniya-v-vuz-v-belarusi-07-24[/url]
[url=http://telegra.ph/kakih-vrachej-nuzhno-prohodit-pered-postupleniem-v-vuz-07-24/]telegra.ph/kakih-vrachej-nuzhno-prohodit-pered-postupleniem-v-vuz-07-24[/url]
Sazrnjk
August 28, 2024 @ 8:26 pm
Стоимость дипломов высшего и среднего образования и процесс их получения
Cazrdat
August 28, 2024 @ 8:32 pm
Купить диплом о среднем образовании в Москве и любом другом городе
Mazrknf
August 28, 2024 @ 8:56 pm
Официальная покупка диплома вуза с сокращенной программой обучения в Москве
Vivod iz zapoya v stacionare_nupr
August 28, 2024 @ 11:19 pm
лечение наркозависимости в стационаре [url=https://vyvod-iz-zapoya-v-stacionare.ru]лечение наркозависимости в стационаре[/url] .
Bryonemaix
August 28, 2024 @ 11:19 pm
[url=https://rashford-marcus-cz.biz]http://www.rashford-marcus-cz.biz[/url]
last news about rashford marcus
https://www.rashford-marcus-cz.biz
ремонт ноутбуков
August 29, 2024 @ 1:31 am
Профессиональный сервисный центр по ремонту ноутбуков и компьютеров.дронов.
Мы предлагаем:ремонт ноутбуков в москве недорого
Наши мастера оперативно устранят неисправности вашего устройства в сервисе или с выездом на дом!
Williamraics
August 29, 2024 @ 2:29 am
[u][b] Добрый день![/b][/u]
Мы готовы предложить дипломы.
[url=http://ingmac.ru/forum/?PAGE_NAME=profile_view&UID=35748/]ingmac.ru/forum/?PAGE_NAME=profile_view&UID=35748[/url]
Eanryja
August 29, 2024 @ 3:57 am
Здравствуйте!
Мы предлагаем дипломы любой профессии по доступным ценам.
[url=http://okna496.ru/rehau/rehau1/]okna496.ru/rehau/rehau1[/url]
Iarioryqw
August 29, 2024 @ 4:21 am
[u][b] Здравствуйте![/b][/u]
Купить документ о получении высшего образования можно в нашей компании в Москве.
[url=http://goalissimo.org/forum/viewtopic.php?f=4&t=695403/]goalissimo.org/forum/viewtopic.php?f=4&t=695403[/url]
[url=http://ya.forum.cool/viewtopic.php?id=5816#p17344/]ya.forum.cool/viewtopic.php?id=5816#p17344[/url]
[url=http://pyha.ru/forum/topic/13499.1#msg216442/]pyha.ru/forum/topic/13499.1#msg216442[/url]
[url=http://ladymystery.ru/forum/topic.asp?TOPIC_ID=9497/]ladymystery.ru/forum/topic.asp?TOPIC_ID=9497[/url]
[url=http://bezone.ru/node/339527/]bezone.ru/node/339527[/url]
Sazrqri
August 29, 2024 @ 6:30 am
Официальная покупка диплома вуза с сокращенной программой обучения в Москве
rudiger-antonio
August 29, 2024 @ 6:34 am
[url=https://rudiger-antonio-cz.biz]http://www.rudiger-antonio-cz.biz[/url]
last news about rudiger antonio
https://www.rudiger-antonio-cz.biz
Cazrxie
August 29, 2024 @ 7:20 am
Купить диплом старого образца, можно ли это сделать по быстрой схеме?
Sazrmpj
August 29, 2024 @ 7:31 am
Как оказалось, купить диплом кандидата наук не так уж и сложно
Trefprs
August 29, 2024 @ 7:39 am
[u][b] Привет, друзья![/b][/u]
Полезная информация как купить диплом о высшем образовании без рисков
[url=http://pedolog-pro.ru/forums/]pedolog-pro.ru/forums[/url]
Окажем помощь!.
Vivod iz zapoya v stacionare_qkSl
August 29, 2024 @ 8:06 am
вывода из запоя в стационаре [url=http://vyvod-iz-zapoya-v-stacionare13.ru]вывода из запоя в стационаре[/url] .
Diplomi_fySi
August 29, 2024 @ 9:04 am
Привет, друзья!
Приобрести документ института можно у нас в столице.
[url=http://w99925ro.bget.ru/main/1-post1/]w99925ro.bget.ru/main/1-post1[/url]
Trefihx
August 29, 2024 @ 10:39 am
[u][b] Добрый день![/b][/u]
Можно ли купить аттестат о среднем образовании, основные моменты и вопросы
[url=http://webmontag.de/doku.php?id=rusddiploms/]webmontag.de/doku.php?id=rusddiploms[/url]
Рады помочь!.
Lazrrqe
August 29, 2024 @ 10:59 am
[u][b] Привет![/b][/u]
[b]Мы предлагаем дипломы[/b] психологов, юристов, экономистов и любых других профессий по приятным тарифам.
[url=http://google.mk/url?sa=i&url=aurus-diploms.com/]google.mk/url?sa=i&url=aurus-diploms.com[/url]
davidalaba
August 29, 2024 @ 11:19 am
[url=https://david-alaba-cz.biz]www.david-alaba-cz.biz[/url]
last news about david alaba
http://www.david-alaba-cz.biz
Mazrznx
August 29, 2024 @ 12:06 pm
[u][b] Привет, друзья![/b][/u]
[b]Быстрая покупка диплома старого образца: возможные риски[/b]
[url=http://lhotkanet.cz/galerie/profile.php?uid=75791/]lhotkanet.cz/galerie/profile.php?uid=75791[/url]
Diplomi_ceSi
August 29, 2024 @ 12:14 pm
Привет!
Купить документ университета вы можете у нас в столице.
[url=http://skybirdint.com/2019/05/31/sky-bird-international-gloves-stitching-hall/#comment-124114/]skybirdint.com/2019/05/31/sky-bird-international-gloves-stitching-hall/#comment-124114[/url]
Lazrcdx
August 29, 2024 @ 12:36 pm
Рекомендации по безопасной покупке диплома о высшем образовании
Domingotes
August 29, 2024 @ 12:48 pm
Авторские джип-туры по Крыму https://м-драйв.рф/tours/edu-kuda-hochu/ из Ялты. Уникальная возможность исследовать самые живописные места Крыма.
Mazrvds
August 29, 2024 @ 1:02 pm
Всё, что нужно знать о покупке аттестата о среднем образовании
Lazrjms
August 29, 2024 @ 2:25 pm
[u][b] Привет, друзья![/b][/u]
[b]Мы изготавливаем дипломы[/b] психологов, юристов, экономистов и других профессий по выгодным ценам.
[url=http://ds-dealer.ru/forum/member.php?u=733168/]ds-dealer.ru/forum/member.php?u=733168[/url]
Xazrrin
August 29, 2024 @ 2:58 pm
Привет, друзья!
Купить документ о получении высшего образования.
Мы готовы предложить дипломы психологов, юристов, экономистов и любых других профессий по приятным тарифам.
[url=http://buycruise.ru/]buycruise.ru[/url]
Рады оказать помощь!.
reiting kapperov_obMn
August 29, 2024 @ 5:43 pm
рейтинг капперов [url=https://rejting-kapperov13.ru/]рейтинг капперов[/url] .
Sazrbnb
August 29, 2024 @ 6:05 pm
Аттестат школы купить официально с упрощенным обучением в Москве
gabriel-jesus
August 29, 2024 @ 7:02 pm
[url=https://gabriel-jesus-cz.biz]https://gabriel-jesus-cz.biz[/url]
last news about gabriel jesus
https://www.gabriel-jesus-cz.biz
jesusgabriel
August 29, 2024 @ 7:36 pm
[url=https://jesus-gabriel-cz.biz]https://www.jesus-gabriel-cz.biz[/url]
last news about jesus gabriel
https://jesus-gabriel-cz.biz
JamesCreew
August 29, 2024 @ 8:40 pm
[url=https://alabadavid-cz.biz]http://www.alabadavid-cz.biz[/url]
last news about alaba david
http://www.alabadavid-cz.biz
Sazrqid
August 29, 2024 @ 8:40 pm
[u][b] Добрый день![/b][/u]
Мы предлагаем [b]дипломы[/b] любой профессии.
Приобретение [b]документа[/b], подтверждающего окончание университета, – это грамотное решение.
[url=http://stalzashita.ru/index.php?subaction=userinfo&user=ugetiq/]stalzashita.ru/index.php?subaction=userinfo&user=ugetiq[/url]
[url=http://b24activities.ru/club/forum/messages/forum4/topic76/message184/?result=reply#message184/]b24activities.ru/club/forum/messages/forum4/topic76/message184/?result=reply#message184[/url]
[url=http://wiki.natlife.ru/index.php?title=radiplom/]wiki.natlife.ru/index.php?title=radiplom[/url]
[url=http://tamklad.ru/viewtopic.php?f=7&t=8899/]tamklad.ru/viewtopic.php?f=7&t=8899[/url]
[url=http://cloudlab.tw/javascript-??????????/comment-page-35080/#comment-3294607/]cloudlab.tw/javascript-??????????/comment-page-35080/#comment-3294607[/url]
gabrieljesus
August 29, 2024 @ 9:04 pm
[url=https://gabrieljesuscz.biz]http://gabrieljesuscz.biz[/url]
last news about gabriel jesus
http://www.gabrieljesuscz.biz
MichaelDar
August 29, 2024 @ 9:25 pm
[url=https://marcusrashford-cz.biz]www.marcusrashford-cz.biz[/url]
last news about marcus rashford
http://www.marcusrashford-cz.biz
Stephennewly
August 29, 2024 @ 9:32 pm
[url=https://romelulukaku-cz.biz/]www.romelulukaku-cz.biz[/url]
last news about romelu lukaku
https://romelulukaku-cz.biz/
Mazrjil
August 29, 2024 @ 10:06 pm
Как приобрести диплом техникума с минимальными рисками
rudigerantoniocz
August 29, 2024 @ 10:59 pm
[url=https://rudiger-antoniocz.biz]www.rudiger-antoniocz.biz[/url]
last news about rudiger antonio
http://rudiger-antoniocz.biz
davidalaba
August 29, 2024 @ 11:01 pm
[url=https://davidalabacz.biz]https://davidalabacz.biz[/url]
last news about david alaba
http://www.davidalabacz.biz
RobertRer
August 29, 2024 @ 11:10 pm
[url=https://rashfordmarcus-cz.biz]http://rashfordmarcus-cz.biz[/url]
last news about rashford marcus
https://www.rashfordmarcus-cz.biz
Mazreal
August 29, 2024 @ 11:15 pm
[u][b] Добрый день![/b][/u]
[b]Как приобрести диплом техникума с минимальными рисками[/b]
[url=http://lada-4×4.net/member.php?u=23337/]lada-4×4.net/member.php?u=23337[/url]
Derrickjoisy
August 29, 2024 @ 11:19 pm
[url=https://marcusrashfordcz.biz]https://marcusrashfordcz.biz[/url]
last news about marcus rashford
https://marcusrashfordcz.biz
gabrieljesus
August 29, 2024 @ 11:26 pm
[url=https://gabrieljesus-cz.biz]www.gabrieljesus-cz.biz[/url]
last news about gabrie ljesus
https://www.gabrieljesus-cz.biz
Sazrqym
August 29, 2024 @ 11:26 pm
[u][b] Добрый день![/b][/u]
Заказать документ о получении высшего образования
[url=http://fbadult.com/blogs/3297/Оформите-диплом-без-ожидания/]fbadult.com/blogs/3297/Оформите-диплом-без-ожидания[/url]
[url=http://sustalks.com/blogs/2877/Решили-узнать-как-приобрести-диплом-в-интернете-недорого-Заходите/]sustalks.com/blogs/2877/Решили-узнать-как-приобрести-диплом-в-интернете-недорого-Заходите[/url]
[url=http://lossantosbattalion17.listbb.ru/viewtopic.php?f=3&t=416/]lossantosbattalion17.listbb.ru/viewtopic.php?f=3&t=416[/url]
[url=http://telegra.ph/10-tonkostej-postupleniya-v-lyuboj-vuz-07-24/]telegra.ph/10-tonkostej-postupleniya-v-lyuboj-vuz-07-24[/url]
[url=http://c90226sl.beget.tech/2024/07/18/oformlenie-diplomov-bez-posrednikov/]c90226sl.beget.tech/2024/07/18/oformlenie-diplomov-bez-posrednikov[/url]
Lazrwgp
August 29, 2024 @ 11:42 pm
Как официально приобрести аттестат 11 класса с минимальными затратами времени
Sazrgag
August 30, 2024 @ 12:58 am
Диплом пту купить официально с упрощенным обучением в Москве
Cazranx
August 30, 2024 @ 12:58 am
[u][b] Привет, друзья![/b][/u]
Купить диплом академии
[url=http://chinchillaorozco.com/asistencia-legal/]купить диплом в соликамске[/url]
[url=http://rockcitytrustcompany.com/2020/01/a-simple-guide-to-design-thinking#comment-27471/]купить диплом в белово[/url]
[url=http://aguasdearuanda.org.br/2014-01-11-reflexao-sobre-a-religiao-umbanda/#comment-628204/]купить диплом в ялте[/url]
[url=http://dnacumaru.com.br/2021/04/07/ola-mundo/#comment-7780/]купить диплом бурильщика[/url]
[url=http://ajcc-conf.net/2020/07/05/ajcc2021-call-for-papers/]купить диплом железнодорожника[/url]
Консультации
August 30, 2024 @ 1:05 am
Faber est suae quisque fortunae
Uazrgsk
August 30, 2024 @ 1:24 am
[u][b]Здравствуйте![/b][/u]
[b]Приобрести диплом любого университета.[/b]
[url=http://telegra.ph/kupit-diplom-vuza-08-13-6/]telegra.ph/kupit-diplom-vuza-08-13-6[/url]
Xazrxnq
August 30, 2024 @ 2:09 am
Привет, друзья!
Заказать документ о получении высшего образования.
Мы изготавливаем дипломы любой профессии по приятным ценам.
[url=http://chento.ru/]chento.ru[/url]
Поможем вам всегда!.
Sazrnnn
August 30, 2024 @ 2:41 am
Приобретение школьного аттестата с официальным упрощенным обучением в Москве
Sazrtfn
August 30, 2024 @ 3:28 am
[u][b] Добрый день![/b][/u]
Купить документ о получении высшего образования
[url=http://forum.aionclassic.net/member.php?u=12170/]forum.aionclassic.net/member.php?u=12170[/url]
Красивый секс
August 30, 2024 @ 3:47 am
Красивый секс https://bit.ly/krasiviy-seks-video-krasiviy-seks
Sazrllq
August 30, 2024 @ 3:56 am
Официальная покупка школьного аттестата с упрощенным обучением в Москве
jesusgabrielcz
August 30, 2024 @ 4:26 am
[url=https://jesusgabriel-cz.biz]www.jesusgabriel-cz.biz[/url]
last news about jesus gabriel
http://jesusgabriel-cz.biz
Mazrxpj
August 30, 2024 @ 4:36 am
Как официально купить диплом вуза с упрощенным обучением в Москве
Lazrmnb
August 30, 2024 @ 6:03 am
[u][b] Привет, друзья![/b][/u]
Приобрести [b]диплом[/b] о высшем образовании.
[url=http://ok-vmeste.ru/toget/1459-zhelaete-zakazat-podlinnyi-diplom/]ok-vmeste.ru/toget/1459-zhelaete-zakazat-podlinnyi-diplom[/url]
Lazruru
August 30, 2024 @ 6:14 am
[u][b] Здравствуйте![/b][/u]
[b]Мы изготавливаем дипломы[/b] психологов, юристов, экономистов и прочих профессий по невысоким тарифам.
[url=http://bn.tobase.ru/viewtopic.php?f=30&t=2737/]bn.tobase.ru/viewtopic.php?f=30&t=2737[/url]
Sazrfct
August 30, 2024 @ 7:45 am
[u][b] Добрый день![/b][/u]
Мы можем предложить [b]дипломы[/b] любой профессии.
Покупка [b]документа[/b], подтверждающего окончание ВУЗа, – это выгодное решение.
[url=http://pechorinase.blogspot.com/2013/04/dartagnan-et-les-trois-mousquetaires/]pechorinase.blogspot.com/2013/04/dartagnan-et-les-trois-mousquetaires[/url]
[url=http://amatagroup.ru/forum/messages/forum1/topic59/message471258/?result=reply#message471258/]amatagroup.ru/forum/messages/forum1/topic59/message471258/?result=reply#message471258[/url]
[url=http://youngwooapt.co.kr/bbs/board.php?bo_table=free&wr_id=382506/]youngwooapt.co.kr/bbs/board.php?bo_table=free&wr_id=382506[/url]
[url=http://krutovo.ru/index.php?name=Account&op=info&uname=ehyniz/]krutovo.ru/index.php?name=Account&op=info&uname=ehyniz[/url]
[url=http://debtcareconsulting.it/2021/04/14/hello-world/#comment-23944/]debtcareconsulting.it/2021/04/14/hello-world/#comment-23944[/url]
ремонту iPhone
August 30, 2024 @ 8:58 am
сервисный центр по ремонту айфонов в москве
Lazrgdv
August 30, 2024 @ 9:49 am
[u][b] Привет, друзья![/b][/u]
[b]Мы можем предложить дипломы[/b] любых профессий по приятным тарифам.
[url=http://2718281828.com/blog/index.php/]2718281828.com/blog/index.php[/url]
Sazrovj
August 30, 2024 @ 9:58 am
[u][b] Привет![/b][/u]
Купить документ института
[url=http://rrsclub.ru/member.php?u=1164/]rrsclub.ru/member.php?u=1164[/url]
[url=http://gemawiraclub.com/blogs/42170/болучите-диплом-без-лишних-хлопот/]gemawiraclub.com/blogs/42170/болучите-диплом-без-лишних-хлопот[/url]
[url=http://telegra.ph/itogi-postupleniya-v-vuzy-2018-07-24/]telegra.ph/itogi-postupleniya-v-vuzy-2018-07-24[/url]
[url=http://pyha.ru/forum/topic/13527.1#msg216474/]pyha.ru/forum/topic/13527.1#msg216474[/url]
[url=http://finttech.ru/vash-diplom-vashe-budushhee/]finttech.ru/vash-diplom-vashe-budushhee[/url]
Kvartirnii pereezd_uwMn
August 30, 2024 @ 10:05 am
заказать машину для переезда [url=www.kvartirnyj-pereezd11.ru/]заказать машину для переезда[/url] .
Germanrip
August 30, 2024 @ 10:20 am
[url=][/url]
Строительство, ремон своими руками. Советы мастеров и лучших специалстов в своем деле.
Каким вы видите свой будущий дом? Выполненный в духе уютного загородного шале или со сложными геометрическими формами и яркими элементами?
<a href=https://www.vuz-chursin.ru/stroitelstvo]Строительство
<a href=https://www.vuz-chursin.ru/stroymaterialy-0]Стройматериалы
<a href=https://www.vuz-chursin.ru/remont-i-otdelka]Отделка
Для большинства людей своя, пускай, даже крохотная, но отдельная однокомнатная квартира, намного слаще, чем аренда 5-комнтантного пентхауса за смешные деньги. Здесь можно устанавливать свои правила, планировать ремонт, делать перестановку, а единственное ограничение – это пространство.
Каталог полезных статей о стройке и ремонте <a href=https://vuz-chursin.ru/.
[url=][/url]
Sazrnhf
August 30, 2024 @ 10:38 am
Полезные советы по безопасной покупке диплома о высшем образовании
Cazrpgm
August 30, 2024 @ 10:40 am
[u][b] Привет![/b][/u]
Купить диплом о высшем образовании
[url=http://c953879x.bget.ru/index.php?newsid=6/]c953879x.bget.ru/index.php?newsid=6[/url]
Sazrbcu
August 30, 2024 @ 11:17 am
Рекомендации по безопасной покупке диплома о высшем образовании
Sazrzti
August 30, 2024 @ 11:21 am
Диплом пту купить официально с упрощенным обучением в Москве
Cazrnld
August 30, 2024 @ 11:24 am
[u][b] Привет![/b][/u]
Получить диплом о высшем образовании
[url=http://synthonia.com/index.php?option=com_k2&view=itemlist&task=user&id=625356/]купить технический диплом[/url]
[url=http://lada-xray.net/showthread.php?p=32961#post32961/]купить диплом в коврове[/url]
[url=http://igram.net/index.php?name=Account&op=userinfo&user_name=ewakow/]купить диплом в новочеркасске[/url]
[url=http://forumru.uniutilis.com/profile.php?action=show&member=6614/]купить диплом в муроме[/url]
[url=http://moolookoo.ru/content/aurusdiplomscom/]купить диплом в сыктывкаре[/url]
Uazrwiu
August 30, 2024 @ 12:17 pm
[u][b]Добрыйдень![/b][/u]
[b]Приобрести диплом о высшем образовании.[/b]
[url=http://telegra.ph/kupit-diplom-deshevo-08-13-10/]telegra.ph/kupit-diplom-deshevo-08-13-10[/url]
Diplomi_stSi
August 30, 2024 @ 12:36 pm
Здравствуйте!
Заказать документ института вы имеете возможность у нас.
[url=http://mathematics-time.blogspot.com/2013/02/blog-post_9352/]mathematics-time.blogspot.com/2013/02/blog-post_9352[/url]
Sazrcuf
August 30, 2024 @ 1:16 pm
Рекомендации по безопасной покупке диплома о высшем образовании
Sazrqpc
August 30, 2024 @ 2:38 pm
Купить диплом о среднем полном образовании, в чем подвох и как избежать обмана?
Mazrlgd
August 30, 2024 @ 2:53 pm
Всё, что нужно знать о покупке аттестата о среднем образовании без рисков
Sazrszd
August 30, 2024 @ 3:00 pm
Рекомендации по безопасной покупке диплома о высшем образовании
Diplomi_roSi
August 30, 2024 @ 4:38 pm
Добрый день!
Заказать документ университета вы можете у нас в Москве.
[url=http://likt590.blogspot.com/2014/01/blog-post/]likt590.blogspot.com/2014/01/blog-post[/url]
Lazrced
August 30, 2024 @ 5:08 pm
[u][b] Добрый день![/b][/u]
Купить [b]диплом[/b] любого ВУЗа.
[url=http://forumrabota.0pk.me/viewtopic.php?id=7320#p16792/]forumrabota.0pk.me/viewtopic.php?id=7320#p16792[/url]
JamesSiz
August 30, 2024 @ 6:26 pm
[url=https://lukaku-romelu-cz.biz]www.lukaku-romelu-cz.biz[/url]
last news about lukaku romelu
http://www.lukaku-romelu-cz.biz
Bryonemaix
August 30, 2024 @ 6:46 pm
[url=https://romelulukakucz.biz]http://romelulukakucz.biz[/url]
last news about romelu lukaku
http://www.romelulukakucz.biz
Douglasbep
August 30, 2024 @ 7:40 pm
[url=https://lukakuromelu-cz.biz]lukakuromelu-cz.biz[/url]
last news about lukaku romelu
http://www.lukakuromelu-cz.biz
vivod iz zapoya sochi_xeon
August 30, 2024 @ 8:57 pm
вывод из запоя стационар (вывода из запоя в стационаре) [url=http://www.vyvod-iz-zapoya-sochi11.ru]вывод из запоя стационар (вывода из запоя в стационаре)[/url] .
Sazrult
August 30, 2024 @ 9:32 pm
Приобретение школьного аттестата с официальным упрощенным обучением в Москве
Cazrqax
August 30, 2024 @ 9:35 pm
[u][b] Здравствуйте![/b][/u]
Получить диплом института
[url=http://romb4x4.ru/forum/threads/kupit-diplom-o-vysshem-obrazovanii-nedorogo-c434l.12253/]romb4x4.ru/forum/threads/kupit-diplom-o-vysshem-obrazovanii-nedorogo-c434l.12253[/url]
Mazriol
August 30, 2024 @ 10:30 pm
Как получить диплом стоматолога быстро и официально
Sazrjie
August 30, 2024 @ 11:05 pm
Приобретение диплома ПТУ с сокращенной программой обучения в Москве
Iariordie
August 30, 2024 @ 11:21 pm
[u][b] Добрый день![/b][/u]
Приобрести документ о получении высшего образования вы имеете возможность в нашей компании в Москве.
[url=http://financeokey.ru/ekspress-dostavka-diplomov-po-vsey-strane/]financeokey.ru/ekspress-dostavka-diplomov-po-vsey-strane[/url]
[url=http://newspromworld.ru/ekspress-diplomyi-po-dostupnyim-tsenam/]newspromworld.ru/ekspress-diplomyi-po-dostupnyim-tsenam[/url]
[url=http://worldavtonew.ru/oformlenie-diplomov-bez-ozhidaniya/]worldavtonew.ru/oformlenie-diplomov-bez-ozhidaniya[/url]
[url=http://animalplanetnews.ru/oformlenie-diplomov-bez-ozhidaniya/]animalplanetnews.ru/oformlenie-diplomov-bez-ozhidaniya[/url]
[url=http://mynewsport.ru/ekspress-dostavka-diplomov-po-vsey-strane/]mynewsport.ru/ekspress-dostavka-diplomov-po-vsey-strane[/url]
Eanryco
August 30, 2024 @ 11:23 pm
Здравствуйте!
Мы предлагаем дипломы любой профессии по выгодным ценам.
[url=http://dolcescents.com/detailS/09/]dolcescents.com/detailS/09[/url]
Charlesnidly
August 30, 2024 @ 11:46 pm
[url=https://romelu-lukaku-cz.biz]http://romelu-lukaku-cz.biz[/url]
last news about romelu lukaku
http://www.romelu-lukaku-cz.biz
Oariorqtu
August 31, 2024 @ 12:42 am
[u][b] Добрый день![/b][/u]
[b]Где заказать диплом специалиста?[/b]
[url=http://telegra.ph/kak-oplatit-obuchenie-v-vuze-07-24/]telegra.ph/kak-oplatit-obuchenie-v-vuze-07-24[/url]
[url=http://telegra.ph/kak-oplatit-obuchenie-v-vuze-cherez-sberbank-07-24/]telegra.ph/kak-oplatit-obuchenie-v-vuze-cherez-sberbank-07-24[/url]
[url=http://telegra.ph/kak-poluchit-kredit-na-obuchenie-v-vuze-07-24/]telegra.ph/kak-poluchit-kredit-na-obuchenie-v-vuze-07-24[/url]
[url=http://telegra.ph/kak-poluchit-nalogovyj-vychet-za-obuchenie-rebenka-v-vuze-07-24/]telegra.ph/kak-poluchit-nalogovyj-vychet-za-obuchenie-rebenka-v-vuze-07-24[/url]
[url=http://telegra.ph/kak-postupit-na-platnoe-obuchenie-v-vuz-07-24/]telegra.ph/kak-postupit-na-platnoe-obuchenie-v-vuz-07-24[/url]
Micahham
August 31, 2024 @ 1:32 am
[url=https://alaba-david-cz.biz]http://alaba-david-cz.biz[/url]
last news about alaba david
https://www.alaba-david-cz.biz
BarryOvete
August 31, 2024 @ 1:47 am
[url=https://riyadmahrezcz.biz]https://www.riyadmahrezcz.biz[/url]
last news about riyad mahrez
http://www.riyadmahrezcz.biz
Robertdup
August 31, 2024 @ 2:00 am
[url=https://marcus-rashford-cz.biz]www.marcus-rashford-cz.biz[/url]
last news about marcus rashford
https://www.marcus-rashford-cz.biz
Sazrrjt
August 31, 2024 @ 2:32 am
Покупка диплома о среднем полном образовании: как избежать мошенничества?
Lazrwdv
August 31, 2024 @ 3:28 am
[u][b] Добрый день![/b][/u]
[b]Мы готовы предложить дипломы[/b] любой профессии по доступным тарифам.
[url=http://tnmiass.ru/programms/besedy-v-shkolah-i-kolledzhah-tvoy-vybor/]tnmiass.ru/programms/besedy-v-shkolah-i-kolledzhah-tvoy-vybor[/url]
antoniorudigercz
August 31, 2024 @ 3:35 am
[url=https://antoniorudigercz.biz]http://www.antoniorudigercz.biz[/url]
last news about antonio rudiger
http://www.antoniorudigercz.biz
do my online accounting class for me
August 31, 2024 @ 6:07 am
In mathematics education, the ultimate goal is to teach students how to solve real-world problems by using mathematical concepts intuitively. Regardless of what field you study, from economics to engineering to medicine to social and behavioral sciences, the ability to think mathematically is fundamental. In algebraic geometry, expressions can be as ancient as rings, groups, and areas, or as contemporary as anything else. In addition to online class assistants, mathematics statistics incorporates concepts from probability theory, mathematical analysis, differential equations, and linear algebra. It may be difficult to understand these ideas if you are short on time or your teacher is unable to assist you. If you want a deeper understanding of them, turn to me. do my online stats class for me will simplify the ideas of statistics.
Sazrrqe
August 31, 2024 @ 6:45 am
Приобретение диплома ПТУ с сокращенной программой обучения в Москве
Lazrohx
August 31, 2024 @ 7:39 am
[u][b] Здравствуйте![/b][/u]
[b]Мы предлагаем дипломы[/b] психологов, юристов, экономистов и других профессий по выгодным тарифам.
[url=http://72reg.ru/]72reg.ru[/url]
Williamraics
August 31, 2024 @ 8:48 am
[u][b] Привет, друзья![/b][/u]
Мы изготавливаем дипломы.
[url=http://alternativaprofi.com/forum/viewthread.php?thread_id=30893/]alternativaprofi.com/forum/viewthread.php?thread_id=30893[/url]
Felixmon
August 31, 2024 @ 9:29 am
[url=][/url]
[url=https://torgdom1.ru/novoe-oborudovanie/]Новое оборудование для ресторанов и кафе[/url]
Предлагаем Новое оборудование для магазинов, ресторанов, гостиниц и других мест общественного питания и обслуживания. В нашем каталоге представлены модели различных габаритов и технических характеристик. На любой товар мы предоставляем гарантийный талон. Доставку осуществляем в течение 1-2 рабочих дней с момента заказа, либо в согласованный день. Предусмотрена оплата для юр.лиц по счету. Чтобы сделать заказ в нашей компании «ТоргХаус», соберите список товаров в корзине, и менеджер свяжется для уточнения способа оплаты и удобного времени доставки.
[url=https://torgdom1.ru/oborudovanie-bu/]БУ оборудование для ресторанов и кафе[/url]
Предлагаем Оборудование для общепита Б/У для магазинов, ресторанов, гостиниц и других мест общественного питания и обслуживания. В нашем каталоге представлены модели различных габаритов и технических характеристик. На любой товар мы предоставляем гарантийный талон.
[url=https://torgdom1.ru/proektirovanie-restoranov/]Проектирование ресторанов и кафе в Екатеринбурге[/url]
Компания «Торгхаус» успешно занимается проектированием предприятий общественного питания.
Заказав проект ресторана у нас, Вы получите:
Отдавая разработку проекта в руки профессионалов, можно быть уверенным в оптимизации и привязке всех систем. Каждое решение органично впишется в общую технологию, снижая затраты, касаемо:
Без соответствующей нормам проектной документации, невозможен запуск ресторана. Схема подводки всех систем обеспечивает безопасность работы в будущем. Соблюдение противопожарных и санитарных норм создаст почву для быстрого прохождения проверок.
Ориентировочная стоимость проектирования
– удачное планировочные решения;
– высокотехнологичную расстановку оборудования;
– верный расчет систем вентиляции и кондиционирования;
– индивидуальный подход к разработке дизайна;
– первоначальных вложений в строительство и перепланировку;
– обслуживание оборудования;
– согласование документации и прочих вопросов.
Цены на всю продукцию [url=https://torgdom1.ru/].[/url]
[url=][/url]
Sazrlkp
August 31, 2024 @ 9:42 am
Как оказалось, купить диплом кандидата наук не так уж и сложно
Sazrowq
August 31, 2024 @ 9:44 am
Официальная покупка школьного аттестата с упрощенным обучением в Москве
Mazrxsq
August 31, 2024 @ 9:48 am
Всё, что нужно знать о покупке аттестата о среднем образовании без рисков
Iariorzjj
August 31, 2024 @ 10:08 am
[u][b] Привет![/b][/u]
Заказать документ ВУЗа можно у нас в столице.
[url=http://obektivcentr.ru/forum/user/17584/]obektivcentr.ru/forum/user/17584[/url]
[url=http://school97.ru/vesti20/index.php?PAGE_NAME=profile_view&UID=210678/]school97.ru/vesti20/index.php?PAGE_NAME=profile_view&UID=210678[/url]
[url=http://sklad-slabov.ru/forum/user/8459/]sklad-slabov.ru/forum/user/8459[/url]
[url=http://mnogozip.ru/forum/user/23890/]mnogozip.ru/forum/user/23890[/url]
[url=http://4mark.net/story/12411296/купить-диплом-о-высшем-образовании-гознак-любого-вуза-россии/]4mark.net/story/12411296/купить-диплом-о-высшем-образовании-гознак-любого-вуза-россии[/url]
Eanrkln
August 31, 2024 @ 10:18 am
Привет, друзья!
Мы изготавливаем дипломы психологов, юристов, экономистов и любых других профессий по приятным ценам.
[url=http://mdr7.ru/topic6433?&p=94269/]mdr7.ru/topic6433?&p=94269[/url]
Sazrofc
August 31, 2024 @ 1:29 pm
Быстрое обучение и получение диплома магистра – возможно ли это?
Cazrxiy
August 31, 2024 @ 1:43 pm
Приобретение школьного аттестата с официальным упрощенным обучением в Москве
gimnas3.ru/d/articles/?gde_kupit_diplom_vuza_sovetu_i_rekomendacii
Mazrblb
August 31, 2024 @ 6:10 pm
Легальная покупка диплома о среднем образовании в Москве и регионах
Lazrkmy
August 31, 2024 @ 7:23 pm
Как приобрести аттестат о среднем образовании в Москве и других городах
Mazrzma
August 31, 2024 @ 7:33 pm
[u][b] Привет![/b][/u]
[b]Как правильно купить диплом колледжа и пту в России, подводные камни[/b]
[url=http://getamped.yxhi.com/doku.php?id=diplomandoks/]getamped.yxhi.com/doku.php?id=diplomandoks[/url]
Diplomi_zuSi
August 31, 2024 @ 9:34 pm
Добрый день!
Приобрести документ о получении высшего образования можно у нас в Москве.
[url=http://matripedia.de/doku.php?id=luxdiplom/]matripedia.de/doku.php?id=luxdiplom[/url]
Xazrzyh
August 31, 2024 @ 10:06 pm
Привет!
Приобрести документ института.
Мы изготавливаем дипломы психологов, юристов, экономистов и любых других профессий по невысоким ценам.
[url=http://avb-service.ru/]avb-service.ru[/url]
Окажем помощь!.
Stephenswogs
September 1, 2024 @ 12:04 am
[url=https://mahrez-riyad-cz.biz]https://mahrez-riyad-cz.biz[/url]
last news about mahrez riyad
https://mahrez-riyad-cz.biz
Cazrohw
September 1, 2024 @ 12:16 am
Узнайте стоимость диплома высшего и среднего образования и процесс получения
uralshooter.ru/wp-content/pages/?kak_kupit_diplom_s_garantiey_provodki_bez_riska
Sazrbbl
September 1, 2024 @ 12:21 am
Как получить диплом техникума с упрощенным обучением в Москве официально
vivod iz zapoya sochi_xoSr
September 1, 2024 @ 12:31 am
вывод из запоя цены сочи [url=vyvod-iz-zapoya-sochi12.ru]vyvod-iz-zapoya-sochi12.ru[/url] .
snyatie zapoya na domy_ifOl
September 1, 2024 @ 2:36 am
лечение алкоголизма на дому [url=snyatie-zapoya-na-domu11.ru]лечение алкоголизма на дому[/url] .
ThomasSot
September 1, 2024 @ 3:05 am
Фонбет промокод промокод не найден
Фонбет регулярно предлагает своим пользователям промокоды, которые позволяют получить дополнительные бонусы и привилегии. Например, промокод ‘GIFT200’ предоставляет новым клиентам бесплатную ставку или другие бонусы при регистрации. Промокоды могут распространяться через рекламные кампании, партнерские программы и специальные акции. Использование промокодов помогает пользователям увеличивать свои шансы на выигрыш и делает процесс игры более увлекательным и прибыльным.
Sazrpco
September 1, 2024 @ 4:06 am
Диплом пту купить официально с упрощенным обучением в Москве
Mazrfis
September 1, 2024 @ 6:12 am
[u][b] Привет![/b][/u]
[b]Покупка диплома о среднем полном образовании: как избежать мошенничества?[/b]
[url=http://gidro2000.com/forum-gidro/user/27779/]gidro2000.com/forum-gidro/user/27779[/url]
Sazrtjr
September 1, 2024 @ 6:32 am
Быстрое обучение и получение диплома магистра – возможно ли это?
Uazrgtv
September 1, 2024 @ 6:42 am
[u][b]Привет,друзья![/b][/u]
[b]Купить диплом университета .[/b]
[url=http://telegra.ph/diplom-magistra-kupit-08-13-2/]telegra.ph/diplom-magistra-kupit-08-13-2[/url]
Lazrtiw
September 1, 2024 @ 6:54 am
Вопросы и ответы: можно ли быстро купить диплом старого образца?
JamesSiz
September 1, 2024 @ 7:09 am
[url=https://tonikroos-cz.biz]http://tonikroos-cz.biz[/url]
last news about toni kroos
tonikroos-cz.biz
david-alaba
September 1, 2024 @ 7:41 am
[url=https://davidalaba-cz.biz]https://davidalaba-cz.biz[/url]
last news about david alaba
https://davidalaba-cz.biz
BarryOvete
September 1, 2024 @ 8:07 am
[url=https://brunoguimaraescz.biz]https://brunoguimaraescz.biz[/url]
last news about bruno guimaraes
https://www.brunoguimaraescz.biz
Bryonemaix
September 1, 2024 @ 8:23 am
[url=https://guimaraes-bruno-cz.biz]http://www.guimaraes-bruno-cz.biz[/url]
last news about guimaraes bruno
http://www.guimaraes-bruno-cz.biz
food
September 1, 2024 @ 8:38 am
[url=https://food.news161.ru]еда на заказ[/url]
обзор еды на заказ в ростовской области
https://food.news161.ru
Sazrwav
September 1, 2024 @ 8:52 am
[u][b] Здравствуйте![/b][/u]
Приобрести документ института
[url=http://twobook.ru/news/akter-seriala-fizryk-egor-klinaev-pogib-v-dtp/]twobook.ru/news/akter-seriala-fizryk-egor-klinaev-pogib-v-dtp[/url]
Darrelnog
September 1, 2024 @ 9:29 am
bonus veren slot siteleri: slot siteleri bonus veren – yeni slot siteleri
Xazrqbx
September 1, 2024 @ 9:37 am
Здравствуйте!
Заказать документ о получении высшего образования.
Мы изготавливаем дипломы любой профессии по выгодным тарифам.
[url=http://ftbteam.ru/]ftbteam.ru[/url]
Будем рады вам помочь!.
Kevinnut
September 1, 2024 @ 9:46 am
http://slotsiteleri.bid/# en iyi slot siteleri
partners
September 1, 2024 @ 10:02 am
[url=https://partners.news161.ru]партнер[/url]
Новости партнеров информационного портала Ростовской области – News161.ru
http://www.partners.news161.ru
Sazrrgb
September 1, 2024 @ 10:03 am
Официальная покупка аттестата о среднем образовании в Москве и других городах
drumsk.ru/bitrix/redirect.php?event1=&event2=&event3=&goto=diplomsagroups.com
Cazrfdy
September 1, 2024 @ 10:14 am
[u][b] Здравствуйте![/b][/u]
Приобрести диплом о высшем образовании
[url=http://studio-pd.ru/index.php?subaction=userinfo&user=irycaze/]куплю диплом[/url]
[url=http://mkala-koncert.ru/users/emoti/]купить аттестат за 9 классов[/url]
[url=http://gdz-fizika.ru/index.php?subaction=userinfo&user=axireh/]купить диплом моториста[/url]
[url=http://kioskindustry.ru/index.php?subaction=userinfo&user=opemiry/]купить диплом судоводителя[/url]
[url=http://mariland.org/index.php?subaction=userinfo&user=obytu/]купить сертификат специалиста[/url]
Sazreis
September 1, 2024 @ 11:50 am
[u][b] Здравствуйте![/b][/u]
Мы изготавливаем [b]дипломы[/b] любых профессий.
Покупка [b]диплома[/b], который подтверждает обучение в ВУЗе, – это грамотное решение.
[url=http://npcenter.blogspot.com/2014/01/blog-post_18?m=1/]npcenter.blogspot.com/2014/01/blog-post_18?m=1[/url]
[url=http://scorers.org/arsenal-re-sign-real-madrids-dani-ceballos-on-loan/#comment-427527/]scorers.org/arsenal-re-sign-real-madrids-dani-ceballos-on-loan/#comment-427527[/url]
[url=http://города-россия.рф/sity_id.php?id=39&comment=success&comment=success&comment=success&comment=success&comment=success&comment=success&comment=success&comment=success&comment=success#comment-24812/]города-россия.рф/sity_id.php?id=39&comment=success&comment=success&comment=success&comment=success&comment=success&comment=success&comment=success&comment=success&comment=success#comment-24812[/url]
[url=http://v208870d.bget.ru/index.php?subaction=userinfo&user=yjikysuc/]v208870d.bget.ru/index.php?subaction=userinfo&user=yjikysuc[/url]
[url=http://polimer42.ru/communication/forum/messages/forum5/topic49/message378486/?result=reply#message378486/]polimer42.ru/communication/forum/messages/forum5/topic49/message378486/?result=reply#message378486[/url]
Darrelnog
September 1, 2024 @ 12:07 pm
yasal slot siteleri: yasal slot siteleri – en iyi slot siteleri 2024
Lazrewq
September 1, 2024 @ 12:20 pm
[u][b] Привет, друзья![/b][/u]
Приобрести [b]диплом[/b] университета.
[url=http://fabnews.ru/blog/6726/]fabnews.ru/blog/6726[/url]
Kevinnut
September 1, 2024 @ 12:45 pm
https://slotsiteleri.bid/# canl? slot siteleri
Sazrbtf
September 1, 2024 @ 1:26 pm
[u][b] Добрый день![/b][/u]
Купить документ о получении высшего образования
[url=http://uktuliza.ru/forum/?PAGE_NAME=profile_view&UID=17802/]uktuliza.ru/forum/?PAGE_NAME=profile_view&UID=17802[/url]
[url=http://leadrp.listbb.ru/viewtopic.php?f=3&t=415/]leadrp.listbb.ru/viewtopic.php?f=3&t=415[/url]
[url=http://sovetushka.forum2x2.ru/t28931-topic#63708/]sovetushka.forum2x2.ru/t28931-topic#63708[/url]
[url=http://ya.5bb.ru/viewtopic.php?id=20035#p60525/]ya.5bb.ru/viewtopic.php?id=20035#p60525[/url]
[url=http://telegra.ph/bally-na-postuplenie-v-vuzy-07-24/]telegra.ph/bally-na-postuplenie-v-vuzy-07-24[/url]
Mazryek
September 1, 2024 @ 2:02 pm
Покупка школьного аттестата с упрощенной программой: что важно знать
Sazrxom
September 1, 2024 @ 2:37 pm
Быстрая схема покупки диплома старого образца: что важно знать?
GregorySeK
September 1, 2024 @ 3:21 pm
[url=][/url]
Лучшая сантехника по низким ценам в Минске
Если вы ищете, где купить сантехнику в Минске, то наш интернет-магазин — для вас.
Большой ассортимент товаров представлен в различных ценовых категориях, а благодаря разнообразию размеров,
форм и других параметров, вы точно найдёте наиболее подходящую модель.
Создайте свой уникальный дизайн ванной комнаты вместе с нами!
Почему Almoni?
Высокое качество: мы работаем с ведущими производителями и брендами с мировым именем,
например: Orans, Ravak, Vitra, Kaldewey, Roca и другие. Все изделия сертифицированы.
Доступные цены: благодаря нашему сотрудничеству напрямую с заводами-изготовителями,
у вас появляется возможность купить сантехнику недорого.
Широкий ассортимент: большой выбор точно позволит вам найти подходящую модель или цвет товара,
чтобы наиболее гармонично вписать его в общий интерьер.
Удобный сервис: заказы обрабатываются в кратчайшие сроки, есть возможность выбрать
подходящий для вас вариант уведомлений (смс/звонок/почта), а оповещения о передвижении
товара позволят вам всегда быть в курсе его местоположения. Оставить онлайн-заказ вы можете
круглосуточно через корзину на сайте, а с 09:00 — 21:00 это можно также сделать через оператора.
Мы любим наших клиентов: при покупке от 3 000 р. вы получаете приятные подарки,
а для наших постоянных покупателей действуют специальные скидки.
Аккуратная доставка в срок курьером доступна по всей Республике Беларусь,
а также более чем в 1000 пунктах самовывоза в России.
Если у Вас возникли вопросы или сложности с заказом, обратитесь к оператору,
который предоставит актуальную информацию, а также проконсультирует по товарам.
Сделать это можно по телефону: +375 (33) 398 66 77.
Интернет-магазин сантехники [url=https://almoni.by/]almoni.by[/url]
[url=][/url]
Sazrxix
September 1, 2024 @ 4:38 pm
Как не стать жертвой мошенников при покупке диплома о среднем полном образовании
Cazrtbr
September 1, 2024 @ 4:40 pm
[u][b] Добрый день![/b][/u]
Купить диплом о высшем образовании
[url=http://чпуремонт.рф/showthread.php?tid=57199/]чпуремонт.рф/showthread.php?tid=57199[/url]
Sazrmhe
September 1, 2024 @ 4:53 pm
Как избежать рисков при покупке диплома колледжа или ПТУ в России
Uazrrlt
September 1, 2024 @ 5:25 pm
Здравствуйте!
Приобрести диплом любого ВУЗа.
telegra.ph/kupit-diplom-v-kieve-08-13-9
RobertSot
September 1, 2024 @ 5:49 pm
Промокод Фонбет на сегодня бесплатно бонусные промокоды на
Для получения бесплатного промокода на Фонбет, действующего сегодня, рекомендуется проверять обновления на официальном сайте или подписаться на рассылку новостей. Промокод ‘GIFT200’ предоставляет бесплатные ставки для новых пользователей. Эти промокоды делают игру более привлекательной и выгодной, предлагая пользователям дополнительные возможности для выигрыша.
Sazrcmw
September 1, 2024 @ 6:33 pm
Как избежать рисков при покупке диплома колледжа или ПТУ в России
Sazrynz
September 1, 2024 @ 7:52 pm
[u][b] Привет, друзья![/b][/u]
Заказать документ о получении высшего образования
[url=http://chl.kiev.ua/cgi-bin/sp/part/index.php?razdel=23a&page=509/]chl.kiev.ua/cgi-bin/sp/part/index.php?razdel=23a&page=509[/url]
Sazrorq
September 1, 2024 @ 9:08 pm
Всё, что нужно знать о покупке аттестата о среднем образовании без рисков
direkt-einkauf.de/includes/refer.php?&id=2&url=diplomsagroups.com
Cazrxwz
September 1, 2024 @ 9:08 pm
[u][b] Привет![/b][/u]
Купить диплом университета
[url=http://octaniumsw.site/2018/01/ssd-m2-acer-nitro-5-an515-41-f9wl/]купить диплом в донском[/url]
[url=http://ks1r1504.blogspot.com/2014/05/blog-post/]купить диплом косметолога[/url]
[url=http://ifreemax.ru/2015/06/forum-madrid-startup-weekend-2015/]купить диплом в мытищах[/url]
[url=http://blagodarova.blogspot.com/2016/03/blog-post_7/]купить диплом в бийске[/url]
[url=http://merendinadatiffani.blogspot.ru/2016/03/aaa-addio-al-nubilato-cercasi/]купить диплом в железногорске[/url]
fanera kypit_lcot
September 1, 2024 @ 10:20 pm
мдф или фанера [url=https://fanera-kupit11.ru/]мдф или фанера[/url] .
Sazrczy
September 1, 2024 @ 11:14 pm
[u][b] Привет![/b][/u]
Мы изготавливаем [b]дипломы[/b] любой профессии.
Заказ [b]документа[/b], подтверждающего обучение в университете, – это разумное решение.
[url=http://nipponsword.ru/profile.php?lookup=24959/]nipponsword.ru/profile.php?lookup=24959[/url]
[url=http://aist-mebel.ru/product/nika-427-t-krovat-cherdak-so-stolom-i-shkafom/#comment-6468/]aist-mebel.ru/product/nika-427-t-krovat-cherdak-so-stolom-i-shkafom/#comment-6468[/url]
[url=http://mafia-game.ru/forum/newthread.php?do=newthread&f=3/]mafia-game.ru/forum/newthread.php?do=newthread&f=3[/url]
[url=http://kiopro.ru/support/forum/view_profile.php?UID=75636/]kiopro.ru/support/forum/view_profile.php?UID=75636[/url]
[url=http://mp3-zone.ru/user/hnnahmaarley9808/]mp3-zone.ru/user/hnnahmaarley9808[/url]
Lazradi
September 1, 2024 @ 11:31 pm
[u][b] Здравствуйте![/b][/u]
Приобрести [b]диплом[/b] о высшем образовании.
[url=http://bajajrussia.club/viewtopic.php?f=33&t=81776/]bajajrussia.club/viewtopic.php?f=33&t=81776[/url]
Darrelnog
September 2, 2024 @ 1:50 am
en guvenilir slot siteleri: bonus veren casino slot siteleri – slot siteleri bonus veren
Sazrlkv
September 2, 2024 @ 2:30 am
Всё, что нужно знать о покупке аттестата о среднем образовании без рисков
Kevinrisse
September 2, 2024 @ 2:36 am
[url=https://darwinnunez-cz.biz]https://www.darwinnunez-cz.biz[/url]
last news about darwin nunez
https://www.darwinnunez-cz.biz
ThomasDok
September 2, 2024 @ 2:43 am
[url=https://darwinnunezcz.biz]http://darwinnunezcz.biz[/url]
last news about darwin nunez
https://darwinnunezcz.biz
JamesCreew
September 2, 2024 @ 3:21 am
[url=https://kroos-toni-cz.biz]http://www.kroos-toni-cz.biz[/url]
last news about kroos toni
kroos-toni-cz.biz
Michaelhox
September 2, 2024 @ 3:32 am
[url=https://toni-kroos-cz.biz]https://www.toni-kroos-cz.biz[/url]
last news about toni kroos
http://toni-kroos-cz.biz
Derrickjoisy
September 2, 2024 @ 3:35 am
[url=https://bruno-guimaraes-cz.biz]http://www.bruno-guimaraes-cz.biz[/url]
last news about bruno guimaraes
http://www.bruno-guimaraes-cz.biz
Sazridd
September 2, 2024 @ 3:38 am
Официальная покупка школьного аттестата с упрощенным обучением в Москве
Cazrgvy
September 2, 2024 @ 3:43 am
[u][b] Добрый день![/b][/u]
Купить диплом о высшем образовании
[url=http://altajska.cz/forum/user/2544-ebejesu/]altajska.cz/forum/user/2544-ebejesu[/url]
Darrelnog
September 2, 2024 @ 3:45 am
en iyi slot siteleri: guvenilir slot siteleri 2024 – slot siteleri
kypit JBI_dmKt
September 2, 2024 @ 3:46 am
железобетонные изделия купить [url=http://www.kupit-zhbi.ru]железобетонные изделия купить[/url] .
RobertRer
September 2, 2024 @ 3:55 am
[url=https://brunoguimaraes-cz.biz]http://brunoguimaraes-cz.biz[/url]
last news about bruno guimaraes
brunoguimaraes-cz.biz
Stephennewly
September 2, 2024 @ 4:04 am
[url=https://guimaraesbruno-cz.biz/]guimaraesbruno-cz.biz[/url]
last news about guimaraes bruno
http://guimaraesbruno-cz.biz/
Lazrlld
September 2, 2024 @ 4:08 am
[u][b] Привет, друзья![/b][/u]
[b]Мы готовы предложить дипломы[/b] любой профессии по разумным ценам.
[url=http://michiya-cs.com/userinfo.php?uid=31162/]michiya-cs.com/userinfo.php?uid=31162[/url]
StevenRaica
September 2, 2024 @ 4:13 am
[url=https://tonikrooscz.biz]https://www.tonikrooscz.biz[/url]
last news about toni kroos
http://tonikrooscz.biz
Kevinnut
September 2, 2024 @ 4:33 am
https://slotsiteleri.bid/# en cok kazandiran slot siteleri
Iariornvh
September 2, 2024 @ 4:49 am
[u][b] Здравствуйте![/b][/u]
Приобрести документ ВУЗа можно в нашей компании в столице.
[url=http://uscgq.com/forum/posts.php?forum=general&id=259709/]uscgq.com/forum/posts.php?forum=general&id=259709[/url]
[url=http://pyha.ru/forum/topic/13527.1#msg216474/]pyha.ru/forum/topic/13527.1#msg216474[/url]
[url=http://avtobestnews.ru/vash-put-k-uspehu-diplom-za-dengi/]avtobestnews.ru/vash-put-k-uspehu-diplom-za-dengi[/url]
[url=http://bestcoolfun.ru/oformlenie-diploma-lyuboy-spetsialnosti/]bestcoolfun.ru/oformlenie-diploma-lyuboy-spetsialnosti[/url]
[url=http://dukan.lovelytutorials.com/member.php?u=5648/]dukan.lovelytutorials.com/member.php?u=5648[/url]
Eanrbra
September 2, 2024 @ 5:39 am
Привет, друзья!
Мы готовы предложить дипломы любой профессии по разумным тарифам.
[url=http://ukrevents.ru/page/4/]ukrevents.ru/page/4[/url]
MichaelDar
September 2, 2024 @ 5:50 am
[url=https://kroostoni-cz.biz]https://kroostoni-cz.biz[/url]
last news about kroos toni
https://kroostoni-cz.biz
LarryHaili
September 2, 2024 @ 6:15 am
[url=https://nunez-darwin-cz.biz]http://www.nunez-darwin-cz.biz[/url]
last news about nunez darwin
nunez-darwin-cz.biz
Diplomi_xsSi
September 2, 2024 @ 6:32 am
Привет, друзья!
Заказать документ о получении высшего образования можно в нашей компании.
[url=http://b24activities.ru/club/forum/messages/forum4/topic76/message105/?result=reply#message105/]b24activities.ru/club/forum/messages/forum4/topic76/message105/?result=reply#message105[/url]
Mazrkjy
September 2, 2024 @ 6:33 am
Купить диплом о среднем образовании в Москве и любом другом городе
Douglasbep
September 2, 2024 @ 6:40 am
[url=https://nunezdarwin-cz.biz]https://www.nunezdarwin-cz.biz[/url]
last news about nunez darwin
http://www.nunezdarwin-cz.biz
StevenSok
September 2, 2024 @ 7:38 am
[url=https://mahrezriyad-cz.biz]www.mahrezriyad-cz.biz[/url]
last news about mahrez riyad
mahrezriyad-cz.biz
Sazruxw
September 2, 2024 @ 8:22 am
Приобретение школьного аттестата с официальным упрощенным обучением в Москве
Zaimlinadow
September 2, 2024 @ 9:16 am
Начинай решать свои финансовые вопросы прямо сейчас! В Telegram канале [url=https://t.me/s/zaim_s_plohoy_ki]Микрозаймы по паспорту мгновенно[/url] ты найдёшь срочные займы от 18 лет под выгодную ставку — всего 0.8% в день. Это твоя возможность получить деньги быстро и без лишних хлопот. Просто выбери подходящее предложение, подай заявку и получи деньги на карту. С нами всё просто: минимум требований, максимум возможностей!
Diplomi_vwSi
September 2, 2024 @ 9:27 am
Здравствуйте!
Заказать документ ВУЗа вы можете в нашей компании.
[url=http://reporteam.ru/2017/02/02/запретный-хвост/]reporteam.ru/2017/02/02/запретный-хвост[/url]
Sazrzea
September 2, 2024 @ 11:01 am
Сколько стоит получить диплом высшего и среднего образования легально?
Mazrbwq
September 2, 2024 @ 1:03 pm
Легальная покупка диплома ПТУ с сокращенной программой обучения
Sazrzde
September 2, 2024 @ 1:07 pm
Приобретение школьного аттестата с официальным упрощенным обучением в Москве
Davidhof
September 2, 2024 @ 1:33 pm
[url=https://darwin-nunez-cz.biz]http://www.darwin-nunez-cz.biz[/url]
last news about darwin nunez
darwin-nunez-cz.biz
Sazrume
September 2, 2024 @ 1:50 pm
Как официально купить аттестат 11 класса с упрощенным обучением в Москве
mazda-demio.ru/forums/index.php?autocom=gallery&req=si&img=4951
Robertdup
September 2, 2024 @ 2:58 pm
[url=https://dijk-virgil-van-cz.biz]https://dijk-virgil-van-cz.biz[/url]
last news about dijk virgil van
https://dijk-virgil-van-cz.biz
Williamraics
September 2, 2024 @ 3:00 pm
[u][b] Добрый день![/b][/u]
Мы предлагаем дипломы.
[url=http://vk.com/unikvseru?w=wall414329581_6128/]vk.com/unikvseru?w=wall414329581_6128[/url]
Darrelnog
September 2, 2024 @ 3:12 pm
slot kumar siteleri: slot kumar siteleri – guvenilir slot siteleri
sightcare reviews
September 2, 2024 @ 3:29 pm
Sight Care, with its natural ingredients and effectiveness, is quickly gaining popularity among those looking to improve their vision.
Iarioraeg
September 2, 2024 @ 3:45 pm
[u][b] Здравствуйте![/b][/u]
Приобрести документ ВУЗа вы можете в нашем сервисе.
[url=http://t67747az.beget.tech/2024/07/23/vash-diplom-vash-put-k-karernomu-rostu/]t67747az.beget.tech/2024/07/23/vash-diplom-vash-put-k-karernomu-rostu[/url]
[url=http://t98223u0.beget.tech/2024/07/23/vysokokachestvennye-diplomy-po-dostupnoy-cene/]t98223u0.beget.tech/2024/07/23/vysokokachestvennye-diplomy-po-dostupnoy-cene[/url]
[url=http://true.pahom.su/2024/07/23/vysokokachestvennye-diplomy-po-dostupnoy-cene/]true.pahom.su/2024/07/23/vysokokachestvennye-diplomy-po-dostupnoy-cene[/url]
[url=http://u90517ol.beget.tech/2024/07/23/nadezhnye-diplomy-dlya-vashego-buduschego/]u90517ol.beget.tech/2024/07/23/nadezhnye-diplomy-dlya-vashego-buduschego[/url]
[url=http://w98804oc.beget.tech/2024/07/23/legko-i-bystro-kupite-diplom/]w98804oc.beget.tech/2024/07/23/legko-i-bystro-kupite-diplom[/url]
Jimmyerura
September 2, 2024 @ 3:52 pm
Спасибо за то, что поделились своим опытом в [направление]. Я ценю честные и содержательные советы! Посетите наш сайт для получения дополнительной информации о Доставка авто из Европы под заказ
Jimmyerura
September 2, 2024 @ 3:53 pm
Спасибо, что поделились с нами своим путешествием. Ваши слова всегда так воодушевляют и воодушевляют. Посетите наш сайт для получения дополнительной информации о Купить автомобиль из Америки в Минске
Cazrrdm
September 2, 2024 @ 4:22 pm
Как безопасно купить диплом колледжа или ВУЗа в России, что важно знать
pedolymp.ru/common/articles/kak_legalno_kupit_diplom_v_2023_godu_sovetu_i_rekomendacii
Eanrdwm
September 2, 2024 @ 4:30 pm
Привет!
Мы предлагаем дипломы любой профессии по доступным ценам.
[url=http://ntsr.info/forum/user/92604/]ntsr.info/forum/user/92604[/url]
Darrelnog
September 2, 2024 @ 5:45 pm
yasal slot siteleri: en yeni slot siteleri – slot siteleri guvenilir
Charlesnidly
September 2, 2024 @ 5:49 pm
[url=https://virgil-van-dijk-cz.biz]https://virgil-van-dijk-cz.biz[/url]
last news about virgil van dijk
http://www.virgil-van-dijk-cz.biz
Ремонт Apple Watch
September 2, 2024 @ 6:27 pm
ремонт эппл вотч
ThomasAlmot
September 2, 2024 @ 6:38 pm
vavada вавада регистрация вавада зеркало сайта
Randallpreal
September 2, 2024 @ 6:43 pm
вавада бездепозитные https://pathiaf.com
Ремонт холодильников
September 2, 2024 @ 6:57 pm
Профессиональный сервисный центр по ремонту холодильников и морозильных камер.
Мы предлагаем: ремонт холодильников на дому
Наши мастера оперативно устранят неисправности вашего устройства в сервисе или с выездом на дом!
Micahham
September 2, 2024 @ 8:44 pm
[url=https://van-dijk-virgil-cz.biz]https://van-dijk-virgil-cz.biz[/url]
last news about van dijk virgil
van-dijk-virgil-cz.biz
Lazrfco
September 2, 2024 @ 8:46 pm
[u][b] Добрый день![/b][/u]
[b]Мы можем предложить дипломы[/b] любой профессии по приятным ценам.
[url=http://forum.ll2.ru/member.php?1255487-donnamiduath/]forum.ll2.ru/member.php?1255487-donnamiduath[/url]
Kevinnut
September 2, 2024 @ 10:07 pm
https://sweetbonanza.network/# sweet bonanza free spin demo
Mazrqmr
September 2, 2024 @ 10:26 pm
Полезные советы по безопасной покупке диплома о высшем образовании
Lazroqy
September 3, 2024 @ 12:10 am
[u][b] Здравствуйте![/b][/u]
[b]Мы изготавливаем дипломы[/b] любой профессии по выгодным ценам.
[url=http://mail.inda-car-audio.tw/home.php?mod=space&username=ybahypihu/]mail.inda-car-audio.tw/home.php?mod=space&username=ybahypihu[/url]
ThomasAlmot
September 3, 2024 @ 12:24 am
сайт vavada тг вавады
Randallpreal
September 3, 2024 @ 12:39 am
игра вавада https://pathiaf.com
Kevinnut
September 3, 2024 @ 1:19 am
https://sweetbonanza.network/# sweet bonanza 90 tl
ремонт ноутбуков
September 3, 2024 @ 2:13 am
Профессиональный сервисный центр по ремонту ноутбуков и компьютеров.дронов.
Мы предлагаем:сервисный центр по ремонту ноутбуков в москве
Наши мастера оперативно устранят неисправности вашего устройства в сервисе или с выездом на дом!
сервис центры в петербурге
September 3, 2024 @ 2:28 am
Профессиональный сервисный центр по ремонту бытовой техники с выездом на дом.
Мы предлагаем:ремонт крупногабаритной техники в петрбурге
Наши мастера оперативно устранят неисправности вашего устройства в сервисе или с выездом на дом!
Cazrrnw
September 3, 2024 @ 2:39 am
Пошаговая инструкция по официальной покупке диплома о высшем образовании
vniro.ru/pages/pgs/?gde_kupit_diplom_o_vusshem_obrazovanii_bustro_i_nadezghno
Lazrsen
September 3, 2024 @ 3:17 am
Приобретение диплома ПТУ с сокращенной программой обучения в Москве
Sazrhqa
September 3, 2024 @ 3:47 am
Аттестат 11 класса купить официально с упрощенным обучением в Москве
moneyschoolinc.mn.co/posts/65295058
Mazrodt
September 3, 2024 @ 5:11 am
Приобретение диплома ПТУ с сокращенной программой обучения в Москве
Sazrgip
September 3, 2024 @ 6:09 am
[u][b] Привет, друзья![/b][/u]
Заказать документ института
[url=http://cdposz.ru/club/user/802/forum/message/1489/1548/]cdposz.ru/club/user/802/forum/message/1489/1548[/url]
[url=http://forum.omnicomm.pro/index.php/topic,25758.0/]forum.omnicomm.pro/index.php/topic,25758.0[/url]
[url=http://together-19.com/read-blog/30571/]together-19.com/read-blog/30571[/url]
[url=http://g95334gq.beget.tech/2024/07/21/diplomy-dlya-uspeshnyh-karernyh-izmeneniy/]g95334gq.beget.tech/2024/07/21/diplomy-dlya-uspeshnyh-karernyh-izmeneniy[/url]
[url=http://hebergementweb.org/threads/diplom-ljuboj-specialnosti-za-paru-dnej.1520813/]hebergementweb.org/threads/diplom-ljuboj-specialnosti-za-paru-dnej.1520813[/url]
Sazrvbk
September 3, 2024 @ 6:37 am
Как быстро получить диплом магистра? Легальные способы
shaman-leaders.mn.co/posts/65295826
Darrelnog
September 3, 2024 @ 7:01 am
en yeni slot siteleri: slot bahis siteleri – en guvenilir slot siteleri
Sazrlbq
September 3, 2024 @ 9:07 am
Как оказалось, купить диплом кандидата наук не так уж и сложно
Darrelnog
September 3, 2024 @ 9:24 am
deneme veren slot siteleri: slot kumar siteleri – 2024 en iyi slot siteleri
ремонт ipad в москве
September 3, 2024 @ 9:38 am
Профессиональный сервисный центр по ремонту планетов в том числе Apple iPad.
Мы предлагаем: ipad ремонт москва
Наши мастера оперативно устранят неисправности вашего устройства в сервисе или с выездом на дом!
can someone write my assignment for me
September 3, 2024 @ 10:01 am
If you’re worried about running out of time, can someone write my assignment for me online tool can assist you. It implies that if you complete your tasks in one or two days, this program will help you in doing so accurately and flawlessly in the bare minimum of time. It’s among the greatest methods to make your educational path easy and stress-free. It is among the primary benefits of this tool. For example, you can use this program to solve your issues if you’ve been given complicated assignments quickly and no other choice. We understand that you always find yourself stuck trying to accomplish homework, putting off all of your different interests. It places you in an unstable position where you are unable to devote enough time to pursuing your interests and finding fulfillment in life. The wonderful thing about using an online tool is that you can finish the assignment faster and devote more time to your other interests.
Uazrrfu
September 3, 2024 @ 10:03 am
[u][b]Здравствуйте![/b][/u]
[b]Приобрести диплом любого университета.[/b]
[url=http://telegra.ph/kupit-diplom-magistra-spb-08-13-10/]telegra.ph/kupit-diplom-magistra-spb-08-13-10[/url]
LloydHew
September 3, 2024 @ 1:11 pm
buy balloons dubai balloons price
Ремонт Apple Watch
September 3, 2024 @ 1:33 pm
apple watch ремонт
Sazrmwo
September 3, 2024 @ 1:58 pm
[u][b] Привет![/b][/u]
Купить документ о получении высшего образования
[url=http://sohapay.com/info/vi/help/huong-dan-thanh-toan/fast-i-bank-techcombank/]sohapay.com/info/vi/help/huong-dan-thanh-toan/fast-i-bank-techcombank[/url]
Lazrgtz
September 3, 2024 @ 2:35 pm
Как официально купить диплом вуза с упрощенным обучением в Москве
Mazrgyy
September 3, 2024 @ 3:05 pm
Можно ли быстро купить диплом старого образца и в чем подвох?
Sazrfib
September 3, 2024 @ 3:43 pm
Как не стать жертвой мошенников при покупке диплома о среднем полном образовании
Sazrrdp
September 3, 2024 @ 4:11 pm
Диплом техникума купить официально с упрощенным обучением в Москве
ds-dealer.ru/forum/member.php?u=733126
Sazrnef
September 3, 2024 @ 4:35 pm
[u][b] Привет![/b][/u]
Купить документ о получении высшего образования
[url=http://kozhuhovo.forum2x2.ru/t6439-topic#13366/]kozhuhovo.forum2x2.ru/t6439-topic#13366[/url]
[url=http://ffxiv.one/blogs/192/Быстрый-и-надежный-способ-получить-диплом/]ffxiv.one/blogs/192/Быстрый-и-надежный-способ-получить-диплом[/url]
[url=http://telegra.ph/kak-oplatit-obuchenie-v-vuze-cherez-sberbank-07-24/]telegra.ph/kak-oplatit-obuchenie-v-vuze-cherez-sberbank-07-24[/url]
[url=http://smd.mybb.ru/viewtopic.php?id=4188#p23371/]smd.mybb.ru/viewtopic.php?id=4188#p23371[/url]
[url=http://g95334gq.beget.tech/2024/06/18/pochemu-diplom-trebuetsya-dlya-professionalnogo-rosta/]g95334gq.beget.tech/2024/06/18/pochemu-diplom-trebuetsya-dlya-professionalnogo-rosta[/url]
Lazrnat
September 3, 2024 @ 5:06 pm
[u][b] Привет![/b][/u]
[b]Мы изготавливаем дипломы[/b] любой профессии по приятным ценам.
[url=http://amongus.begandigital.com/diplom-672532qnarl/]amongus.begandigital.com/diplom-672532qnarl[/url]
ремонт дронов
September 3, 2024 @ 5:31 pm
Профессиональный сервисный центр по ремонту радиоуправляемых устройства – квадрокоптеры, дроны, беспилостники в том числе Apple iPad.
Мы предлагаем: ремонт дрона
Наши мастера оперативно устранят неисправности вашего устройства в сервисе или с выездом на дом!
сервис профи
September 3, 2024 @ 5:50 pm
Если вы искали где отремонтировать сломаную технику, обратите внимание – ремонт бытовой техники
сервис профи петербург
September 3, 2024 @ 5:52 pm
Если вы искали где отремонтировать сломаную технику, обратите внимание – сервисный центр в спб
Sazruru
September 3, 2024 @ 7:29 pm
Полезная информация как купить диплом о высшем образовании без рисков
Lazrkro
September 3, 2024 @ 8:30 pm
[u][b] Добрый день![/b][/u]
[b]Мы можем предложить дипломы[/b] любых профессий по приятным тарифам.
[url=http://autogroupe.ru/poluchite-diplom-i-dostignite-svoih-tseley/]autogroupe.ru/poluchite-diplom-i-dostignite-svoih-tseley[/url]
Uazrfis
September 3, 2024 @ 9:09 pm
[u][b]Привет![/b][/u]
[b]Купить диплом университета .[/b]
[url=http://telegra.ph/kupit-diplom-v-novosibirske-08-13-6/]telegra.ph/kupit-diplom-v-novosibirske-08-13-6[/url]
ремонт техники профи в екб
September 3, 2024 @ 9:22 pm
Если вы искали где отремонтировать сломаную технику, обратите внимание – профи екб
Mazrxbq
September 3, 2024 @ 9:36 pm
Приобретение школьного аттестата с официальным упрощенным обучением в Москве
Darrelnog
September 3, 2024 @ 10:03 pm
en iyi slot siteler: guvenilir slot siteleri – guvenilir slot siteleri
Sazrwuy
September 3, 2024 @ 10:23 pm
Быстрая покупка диплома старого образца: возможные риски
Kak zarabotat v internete_cpOa
September 3, 2024 @ 11:57 pm
заработок онлайн [url=www.kak-zarabotat-v-internete12.ru/]заработок онлайн[/url] .
snyatie zapoya na domy_vkMt
September 4, 2024 @ 12:03 am
снятие запоя на дому [url=http://www.snyatie-zapoya-na-domu13.ru]снятие запоя на дому[/url] .
Darrelnog
September 4, 2024 @ 12:25 am
en yeni slot siteleri: en cok kazandiran slot siteleri – guvenilir slot siteleri
Cazrcmk
September 4, 2024 @ 1:27 am
[u][b] Привет, друзья![/b][/u]
Получить диплом о высшем образовании
[url=http://pawsarl.es/blog/амрах/]купить диплом в бердске[/url]
Sazrjtz
September 4, 2024 @ 1:55 am
[u][b] Добрый день![/b][/u]
Приобрести документ института
[url=http://mail.bike.by/forum/viewtopic.php?f=81&t=27279/]mail.bike.by/forum/viewtopic.php?f=81&t=27279[/url]
Sazrgzs
September 4, 2024 @ 2:23 am
Официальная покупка диплома вуза с сокращенной программой обучения в Москве
Sazrloc
September 4, 2024 @ 3:41 am
Всё, что нужно знать о покупке аттестата о среднем образовании без рисков
k-grant.com/bbs/bbs2.cgi?mode=res&reno=91531
JerryEffon
September 4, 2024 @ 5:18 am
[url=][/url]
Интернет-магазин климатической техники «СКСэйл»
Канальные, кассетные кондиционеры и сплит-системы, а также все, что вы хотите знать о кондиционерах, есть на нашем сайте. Подробно рассказано об оконных и канальных кондиционерах, тепловых завесах, системах вентиляции, очистителях, увлажнителях и осушителях воздуха. Цены на продукцию можно посмотреть в каталоге.
Продажа, монтаж и установка кондиционеров — это наша работа, которую мы выполняем профессионально и с огромным удовольствием. Мы предлагаем только лучшее оборудование, которое хорошо зарекомендовало себя в России и признанно во всем мире.
Мы продаём не только бытовые сплит системы, но и колонные конционеры, канальные кондиционеры, потолочные и центральные системы кондиционирования.
Центральные кондиционеры отлично подойдут для средних и больших загородных коттеджей, поскольку внешний блок будет только один. Установка кондиционеров любого типа производится нами в максимально сжатые сроки.
Официальный сайт «СКСэйл».
[url=][/url]
pay someone to take my online english class
September 4, 2024 @ 5:47 am
Nursing degrees have a large number of assignments, tests, and assessments that put a lot of mental pressure on online nursing students. You can easily pay someone to do my online nursing class and remove all the stress and pressure from it. Let our experts handle your nursing degree while you relax while we provide solutions to your do my online nursing class request. With our help for ‘ do my online nursing class for me’ questions, you can achieve good grades in online nursing classes. It is our goal to improve the grades and academic performance of nursing students at the BSN, MSN, and DNP levels. Are you considering paying someone to take my class online? Our class-takers can assist you in showing the best academic performance. Let our intellectuals assist you with all your nursing courses. We guarantee academic excellence with our online class assistance services.
WarrenByday
September 4, 2024 @ 6:30 am
http://pin-up.diy/# пин ап казино
Расстановки Хеллингер
September 4, 2024 @ 7:49 am
Семейные расстановки и шаманизм. https://rasstanovkiural.ru
WarrenByday
September 4, 2024 @ 7:54 am
https://pin-up.diy/# пинап казино
Scottfup
September 4, 2024 @ 8:00 am
вавада: vavada online casino – вавада рабочее зеркало
Scottger
September 4, 2024 @ 8:53 am
pin up: pin up казино – пин ап вход
Sazrtyo
September 4, 2024 @ 9:42 am
Пошаговая инструкция по безопасной покупке диплома о высшем образовании
сервис профи
September 4, 2024 @ 9:56 am
Если вы искали где отремонтировать сломаную технику, обратите внимание – тех профи
Oariorqqd
September 4, 2024 @ 10:27 am
[u][b] Привет![/b][/u]
[b]Где заказать диплом по актуальной специальности?[/b]
[url=http://thesleepinghusband.rolka.me/viewtopic.php?id=22081#p53887/]thesleepinghusband.rolka.me/viewtopic.php?id=22081#p53887[/url]
[url=http://markusragger.at/chess/index.php/kforum/jm-news-pro-module/651333/]markusragger.at/chess/index.php/kforum/jm-news-pro-module/651333[/url]
[url=http://soad.msk.ru/forum/index.php?sid=762e4b94a9d4a2c9734fb98e4b8842ae/]soad.msk.ru/forum/index.php?sid=762e4b94a9d4a2c9734fb98e4b8842ae[/url]
[url=http://chayka.ixbb.ru/viewtopic.php?id=138#p148/]chayka.ixbb.ru/viewtopic.php?id=138#p148[/url]
[url=http://n-staff.ru/forum?mode=edit&type=thread&forum_id=74041/]n-staff.ru/forum?mode=edit&type=thread&forum_id=74041[/url]
Iariorprx
September 4, 2024 @ 10:28 am
[u][b] Здравствуйте![/b][/u]
Приобрести документ о получении высшего образования вы сможете в нашей компании в столице.
[url=http://b-stringer.ru/cat/publikacii/universalnoe/]b-stringer.ru/cat/publikacii/universalnoe[/url]
[url=http://club.sabaylok.com/blogs/5791/Купите-диплом-быстро-и-легко/]club.sabaylok.com/blogs/5791/Купите-диплом-быстро-и-легко[/url]
[url=http://dailygram.com/blog/1307740/купите-диплом-быстро-и-легко/]dailygram.com/blog/1307740/купите-диплом-быстро-и-легко[/url]
[url=http://epistles.ru/blogs/1719/Купите-диплом-быстро-и-легко/]epistles.ru/blogs/1719/Купите-диплом-быстро-и-легко[/url]
[url=http://faithstreamer.com/blogs/3347/Купите-диплом-быстро-и-легко/]faithstreamer.com/blogs/3347/Купите-диплом-быстро-и-легко[/url]
ремонт айпадов в москве
September 4, 2024 @ 12:22 pm
Профессиональный сервисный центр по ремонту планетов в том числе Apple iPad.
Мы предлагаем: срочный ремонт ipad
Наши мастера оперативно устранят неисправности вашего устройства в сервисе или с выездом на дом!
Lazrrgk
September 4, 2024 @ 12:22 pm
[u][b] Добрый день![/b][/u]
[b]Мы изготавливаем дипломы[/b] психологов, юристов, экономистов и любых других профессий по приятным ценам.
[url=http://littleyaksa.yodev.net/bbs/board.php?bo_table=free&wr_id=4338574&c_id=4491619&w=c/]littleyaksa.yodev.net/bbs/board.php?bo_table=free&wr_id=4338574&c_id=4491619&w=c[/url]
Scottfup
September 4, 2024 @ 12:46 pm
vavada зеркало: вавада зеркало – вавада казино
Williamraics
September 4, 2024 @ 12:53 pm
[u][b] Привет, друзья![/b][/u]
Мы изготавливаем дипломы.
[url=http://army.clanfm.ru/viewtopic.php?f=2&t=35340/]army.clanfm.ru/viewtopic.php?f=2&t=35340[/url]
ремонт бытовой техники в спб
September 4, 2024 @ 1:27 pm
Если вы искали где отремонтировать сломаную технику, обратите внимание – профи ремонт
Sazrcdu
September 4, 2024 @ 2:03 pm
[u][b] Привет![/b][/u]
Мы изготавливаем [b]дипломы[/b] любой профессии.
Заказ [b]документа[/b], подтверждающего обучение в университете, – это выгодное решение.
[url=http://kroosuriya.com/krusuriya/modules.php?name=Journal&file=display&jid=10848/]kroosuriya.com/krusuriya/modules.php?name=Journal&file=display&jid=10848[/url]
[url=http://forum.stpku.ru/viewtopic.php?f=2&t=79483/]forum.stpku.ru/viewtopic.php?f=2&t=79483[/url]
[url=http://laflore.ru/sale/urinastop#comment-213696/]laflore.ru/sale/urinastop#comment-213696[/url]
[url=http://videopro.ru/xenforo/index.php?members/chriswhjak.1544/]videopro.ru/xenforo/index.php?members/chriswhjak.1544[/url]
[url=http://lada-xray.net/showthread.php?p=32970#post32970/]lada-xray.net/showthread.php?p=32970#post32970[/url]
ремонт квадрокоптеров москве
September 4, 2024 @ 2:04 pm
Профессиональный сервисный центр по ремонту радиоуправляемых устройства – квадрокоптеры, дроны, беспилостники в том числе Apple iPad.
Мы предлагаем: ремонт квадрокоптеров москва
Наши мастера оперативно устранят неисправности вашего устройства в сервисе или с выездом на дом!
Lazrigt
September 4, 2024 @ 3:51 pm
[u][b] Здравствуйте![/b][/u]
[b]Мы предлагаем дипломы[/b] любой профессии по доступным тарифам.
[url=http://make-coin.ru/vash-diplom-vash-uspeh/]make-coin.ru/vash-diplom-vash-uspeh[/url]
vivod iz zapoya v stacionare_mjol
September 4, 2024 @ 5:45 pm
вывод из запоя самара стационар [url=www.vyvod-iz-zapoya-v-stacionare-samara.ru]вывод из запоя самара стационар[/url] .
Scottfup
September 4, 2024 @ 6:15 pm
pin up: пин ап казино – pin up
Douglasjam
September 4, 2024 @ 6:29 pm
Fonbet промокод на фрибет при регистрации https://kmural.ru/news_importer/inc/aktualnue_promokodu_bukmekerskoy_kontoru_fonbet.html
Промокод на фрибет при регистрации на Fonbet предоставляет новым пользователям бесплатные ставки. Примером такого промокода является ‘GIFT200’, который активирует фрибеты для новых игроков. Ввод этого промокода в специальное поле при регистрации позволяет сделать ставки без использования собственных средств, увеличивая шансы на выигрыш.
Cazrgcg
September 4, 2024 @ 7:07 pm
Реально ли приобрести диплом стоматолога? Основные этапы
apteka.podolsk.ru/dis/pgs/gde_kupit_diplom_s_vkladushem_bustro_i_nadezghno
LeslieFon
September 4, 2024 @ 7:51 pm
вавада казино [url=https://vavada.auction/#]vavada казино[/url] вавада зеркало
MarioCig
September 4, 2024 @ 8:08 pm
комплексное продвижение сайта seo заказать продвижение сайтов в москве
Scottger
September 4, 2024 @ 8:12 pm
пин ап казино вход: пин ап вход – пин ап казино
Oarioryeh
September 4, 2024 @ 8:14 pm
[u][b] Привет, друзья![/b][/u]
[b]Где купить диплом по необходимой специальности?[/b]
[url=http://cittaviva.net/read-blog/89/]cittaviva.net/read-blog/89[/url]
[url=http://followingbook.com/create-page/]followingbook.com/create-page[/url]
[url=http://gerosland.com/read-blog/200/]gerosland.com/read-blog/200[/url]
[url=http://itstagram.ru/read-blog/970/]itstagram.ru/read-blog/970[/url]
[url=http://local-urban-eats.mn.co/posts/62883286/]local-urban-eats.mn.co/posts/62883286[/url]
Iariorspu
September 4, 2024 @ 8:27 pm
[u][b] Привет, друзья![/b][/u]
Приобрести документ о получении высшего образования вы имеете возможность в нашем сервисе.
[url=http://telegra.ph/do-kakogo-chisla-mozhno-oplatit-obuchenie-v-vuze-07-24/]telegra.ph/do-kakogo-chisla-mozhno-oplatit-obuchenie-v-vuze-07-24[/url]
[url=http://telegra.ph/dogovor-na-obuchenie-v-vuze-07-24/]telegra.ph/dogovor-na-obuchenie-v-vuze-07-24[/url]
[url=http://telegra.ph/dogovor-ob-obuchenii-v-vuze-07-24/]telegra.ph/dogovor-ob-obuchenii-v-vuze-07-24[/url]
[url=http://telegra.ph/dokumenty-na-vozvrat-naloga-za-obuchenie-v-vuze-07-24/]telegra.ph/dokumenty-na-vozvrat-naloga-za-obuchenie-v-vuze-07-24[/url]
[url=http://telegra.ph/zakazat-spravku-ob-obuchenii-v-vuze-07-24/]telegra.ph/zakazat-spravku-ob-obuchenii-v-vuze-07-24[/url]
WarrenByday
September 4, 2024 @ 9:17 pm
https://vavada.auction/# vavada зеркало
сервис профи екатеринбург
September 4, 2024 @ 9:51 pm
Если вы искали где отремонтировать сломаную технику, обратите внимание – профи ремонт
Scottger
September 4, 2024 @ 10:04 pm
пин ап зеркало: пинап казино – pin up
Kak zarabotat v internete_knml
September 4, 2024 @ 10:27 pm
как начать зарабатывать в интернете [url=www.kak-zarabotat-v-internete11.ru/]как начать зарабатывать в интернете[/url] .
Eanrrbx
September 4, 2024 @ 10:55 pm
Добрый день!
Мы изготавливаем дипломы любой профессии по разумным ценам.
[url=http://textualheritage.org/en/the-materials-of-the-conference-el-manusctipt-2014/82/]textualheritage.org/en/the-materials-of-the-conference-el-manusctipt-2014/82[/url]
Scottfup
September 4, 2024 @ 11:10 pm
вавада: vavada online casino – вавада казино
Williamraics
September 4, 2024 @ 11:11 pm
[u][b] Привет![/b][/u]
Мы изготавливаем дипломы.
[url=http://raznoe.rolka.me/viewtopic.php?id=5186#p15229/]raznoe.rolka.me/viewtopic.php?id=5186#p15229[/url]
Sazrarw
September 5, 2024 @ 12:13 am
[u][b] Привет, друзья![/b][/u]
Мы можем предложить [b]дипломы[/b] любых профессий.
Приобретение [b]документа[/b], подтверждающего обучение в университете, – это грамотное решение.
[url=http://выкуп-ноутбуков.рф/user/ellpetrovz2489/]выкуп-ноутбуков.рф/user/ellpetrovz2489[/url]
[url=http://zamok.druzya.org/index.php?/register/]zamok.druzya.org/index.php?/register[/url]
[url=http://sweetburg.ru/course/paket-reczepturnyh-kart-na-mussovye-torty-koktejlnaya-vecherinka/#comment-5806/]sweetburg.ru/course/paket-reczepturnyh-kart-na-mussovye-torty-koktejlnaya-vecherinka/#comment-5806[/url]
[url=http://автосервис-ковров.рф/index.php?subaction=userinfo&user=inerevyq/]автосервис-ковров.рф/index.php?subaction=userinfo&user=inerevyq[/url]
[url=http://myskupera.ru/forum/messages/forum1/topic12/message116249/?result=reply#message116249/]myskupera.ru/forum/messages/forum1/topic12/message116249/?result=reply#message116249[/url]
Sazruvr
September 5, 2024 @ 12:22 am
Приобретение школьного аттестата с официальным упрощенным обучением в Москве
santa4.su/gallery/image/349-nuzhno-priobresti-diplom-posetite-luchshiy-onlayn-magazin
MarioCig
September 5, 2024 @ 1:37 am
комплексное продвижение в интернете в москве продвижение сайта москва
instagram viewer_pwsi
September 5, 2024 @ 2:41 am
view instagram stories anonymously [url=inviewanon.com]view instagram stories anonymously[/url] .
vivod iz zapoya v stacionare_kzKi
September 5, 2024 @ 2:54 am
вывод из запоя в стационаре самара [url=https://vyvod-iz-zapoya-v-stacionare-samara11.ru]https://vyvod-iz-zapoya-v-stacionare-samara11.ru[/url] .
Sazrwqd
September 5, 2024 @ 3:17 am
Полезная информация как купить диплом о высшем образовании без рисков
extinct-networks.mn.co/posts/65295853
Scottfup
September 5, 2024 @ 4:41 am
1xbet зеркало рабочее на сегодня: 1xbet официальный сайт – 1xbet зеркало
Cazrczq
September 5, 2024 @ 4:47 am
Официальная покупка диплома вуза с упрощенной программой обучения
sbmk.org/css/pgs/kupit_diplom_universiteta__gde_nayti_i_na_chto_obratit_vnimanie
EliseoMuG
September 5, 2024 @ 6:00 am
[url=][/url]
Многие из нас с волнением и беспокойством идут на прием к врачу акушеру-гинекологу, часто откладывая посещение.
А ведь к женскому врачу приходится обращаться в течение всей жизни, особенно в такие важные периоды, как беременность и роды.
Естественно, что каждая женщина хочет чувствовать себя уверенной и защищенной, когда речь идет о здоровье.
Понимая ваше волнение, чувствуя ответственность за вашу жизнь и жизнь ваших детей, был создан ознакомительный сайт «Женский Доктор».
На сайте будут освещены основные темы в области акушерства и гинекологии в доступной широкому кругу читателей форме.
По мере поступления откликов, будут выходить новые статьи, чтобы вы смогли найти ответы на волнующие вас вопросы.
Зная, как важно общение с доктором на родном языке, я буду рада помочь вам в моем частном кабинете расположенном рядом с клиникой Medical Park,
Турция, где вы найдете квалифицированную помощь специалиста акушера-гинеколога, а будущие мамы смогут в полной мере подготовиться к родам, оставив позади свои тревоги.
Мой сайт rusginekolog.com
Частный кабинет, расположенный в центре Европейской части г. Стамбул, у автотрассы E-5 и в 5 минутах езды от аэропорта Ататюрк.
Кабинет No 1009, 10 этаж.
Bahcelievler mah., E-5 Yanyol No 14/B Metroport Busidence /Bahcelievler/Istanbul
[url=][/url]
ремонт телевизора
September 5, 2024 @ 6:51 am
ремонт телевизоров в москве
Lazrzox
September 5, 2024 @ 7:04 am
[u][b] Привет, друзья![/b][/u]
[b]Мы изготавливаем дипломы[/b] любых профессий по приятным ценам.
[url=http://occ24.ru/preimushhestva-pokupki-diploma-3/]occ24.ru/preimushhestva-pokupki-diploma-3[/url]
сервис профи
September 5, 2024 @ 7:31 am
Если вы искали где отремонтировать сломаную технику, обратите внимание – профи тех сервис москва
Sazrtrh
September 5, 2024 @ 8:05 am
[u][b] Привет![/b][/u]
Заказать документ института
[url=http://ya.0bb.ru/viewtopic.php?id=12136#p32290/]ya.0bb.ru/viewtopic.php?id=12136#p32290[/url]
[url=http://probox-club.ru/forums/index.php?autocom=gallery&req=si&img=5100/]probox-club.ru/forums/index.php?autocom=gallery&req=si&img=5100[/url]
[url=http://telegra.ph/lgoty-po-potere-kormilca-pri-postuplenii-v-vuz-2018-07-24/]telegra.ph/lgoty-po-potere-kormilca-pri-postuplenii-v-vuz-2018-07-24[/url]
[url=http://master39.net/forum/user/5288/]master39.net/forum/user/5288[/url]
[url=http://telegra.ph/igrovye-metody-obucheniya-v-vuze-07-24/]telegra.ph/igrovye-metody-obucheniya-v-vuze-07-24[/url]
Lazrbre
September 5, 2024 @ 8:30 am
Официальная покупка диплома вуза с сокращенной программой обучения в Москве
ремонт ноутбуков
September 5, 2024 @ 8:34 am
Профессиональный сервисный центр по ремонту ноутбуков и компьютеров.дронов.
Мы предлагаем:ноутбук ремонт сервисный
Наши мастера оперативно устранят неисправности вашего устройства в сервисе или с выездом на дом!
ремонт бытовой техники в петербурге
September 5, 2024 @ 8:58 am
Профессиональный сервисный центр по ремонту бытовой техники с выездом на дом.
Мы предлагаем:сервисные центры по ремонту техники в спб
Наши мастера оперативно устранят неисправности вашего устройства в сервисе или с выездом на дом!
Scottger
September 5, 2024 @ 9:01 am
пин ап зеркало: пин ап – pin up casino
Scottfup
September 5, 2024 @ 9:32 am
ван вин: ван вин – 1win официальный сайт
Lazrpds
September 5, 2024 @ 10:39 am
[u][b] Здравствуйте![/b][/u]
[b]Мы изготавливаем дипломы[/b] любой профессии по приятным ценам.
[url=http://vezuviy.my1.ru/load/14/]vezuviy.my1.ru/load/14[/url]
Scottger
September 5, 2024 @ 11:01 am
пин ап казино вход: пин ап казино – pin up
Sazrejp
September 5, 2024 @ 11:34 am
Аттестат школы купить официально с упрощенным обучением в Москве
nine casino app
September 5, 2024 @ 12:00 pm
Se stai cercando un’esperienza di gioco emozionante e sicura, ninecasino e la scelta giusta per te. Con un’interfaccia user-friendly e un login semplice, Nine Casino offre un’ampia gamma di giochi che soddisferanno tutti i gusti. Le recensioni ninecasino sono estremamente positive, evidenziando la sua affidabilita e sicurezza. Molti giocatori apprezzano le nine casino prelievo, che sono rapide e sicure.
Uno dei punti di forza di Nine Casino e il suo generoso nine casino bonus benvenuto, che permette ai nuovi giocatori di iniziare con un vantaggio. Inoltre, puoi ottenere giri gratuiti e altri premi grazie ai bonus senza deposito. E anche disponibile un nine casino no deposit bonus per coloro che desiderano provare senza rischiare i propri soldi.
Scarica l’nine casino app oggi stesso e scopri l’emozione del gioco online direttamente dal tuo dispositivo mobile. Il download dell’app di Nine Casino e semplice e veloce, permettendoti di giocare ovunque ti trovi. Molti si chiedono, “nine casino e sicuro?” La risposta e si: ninecasino e completamente legale in Italia e garantisce un ambiente di gioco sicuro e regolamentato. Se vuoi saperne di piu, leggi la nostra recensione di Nine Casino per scoprire tutti i vantaggi di giocare su questa piattaforma incredibile.
ninecasino [url=https://casinonine-bonus.com/]https://casinonine-bonus.com/[/url] .
LloydMem
September 5, 2024 @ 12:03 pm
sample resume of an engineer cv format for engineers
WarrenByday
September 5, 2024 @ 12:46 pm
http://vavada.auction/# вавада рабочее зеркало
ремонт бытовой техники новосибирск
September 5, 2024 @ 12:54 pm
Если вы искали где отремонтировать сломаную технику, обратите внимание – сервисный центр в новосибирск
Ремонт iPhone
September 5, 2024 @ 12:59 pm
Профессиональный сервисный центр по ремонту Apple iPhone в Москве.
Мы предлагаем: ремонт iphone на дому в москве
Наши мастера оперативно устранят неисправности вашего устройства в сервисе или с выездом на дом!
Ремонт сотовых телефонов в Москве
September 5, 2024 @ 1:03 pm
ремонт телефонов в москве адреса
Ремонт ИБП в Москве
September 5, 2024 @ 1:16 pm
Профессиональный сервисный центр по ремонту источников бесперебойного питания.
Мы предлагаем: сервис ибп
Наши мастера оперативно устранят неисправности вашего устройства в сервисе или с выездом на дом!
WarrenByday
September 5, 2024 @ 2:14 pm
https://pin-up.diy/# pin up
Uazrggk
September 5, 2024 @ 2:48 pm
[u][b]Привет![/b][/u]
[b]Заказать диплом ВУЗа .[/b]
[url=http://telegra.ph/kupit-diplom-o-vysshem-obrazovanii-s-zaneseniem-v-reestr-moskva-08-13-7/]telegra.ph/kupit-diplom-o-vysshem-obrazovanii-s-zaneseniem-v-reestr-moskva-08-13-7[/url]
Scottfup
September 5, 2024 @ 2:57 pm
1xbet зеркало рабочее на сегодня: 1xbet официальный сайт – 1хбет
Sazrcjb
September 5, 2024 @ 5:25 pm
Пошаговая инструкция по официальной покупке диплома о высшем образовании
ingprint.ru/club/user/58725/forum/message/7206/203580
nine casino app download
September 5, 2024 @ 5:26 pm
Se stai cercando un’esperienza di gioco emozionante e sicura, ninecasino e la scelta giusta per te. Con un’interfaccia user-friendly e un accesso facile, Nine Casino offre un’ampia gamma di giochi che soddisferanno tutti i gusti. Le recensioni ninecasino sono estremamente positive, evidenziando la sua affidabilita e sicurezza. Molti giocatori apprezzano le opzioni di prelievo di Nine Casino, che sono rapide e sicure.
Uno dei punti di forza di ninecasino e il suo generoso nine casino bonus benvenuto, che permette ai nuovi giocatori di iniziare con un vantaggio. Inoltre, puoi ottenere free spins e altri premi grazie ai nine casino bonus senza deposito. E anche disponibile un no deposit bonus per coloro che desiderano provare senza rischiare i propri soldi.
Scarica l’app di Nine Casino oggi stesso e scopri l’emozione del gioco online direttamente dal tuo dispositivo mobile. Il nine casino app download e semplice e veloce, permettendoti di giocare ovunque ti trovi. Molti si chiedono, “nine casino e sicuro?” La risposta e si: Nine Casino e completamente legale in Italia e garantisce un ambiente di gioco sicuro e regolamentato. Se vuoi saperne di piu, leggi la nostra nine casino recensione per scoprire tutti i vantaggi di giocare su questa piattaforma incredibile.
nine casino bonus senza deposito [url=https://casinonine-it.com/]https://casinonine-it.com/[/url] .
Sazrosz
September 5, 2024 @ 6:04 pm
[u][b] Здравствуйте![/b][/u]
Заказать документ института
[url=http://telegra.ph/v-kakih-voennyh-vuzah-est-zaochnoe-obuchenie-07-24-2/]telegra.ph/v-kakih-voennyh-vuzah-est-zaochnoe-obuchenie-07-24-2[/url]
[url=http://mindclub.online/wiki/index.php/Купите_диплом_и_начните_новую_жизнь/]mindclub.online/wiki/index.php/Купите_диплом_и_начните_новую_жизнь[/url]
[url=http://rrsclub.ru/member.php?u=1164/]rrsclub.ru/member.php?u=1164[/url]
[url=http://flerus-shop.hcp.dilhost.ru/club/user/4/forum/message/938/934/#message934/]flerus-shop.hcp.dilhost.ru/club/user/4/forum/message/938/934/#message934[/url]
[url=http://cslregister.com/forum/member.php?u=6225/]cslregister.com/forum/member.php?u=6225[/url]
Lazrphv
September 5, 2024 @ 6:55 pm
Приобретение школьного аттестата с официальным упрощенным обучением в Москве
Sazrwzi
September 5, 2024 @ 7:38 pm
Как приобрести аттестат о среднем образовании в Москве и других городах
Scottfup
September 5, 2024 @ 7:41 pm
1вин зеркало: 1вин официальный сайт – 1win вход
Sazrrin
September 5, 2024 @ 7:45 pm
[u][b] Добрый день![/b][/u]
Купить документ университета
[url=http://google.com.vc/url?q=aurus-diploms.com/]google.com.vc/url?q=aurus-diploms.com[/url]
Sazrwsi
September 5, 2024 @ 8:23 pm
Узнайте стоимость диплома высшего и среднего образования и процесс получения
infobidz.fun/read-blog/535
Sazrdkx
September 5, 2024 @ 9:15 pm
Узнайте, как приобрести диплом о высшем образовании без рисков
Sazrkxj
September 5, 2024 @ 9:34 pm
Как правильно приобрести диплом колледжа или ПТУ в России, важные моменты
ninecasino
September 5, 2024 @ 10:49 pm
Se stai cercando un’esperienza di gioco emozionante e sicura, ninecasino e la scelta giusta per te. Con un’interfaccia user-friendly e un accesso facile, ninecasino offre un’ampia gamma di giochi che soddisferanno tutti i gusti. Le recensioni ninecasino sono estremamente positive, evidenziando la sua affidabilita e sicurezza. Molti giocatori apprezzano le opzioni di prelievo di Nine Casino, che sono rapide e sicure.
Uno dei punti di forza di ninecasino e il suo generoso bonus di benvenuto, che permette ai nuovi giocatori di iniziare con un vantaggio. Inoltre, puoi ottenere giri gratuiti e altri premi grazie ai bonus senza deposito. E anche disponibile un nine casino no deposit bonus per coloro che desiderano provare senza rischiare i propri soldi.
Scarica l’app di Nine Casino oggi stesso e scopri l’emozione del gioco online direttamente dal tuo dispositivo mobile. Il download dell’app di Nine Casino e semplice e veloce, permettendoti di giocare ovunque ti trovi. Molti si chiedono, “nine casino e sicuro?” La risposta e si: Nine Casino e completamente legale in Italia e garantisce un ambiente di gioco sicuro e regolamentato. Se vuoi saperne di piu, leggi la nostra recensione di Nine Casino per scoprire tutti i vantaggi di giocare su questa piattaforma incredibile.
nine casino [url=https://nine-casino-italia.com/]https://nine-casino-italia.com/[/url] .
Scottfup
September 6, 2024 @ 12:32 am
казино вавада: vavada зеркало – вавада
Uazrefu
September 6, 2024 @ 12:58 am
[u][b]Здравствуйте![/b][/u]
[b]Приобрести диплом любого университета.[/b]
[url=http://telegra.ph/kupit-diplom-chita-08-13-7/]telegra.ph/kupit-diplom-chita-08-13-7[/url]
Lazruug
September 6, 2024 @ 1:20 am
[u][b] Привет![/b][/u]
[b]Мы изготавливаем дипломы[/b] психологов, юристов, экономистов и любых других профессий по невысоким тарифам.
[url=http://qigong.com/diplom-347964mhaon/]qigong.com/diplom-347964mhaon[/url]
KyleSot
September 6, 2024 @ 2:14 am
Букмекерская контора «Мелбет» – это уникальная возможность для всех желающих получить дополнительный доход. Здесь можно делать ставки на самые разные спортивные события по всему миру. Высокие коэффициенты, круглосуточная техническая поддержка, удобное мобильное приложение и многие другие преимущества проекта делают его весьма популярным среди любителей азартных игр. Доступ на виртуальную площадку открыт лишь для зарегистрированных пользователей. Melbet промокод бонус помогут сэкономить как начинающим, так и опытным игрокам. В такой надежной и стабильной букмекерской конторе вы всегда можете быть уверены.
WarrenByday
September 6, 2024 @ 3:21 am
http://1win.directory/# 1вин сайт
Sazrkji
September 6, 2024 @ 3:39 am
Купить диплом магистра оказалось возможно, быстрое обучение и диплом на руки
nine casino e legale in italia
September 6, 2024 @ 4:12 am
Se stai cercando un’esperienza di gioco emozionante e sicura, ninecasino e la scelta giusta per te. Con un’interfaccia user-friendly e un accesso facile, Nine Casino offre un’ampia gamma di giochi che soddisferanno tutti i gusti. Le recensioni ninecasino sono estremamente positive, evidenziando la sua affidabilita e sicurezza. Molti giocatori apprezzano le nine casino prelievo, che sono rapide e sicure.
Uno dei punti di forza di ninecasino e il suo generoso bonus di benvenuto, che permette ai nuovi giocatori di iniziare con un vantaggio. Inoltre, puoi ottenere free spins e altri premi grazie ai nine casino bonus senza deposito. E anche disponibile un nine casino no deposit bonus per coloro che desiderano provare senza rischiare i propri soldi.
Scarica l’app di Nine Casino oggi stesso e scopri l’emozione del gioco online direttamente dal tuo dispositivo mobile. Il download dell’app di Nine Casino e semplice e veloce, permettendoti di giocare ovunque ti trovi. Molti si chiedono, “nine casino e sicuro?” La risposta e si: Nine Casino e completamente legale in Italia e garantisce un ambiente di gioco sicuro e regolamentato. Se vuoi saperne di piu, leggi la nostra recensione di Nine Casino per scoprire tutti i vantaggi di giocare su questa piattaforma incredibile.
nine casino free spins [url=https://nine-casino-italy.com/]https://nine-casino-italy.com/[/url] .
LloydMem
September 6, 2024 @ 5:06 am
professional engineer resume format https://resume-engineering-builder.com
Scottfup
September 6, 2024 @ 5:11 am
казино вавада: vavada зеркало – вавада рабочее зеркало
Cazrpnr
September 6, 2024 @ 6:16 am
[u][b] Привет, друзья![/b][/u]
Заказать диплом академии
[url=http://yartube.ru/stati-v/456-nakrutka-vk-kak-mozhno-prodvinut-gruppu-vkontakte/]купить диплом юриста[/url]
Lazrhdq
September 6, 2024 @ 6:52 am
[u][b] Здравствуйте![/b][/u]
[b]Мы изготавливаем дипломы[/b] любой профессии по разумным тарифам.
[url=http://superdiplomnik.ru/]superdiplomnik.ru[/url]
Sazrhsd
September 6, 2024 @ 7:27 am
[u][b] Привет![/b][/u]
Приобрести документ ВУЗа
[url=http://lasfloresboutique.ru/product-page/букет-из-роз-и-астральмерии/]lasfloresboutique.ru/product-page/букет-из-роз-и-астральмерии[/url]
сервис профи барнаул
September 6, 2024 @ 7:32 am
Если вы искали где отремонтировать сломаную технику, обратите внимание – ремонт бытовой техники
Vivod iz zapoya v sankt peterbyrge_acka
September 6, 2024 @ 8:28 am
выведение из запоя санкт петербург [url=http://vyvod-iz-zapoya-v-sankt-peterburge.ru/]http://vyvod-iz-zapoya-v-sankt-peterburge.ru/[/url] .
Snyatie lomki narkolog_mzOn
September 6, 2024 @ 8:30 am
снятие ломки на дому цена [url=http://snyatie-lomki-narkolog.ru]http://snyatie-lomki-narkolog.ru[/url] .
ремонт техники профи в новосибирск
September 6, 2024 @ 9:31 am
Если вы искали где отремонтировать сломаную технику, обратите внимание – ремонт бытовой техники
сервис профи челябинск
September 6, 2024 @ 9:39 am
Если вы искали где отремонтировать сломаную технику, обратите внимание – техпрофи
Sazrwfa
September 6, 2024 @ 9:54 am
Пошаговая инструкция по безопасной покупке диплома о высшем образовании
Ремонт iPhone
September 6, 2024 @ 10:23 am
Профессиональный сервисный центр по ремонту Apple iPhone в Москве.
Мы предлагаем: качественный ремонт айфонов в москве
Наши мастера оперативно устранят неисправности вашего устройства в сервисе или с выездом на дом!
Scottfup
September 6, 2024 @ 10:25 am
pin up: пин ап вход – pin up casino
slot gacor
September 6, 2024 @ 10:28 am
I was extremely pleased to find this site. I want to to
thank you for ones time just for this fantastic read!!
I definitely liked every part of it and i also have you saved as a favorite to check out new things in your website.
Ремонт ИБП в Москве
September 6, 2024 @ 11:11 am
Профессиональный сервисный центр по ремонту источников бесперебойного питания.
Мы предлагаем: ремонт плат ибп
Наши мастера оперативно устранят неисправности вашего устройства в сервисе или с выездом на дом!
Oariordzv
September 6, 2024 @ 1:18 pm
[u][b] Здравствуйте![/b][/u]
[b]Где заказать диплом специалиста?[/b]
[url=http://telegra.ph/lgoty-detyam-voennosluzhashchih-pri-postuplenii-v-vuz-v-uzbekistane-07-24/]telegra.ph/lgoty-detyam-voennosluzhashchih-pri-postuplenii-v-vuz-v-uzbekistane-07-24[/url]
[url=http://telegra.ph/lgoty-detyam-iz-mnogodetnyh-semej-pri-postuplenii-v-vuz-ukraina-07-24/]telegra.ph/lgoty-detyam-iz-mnogodetnyh-semej-pri-postuplenii-v-vuz-ukraina-07-24[/url]
[url=http://telegra.ph/minimalnye-bally-dlya-postupleniya-v-vuz-2017-07-24/]telegra.ph/minimalnye-bally-dlya-postupleniya-v-vuz-2017-07-24[/url]
[url=http://telegra.ph/molitva-o-postuplenii-syna-v-vuz-na-byudzhet-07-24/]telegra.ph/molitva-o-postuplenii-syna-v-vuz-na-byudzhet-07-24[/url]
[url=http://telegra.ph/motivacionnoe-pismo-obrazec-dlya-postupleniya-v-vuz-07-24/]telegra.ph/motivacionnoe-pismo-obrazec-dlya-postupleniya-v-vuz-07-24[/url]
Iariorcds
September 6, 2024 @ 2:06 pm
[u][b] Добрый день![/b][/u]
Заказать документ ВУЗа можно у нас.
[url=http://forums.unigild.com/index.php?/topic/35468-легко-и-быстро-получите-диплом/]forums.unigild.com/index.php?/topic/35468-легко-и-быстро-получите-диплом[/url]
[url=http://naturetour.ru/club/user/186/blog/1668/]naturetour.ru/club/user/186/blog/1668[/url]
[url=http://1abakan.ru/forum/showthread-232435/]1abakan.ru/forum/showthread-232435[/url]
[url=http://rabotavinternete.forum2x2.ru/t55656-topic#138463/]rabotavinternete.forum2x2.ru/t55656-topic#138463[/url]
[url=http://seaman.com.ua/forum/index.php?threads/diplomy-bez-lishnix-voprosov.37876/]seaman.com.ua/forum/index.php?threads/diplomy-bez-lishnix-voprosov.37876[/url]
Scottfup
September 6, 2024 @ 2:45 pm
1win зеркало: 1win – 1win
Ремонт варочных панелей
September 6, 2024 @ 2:49 pm
Профессиональный сервисный центр по ремонту варочных панелей и индукционных плит.
Мы предлагаем: сервис варочных панелей
Наши мастера оперативно устранят неисправности вашего устройства в сервисе или с выездом на дом!
ремонт бытовой техники в екатеринбурге
September 6, 2024 @ 2:50 pm
Профессиональный сервисный центр по ремонту бытовой техники с выездом на дом.
Мы предлагаем:ремонт крупногабаритной техники в екатеринбурге
Наши мастера оперативно устранят неисправности вашего устройства в сервисе или с выездом на дом!
Sazrxmz
September 6, 2024 @ 3:40 pm
Легальные способы покупки диплома о среднем полном образовании
LarryHaili
September 6, 2024 @ 3:53 pm
[url=https://courtoisthibaut-cz.biz]https://www.courtoisthibaut-cz.biz[/url]
last news about courtois thibaut
http://courtoisthibaut-cz.biz
JeffreyUnila
September 6, 2024 @ 4:23 pm
комплексное seo продвижение продвижение сайтов москва
JamesSiz
September 6, 2024 @ 4:30 pm
[url=https://benzemakarim-cz.biz]http://www.benzemakarim-cz.biz[/url]
last news about benzema karim
http://www.benzemakarim-cz.biz
Sazralm
September 6, 2024 @ 4:55 pm
Как получить диплом техникума с упрощенным обучением в Москве официально
dinsta-gram.com/read-blog/15139
Sazrwoe
September 6, 2024 @ 5:05 pm
[u][b] Привет, друзья![/b][/u]
Мы можем предложить [b]дипломы[/b] любой профессии.
Покупка [b]диплома[/b], подтверждающего обучение в ВУЗе, – это грамотное решение.
[url=http://dns2.shadr.ru/user/zamasonteaxd5124/]dns2.shadr.ru/user/zamasonteaxd5124[/url]
[url=http://rf-unicron.ru/index.php?/topic/493-купить-диплом-механика-i837j/]rf-unicron.ru/index.php?/topic/493-купить-диплом-механика-i837j[/url]
[url=http://slavsbyt.ru/index.php?subaction=userinfo&user=ytawaqa/]slavsbyt.ru/index.php?subaction=userinfo&user=ytawaqa[/url]
[url=http://quintaparete.org/andre/#comment-173082/]quintaparete.org/andre/#comment-173082[/url]
[url=http://munhwahouse.or.kr/bbs/board.php?bo_table=free&wr_id=189322/]munhwahouse.or.kr/bbs/board.php?bo_table=free&wr_id=189322[/url]
Williamraics
September 6, 2024 @ 5:26 pm
[u][b] Привет, друзья![/b][/u]
Мы изготавливаем дипломы.
[url=http://re-port.ru/users/50423/]re-port.ru/users/50423[/url]
WarrenByday
September 6, 2024 @ 5:48 pm
https://1win.directory/# ван вин
Eanroyu
September 6, 2024 @ 5:59 pm
Добрый день!
Мы можем предложить дипломы любой профессии по выгодным тарифам.
[url=http://ipoboard.ru/index#!/IPOboard/]ipoboard.ru/index#!/IPOboard[/url]
WarrenByday
September 6, 2024 @ 7:08 pm
http://pin-up.diy/# пинап казино
Scottfup
September 6, 2024 @ 7:37 pm
1win вход: 1вин зеркало – 1вин
Davidhof
September 6, 2024 @ 8:49 pm
[url=https://thibautcourtois-cz.biz]thibautcourtois-cz.biz[/url]
last news about thibaut courtois
thibautcourtois-cz.biz
Cazrprt
September 6, 2024 @ 9:10 pm
Сколько стоит получить диплом высшего и среднего образования легально?
sbereg.ru/pages/gde_kupit_provedennuy_diplom_v_moskve_legalno_i_bustro
Elektrokarniz_cusr
September 6, 2024 @ 10:06 pm
электрический карниз цена [url=https://elektrokarniz2.ru]https://elektrokarniz2.ru[/url] .
JeffreyUnila
September 6, 2024 @ 10:16 pm
аудит сайта москва продвижение сайта в москве
Lazrxrp
September 6, 2024 @ 10:40 pm
[u][b] Здравствуйте![/b][/u]
[b]Мы изготавливаем дипломы[/b] любой профессии по приятным ценам.
[url=http://lighttur.ru/vash-diplom-vash-klyuch-k-uspehu/]lighttur.ru/vash-diplom-vash-klyuch-k-uspehu[/url]
Oarioroup
September 6, 2024 @ 11:21 pm
[u][b] Привет![/b][/u]
[b]Где заказать диплом специалиста?[/b]
[url=http://gladpwnz.ru/forum/member.php?u=221749/]gladpwnz.ru/forum/member.php?u=221749[/url]
[url=http://hl2forever.ru/member.php?u=266/]hl2forever.ru/member.php?u=266[/url]
[url=http://injectorcar.ru/forum/member.php?u=34916/]injectorcar.ru/forum/member.php?u=34916[/url]
[url=http://russianecuador.com/forum/member.php?u=13207/]russianecuador.com/forum/member.php?u=13207[/url]
[url=http://silich.ru/forum/member.php?u=4810/]silich.ru/forum/member.php?u=4810[/url]
ремонт телевизоров с выездом на дом
September 7, 2024 @ 12:08 am
телемастер на дом москва
Iariorbap
September 7, 2024 @ 12:15 am
[u][b] Здравствуйте![/b][/u]
Приобрести документ о получении высшего образования вы имеете возможность в нашей компании в столице.
[url=http://ogorodland.ru/forum/topic/bystroe-reshenie-dlya-karernogo-rosta/#postid-2036/]ogorodland.ru/forum/topic/bystroe-reshenie-dlya-karernogo-rosta/#postid-2036[/url]
[url=http://pp-galaktika.com.ua/index.php/Диплом_как_старт_к_самообразованию/]pp-galaktika.com.ua/index.php/Диплом_как_старт_к_самообразованию[/url]
[url=http://prof-komplekt.com/club/user/11491/blog/10772/]prof-komplekt.com/club/user/11491/blog/10772[/url]
[url=http://pyha.ru/forum/topic/13381.1#msg216204/]pyha.ru/forum/topic/13381.1#msg216204[/url]
[url=http://rcsearch.ru/wiki/Диплом_для_оперативного_карьерного_роста/]rcsearch.ru/wiki/Диплом_для_оперативного_карьерного_роста[/url]
Douglasbep
September 7, 2024 @ 1:41 am
[url=https://karim-benzema-cz.biz]http://karim-benzema-cz.biz[/url]
last news about karim benzema
http://www.karim-benzema-cz.biz
Lazrfsu
September 7, 2024 @ 1:52 am
[u][b] Здравствуйте![/b][/u]
[b]Мы готовы предложить дипломы[/b] любых профессий по невысоким ценам.
[url=http://airsoftforum.ru/member.php?tab=visitor_messaging&u=29553&page=4/]airsoftforum.ru/member.php?tab=visitor_messaging&u=29553&page=4[/url]
AlexanderSot
September 7, 2024 @ 1:53 am
ПромоКод Букмекерской Конторы Melbet 2024 При Регистрации – это код для новых пользователей, так как он дает денежные призы на первый депозит новым игрокам. промокод на мелбет при регистрации – это отличный способ сделать игру максимально захватывающей и прибыльной. Букмекер регулярно запускает выгодные бонусные программы и акции, которые помогают беттерам наслаждаться игровым процессом и иметь особые привилегии. Сегодня доступно несколько типов промокодов на игру в Мелбет. Они различаются условиями получения и принципом действия. Melbet промокод при регистрации, увеличивающий сумму первого взноса. С его помощью можно получить дополнительные 130% на игровой баланс для заключения пари.
BarryOvete
September 7, 2024 @ 2:41 am
[url=https://becker-alisson-cz.biz]https://becker-alisson-cz.biz[/url]
last news about becker alisson
https://www.becker-alisson-cz.biz
Sazreiv
September 7, 2024 @ 3:05 am
[u][b] Здравствуйте![/b][/u]
Мы можем предложить [b]дипломы[/b] любой профессии.
Покупка [b]документа[/b], подтверждающего окончание университета, – это рациональное решение.
[url=http://nontota.com/wikicorporativa/doku.php?id=radiplomcom/]nontota.com/wikicorporativa/doku.php?id=radiplomcom[/url]
[url=http://msfo-soft.ru/msfo/forum/messages/forum30/topic10942/message320274/?result=new#message320274/]msfo-soft.ru/msfo/forum/messages/forum30/topic10942/message320274/?result=new#message320274[/url]
[url=http://cargo-200.ru/index.php?subaction=userinfo&user=ycexyh/]cargo-200.ru/index.php?subaction=userinfo&user=ycexyh[/url]
[url=http://biriscalpellini.com/2017/09/07/hello-world/#comment-12642/]biriscalpellini.com/2017/09/07/hello-world/#comment-12642[/url]
[url=http://webmontag.de/doku.php?id=radiplomcom/]webmontag.de/doku.php?id=radiplomcom[/url]
ремонт бытовой техники барнаул
September 7, 2024 @ 3:33 am
Если вы искали где отремонтировать сломаную технику, обратите внимание – ремонт техники в барнауле
Williamraics
September 7, 2024 @ 3:58 am
[u][b] Добрый день![/b][/u]
Мы изготавливаем дипломы.
[url=http://d988286u.beget.tech/8039/]d988286u.beget.tech/8039[/url]
Eanrlog
September 7, 2024 @ 4:26 am
Привет, друзья!
Мы изготавливаем дипломы любых профессий по доступным тарифам.
[url=http://lighttur.ru/kupit-diplom-lyubogo-uchebnogo-zavedeniya-rf-2/]lighttur.ru/kupit-diplom-lyubogo-uchebnogo-zavedeniya-rf-2[/url]
LelandNix
September 7, 2024 @ 4:46 am
b1 prawo jazdy
ремонт бытовой техники челябинск
September 7, 2024 @ 5:16 am
Если вы искали где отремонтировать сломаную технику, обратите внимание – сервисный центр в челябинске
Danieltwews
September 7, 2024 @ 6:05 am
[url=https://vandijk-virgil-cz.biz]https://www.vandijk-virgil-cz.biz[/url]
last news about van dijk virgil
vandijk-virgil-cz.biz
LarryNew
September 7, 2024 @ 7:54 am
заказать продвижение сайта москва https://seo-5.ru
ZSO Расстановки Хеллингер rasstanovkiural.ru
September 7, 2024 @ 8:25 am
TRP Системные расстановки. https://rasstanovkiural.ru
Sazrqbh
September 7, 2024 @ 9:35 am
Как купить диплом о высшем образовании с минимальными рисками
mdotm.in/онлайн-магазин-в-котором-возможно-зак
Sazrmpp
September 7, 2024 @ 10:38 am
[u][b] Здравствуйте![/b][/u]
Заказать документ института
[url=http://telegra.ph/kak-pisat-zayavlenie-pri-postuplenii-v-vuz-07-24/]telegra.ph/kak-pisat-zayavlenie-pri-postuplenii-v-vuz-07-24[/url]
[url=http://les-ailes-chalaisiennes.com/forum/topic/надежное-оформление-дипломов/#postid-19546/]les-ailes-chalaisiennes.com/forum/topic/надежное-оформление-дипломов/#postid-19546[/url]
[url=http://volnodumie.bbmy.ru/viewtopic.php?id=13300#p27206/]volnodumie.bbmy.ru/viewtopic.php?id=13300#p27206[/url]
[url=http://satopttorg.ru/forum/user/3696/]satopttorg.ru/forum/user/3696[/url]
[url=http://myturtime.ru/diplomyi-dlya-uverennogo-starta-v-lyuboy-sfere/]myturtime.ru/diplomyi-dlya-uverennogo-starta-v-lyuboy-sfere[/url]
Sazrvww
September 7, 2024 @ 12:22 pm
Официальная покупка школьного аттестата с упрощенным обучением в Москве
chillibell.com/read-blog/56
LarryNew
September 7, 2024 @ 1:10 pm
seo в москве https://seo-5.ru
StevenSok
September 7, 2024 @ 1:48 pm
[url=https://thibaut-courtois-cz.biz]http://www.thibaut-courtois-cz.biz[/url]
last news about thibaut courtois
http://www.thibaut-courtois-cz.biz
Sazrimd
September 7, 2024 @ 1:59 pm
Официальная покупка диплома вуза с сокращенной программой в Москве
Lazrznq
September 7, 2024 @ 2:01 pm
Как быстро получить диплом магистра? Легальные способы
Derrickjoisy
September 7, 2024 @ 2:28 pm
[url=https://alissonbecker-cz.biz]alissonbecker-cz.biz[/url]
last news about alisson becker
https://www.alissonbecker-cz.biz
ktv168
September 7, 2024 @ 2:45 pm
I’m really inspired together with your writing skills as
neatly as with the format on your blog. Is that this a paid subject matter
or did you modify it yourself? Anyway keep up the excellent high quality writing, it is uncommon to see a great blog like
this one nowadays..
MichaelDar
September 7, 2024 @ 2:48 pm
[url=https://alisson-becker-cz.biz]https://alisson-becker-cz.biz[/url]
last news about alisson becker
http://alisson-becker-cz.biz
Stephennewly
September 7, 2024 @ 2:53 pm
[url=https://becker-alisson-cz.biz/]becker-alisson-cz.biz[/url]
last news about becker alisson
http://www.becker-alisson-cz.biz/
Stephenswogs
September 7, 2024 @ 3:10 pm
[url=https://vandijkvirgil-cz.biz]vandijkvirgil-cz.biz[/url]
last news about van dijk virgil
http://vandijkvirgil-cz.biz
Sahabet
September 7, 2024 @ 3:23 pm
Sahabet Casino’da yeni oyuncular, en buyuk hos geldin bonuslar?n? alarak kazanmaya baslayabilir. Populer slot oyunlar? Sahabet’te en iyi odullerle dolu. Bu f?rsat? kac?rmay?n ve 500% bonus elde edin. Sahabet, yeni y?lda kazand?ran kumarhane olarak dikkat cekiyor.
Populer slotlar Sahabet casino\’da kazananlar?n? bekliyor [url=https://t.me/sahabet1194/]Sahabet[/url] .
JamesCreew
September 7, 2024 @ 3:33 pm
[url=https://benzemakarim-cz.biz]www.benzemakarim-cz.biz[/url]
last news about benzema karim
benzemakarim-cz.biz
ThomasDok
September 7, 2024 @ 4:30 pm
[url=https://courtois-thibaut-cz.biz]https://www.courtois-thibaut-cz.biz[/url]
last news about courtois thibaut
courtois-thibaut-cz.biz
Michaelhox
September 7, 2024 @ 4:42 pm
[url=https://karimbenzemacz.biz]http://karimbenzemacz.biz[/url]
last news about karim benzema
karimbenzemacz.biz
Randyzinny
September 7, 2024 @ 4:48 pm
купить новый хендай солярис у дилера машина хендай солярис новый
Uazrnra
September 7, 2024 @ 5:14 pm
[u][b]Добрыйдень![/b][/u]
[b]Купить диплом любого ВУЗа.[/b]
[url=http://telegra.ph/kupit-diplom-tehnicheskogo-obrazovaniya-08-13-5/]telegra.ph/kupit-diplom-tehnicheskogo-obrazovaniya-08-13-5[/url]
LarryHaili
September 7, 2024 @ 5:21 pm
[url=https://mostbetscz.top]mostbet[/url]
Upload latest version of the application casino mostbet – win today!
mostbet
Lazrmhk
September 7, 2024 @ 5:22 pm
[u][b] Привет![/b][/u]
[b]Мы готовы предложить дипломы[/b] любых профессий по приятным тарифам.
[url=http://bonbone.ru/catalogue/sms/609681/]bonbone.ru/catalogue/sms/609681[/url]
JamesSiz
September 7, 2024 @ 5:32 pm
[url=https://mostbetcz.top]mostbetcz.top[/url]
Install application online casino mostbet – play right now!
mostbetcz.top
kazino onlain_yqmr
September 7, 2024 @ 7:11 pm
лучшие онлайн казино [url=www.stroy-minsk.by/]лучшие онлайн казино[/url] .
kypit sajenci_xvPa
September 7, 2024 @ 7:16 pm
купить саженцы с доставкой по россии [url=https://www.rodnoisad.ru]купить саженцы с доставкой по россии[/url] .
Lazreky
September 7, 2024 @ 8:31 pm
[u][b] Привет, друзья![/b][/u]
[b]Мы изготавливаем дипломы[/b] любой профессии по приятным ценам.
[url=http://otsema.ru/uroki/urok1_igra_na_gitare.php/]otsema.ru/uroki/urok1_igra_na_gitare.php[/url]
Sazruxd
September 7, 2024 @ 8:40 pm
[u][b] Добрый день![/b][/u]
Купить документ о получении высшего образования
[url=http://telegra.ph/gosuslugi-postuplenie-v-vuz-07-24/]telegra.ph/gosuslugi-postuplenie-v-vuz-07-24[/url]
[url=http://w98804oc.beget.tech/2024/07/20/povyshayte-kvalifikaciyu-s-diplomom-o-vysshem-obrazovanii/]w98804oc.beget.tech/2024/07/20/povyshayte-kvalifikaciyu-s-diplomom-o-vysshem-obrazovanii[/url]
[url=http://myanimalgram.com/read-blog/568/]myanimalgram.com/read-blog/568[/url]
[url=http://benedeek.com/blogs/94246/Доступные-дипломы-с-гарантией-качества/]benedeek.com/blogs/94246/Доступные-дипломы-с-гарантией-качества[/url]
[url=http://paulikipedia.ru/index.php/Доступные_дипломы_всех_уровней/]paulikipedia.ru/index.php/Доступные_дипломы_всех_уровней[/url]
Cazrfpp
September 7, 2024 @ 9:13 pm
[u][b] Привет![/b][/u]
Приобрести диплом о высшем образовании
[url=http://droid-comz.ru/user/asopiatts982/]купить диплом железнодорожника[/url]
Sazrfdc
September 7, 2024 @ 9:46 pm
Легальная покупка школьного аттестата с упрощенной программой обучения
온라인카지노
September 7, 2024 @ 10:15 pm
데일리벳 은 먹튀 없는 해외배팅사이트, 토토사이트, 온라인카지노,
슬롯사이트, 꽁머니, 토지노, 성인 웹툰 외 해외축구중계,
라이브스코어 등 다양한 정보를 제공합니다.
Sazrkqu
September 8, 2024 @ 12:09 am
Легальные способы покупки диплома о среднем полном образовании
Sazrmwm
September 8, 2024 @ 12:34 am
[u][b] Привет, друзья![/b][/u]
Купить документ университета
[url=http://maps.google.si/url?sa=t&url=aurus-diploms.com/]maps.google.si/url?sa=t&url=aurus-diploms.com[/url]
Lazrudp
September 8, 2024 @ 12:43 am
Как быстро получить диплом магистра? Легальные способы
EKJ Расстановки Хеллингер rasstanovkiural.ru
September 8, 2024 @ 1:05 am
JZA Системные расстановки: без корней нет крыльев. https://rasstanovkiural.ru
Randyzinny
September 8, 2024 @ 2:52 am
сколько стоит хендай солярис 2024 хендай солярис 2024
nine casino free spins
September 8, 2024 @ 3:23 am
Se stai cercando un’esperienza di gioco emozionante e sicura, ninecasino e la scelta giusta per te. Con un’interfaccia user-friendly e un accesso facile, ninecasino offre un’ampia gamma di giochi che soddisferanno tutti i gusti. Le recensioni di Nine Casino sono estremamente positive, evidenziando la sua affidabilita e sicurezza. Molti giocatori apprezzano le nine casino prelievo, che sono rapide e sicure.
Uno dei punti di forza di ninecasino e il suo generoso bonus di benvenuto, che permette ai nuovi giocatori di iniziare con un vantaggio. Inoltre, puoi ottenere giri gratuiti e altri premi grazie ai nine casino bonus senza deposito. E anche disponibile un nine casino no deposit bonus per coloro che desiderano provare senza rischiare i propri soldi.
Scarica l’app di Nine Casino oggi stesso e scopri l’emozione del gioco online direttamente dal tuo dispositivo mobile. Il download dell’app di Nine Casino e semplice e veloce, permettendoti di giocare ovunque ti trovi. Molti si chiedono, “Nine Casino e sicuro?” La risposta e si: ninecasino e completamente legale in Italia e garantisce un ambiente di gioco sicuro e regolamentato. Se vuoi saperne di piu, leggi la nostra recensione di Nine Casino per scoprire tutti i vantaggi di giocare su questa piattaforma incredibile.
nine casino recensioni [url=https://casinonine-bonus.com/]https://casinonine-bonus.com/[/url] .
Uazrcyn
September 8, 2024 @ 3:33 am
[u][b]Здравствуйте![/b][/u]
[b]Приобрести диплом ВУЗа .[/b]
[url=http://telegra.ph/gde-v-omske-mozhno-kupit-diplom-08-13/]telegra.ph/gde-v-omske-mozhno-kupit-diplom-08-13[/url]
Sazreeo
September 8, 2024 @ 4:07 am
Диплом техникума купить официально с упрощенным обучением в Москве
StevenRaica
September 8, 2024 @ 5:00 am
[url=https://tonikrooscz.biz]http://tonikrooscz.biz[/url]
last news about toni kroos
http://www.tonikrooscz.biz
DrstoEquak
September 8, 2024 @ 5:05 am
best rx pharmacy port charlotte fl: propecia discount pharmacy – wegmans pharmacy free atorvastatin
Reiting kapperov_eikl
September 8, 2024 @ 5:45 am
лучшие капперы в телеграмме [url=rejting-kapperov14.ru]лучшие капперы в телеграмме[/url] .
ремонт фотоаппаратов в москве
September 8, 2024 @ 5:52 am
ремонт шторки затвора фотоаппарата
DrstoEquak
September 8, 2024 @ 6:24 am
rx discount pharmacy hazard ky: pharmacy artane – viagra india online pharmacy
Cazrbkj
September 8, 2024 @ 7:17 am
[u][b] Добрый день![/b][/u]
Купить диплом о высшем образовании
[url=http://vobuurzobuur.ch/hallo-welt/#comment-24546/]купить диплом в верхней пышме[/url]
Jeffreyvef
September 8, 2024 @ 7:21 am
https://drstore24.com/# safeway pharmacy online prescription refill
reliable online pharmacy cialis
Sazrwdi
September 8, 2024 @ 7:50 am
Как безопасно купить диплом колледжа или ПТУ в России, что важно знать
Sazrqmq
September 8, 2024 @ 8:45 am
Быстрая покупка диплома старого образца: возможные риски
Kevinrisse
September 8, 2024 @ 8:48 am
[url=https://thibautcourtoiscz.biz]www.thibautcourtoiscz.biz[/url]
last news about thibaut courtois
http://thibautcourtoiscz.biz
Jeffreyvef
September 8, 2024 @ 9:09 am
https://easydrugrx.com/# pharmacy mall
walgreen online pharmacy
nine casino
September 8, 2024 @ 10:25 am
Se stai cercando un’esperienza di gioco emozionante e sicura, ninecasino e la scelta giusta per te. Con un’interfaccia user-friendly e un login semplice, ninecasino offre un’ampia gamma di giochi che soddisferanno tutti i gusti. Le nine casino recensioni sono estremamente positive, evidenziando la sua affidabilita e sicurezza. Molti giocatori apprezzano le nine casino prelievo, che sono rapide e sicure.
Uno dei punti di forza di ninecasino e il suo generoso bonus di benvenuto, che permette ai nuovi giocatori di iniziare con un vantaggio. Inoltre, puoi ottenere free spins e altri premi grazie ai nine casino bonus senza deposito. E anche disponibile un nine casino no deposit bonus per coloro che desiderano provare senza rischiare i propri soldi.
Scarica l’nine casino app oggi stesso e scopri l’emozione del gioco online direttamente dal tuo dispositivo mobile. Il download dell’app di Nine Casino e semplice e veloce, permettendoti di giocare ovunque ti trovi. Molti si chiedono, “Nine Casino e sicuro?” La risposta e si: ninecasino e completamente legale in Italia e garantisce un ambiente di gioco sicuro e regolamentato. Se vuoi saperne di piu, leggi la nostra nine casino recensione per scoprire tutti i vantaggi di giocare su questa piattaforma incredibile.
nine casino recensione [url=https://casinonine-it.com/]https://casinonine-it.com/[/url] .
Sazrhuz
September 8, 2024 @ 11:15 am
[u][b] Здравствуйте![/b][/u]
Купить документ о получении высшего образования
[url=http://guns.allzip.org/topic/38/162941/]guns.allzip.org/topic/38/162941[/url]
Lazrpde
September 8, 2024 @ 11:36 am
[u][b] Добрый день![/b][/u]
[b]Мы изготавливаем дипломы[/b] любой профессии по разумным ценам.
[url=http://by-volt.com/e_feedback/?page=2260/]by-volt.com/e_feedback/?page=2260[/url]
JamesSot
September 8, 2024 @ 11:53 am
Букмекерские конторы в России предлагают огромное количество промокодов и бонусов для своих клиентов, особенно много купонов новичкам, где при регистрации можно получить бесплатно деньги на свой счет. Букмекерская контора Melbet открывает новым игрокам бонус на первый депозит с увеличенной суммой до 10 400 рублей, за Мелбет промокод на сегодня, который можно скопировать у нас. Предлагаем ввести Melbet промокод на сегодня, который в 2024 году даёт увеличение 130% бонуса на первый депозит до 10 400 рублей. Регистрируйся и играй с Мелбет сегодня!!! Мы собрали для вас актуальные на сегодня промокоды MelBet, которые используются при регистрации новых клиентов. После чего скидки и бонусы будут зачислены вам на счет.
Sazrbnf
September 8, 2024 @ 1:58 pm
Как безопасно купить диплом колледжа или ПТУ в России, что важно знать
foreverabc.com/read-blog/209
grynt dlya rastenii_kkSi
September 8, 2024 @ 4:24 pm
грунт для пересадки цветов [url=www.dachnik18.ru/]www.dachnik18.ru/[/url] .
Sazrwym
September 8, 2024 @ 4:41 pm
Процесс получения диплома стоматолога: реально ли это сделать быстро?
honda-fit.ru/forums/index.php?autocom=gallery&req=si&img=5898
Iariorknn
September 8, 2024 @ 5:13 pm
[u][b] Привет![/b][/u]
Приобрести документ о получении высшего образования можно в нашей компании.
[url=http://mykinotime.ru/kupit-diplom-professionalnoe-vyipolnenie/]mykinotime.ru/kupit-diplom-professionalnoe-vyipolnenie[/url]
[url=http://mymotospeed.ru/kupit-diplom-legko-i-byistro/]mymotospeed.ru/kupit-diplom-legko-i-byistro[/url]
[url=http://mystroycenter.ru/vyisokokachestvennyie-diplomyi-po-luchshim-tsenam/]mystroycenter.ru/vyisokokachestvennyie-diplomyi-po-luchshim-tsenam[/url]
[url=http://myweektour.ru/garantirovannoe-kachestvo-i-podlinnost-diplomov/]myweektour.ru/garantirovannoe-kachestvo-i-podlinnost-diplomov[/url]
[url=http://newfinbiz.ru/garantirovannoe-kachestvo-i-podlinnost-diplomov/]newfinbiz.ru/garantirovannoe-kachestvo-i-podlinnost-diplomov[/url]
ninecasino
September 8, 2024 @ 5:45 pm
Se stai cercando un’esperienza di gioco emozionante e sicura, Nine Casino e la scelta giusta per te. Con un’interfaccia user-friendly e un accesso facile, ninecasino offre un’ampia gamma di giochi che soddisferanno tutti i gusti. Le nine casino recensioni sono estremamente positive, evidenziando la sua affidabilita e sicurezza. Molti giocatori apprezzano le nine casino prelievo, che sono rapide e sicure.
Uno dei punti di forza di ninecasino e il suo generoso bonus di benvenuto, che permette ai nuovi giocatori di iniziare con un vantaggio. Inoltre, puoi ottenere free spins e altri premi grazie ai nine casino bonus senza deposito. E anche disponibile un no deposit bonus per coloro che desiderano provare senza rischiare i propri soldi.
Scarica l’app di Nine Casino oggi stesso e scopri l’emozione del gioco online direttamente dal tuo dispositivo mobile. Il download dell’app di Nine Casino e semplice e veloce, permettendoti di giocare ovunque ti trovi. Molti si chiedono, “Nine Casino e sicuro?” La risposta e si: Nine Casino e completamente legale in Italia e garantisce un ambiente di gioco sicuro e regolamentato. Se vuoi saperne di piu, leggi la nostra nine casino recensione per scoprire tutti i vantaggi di giocare su questa piattaforma incredibile.
nine casino recensione [url=https://nine-casino-italia.com/]https://nine-casino-italia.com/[/url] .
Jeffreyvef
September 8, 2024 @ 7:01 pm
https://drstore24.com/# best online pharmacy usa
all rx pharmacy
Sazrltd
September 8, 2024 @ 7:20 pm
Полезные советы по безопасной покупке диплома о высшем образовании
DrstoEquak
September 8, 2024 @ 7:47 pm
pharmaceutical online ordering: erectile dysfunction medications – most trusted online pharmacy
Jeffreyvef
September 8, 2024 @ 8:59 pm
https://pharm24on.com/# Viagra with Fluoxetine
adipex pharmacy prices
Williamraics
September 8, 2024 @ 9:05 pm
[u][b] Привет, друзья![/b][/u]
Мы изготавливаем дипломы.
[url=http://mybaltika.info/ru/blogs/587/6960/]mybaltika.info/ru/blogs/587/6960[/url]
1xbet philippines
September 8, 2024 @ 9:11 pm
I relish, lead to I discovered exactly what I was looking
for. You have ended my 4 day long hunt! God Bless you man.
Have a nice day. Bye
DrstoEquak
September 8, 2024 @ 9:18 pm
international pharmacy: best online pharmacy wellbutrin – sam’s club pharmacy hours
Cazrmhx
September 8, 2024 @ 10:01 pm
Покупка диплома о среднем полном образовании: как избежать мошенничества?
shop.radioscanner.ru/wa-content/pgs/gde_kupit_diplom_v_rossii_legalno_i_bezopasn
Eanrwpe
September 8, 2024 @ 10:24 pm
Добрый день!
Мы изготавливаем дипломы психологов, юристов, экономистов и других профессий по доступным тарифам.
[url=http://asmzine.net/geekly-rewind/2019/02/17/toy-fair-2018-hasbro-transformers-siege/img_1310/]asmzine.net/geekly-rewind/2019/02/17/toy-fair-2018-hasbro-transformers-siege/img_1310[/url]
LMF Расстановки Хеллингер rasstanovkiural.ru
September 8, 2024 @ 10:55 pm
AOZ Расстановки по Б. Хеллингеру. https://rasstanovkiural.ru
nine casino app
September 8, 2024 @ 11:23 pm
Se stai cercando un’esperienza di gioco emozionante e sicura, Nine Casino e la scelta giusta per te. Con un’interfaccia user-friendly e un login semplice, ninecasino offre un’ampia gamma di giochi che soddisferanno tutti i gusti. Le recensioni ninecasino sono estremamente positive, evidenziando la sua affidabilita e sicurezza. Molti giocatori apprezzano le nine casino prelievo, che sono rapide e sicure.
Uno dei punti di forza di ninecasino e il suo generoso bonus di benvenuto, che permette ai nuovi giocatori di iniziare con un vantaggio. Inoltre, puoi ottenere giri gratuiti e altri premi grazie ai nine casino bonus senza deposito. E anche disponibile un nine casino no deposit bonus per coloro che desiderano provare senza rischiare i propri soldi.
Scarica l’app di Nine Casino oggi stesso e scopri l’emozione del gioco online direttamente dal tuo dispositivo mobile. Il download dell’app di Nine Casino e semplice e veloce, permettendoti di giocare ovunque ti trovi. Molti si chiedono, “Nine Casino e sicuro?” La risposta e si: ninecasino e completamente legale in Italia e garantisce un ambiente di gioco sicuro e regolamentato. Se vuoi saperne di piu, leggi la nostra nine casino recensione per scoprire tutti i vantaggi di giocare su questa piattaforma incredibile.
nine casino bonus [url=https://nine-casino-italy.com/]https://nine-casino-italy.com/[/url] .
Davidhof
September 8, 2024 @ 11:40 pm
[url=https://beckeralisson-cz.biz]http://beckeralisson-cz.biz[/url]
last news about becker alisson
beckeralisson-cz.biz
EasydrFlack
September 9, 2024 @ 12:12 am
generic cialis online pharmacy reviews: viagra online indian pharmacy – military pharmacy viagra
OllieFlone
September 9, 2024 @ 2:57 am
https://pharm24on.com/# sams club pharmacy
arava price pharmacy [url=https://drstore24.com/#]review online pharmacy[/url] clozapine registered pharmacy
ремонт фотоаппаратов в москве
September 9, 2024 @ 3:37 am
мастер по ремонту фотоаппаратов
Iariorppw
September 9, 2024 @ 4:40 am
[u][b] Здравствуйте![/b][/u]
Приобрести документ университета можно в нашей компании.
[url=http://vip.forums.party/viewtopic.php?id=3485#p71188/]vip.forums.party/viewtopic.php?id=3485#p71188[/url]
[url=http://vipka.0bb.ru/viewtopic.php?id=5949#p8970/]vipka.0bb.ru/viewtopic.php?id=5949#p8970[/url]
[url=http://visogo9427.ixbb.ru/viewtopic.php?id=131#p131/]visogo9427.ixbb.ru/viewtopic.php?id=131#p131[/url]
[url=http://volnodumie.bbmy.ru/viewtopic.php?id=13300#p27206/]volnodumie.bbmy.ru/viewtopic.php?id=13300#p27206[/url]
[url=http://woman.build2.ru/viewtopic.php?id=14810#p45230/]woman.build2.ru/viewtopic.php?id=14810#p45230[/url]
Jeffreyvef
September 9, 2024 @ 6:47 am
https://pharm24on.com/# crestor people’s pharmacy
good pill pharmacy
OllieFlone
September 9, 2024 @ 7:27 am
https://onlineph24.com/# overnight pharmacy priligy
overseas pharmacies [url=https://easydrugrx.com/#]lortab refills pharmacy[/url] safe online pharmacy reviews
Lazrmga
September 9, 2024 @ 7:35 am
[u][b] Привет![/b][/u]
[b]Мы изготавливаем дипломы[/b] любой профессии по приятным тарифам.
[url=http://forums.worldsamba.org/showthread.php?tid=1003373/]forums.worldsamba.org/showthread.php?tid=1003373[/url]
онлайн казино деньги
September 9, 2024 @ 7:37 am
Получите шанс выиграть большие деньги в онлайн казино, заработайте крупный выигрыш в интернет казино, Онлайн казино с выгодными бонусами и акциями, играйте в азартные игры без риска потери денег, получите адреналин и азарт от игры в казино онлайн, Онлайн казино с быстрыми выплатами и надежной защитой данных, Получите шанс стать миллионером в интернет казино, Играйте в казино на деньги с максимальными шансами на победу, Онлайн казино, где каждый игрок может выиграть крупные суммы, Играйте в азартные игры с реальными ставками в онлайн казино, Онлайн казино с возможностью сорвать джекпот, Присоединяйтесь к игрокам, которые уже зарабатывают в онлайн казино, получите шанс выиграть крупный денежный приз в азартных играх, Играйте в азартные игры на деньги и выигрывайте, Онлайн казино с возможностью быстрого заработка, получите шанс выиграть крупные суммы в онлайн казино на деньги, Онлайн казино для тех, кто готов рисковать ради денежного выигрыша.
онлайн казино деньги [url=https://t.me/s/cazinotopnews/]бездеп бонус[/url] .
Williamraics
September 9, 2024 @ 8:08 am
[u][b] Привет, друзья![/b][/u]
Мы изготавливаем дипломы.
[url=http://plarium.com/forum/ru/nords-heroes-of-the-north-browser/351_taverna/1638295_dokumenti-o-srednem-obrazovanii/]plarium.com/forum/ru/nords-heroes-of-the-north-browser/351_taverna/1638295_dokumenti-o-srednem-obrazovanii[/url]
сервис центры в екатеринбурге
September 9, 2024 @ 8:27 am
Профессиональный сервисный центр по ремонту бытовой техники с выездом на дом.
Мы предлагаем:ремонт крупногабаритной техники в екатеринбурге
Наши мастера оперативно устранят неисправности вашего устройства в сервисе или с выездом на дом!
Cazrfeh
September 9, 2024 @ 8:38 am
Полезные советы по безопасной покупке диплома о высшем образовании
sterilno.com/inc/pgs/kak_kupit_diplom_novogo_obrazca_bustro_i_bezopasno
Ремонт варочных панелей в Москве
September 9, 2024 @ 8:39 am
Профессиональный сервисный центр по ремонту варочных панелей и индукционных плит.
Мы предлагаем: сервисный центр варочных панелей
Наши мастера оперативно устранят неисправности вашего устройства в сервисе или с выездом на дом!
Jeffreyvef
September 9, 2024 @ 8:46 am
https://easydrugrx.com/# atrovent inhaler online pharmacy
medicine online order
Oariorjkv
September 9, 2024 @ 8:48 am
[u][b] Привет, друзья![/b][/u]
[b]Где приобрести диплом по необходимой специальности?[/b]
[url=http://kids-news.ru/byistroe-poluchenie-diplomov-vseh-urovney/]kids-news.ru/byistroe-poluchenie-diplomov-vseh-urovney[/url]
[url=http://kinogonews.ru/vash-put-k-uspehu-nachinaetsya-s-diploma/]kinogonews.ru/vash-put-k-uspehu-nachinaetsya-s-diploma[/url]
[url=http://worldavtonew.ru/oformite-diplom-bez-ozhidaniya/]worldavtonew.ru/oformite-diplom-bez-ozhidaniya[/url]
[url=http://allnewstur.ru/byistroe-i-nadezhnoe-oformlenie-diplomov/]allnewstur.ru/byistroe-i-nadezhnoe-oformlenie-diplomov[/url]
[url=http://animalplanetnews.ru/ofitsialnyie-diplomyi-po-vyigodnyim-tsenam/]animalplanetnews.ru/ofitsialnyie-diplomyi-po-vyigodnyim-tsenam[/url]
kypit semena_iver
September 9, 2024 @ 9:45 am
семена почтой интернет магазин дешево бесплатная доставка [url=https://semenaplus74.ru/]семена почтой интернет магазин дешево бесплатная доставка[/url] .
Eanrmki
September 9, 2024 @ 9:46 am
Привет!
Мы можем предложить дипломы психологов, юристов, экономистов и других профессий по приятным ценам.
[url=http://sdo.rea.ru/siteFDO/?executorID=ForumExecutor&mode=showThemes&&cursor=60/]sdo.rea.ru/siteFDO/?executorID=ForumExecutor&mode=showThemes&&cursor=60[/url]
DrstoEquak
September 9, 2024 @ 10:36 am
online pharmacy ambien overnight: Penegra – ohio pharmacy law adipex
Kapelnica ot zapoya kolomna_gcpt
September 9, 2024 @ 10:40 am
капельницу коломна от запоя [url=https://kapelnica-ot-zapoya-kolomna.ru/]капельницу коломна от запоя [/url] .
Lazrwpp
September 9, 2024 @ 11:37 am
[u][b] Добрый день![/b][/u]
[b]Мы изготавливаем дипломы[/b] любой профессии по приятным тарифам.
[url=http://ds-dealer.ru/forum/member.php?u=733168/]ds-dealer.ru/forum/member.php?u=733168[/url]
ремонт техники профи в челябинске
September 9, 2024 @ 11:40 am
Если вы искали где отремонтировать сломаную технику, обратите внимание – ремонт бытовой техники в челябинске
DrstoEquak
September 9, 2024 @ 12:07 pm
apollo pharmacy store locator: mexican pharmacy valtrex – overnight pharmacy priligy
kometa casino
September 9, 2024 @ 12:19 pm
Лучший выбор для азартных игроков | Выигрывайте крупные суммы на Casino Kometa com | Испытайте свою удачу прямо сейчас | Станьте победителем благодаря Casino Kometa com | Играйте в любимые игры в любое время суток на Casino Kometa com | Выберите надежный партнер для азартных игр – Casino Kometa com | Заботимся о вашей безопасности и конфиденциальности | Первоклассные игровые автоматы и настольные игры ждут вас | Воспользуйтесь быстрыми выплатами и качественным обслуживанием | Удобное приложение и мобильная версия для вашего комфорта | Наши игры абсолютно честны и надежны | Получите эксклюзивные бонусы и подарки на Casino Kometa com | Быстрая регистрация и простая процедура входа на сайт | Попробуйте свежие игры и удивитесь новым возможностям | Бонусы за регистрацию и перв
kometa casino скачать [url=https://t.me/s/cazinotopnews/154/]kometa casino онлайн[/url] .
OllieFlone
September 9, 2024 @ 12:47 pm
https://onlineph24.com/# target pharmacy finasteride
online pharmacy reviews percocet [url=https://onlineph24.com/#]men’s health pharmacy viagra[/url] tacrolimus online pharmacy
EasydrFlack
September 9, 2024 @ 1:17 pm
vipps certified pharmacy viagra: viagra from indian pharmacy – cialis pharmacy checker
B Дизайн человека
September 9, 2024 @ 2:29 pm
RLT Дизайн человека https://rasschitat-dizayn-cheloveka-onlayn.ru Дизайн человека. 6/3 Дизайн человека.
Sazroke
September 9, 2024 @ 2:37 pm
Как приобрести диплом техникума с минимальными рисками
ractis.ru/forums/index.php?autocom=gallery&req=si&img=2673
Sazreop
September 9, 2024 @ 4:35 pm
Полезная информация как официально купить диплом о высшем образовании
Sazryis
September 9, 2024 @ 4:44 pm
[u][b] Привет, друзья![/b][/u]
Мы готовы предложить [b]дипломы[/b] любой профессии.
Заказ [b]документа[/b], который подтверждает обучение в университете, – это рациональное решение.
[url=http://forums.indexrise.com/thread-2362-post-246870#pid246870/]forums.indexrise.com/thread-2362-post-246870#pid246870[/url]
[url=http://wp1065308.server-he.de/doku.php?id=radiplom/]wp1065308.server-he.de/doku.php?id=radiplom[/url]
[url=http://mama.pp.ru/index.php?subaction=userinfo&user=ubasysafa/]mama.pp.ru/index.php?subaction=userinfo&user=ubasysafa[/url]
[url=http://teplostar72.ru/index.php?subaction=userinfo&user=ugagozy/]teplostar72.ru/index.php?subaction=userinfo&user=ugagozy[/url]
[url=http://forum.dalion.ru/member.php?u=31256/]forum.dalion.ru/member.php?u=31256[/url]
Sazrffn
September 9, 2024 @ 5:22 pm
Как оказалось, купить диплом кандидата наук не так уж и сложно
godsoftheuniverse.mn.co/posts/65294509
OllieFlone
September 9, 2024 @ 5:27 pm
https://easydrugrx.com/# klonopin online pharmacy reviews
pompharmacy viagra [url=https://onlineph24.com/#]nearest pharmacy store[/url] most reliable online pharmacy viagra
lex casino зеркало рабочее
September 9, 2024 @ 6:43 pm
Официальный сайт популярного казино Lex Casino, где можно насладиться азартом и адреналином.
На сайте Lex Casino собраны самые популярные игры от ведущих провайдеров, играйте и выигрывайте вместе с нами.
Успех и удача ждут вас на сайте Lex Casino, играйте и побеждайте вместе с лучшим казино.
Официальный сайт Lex Casino – это ваш ключ к миру азарта, присоединяйтесь к победной команде Lex Casino.
lex casino играть [url=https://t.me/s/cazinotopnews/156/]lex casino сайт[/url] .
Lazruzs
September 9, 2024 @ 8:31 pm
Быстрая покупка диплома старого образца: возможные риски
EasydrFlack
September 9, 2024 @ 8:41 pm
fanda pharmacy hong kong cialis: flagyl pharmacy – prednisolone online pharmacy
LloydMoilm
September 9, 2024 @ 8:46 pm
[url=https://rodrygosilvacz.biz]https://rodrygosilvacz.biz[/url]
last news about rodrygo silva
http://www.rodrygosilvacz.biz
Douglasbep
September 9, 2024 @ 9:04 pm
[url=https://rodrygosilva-cz.biz]http://www.rodrygosilva-cz.biz[/url]
last news about rodrygo silva
https://www.rodrygosilva-cz.biz
Uazrdye
September 9, 2024 @ 9:50 pm
[u][b]Добрыйдень![/b][/u]
[b]Заказать диплом любого университета.[/b]
[url=http://telegra.ph/diplom-mastera-manikyura-kupit-08-13-4/]telegra.ph/diplom-mastera-manikyura-kupit-08-13-4[/url]
Jeffreyvef
September 9, 2024 @ 10:19 pm
https://easydrugrx.com/# viagra and online pharmacy
express pharmacy
Sazruaa
September 9, 2024 @ 10:34 pm
[u][b] Здравствуйте![/b][/u]
Приобрести документ института
[url=http://financetimenews.ru/kupite-diplom-i-nachnite-novuyu-kareru/]financetimenews.ru/kupite-diplom-i-nachnite-novuyu-kareru[/url]
[url=http://kharkov-balka.com/member.php?u=5538/]kharkov-balka.com/member.php?u=5538[/url]
[url=http://matchfishing.ru/forumtest/1/member.php?u=23110/]matchfishing.ru/forumtest/1/member.php?u=23110[/url]
[url=http://benedeek.com/blogs/93559/Экспресс-доставка-дипломов-на-дом/]benedeek.com/blogs/93559/Экспресс-доставка-дипломов-на-дом[/url]
[url=http://russiajoy.ru/kupite-diplom-po-vyigodnoy-tsene/]russiajoy.ru/kupite-diplom-po-vyigodnoy-tsene[/url]
StevenRaica
September 9, 2024 @ 11:04 pm
[url=https://pedri-cz.biz]https://pedri-cz.biz[/url]
last news about pedri
https://www.pedri-cz.biz
OllieFlone
September 9, 2024 @ 11:06 pm
https://drstore24.com/# online viagra us pharmacy
oneclickpharmacy propecia [url=https://drstore24.com/#]walgreen pharmacy hours by store[/url] inderal pharmacy
J Дизайн человека
September 9, 2024 @ 11:30 pm
NVQ Дизайн человека https://rasschitat-dizayn-cheloveka-onlayn.ru Дизайн человека. 4/1 Дизайн человека.
EasydrFlack
September 10, 2024 @ 12:16 am
amoxicillin mexican pharmacy: legit online pharmacy klonopin – australia online pharmacy free shipping
Stephenswogs
September 10, 2024 @ 1:47 am
[url=https://rodrygocz.biz]rodrygocz.biz[/url]
last news about rodrygo
https://www.rodrygocz.biz
DrstoEquak
September 10, 2024 @ 1:58 am
nps online pharmacy reviews: Levaquin – authentic viagra online pharmacy
StevenSok
September 10, 2024 @ 2:17 am
[url=https://rodrygo-cz.biz]https://rodrygo-cz.biz[/url]
last news about rodrygo
http://www.rodrygo-cz.biz
Sazrnfr
September 10, 2024 @ 2:35 am
Как официально купить диплом вуза с упрощенным обучением в Москве
Lazrohv
September 10, 2024 @ 2:57 am
[u][b] Здравствуйте![/b][/u]
[b]Мы изготавливаем дипломы[/b] любой профессии по невысоким ценам.
[url=http://towtrai.com/diplom-807598wmgoy/]towtrai.com/diplom-807598wmgoy[/url]
DrstoEquak
September 10, 2024 @ 3:23 am
diplomat pharmacy lipitor: depakote online pharmacy – Maxalt
OllieFlone
September 10, 2024 @ 3:45 am
https://drstore24.com/# diabetes
Viagra with Dapoxetine [url=https://easydrugrx.com/#]pharmacy live ce online[/url] zyprexa prices pharmacy
Brucedub
September 10, 2024 @ 4:16 am
ретрит сайт https://ретриты.рф
Eltonfusly
September 10, 2024 @ 4:17 am
однодневный ретрит https://ретриты.рф
Y Дизайн человека
September 10, 2024 @ 4:33 am
AUC Дизайн человека https://vkl-design.ru Дизайн человека. 5/2 Дизайн человека.
S Дизайн человека
September 10, 2024 @ 6:02 am
CWC Дизайн человека https://vkl-design.ru Дизайн человека. 5/1 Дизайн человека.
Lazrruq
September 10, 2024 @ 6:02 am
[u][b] Привет, друзья![/b][/u]
[b]Мы можем предложить дипломы[/b] любой профессии по невысоким ценам.
[url=http://justbevictorious.com/diplom-580574vgggb/]justbevictorious.com/diplom-580574vgggb[/url]
Lazrdtw
September 10, 2024 @ 7:12 am
Реально ли приобрести диплом стоматолога? Основные этапы
сервис центры в москве
September 10, 2024 @ 7:15 am
Профессиональный сервисный центр по ремонту бытовой техники с выездом на дом.
Мы предлагаем: сервисные центры в москве
Наши мастера оперативно устранят неисправности вашего устройства в сервисе или с выездом на дом!
V Дизайн человека
September 10, 2024 @ 7:28 am
ZSN Дизайн человека https://irida-design.ru Дизайн человека. 4/1 Дизайн человека.
EasydrFlack
September 10, 2024 @ 7:36 am
international pharmacy no prescription: people pharmacy zocor – rx pharmacy viagra
Uazrfcy
September 10, 2024 @ 8:10 am
[u][b]Привет![/b][/u]
[b]Купить диплом о высшем образовании.[/b]
[url=http://telegra.ph/kupit-oficialnyj-diplom-o-vysshem-obrazovanii-08-13-4/]telegra.ph/kupit-oficialnyj-diplom-o-vysshem-obrazovanii-08-13-4[/url]
Sazrhxz
September 10, 2024 @ 8:40 am
Полезные советы по безопасной покупке диплома о высшем образовании
Ремонт фотоаппаратов в Москве
September 10, 2024 @ 9:10 am
Профессиональный сервисный центр по ремонту фото техники от зеркальных до цифровых фотоаппаратов.
Мы предлагаем: отремонтировать фотоаппарат
Наши мастера оперативно устранят неисправности вашего устройства в сервисе или с выездом на дом!
OllieFlone
September 10, 2024 @ 9:17 am
https://easydrugrx.com/# get cialis online pharmacy
tylenol pharmacy scholarship [url=https://onlineph24.com/#]topamax pharmacy[/url] boots pharmacy doxycycline
ремонт бытовой техники челябинск
September 10, 2024 @ 9:36 am
Если вы искали где отремонтировать сломаную технику, обратите внимание – ремонт техники в челябинске
spam
September 10, 2024 @ 9:41 am
I really like your blog.. very nice colors & theme.
Did you make this website yourself or did you hire someone to do
it for you? Plz answer back as I’m looking to create my own blog and would like to find
out where u got this from. appreciate it
ReeceSot
September 10, 2024 @ 11:33 am
Ou trouver le code promo 1xbet abidjan?
Les codes promotionnels actuels 1xBet sont distribues sur Internet via des ressources partenaires, via les reseaux sociaux, via des blogueurs populaires. Les clients reguliers du bureau peuvent recevoir des codes promotionnels a la suite de la diffusion par e-mail ou par le biais de la «vitrine des codes promotionnels».
EasydrFlack
September 10, 2024 @ 11:35 am
compounding pharmacy prometrium: abdulhay ali ahmed alawadhiand bahrain pharmacy & general store – order cialis at online pharmacy
Sazriyk
September 10, 2024 @ 12:13 pm
Купить диплом о среднем образовании в Москве и любом другом городе
Cazrkni
September 10, 2024 @ 1:00 pm
[u][b] Здравствуйте![/b][/u]
Получить диплом о высшем образовании
[url=http://therebepipers.com/2015/02/01/first-exposures/#comment-83880/]купить диплом в ельце[/url]
Brucedub
September 10, 2024 @ 1:27 pm
ретриты геленджик https://ретриты.рф
перетяжка мебели
September 10, 2024 @ 2:35 pm
Как выбрать профессионалов по перетяжке мягкой мебели в Минске
Качественная перетяжка мягкой мебели в Минске
Перетяжка мягкой мебели на заказ в Минске
Недорогая перетяжка мягкой мебели в Минске
Сколько стоит перетяжка мягкой мебели в Минске
Как правильно подобрать ткань для мягкой мебели
Расцветка и дизайн мягкой мебели в Минске
Отзывы клиентов о перетяжке мебели в Минске
Экономьте с нами на перетяжке мебели
Уникальные проекты по перетяжке мягкой мебели
Индивидуальный дизайн перетяжки мягкой мебели
Как обновить мебель с минимальными затратами
Онлайн-консультация по перетяжке мягкой мебели
Инновации в процессе перетяжки мебели
Какие виды мебели мы перетягиваем
Разнообразие цветов и оттенков для перетяжки мягкой мебели в Минске
Какие условия гарантии мы предоставляем
Индивидуальный подход к оформлению вашей мебели
перетяжка мебели в Минске [url=https://t.me/s/peretyazhka_mebeli_bel/]перетяжка мебели в Минске[/url] .
Бесплатный забор и доставка мебели для перетяжки в Минске
Этапы и сроки перетяжки мебели
Ткани для мебели: преимущества и недостатки
Идеальный вариант перетяжки мебели в Минске
Оригинальный дизайн для мебели
Эксклюзивные решения для вашей мебели
Оцените стоимость перетяжки мягкой мебели
Онлайн-заказ перетяжки мебели в Минске
Интересные идеи для перетяжки мягкой мебели
Как проверить квалификацию мастеров по перетяжке мягкой мебели
Преимущества заказной перетяжки мягкой мебели
Где можно быстро и качественно перетянуть мягкую мебель в Минске
OllieFlone
September 10, 2024 @ 2:36 pm
https://onlineph24.com/# us pharmacy
viagra uk online pharmacy [url=https://easydrugrx.com/#]pantoprazole online pharmacy[/url] fluconazole mexico pharmacy
Eltonfusly
September 10, 2024 @ 3:33 pm
лучшие ретриты https://ретриты.рф
DrstoEquak
September 10, 2024 @ 5:35 pm
qatar pharmacy cialis: online international pharmacy – online pharmacy australia viagra
DrstoEquak
September 10, 2024 @ 7:05 pm
ed pills that work quickly: vipps pharmacy viagra – medical rx pharmacy
EasydrFlack
September 10, 2024 @ 7:22 pm
no prescription viagra online pharmacy: aetna online pharmacy – lamisil boots pharmacy
LloydMoilm
September 10, 2024 @ 7:27 pm
[url=https://pedri-lopas-cz.biz]www.pedri-lopas-cz.biz[/url]
last news about pedri lopas
https://pedri-lopas-cz.biz
Sazrnfl
September 10, 2024 @ 7:28 pm
Вопросы и ответы: можно ли быстро купить диплом старого образца?
OllieFlone
September 10, 2024 @ 8:28 pm
https://easydrugrx.com/# top rated online pharmacy
overseas pharmacy no prescription [url=https://easydrugrx.com/#]kamagra pharmacy bangkok[/url] cheap pharmacy cialis
Douglasbep
September 10, 2024 @ 8:52 pm
[url=https://lopas-pedri-cz.biz]http://www.lopas-pedri-cz.biz[/url]
last news about lopas pedri
http://www.lopas-pedri-cz.biz
Eltonfusly
September 10, 2024 @ 9:22 pm
ретрит перезагрузка https://ретриты.рф
Iariorvig
September 10, 2024 @ 9:37 pm
[u][b] Здравствуйте![/b][/u]
Купить документ ВУЗа вы сможете у нас.
[url=http://ya.10bb.ru/viewtopic.php?id=4105#p7321/]ya.10bb.ru/viewtopic.php?id=4105#p7321[/url]
[url=http://fauna.0pk.me/viewtopic.php?id=1357#p6277/]fauna.0pk.me/viewtopic.php?id=1357#p6277[/url]
[url=http://bvf.ru/forum/showthread.php?p=25250184#post25250184/]bvf.ru/forum/showthread.php?p=25250184#post25250184[/url]
[url=http://ya.5bb.ru/viewtopic.php?id=19989#p60454/]ya.5bb.ru/viewtopic.php?id=19989#p60454[/url]
[url=http://moscowurban.getbb.ru/viewtopic.php?f=12&t=1696/]moscowurban.getbb.ru/viewtopic.php?f=12&t=1696[/url]
Lazrakg
September 10, 2024 @ 10:06 pm
[u][b] Здравствуйте![/b][/u]
[b]Мы изготавливаем дипломы[/b] любой профессии по доступным ценам.
[url=http://freeboard.com.ua/forum/viewtopic.php?pid=947522/]freeboard.com.ua/forum/viewtopic.php?pid=947522[/url]
Sazrivq
September 10, 2024 @ 10:34 pm
Официальная покупка школьного аттестата с упрощенным обучением в Москве
Cazrwku
September 10, 2024 @ 10:58 pm
[u][b] Здравствуйте![/b][/u]
Купить диплом о высшем образовании
[url=http://rogerfilms.com/index.php?title=premialniediplomix/]купить диплом сантехника[/url]
EasydrFlack
September 10, 2024 @ 11:07 pm
tylenol 3 pharmacy name: rx pharmacy meaning – doxycycline hyclate online pharmacy
Williamraics
September 11, 2024 @ 12:00 am
[u][b] Здравствуйте![/b][/u]
Мы изготавливаем дипломы.
[url=http://poselki.animetalk.ru/viewtopic.php?id=24678#p37316/]poselki.animetalk.ru/viewtopic.php?id=24678#p37316[/url]
Davidhof
September 11, 2024 @ 12:24 am
[url=https://lopaspedri-cz.biz]www.lopaspedri-cz.biz[/url]
last news about lopas pedri
https://lopaspedri-cz.biz
Lazrxnh
September 11, 2024 @ 12:58 am
[u][b] Добрый день![/b][/u]
[b]Мы предлагаем дипломы[/b] любых профессий по приятным тарифам.
[url=http://google.mk/url?sa=i&url=aurus-diploms.com/]google.mk/url?sa=i&url=aurus-diploms.com[/url]
Cazrymo
September 11, 2024 @ 1:29 am
Пошаговая инструкция по официальной покупке диплома о высшем образовании
sbmk.org/css/pgs/kupit_diplom_universiteta__gde_nayti_i_na_chto_obratit_vnimanie
OllieFlone
September 11, 2024 @ 1:39 am
https://easydrugrx.com/# ed meds online without doctor prescription
montelukast online pharmacy [url=https://onlineph24.com/#]azithromycin mexico pharmacy[/url] lotemax online pharmacy
N Дизайн человека
September 11, 2024 @ 1:48 am
BDE Дизайн человека https://designmsu.ru Дизайн человека. 6/3 Дизайн человека.
RobertRer
September 11, 2024 @ 1:54 am
[url=https://brunoguimaraes-cz.biz]http://www.brunoguimaraes-cz.biz[/url]
last news about alisson becker
https://brunoguimaraes-cz.biz
Eanrvcb
September 11, 2024 @ 3:15 am
Привет!
Мы можем предложить дипломы любых профессий по выгодным тарифам.
[url=http://verbanent.hu/elfelejtett-jelszo/]verbanent.hu/elfelejtett-jelszo[/url]
M Дизайн человека
September 11, 2024 @ 3:21 am
LFS Дизайн человека https://vkl-design.ru Дизайн человека. 4/6 Дизайн человека.
Sazruvj
September 11, 2024 @ 3:29 am
Как приобрести диплом о среднем образовании в Москве и других городах
bakinsky-dvorik.ru/club/user/127071/blog/4892
C Дизайн человека
September 11, 2024 @ 4:17 am
UIF Дизайн человека https://designchita.ru Дизайн человека. 6/3 Дизайн человека.
Kevinrisse
September 11, 2024 @ 5:00 am
[url=https://rodrygo-silva-cz.biz]www.rodrygo-silva-cz.biz[/url]
last news about rodrygo silva
https://www.rodrygo-silva-cz.biz
Douglasjam
September 11, 2024 @ 5:40 am
Промокод Fonbet на сегодня при регистрации https://mcv26.ru/news/aktualnuy_promokod_fonbet_pri_registracii.html
Для получения актуального промокода при регистрации на сегодня рекомендуется проверять обновления на сайте Fonbet. Примером такого промокода может быть ‘GIFT200’, который активирует бесплатные ставки или другие бонусы для новых пользователей. Ввод промокода при регистрации делает процесс начала игры более выгодным и интересным.
ArchieHiz
September 11, 2024 @ 6:28 am
Pin up casino official Pil up casino website – login and play online
Derrickjoisy
September 11, 2024 @ 6:58 am
[url=https://pedricz.biz]www.pedricz.biz[/url]
last news about pedri
https://pedricz.biz
Iarioridr
September 11, 2024 @ 7:41 am
[u][b] Привет, друзья![/b][/u]
Приобрести документ о получении высшего образования можно в нашей компании.
[url=http://kuvandyk.ru/forums.php?m=posts&q=1841&n=last#bottom/]kuvandyk.ru/forums.php?m=posts&q=1841&n=last#bottom[/url]
[url=http://l67697qa.beget.tech/2024/06/18/pochemu-diplom-neobhodim-dlya-uspeshnogo-buduschego/]l67697qa.beget.tech/2024/06/18/pochemu-diplom-neobhodim-dlya-uspeshnogo-buduschego[/url]
[url=http://molbiol.ru/forums/index.php?showtopic=1191052/]molbiol.ru/forums/index.php?showtopic=1191052[/url]
[url=http://msfo-soft.ru/msfo/forum/messages/forum27/topic9933/message278468/?result=new#message278468/]msfo-soft.ru/msfo/forum/messages/forum27/topic9933/message278468/?result=new#message278468[/url]
[url=http://mysmart.ru/index.php?/gallery/image/640-диплом-как-вложение-в-будущее/]mysmart.ru/index.php?/gallery/image/640-диплом-как-вложение-в-будущее[/url]
ремонт бытовой техники в краснодаре
September 11, 2024 @ 8:55 am
Если вы искали где отремонтировать сломаную технику, обратите внимание – ремонт бытовой техники
Sazrvea
September 11, 2024 @ 9:11 am
[u][b] Привет![/b][/u]
Мы изготавливаем [b]дипломы[/b] психологов, юристов, экономистов и других профессий.
Приобретение [b]документа[/b], подтверждающего окончание ВУЗа, – это выгодное решение.
[url=http://kafdp.or.kr/bbs/board.php?bo_table=free&wr_id=495918/]kafdp.or.kr/bbs/board.php?bo_table=free&wr_id=495918[/url]
[url=http://wellenergy.by/news/by-htm/?unapproved=459680&moderation-hash=60269bf3958fb7758c7a7c952d5dc3fb#comment-459680/]wellenergy.by/news/by-htm/?unapproved=459680&moderation-hash=60269bf3958fb7758c7a7c952d5dc3fb#comment-459680[/url]
[url=http://propertyclaimspain.co.uk/hello-world/#comment-65754/]propertyclaimspain.co.uk/hello-world/#comment-65754[/url]
[url=http://85sucai.com/member/index.php?uid=ypeloc/]85sucai.com/member/index.php?uid=ypeloc[/url]
[url=http://old.trudcher.ru/users.php?m=details&id=16345/]old.trudcher.ru/users.php?m=details&id=16345[/url]
Ремонт планшетов в Москве
September 11, 2024 @ 9:14 am
Профессиональный сервисный центр по ремонту планшетов в Москве.
Мы предлагаем: сколько стоит ремонт планшета
Наши мастера оперативно устранят неисправности вашего устройства в сервисе или с выездом на дом!
Williamraics
September 11, 2024 @ 9:49 am
[u][b] Привет, друзья![/b][/u]
Мы готовы предложить дипломы.
[url=http://vladimir.ru/forum/forum/thread/39471/]vladimir.ru/forum/forum/thread/39471[/url]
Oariorklf
September 11, 2024 @ 10:31 am
[u][b] Добрый день![/b][/u]
[b]Где приобрести диплом специалиста?[/b]
[url=http://socialamerica.net/blogs/2383/Хотите-узнать-как-приобрести-документы-в-интернете-Ждем-Вас/]socialamerica.net/blogs/2383/Хотите-узнать-как-приобрести-документы-в-интернете-Ждем-Вас[/url]
[url=http://newnormalnetwork.me/read-blog/382/]newnormalnetwork.me/read-blog/382[/url]
[url=http://driversspace.com/blogs/3285/Важные-советы-что-позволят-подыскать-надежный-магазин-с-документами/]driversspace.com/blogs/3285/Важные-советы-что-позволят-подыскать-надежный-магазин-с-документами[/url]
[url=http://sustalks.com/blogs/2877/Решили-узнать-как-приобрести-диплом-в-интернете-недорого-Заходите/]sustalks.com/blogs/2877/Решили-узнать-как-приобрести-диплом-в-интернете-недорого-Заходите[/url]
[url=http://facesocial.demo3.dedicatedhost247.com/read-blog/15036/]facesocial.demo3.dedicatedhost247.com/read-blog/15036[/url]
E Дизайн человека
September 11, 2024 @ 10:58 am
HOQ Дизайн человека https://designchita.ru Дизайн человека. 1/3 Дизайн человека.
Cazrtfo
September 11, 2024 @ 11:07 am
Стоимость дипломов высшего и среднего образования и процесс их получения
cockpit.ru/img/pgs/kak_kupit_diplom_kotoruy_proydet_lubue_proverki_i_garantiruet_uspeh
ArchieHiz
September 11, 2024 @ 12:09 pm
Pin up casino official Slot pick up casino website – login and play online
Z Дизайн человека
September 11, 2024 @ 12:27 pm
OMC Дизайн человека https://designchita.ru Дизайн человека. 2/5 Дизайн человека.
MauriceJap
September 11, 2024 @ 12:53 pm
best australian online pharmacy: online pharmacy fioricet – wellbutrin target pharmacy
Z Дизайн человека
September 11, 2024 @ 2:21 pm
JGM Дизайн человека https://design-human.ru Дизайн человека. 6/3 Дизайн человека.
Arthurpax
September 11, 2024 @ 3:00 pm
http://pharmbig24.com/# Super Kamagra
RobertAmipt
September 11, 2024 @ 3:02 pm
envision rx pharmacy: buying viagra from pharmacy – rx pharmacy symbol
BarryOvete
September 11, 2024 @ 4:10 pm
[url=https://gavi-pablo-cz.biz]https://www.gavi-pablo-cz.biz[/url]
last news about gavi pablo
http://www.gavi-pablo-cz.biz
Bryonemaix
September 11, 2024 @ 5:00 pm
[url=https://gavipablo-cz.biz]www.gavipablo-cz.biz[/url]
last news about gavi pablo
http://www.gavipablo-cz.biz
Ремонт фотоаппаратов
September 11, 2024 @ 5:33 pm
Профессиональный сервисный центр по ремонту фото техники от зеркальных до цифровых фотоаппаратов.
Мы предлагаем: ремонт цифровых фотоаппаратов
Наши мастера оперативно устранят неисправности вашего устройства в сервисе или с выездом на дом!
Josephglype
September 11, 2024 @ 7:27 pm
clomid indian pharmacy [url=https://pharmbig24.online/#]caverta online pharmacy[/url] tijuana pharmacy cialis
RobertAmipt
September 11, 2024 @ 7:33 pm
mexican rx online: mexico drug stores pharmacies – mexican rx online
MauriceJap
September 11, 2024 @ 7:53 pm
cialis online review online pharmacy: pharmacy warfarin counseling – online pharmacy quick delivery
ремонт бытовой техники в новосибирске
September 11, 2024 @ 8:18 pm
Профессиональный сервисный центр по ремонту бытовой техники с выездом на дом.
Мы предлагаем:сервис центры бытовой техники новосибирск
Наши мастера оперативно устранят неисправности вашего устройства в сервисе или с выездом на дом!
Coreysah
September 11, 2024 @ 9:28 pm
Discover the world of excitement at Pin Up Casino, the world’s leading online casino. The official website Game pic up offers more than 4,000 slot machines. Play online for real money or for free using the working link today
mobile erotic massage
September 11, 2024 @ 10:28 pm
Thank you for sharing your thoughts. I truly appreciate your efforts
and I will be waiting for your next post thanks once again.
ArthurGlymn
September 11, 2024 @ 11:45 pm
Ретрит http://ретриты.рф международное обозначение времяпрепровождения, посвящённого духовной практике. Ретриты бывают уединённые и коллективные; на коллективных чаще всего проводится обучение практике медитации.
MauriceJap
September 12, 2024 @ 12:09 am
buying percocet online pharmacy: low dose naltrexone pharmacy – malaysia online pharmacy store
RobertAmipt
September 12, 2024 @ 12:47 am
atacand online pharmacy: bactrim pharmacy – erectile dysfunction drug
JamesSot
September 12, 2024 @ 2:18 am
Quelles methodes puis-je utiliser pour l’entree et la sortie?
Le bookmaker 1xbet met a la disposition des joueurs tous les moyens de paiement disponibles aujourd’hui.промокоды на один 1хбет на сегодня Vous pouvez faire un depot avec des cartes bancaires Visa et Mastercard, avec des portefeuilles electroniques, via des systemes de paiement, via des services bancaires par Internet, en utilisant les services des operateurs mobiles, sur des portefeuilles pour crypto – monnaie. Pour le retrait, il est recommande d’utiliser des cartes bancaires, des portefeuilles electroniques et des systemes de paiement. La liste des methodes de paiement se trouve dans la section «Paiements».
Coreysah
September 12, 2024 @ 2:34 am
Discover the world of excitement at Pin Up Casino, the world’s leading online casino. The official website Game pic up offers more than 4,000 slot machines. Play online for real money or for free using the working link today
Arthurpax
September 12, 2024 @ 4:23 am
https://pharmbig24.com/# kaiser online pharmacy
StevenSok
September 12, 2024 @ 5:01 am
[url=https://gavicz.biz]gavicz.biz[/url]
last news about gavi
http://www.gavicz.biz
RobertAmipt
September 12, 2024 @ 5:16 am
india pharmacy mail order: indian pharmacy paypal – reputable indian online pharmacy
Arthurpax
September 12, 2024 @ 5:55 am
http://pharmbig24.com/# rite aid pharmacy store
ремонт бытовой техники казань
September 12, 2024 @ 6:47 am
Если вы искали где отремонтировать сломаную технику, обратите внимание – ремонт бытовой техники в казани
Josephglype
September 12, 2024 @ 6:52 am
robaxin online pharmacy [url=https://pharmbig24.online/#]schnucks pharmacy buttler hill rd store hours[/url] wal mart pharmacy prices cialis
ремонт бытовой техники в краснодаре
September 12, 2024 @ 6:52 am
Если вы искали где отремонтировать сломаную технику, обратите внимание – ремонт техники в краснодаре
MauriceJap
September 12, 2024 @ 9:49 am
online pharmacy paroxetine: crestor online pharmacy – cheapest pharmacy to get concerta
StevenRaica
September 12, 2024 @ 10:22 am
[url=https://lopaspedri-cz.biz]https://lopaspedri-cz.biz[/url]
last news about pedri
https://lopaspedri-cz.biz
RobertAmipt
September 12, 2024 @ 11:00 am
buy prescription drugs from india: buy prescription drugs from india – reputable indian online pharmacy
Ремонт планшетов
September 12, 2024 @ 11:38 am
Профессиональный сервисный центр по ремонту планшетов в Москве.
Мы предлагаем: ремонт экрана планшета
Наши мастера оперативно устранят неисправности вашего устройства в сервисе или с выездом на дом!
F Дизайн человека
September 12, 2024 @ 12:07 pm
HPR Дизайн человека https://vkl-design.ru Дизайн человека. 1/4 Дизайн человека.
Игорь Тальков Я Вернусь
September 12, 2024 @ 1:06 pm
Поклонникам творчества Игоря Талькова, не пропустите!
По ссылке вы найдете легендарную
композицию “Я Вернусь” Игоря Талькова
Игорь Тальков Я Вернусь.
Окунитесь в мир глубоких эмоций прямо сейчас!
dae JtH
X Дизайн человека
September 12, 2024 @ 2:15 pm
PTO Дизайн человека https://designchita.ru Дизайн человека. 3/6 Дизайн человека.
MauriceJap
September 12, 2024 @ 2:29 pm
buy prescription drugs from india: buy prescription drugs from india – reputable indian online pharmacy
Josephglype
September 12, 2024 @ 2:35 pm
ambien pharmacy [url=https://pharmbig24.online/#]birth control online pharmacy[/url] propecia pharmacy uk
MichaelDar
September 12, 2024 @ 2:46 pm
[url=https://pablogavicz.biz]https://pablogavicz.biz[/url]
last news about pablo gavi
https://pablogavicz.biz
LloydMoilm
September 12, 2024 @ 3:06 pm
[url=https://lukamodric-cz.biz]lukamodric-cz.biz[/url]
last news about luka modric
https://lukamodric-cz.biz
BarryOvete
September 12, 2024 @ 3:18 pm
[url=https://ronaldo-cristiano-cz.biz]www.ronaldo-cristiano-cz.biz[/url]
last news about ronaldo cristiano
http://ronaldo-cristiano-cz.biz
Douglasbep
September 12, 2024 @ 4:02 pm
[url=https://lukamodriccz.biz]lukamodriccz.biz[/url]
last news about luka modric
https://www.lukamodriccz.biz
RobertAmipt
September 12, 2024 @ 4:03 pm
mexican drugstore online: mexican border pharmacies shipping to usa – mexican mail order pharmacies
Bryonemaix
September 12, 2024 @ 4:11 pm
[url=https://cristianoronaldocz.biz]https://www.cristianoronaldocz.biz[/url]
last news about cristiano ronaldo
http://cristianoronaldocz.biz
RobertRer
September 12, 2024 @ 4:44 pm
[url=https://ronaldocristiano-cz.biz]https://ronaldocristiano-cz.biz[/url]
last news about ronaldo cristiano
ronaldocristiano-cz.biz
Josephglype
September 12, 2024 @ 6:12 pm
mexico pharmacies prescription drugs [url=http://mexicopharmacy.cheap/#]buying from online mexican pharmacy[/url] mexican mail order pharmacies
Davidhof
September 12, 2024 @ 6:15 pm
[url=https://modricluka-cz.biz]modricluka-cz.biz[/url]
last news about modric luka
http://modricluka-cz.biz
Ремонт видеокамер в Москве
September 12, 2024 @ 8:18 pm
Профессиональный сервисный центр по ремонту видео техники а именно видеокамер.
Мы предлагаем: ремонт видеокамер
Наши мастера оперативно устранят неисправности вашего устройства в сервисе или с выездом на дом!
Arthurpax
September 12, 2024 @ 8:43 pm
http://pharmbig24.com/# pharmacy bangkok kamagra
RobertAmipt
September 12, 2024 @ 10:05 pm
mail order pharmacy india: buy prescription drugs from india – top 10 online pharmacy in india
Arthurpax
September 12, 2024 @ 10:17 pm
https://indianpharmacy.company/# buy medicines online in india
Stephennewly
September 12, 2024 @ 11:16 pm
[url=https://cristianoronaldo-cz.biz/]http://www.cristianoronaldo-cz.biz[/url]
last news about cristiano ronaldo
http://cristianoronaldo-cz.biz
Charlesnidly
September 12, 2024 @ 11:17 pm
[url=https://cristiano-ronaldo-cz.biz]http://www.cristiano-ronaldo-cz.biz[/url]
last news about cristiano ronaldo
http://www.cristiano-ronaldo-cz.biz
Douglasjam
September 12, 2024 @ 11:32 pm
Промокод в Фонбет https://macroclub.ru/glr/inc/aktualnuy_promokod_fonbet_pri_registracii.html
Промокоды в Фонбет предоставляют различные бонусы и преимущества для пользователей. Примером такого промокода является ‘GIFT200’, который активирует бесплатные ставки или другие бонусы при регистрации. Использование промокодов делает процесс игры на платформе более интересным и выгодным для пользователей.
MauriceJap
September 13, 2024 @ 12:16 am
india pharmacy mail order: indian pharmacy – best india pharmacy
Stephenswogs
September 13, 2024 @ 12:17 am
[url=https://gavi-cz.biz]http://www.gavi-cz.biz[/url]
last news about gavi
http://www.gavi-cz.biz
Josephglype
September 13, 2024 @ 2:01 am
latisse online pharmacy [url=https://pharmbig24.com/#]doxycycline online pharmacy no prescription[/url] dutasteride inhouse pharmacy
RobertAmipt
September 13, 2024 @ 3:29 am
online pharmacy ireland viagra: get prescription online – claritin d pharmacy counter
MauriceJap
September 13, 2024 @ 4:38 am
india pharmacy mail order: indianpharmacy com – india online pharmacy
Josephglype
September 13, 2024 @ 5:41 am
mexico drug stores pharmacies [url=http://mexicopharmacy.cheap/#]best online pharmacies in mexico[/url] reputable mexican pharmacies online
сервис профи красноярск
September 13, 2024 @ 6:55 am
Если вы искали где отремонтировать сломаную технику, обратите внимание – выездной ремонт бытовой техники в красноярске
ремонт бытовой техники в казани
September 13, 2024 @ 7:25 am
Если вы искали где отремонтировать сломаную технику, обратите внимание – ремонт бытовой техники в казани
RobertAmipt
September 13, 2024 @ 9:07 am
medicine in mexico pharmacies: mexican mail order pharmacies – mexican pharmaceuticals online
CharlieSot
September 13, 2024 @ 9:53 am
промокод в 1хбет на сегодня бесплатноt utilisez le code bonus pour obtenir un bonus VIP de 100% jusqu’a 130€ pour les Paris sportifs, ainsi qu’un bonus de casino de 1950 € + 150 tours de machines a sous. En utilisant le code promotionnel, vous pouvez vous attendre a ce que vos gains soient aussi eleves que dans le cas du plus grand bookmaker 1xbet. Tous les joueurs des pays suivants peuvent utiliser le code lors de l’inscription et recevoir un bonus de bienvenue:
Arthurpax
September 13, 2024 @ 12:44 pm
https://mexicopharmacy.cheap/# mexican drugstore online
Josephglype
September 13, 2024 @ 12:51 pm
mexican rx online [url=http://mexicopharmacy.cheap/#]best online pharmacies in mexico[/url] п»їbest mexican online pharmacies
RobertAmipt
September 13, 2024 @ 1:59 pm
buy medicines online in india: india online pharmacy – best india pharmacy
Arthurpax
September 13, 2024 @ 2:10 pm
https://mexicopharmacy.cheap/# best online pharmacies in mexico
LloydMoilm
September 13, 2024 @ 4:00 pm
[url=https://sonheungmincz.biz]https://www.sonheungmincz.biz[/url]
last news about son heung min
http://sonheungmincz.biz
Josephglype
September 13, 2024 @ 4:15 pm
reputable indian pharmacies [url=https://indianpharmacy.company/#]best india pharmacy[/url] india pharmacy
Douglasbep
September 13, 2024 @ 4:37 pm
[url=https://sonheungmin-cz.biz]http://www.sonheungmin-cz.biz[/url]
last news about son heung min
http://sonheungmin-cz.biz
ремонт техники в москве
September 13, 2024 @ 5:29 pm
Профессиональный сервисный центр по ремонту бытовой техники с выездом на дом.
Мы предлагаем: сервис центры бытовой техники москва
Наши мастера оперативно устранят неисправности вашего устройства в сервисе или с выездом на дом!
BarryOvete
September 13, 2024 @ 6:04 pm
[url=https://ronaldo-cristiano-cz.biz]https://www.ronaldo-cristiano-cz.biz[/url]
last news about robert lewandowski
https://www.ronaldo-cristiano-cz.biz
MauriceJap
September 13, 2024 @ 6:14 pm
mexican mail order pharmacies: medicine in mexico pharmacies – mexican pharmaceuticals online
ремонт техники профи в нижнем новгороде
September 13, 2024 @ 6:17 pm
Если вы искали где отремонтировать сломаную технику, обратите внимание – ремонт бытовой техники
Bryonemaix
September 13, 2024 @ 6:31 pm
[url=https://robertlewandowskicz.biz]http://robertlewandowskicz.biz[/url]
last news about robert lewandowski
http://www.robertlewandowskicz.biz
StevenRaica
September 13, 2024 @ 6:50 pm
[url=https://heungmin-son-cz.biz]http://www.heungmin-son-cz.biz[/url]
last news about heung min son
http://heungmin-son-cz.biz
Ремонт видеокамер в Москве
September 13, 2024 @ 8:55 pm
Профессиональный сервисный центр по ремонту видео техники а именно видеокамер.
Мы предлагаем: ремонт видеокамеры
Наши мастера оперативно устранят неисправности вашего устройства в сервисе или с выездом на дом!
Derrickjoisy
September 13, 2024 @ 9:55 pm
[url=https://luka-modric-cz.biz]www.luka-modric-cz.biz[/url]
last news about luka modric
http://www.luka-modric-cz.biz
Kevinrisse
September 13, 2024 @ 10:14 pm
[url=https://modric-luka-cz.biz]modric-luka-cz.biz[/url]
last news about modric luka
https://www.modric-luka-cz.biz
Josephglype
September 13, 2024 @ 11:48 pm
good online pharmacy [url=http://pharmbig24.com/#]nexium pharmacy prices[/url] mail order pharmacy
RobertAmipt
September 14, 2024 @ 12:24 am
best online pharmacies in mexico: mexico drug stores pharmacies – best online pharmacies in mexico
MauriceJap
September 14, 2024 @ 3:33 am
mexican online pharmacies prescription drugs: mexican rx online – buying prescription drugs in mexico online
Arthurpax
September 14, 2024 @ 4:04 am
https://mexicopharmacy.cheap/# medication from mexico pharmacy
ремонт техники профи в новосибирске
September 14, 2024 @ 4:52 am
Если вы искали где отремонтировать сломаную технику, обратите внимание – профи тех сервис новосибирск
StevenSok
September 14, 2024 @ 5:10 am
[url=https://son-heung-min-cz.biz]https://son-heung-min-cz.biz[/url]
last news about son heung min
https://www.son-heung-min-cz.biz
Arthurpax
September 14, 2024 @ 5:29 am
https://mexicopharmacy.cheap/# mexican drugstore online
RobertAmipt
September 14, 2024 @ 5:58 am
purple pharmacy mexico price list: medication from mexico pharmacy – mexico pharmacies prescription drugs
ремонт техники профи в красноярске
September 14, 2024 @ 7:25 am
Если вы искали где отремонтировать сломаную технику, обратите внимание – техпрофи
GeorgeSot
September 14, 2024 @ 7:57 am
Le что дают промокоды 1хбет 2024 pour beneficier d’une offre VIP de 100% jusqu’a 130€ sur les paris sportifs ou de 1950€ + 150 tours. Tous les nouveaux joueurs ayant utilise le code lors de leur inscription recoivent un bonus de bienvenue. Ce code bonus special vous permettra de recevoir un bonus augmente lors de votre inscription sur le site du bookmaker 1xBet.com ! Ne manquez pas cette occasion et inscrivez-vous des maintenant avec le code promo 1xbet pour obtenir un bonus allant jusqu’a 130 €/$.
MauriceJap
September 14, 2024 @ 8:02 am
mexican border pharmacies shipping to usa: medication from mexico pharmacy – mexican border pharmacies shipping to usa
Josephglype
September 14, 2024 @ 10:39 am
buying from online mexican pharmacy [url=http://mexicopharmacy.cheap/#]medication from mexico pharmacy[/url] purple pharmacy mexico price list
RobertAmipt
September 14, 2024 @ 10:40 am
best online pharmacy india: mail order pharmacy india – indian pharmacies safe
ремонт бытовой техники в москве
September 14, 2024 @ 1:16 pm
Профессиональный сервисный центр по ремонту бытовой техники с выездом на дом.
Мы предлагаем: ремонт крупногабаритной техники в москве
Наши мастера оперативно устранят неисправности вашего устройства в сервисе или с выездом на дом!
Josephglype
September 14, 2024 @ 1:58 pm
pharmacies in mexico that ship to usa [url=https://mexicopharmacy.cheap/#]buying prescription drugs in mexico[/url] mexican mail order pharmacies
RobertAmipt
September 14, 2024 @ 3:55 pm
purple pharmacy mexico price list: mexican pharmaceuticals online – buying prescription drugs in mexico online
сервис профи нижний новгород
September 14, 2024 @ 5:28 pm
Если вы искали где отремонтировать сломаную технику, обратите внимание – ремонт бытовой техники
MauriceJap
September 14, 2024 @ 5:35 pm
purple pharmacy mexico price list: mexican mail order pharmacies – mexico pharmacies prescription drugs
Ремонт стиральных машин в Москве
September 14, 2024 @ 6:35 pm
Профессиональный сервисный центр по ремонту стиральных машин с выездом на дом по Москве.
Мы предлагаем: ремонт стиральных машин в москве
Наши мастера оперативно устранят неисправности вашего устройства в сервисе или с выездом на дом!
сервис центры в казани
September 14, 2024 @ 6:42 pm
Профессиональный сервисный центр по ремонту бытовой техники с выездом на дом.
Мы предлагаем: ремонт бытовой техники в казани
Наши мастера оперативно устранят неисправности вашего устройства в сервисе или с выездом на дом!
Arthurpax
September 14, 2024 @ 7:04 pm
https://pharmbig24.com/# discount pharmacy viagra
Douglasbep
September 14, 2024 @ 9:08 pm
[url=https://lewandowskirobert-cz.biz]https://www.lewandowskirobert-cz.biz[/url]
last news about lewandowski robert
lewandowskirobert-cz.biz
Josephglype
September 14, 2024 @ 9:15 pm
best online pharmacy viagra review [url=http://pharmbig24.com/#]online pharmacy that sells viagra[/url] doxycycline people’s pharmacy
LloydMoilm
September 14, 2024 @ 9:17 pm
[url=https://antoine-griezmann-cz.biz]antoine-griezmann-cz.biz[/url]
last news about antoine griezmann
http://www.antoine-griezmann-cz.biz
BarryOvete
September 14, 2024 @ 10:10 pm
[url=https://antoinegriezmanncz.biz]www.antoinegriezmanncz.biz[/url]
last news about antoine griezmann
antoinegriezmanncz.biz
MauriceJap
September 14, 2024 @ 10:12 pm
mexico drug stores pharmacies: medication from mexico pharmacy – medication from mexico pharmacy
Bryonemaix
September 14, 2024 @ 10:28 pm
[url=https://griezmannantoine-cz.biz]http://www.griezmannantoine-cz.biz[/url]
last news about griezmann antoine
griezmannantoine-cz.biz
Davidhof
September 14, 2024 @ 11:08 pm
[url=https://lewandowski-robert-cz.biz]lewandowski-robert-cz.biz[/url]
last news about lewandowski robert
lewandowski-robert-cz.biz
Josephglype
September 15, 2024 @ 12:41 am
reputable mexican pharmacies online [url=https://mexicopharmacy.cheap/#]mexican border pharmacies shipping to usa[/url] purple pharmacy mexico price list
Bonus de parrainage Binance
September 15, 2024 @ 1:14 am
Your point of view caught my eye and was very interesting. Thanks. I have a question for you. https://accounts.binance.com/fr-AF/register-person?ref=JHQQKNKN
RobertAmipt
September 15, 2024 @ 2:01 am
buying from online mexican pharmacy: medicine in mexico pharmacies – pharmacies in mexico that ship to usa
RobertRer
September 15, 2024 @ 2:40 am
[url=https://robertlewandowski-cz.biz]www.robertlewandowski-cz.biz[/url]
last news about robert lewandowski
https://www.robertlewandowski-cz.biz
Douglasjam
September 15, 2024 @ 2:56 am
промокод 1xbet слоты
Free promo codes for 1xBet offer users the opportunity to claim bonuses without making a deposit. These codes may provide free bets or free spins, making them highly sought after by both new and existing users.
сервис профи новосибирск
September 15, 2024 @ 5:01 am
Если вы искали где отремонтировать сломаную технику, обратите внимание – профи ремонт
RobertAmipt
September 15, 2024 @ 6:45 am
cheapest online pharmacy india: pharmacy website india – top 10 pharmacies in india
Josephglype
September 15, 2024 @ 7:48 am
pharmacy website india [url=https://indianpharmacy.company/#]indian pharmacy[/url] pharmacy website india
MauriceJap
September 15, 2024 @ 7:56 am
buy latisse online pharmacy: medication costs – rite aid pharmacy online
Arthurpax
September 15, 2024 @ 10:33 am
http://mexicopharmacy.cheap/# mexican pharmaceuticals online
Josephglype
September 15, 2024 @ 11:04 am
lamotrigine target pharmacy [url=https://pharmbig24.online/#]diovan pharmacy coupons[/url] 24 hour drug store
Arthurpax
September 15, 2024 @ 12:03 pm
https://pharmbig24.online/# fear pharmacy ativan
RobertAmipt
September 15, 2024 @ 12:08 pm
buying prescription drugs in mexico online: pharmacies in mexico that ship to usa – mexican drugstore online
Психолог tmq
September 15, 2024 @ 12:12 pm
Надежный психоаналитик в сети. Консультации доступна 24/7. Приватно, эффективно. Индивидуальный подход. Запишитесь сейчас! https://3723-doctor-psiholog.tds-ka.ru/
MauriceJap
September 15, 2024 @ 12:27 pm
best india pharmacy: indian pharmacies safe – buy prescription drugs from india
ремонт техники профи в перми
September 15, 2024 @ 1:31 pm
Если вы искали где отремонтировать сломаную технику, обратите внимание – ремонт бытовой техники
RobertAmipt
September 15, 2024 @ 4:53 pm
indian pharmacy: indian pharmacies safe – online shopping pharmacy india
Josephglype
September 15, 2024 @ 6:11 pm
india online pharmacy [url=https://indianpharmacy.company/#]cheapest online pharmacy india[/url] india pharmacy mail order
Ремонт стиральных машин в Москве
September 15, 2024 @ 6:50 pm
Профессиональный сервисный центр по ремонту стиральных машин с выездом на дом по Москве.
Мы предлагаем: сервисный ремонт стиральных машин
Наши мастера оперативно устранят неисправности вашего устройства в сервисе или с выездом на дом!
ремонт техники в казани
September 15, 2024 @ 6:59 pm
Профессиональный сервисный центр по ремонту бытовой техники с выездом на дом.
Мы предлагаем: ремонт бытовой техники в казани
Наши мастера оперативно устранят неисправности вашего устройства в сервисе или с выездом на дом!
MichaelDar
September 15, 2024 @ 7:01 pm
[url=https://robert-lewandowski-cz.biz]https://www.robert-lewandowski-cz.biz[/url]
last news about robert lewandowski
http://robert-lewandowski-cz.biz
Josephglype
September 15, 2024 @ 9:40 pm
п»їbest mexican online pharmacies [url=http://mexicopharmacy.cheap/#]mexico drug stores pharmacies[/url] mexico drug stores pharmacies
MauriceJap
September 15, 2024 @ 10:08 pm
tadalafil online us pharmacy: cheap cialis online pharmacy – drug store
RobertAmipt
September 15, 2024 @ 10:26 pm
india pharmacy: top 10 pharmacies in india – buy prescription drugs from india
CharlieSot
September 15, 2024 @ 11:09 pm
Qu’est-ce que la strategie de jeu?
La strategie de jeu est un algorithme d’action que le joueur doit suivre en faisant un pari. Les actions peuvent inclure la taille du pari, le montant des cotes, la duree de la serie de Paris.Haiti L’objectif principal de toute strategie est d’obtenir des gains qui seront en mesure de compenser toutes les depenses precedentes.
Arthurpax
September 16, 2024 @ 2:00 am
http://mexicopharmacy.cheap/# mexican border pharmacies shipping to usa
MauriceJap
September 16, 2024 @ 2:40 am
п»їlegitimate online pharmacies india: top 10 pharmacies in india – buy medicines online in india
Josephglype
September 16, 2024 @ 4:43 am
mexican veterinary pharmacy online [url=https://pharmbig24.online/#]selegiline pharmacy[/url] people pharmacy store
StevenRaica
September 16, 2024 @ 6:15 am
[url=https://antoinegriezmann-cz.biz]https://www.antoinegriezmann-cz.biz[/url]
last news about antoine griezmann
http://www.antoinegriezmann-cz.biz
Charlesnidly
September 16, 2024 @ 6:29 am
[url=https://griezmann-antoine-cz.biz]https://www.griezmann-antoine-cz.biz[/url]
last news about griezmann antoine
griezmann-antoine-cz.biz
Stephennewly
September 16, 2024 @ 6:33 am
[url=https://bernardo-silva-cz.biz/]www.bernardo-silva-cz.biz[/url]
last news about bernardo silva
bernardo-silva-cz.biz
Психолог vzy
September 16, 2024 @ 6:52 am
Квалифицированный психотерапевт дистанционно. Поддержка доступна круглосуточно. Приватно, результативно. Доступные тарифы. Сделайте шаг к улучшению! https://1371-msk-psiholog.tds-ka.ru/
mbxat3
September 16, 2024 @ 7:20 am
ohd 753792985880936 bwvmi
Josephglype
September 16, 2024 @ 7:57 am
Online medicine order [url=http://indianpharmacy.company/#]reputable indian pharmacies[/url] indian pharmacy paypal
Douglasjam
September 16, 2024 @ 8:05 am
1xBet
1xBet provides customer support through various channels, including WhatsApp. In Sri Lanka, users can contact 1xBet’s support team via WhatsApp for assistance with issues like promo codes, bonuses, and account management.
сервис профи ростов на дону
September 16, 2024 @ 8:36 am
Если вы искали где отремонтировать сломаную технику, обратите внимание – выездной ремонт бытовой техники в ростове на дону
RobertAmipt
September 16, 2024 @ 8:48 am
reliable rx pharmacy coupons: mexican pharmacy prednisone – erectile dysfunction causes
криптобосс cryptoboss
September 16, 2024 @ 10:31 am
Увлекательное казино Cryptoboss ждет вас, играйте и выигрывайте вместе с Cryptoboss, возможность выиграть крупный джекпот, выиграйте криптовалюты в казино от Cryptoboss, Cryptoboss casino – ваш путь к успеху, захватывающий азарт с криптовалютным боссом, будьте боссом в мире криптовалютных игр с Cryptoboss casino, эксклюзивное казино для ценителей криптовалют, качественный сервис и безопасность с Cryptoboss casino, особые привилегии для лучших игроков, революция в криптовалютных играх с Cryptoboss casino, большие выигрыши ждут вас в Cryptoboss casino, получите криптовалютные призы в Cryptoboss casino, встречайте криптовалютного короля в казино, Cryptoboss casino – ваше казино удачи, присоединяйтесь к лидерам в мире криптовалютных игр с Cryptoboss casino.
cryptoboss casino [url=https://ikea-expert.ru/]cryptoboss ru[/url] .
MauriceJap
September 16, 2024 @ 12:26 pm
best online pharmacy generic levitra: online pharmacy reviews generic viagra – precision rx specialty pharmacy
Josephglype
September 16, 2024 @ 3:13 pm
mail order pharmacy india [url=http://indianpharmacy.company/#]п»їlegitimate online pharmacies india[/url] reputable indian pharmacies
ремонт бытовой техники пермь
September 16, 2024 @ 3:23 pm
Если вы искали где отремонтировать сломаную технику, обратите внимание – ремонт бытовой техники в перми
Douglasbep
September 16, 2024 @ 3:47 pm
[url=https://lionel-messi-cz.biz]http://www.lionel-messi-cz.biz[/url]
last news about lionel messi
http://lionel-messi-cz.biz
LloydMoilm
September 16, 2024 @ 4:09 pm
[url=https://lionelmessi-cz.biz]lionelmessi-cz.biz[/url]
last news about lionel messi
https://www.lionelmessi-cz.biz
cryptoboss casino зеркало рабочее на сегодня
September 16, 2024 @ 4:56 pm
Новые зеркала Cryptoboss Casino уже здесь!, играйте без проблем!
Самое актуальное зеркало Cryptoboss Casino всегда под рукой, бесперебойный доступ гарантированы.
Самое популярное зеркало Cryptoboss Casino ждет вас прямо сейчас, пропустите другие варианты!
Проводите время с удовольствием на зеркале Cryptoboss Casino!, не пропустите свой шанс!
Самое надежное зеркало Cryptoboss Casino только у нас, получайте удовольствие без лишних хлопот!
криптобосс зеркало рабочее [url=https://motorola-profi.ru/]криптобосс официальное зеркало[/url] .
MauriceJap
September 16, 2024 @ 5:05 pm
rx discount pharmacy middlesboro ky: simvastatin kroger pharmacy – inhouse pharmacy finasteride
Arthurpax
September 16, 2024 @ 5:05 pm
https://pharmbig24.online/# pharmacy program online
BarryOvete
September 16, 2024 @ 6:08 pm
[url=https://messi-lionel-cz.biz]messi-lionel-cz.biz[/url]
last news about messi lionel
messi-lionel-cz.biz
Bryonemaix
September 16, 2024 @ 6:27 pm
[url=https://kevin-de-bruyne-cz.biz]kevin-de-bruyne-cz.biz[/url]
last news about kevin de bruyne
http://www.kevin-de-bruyne-cz.biz
Josephglype
September 16, 2024 @ 6:51 pm
online shopping pharmacy india [url=http://indianpharmacy.company/#]indian pharmacies safe[/url] pharmacy website india
Ремонт игровых приставок
September 16, 2024 @ 7:14 pm
Профессиональный сервисный центр по ремонту игровых консолей Sony Playstation, Xbox, PSP Vita с выездом на дом по Москве.
Мы предлагаем: ремонт игровых приставок
Наши мастера оперативно устранят неисправности вашего устройства в сервисе или с выездом на дом!
Davidhof
September 16, 2024 @ 7:38 pm
[url=https://silvabernardocz.biz]silvabernardocz.biz[/url]
last news about silva bernardo
silvabernardocz.biz
StevenSok
September 16, 2024 @ 7:45 pm
[url=https://bernardosilva-cz.biz]www.bernardosilva-cz.biz[/url]
last news about bernardo silva
https://www.bernardosilva-cz.biz
RobertAmipt
September 16, 2024 @ 7:52 pm
mexico drug stores pharmacies: mexican pharmaceuticals online – mexico drug stores pharmacies
Ремонт видеокарт в Москве
September 16, 2024 @ 8:25 pm
Профессиональный сервисный центр по ремонту компьютерных видеокарт по Москве.
Мы предлагаем: [url=remont-videokart-gar.ru]обслуживание видеокарты цена[/url]
Наши мастера оперативно устранят неисправности вашего устройства в сервисе или с выездом на дом!
MichaelSot
September 16, 2024 @ 10:11 pm
Des que le compte est active et que le premier depot est recu sur le compte courant, le compte bonus recevra un montant de 130$. En consequence, les chances d’un joueur de gagner au premier pari augmentent de 130%. Et cet avantage est rendu disponible grace au Liban.
сервис центры в москве
September 16, 2024 @ 10:33 pm
Профессиональный сервисный центр по ремонту бытовой техники с выездом на дом.
Мы предлагаем: сервис центры бытовой техники москва
Наши мастера оперативно устранят неисправности вашего устройства в сервисе или с выездом на дом!
explainer videos
September 16, 2024 @ 11:22 pm
Great blog you have got here.. It’s difficult to find quality writing like yours these days.
I really appreciate people like you! Take care!!
Feel free to visit my blog post explainer videos
RobertAmipt
September 17, 2024 @ 1:00 am
reputable mexican pharmacies online: medication from mexico pharmacy – mexican online pharmacies prescription drugs
Josephglype
September 17, 2024 @ 2:29 am
can you buy viagra from the pharmacy [url=https://pharmbig24.com/#]best online pharmacy tadalafil[/url] prilosec online pharmacy
ремонт техники профи в ростове на дону
September 17, 2024 @ 8:49 am
Если вы искали где отремонтировать сломаную технику, обратите внимание – ремонт бытовой техники
Психолог etb
September 17, 2024 @ 5:45 pm
Компетентный специалист по психическому здоровью удаленно. Решение проблем доступна круглосуточно. Анонимно, результативно. Первая консультация -50% от цены. Начните менять жизнь сегодня! https://w-495.ru/
LloydMoilm
September 17, 2024 @ 6:45 pm
[url=https://mohamedsalahcz.biz]www.mohamedsalahcz.biz[/url]
last news about mohamed salah
http://www.mohamedsalahcz.biz
BarryOvete
September 17, 2024 @ 7:02 pm
[url=https://de-bruyne-cz.biz]de-bruyne-cz.biz[/url]
last news about de bruyne
de-bruyne-cz.biz
Douglasbep
September 17, 2024 @ 7:11 pm
[url=https://salahmohamedcz.biz]https://salahmohamedcz.biz[/url]
last news about salah mohamed
https://salahmohamedcz.biz
Bryonemaix
September 17, 2024 @ 7:27 pm
[url=https://kevindebruynecz.biz]http://kevindebruynecz.biz[/url]
last news about kevin debruyne
kevindebruynecz.biz
Ремонт игровых консолей
September 17, 2024 @ 7:34 pm
Профессиональный сервисный центр по ремонту игровых консолей Sony Playstation, Xbox, PSP Vita с выездом на дом по Москве.
Мы предлагаем: ремонт игровых приставок
Наши мастера оперативно устранят неисправности вашего устройства в сервисе или с выездом на дом!
Психолог lcf
September 17, 2024 @ 7:47 pm
Квалифицированный психоаналитик удаленно. Помощь в трудных ситуациях возможна круглосуточно. Безопасно, продуктивно. Индивидуальный подход. Сделайте шаг к улучшению! https://w-495.ru/
Kevinrisse
September 17, 2024 @ 8:18 pm
[url=https://silva-bernardo-cz.biz]https://www.silva-bernardo-cz.biz[/url]
last news about silva bernardo
https://www.silva-bernardo-cz.biz
Stephenswogs
September 17, 2024 @ 8:29 pm
[url=https://bernardosilvacz.biz]http://www.bernardosilvacz.biz[/url]
last news about bernardo silva
http://www.bernardosilvacz.biz
RobertRer
September 17, 2024 @ 8:58 pm
[url=https://messilionelcz.biz]https://www.messilionelcz.biz[/url]
last news about messi lionel
https://www.messilionelcz.biz
Derrickjoisy
September 17, 2024 @ 9:24 pm
[url=https://lionelmessicz.biz]https://www.lionelmessicz.biz[/url]
last news about lionel messi
lionelmessicz.biz
Психолог mey
September 18, 2024 @ 1:14 am
Опытный психоаналитик онлайн. Консультации доступна в любое время. Конфиденциально, действенно. Доступные тарифы. Сделайте шаг к улучшению! https://w-495.ru/
JamesCoels
September 18, 2024 @ 1:28 am
starz bet giris [url=https://starzbet.shop/#]starzbet[/url] starzbet giris
Houstonzib
September 18, 2024 @ 4:25 am
starzbet guncel giris: starz bet giris – starzbet guncel giris
betine com guncel giris [url=http://betine.online/#]betine promosyon kodu 2024[/url] betine guncel
Чат mpa
September 18, 2024 @ 5:14 am
Telegram как инструмент психотерапии: инновационные методы в психологии Чат mrl
Аптечка Онлайн
September 18, 2024 @ 6:46 am
Аптечка Онлайн – это уникальный справочник, предоставляющий полную информацию о лекарствах, таких как Ксефокам, и популярных лекарственных средствах. На Aptechka.Online пользователи могут ознакомиться с подробной информацией о таких средствах, как Урсодез, побочных эффектах и сравнить их для лечения.
Аптечка Онлайн также предоставляет пользователям возможность ознакомиться с рекомендациями о различных препаратах, таких как Ксефокам. Эти отзывы помогают определиться, какое лекарство будет подходящим в конкретном случае. Кроме того, на сайте представлено сравнение аналогов, что облегчает выбор альтернативных вариантов.
Благодаря удобной структуре на Aptechka Online, пользователи могут быстро найти интересующий их препарат, будь то описание действия или побочные эффекты. Это делает ресурс полезным помощником для тех, кто заботится о своем здоровье.
[b]Aptechka Online[/b] предлагает подробные инструкции по применению препаратов, таких как Адаптол, что помогает пользователям лучше понять, как использовать средства для профилактики различных заболеваний. На сайте также можно найти обновленные данные о противопоказаниях и возможных реакциях, что важно для эффективного применения.
Дополнительно, Aptechka Online предлагает обзоры по сравнению аналогов, таких как Урсосан. Это помогает пользователям принимать информированные решения и находить более доступные варианты лекарственных препаратов, не теряя при этом в качестве.
Houstonzib
September 18, 2024 @ 8:50 am
betine guncel: betine giris – betine giris
gates of olympus demo turkce oyna [url=http://gatesofolympusoyna.online/#]gates of olympus slot[/url] Gates of Olympus
Ремонт фотовспышек
September 18, 2024 @ 9:42 am
Профессиональный сервисный центр по ремонту фототехники в Москве.
Мы предлагаем: купить в москве фотовспышку дешево
Наши мастера оперативно устранят неисправности вашего устройства в сервисе или с выездом на дом!
Подробнее на сайте сервисного центра remont-vspyshek-realm.ru
Georgetax
September 18, 2024 @ 10:00 am
https://gatesofolympusoyna.online/# gates of olympus giris
Ремонт видео карты
September 18, 2024 @ 10:07 am
Профессиональный сервисный центр по ремонту компьютерных видеокарт по Москве.
Мы предлагаем: [url=remont-videokart-gar.ru]ремонт видеокарты компьютера[/url]
Наши мастера оперативно устранят неисправности вашего устройства в сервисе или с выездом на дом!
Georgetax
September 18, 2024 @ 11:27 am
https://gatesofolympusoyna.online/# gates of olympus demo turkce
Houstonzib
September 18, 2024 @ 2:17 pm
gates of olympus demo turkce oyna: gates of olympus giris – gates of olympus giris
betine guncel [url=https://betine.online/#]betine promosyon kodu[/url] betine promosyon kodu
Louisamark
September 18, 2024 @ 2:25 pm
эскорт услуга в москве работа эскорт услуги девочки москва
Ремонт компьютеров
September 18, 2024 @ 2:48 pm
Профессиональный сервисный центр по ремонту компьютероной техники в Москве.
Мы предлагаем: ремонт системного блока
Наши мастера оперативно устранят неисправности вашего устройства в сервисе или с выездом на дом!
JosephGrate
September 18, 2024 @ 3:21 pm
эскорт услуги москва фото эскорт москва услуги девочки
Davidhof
September 18, 2024 @ 5:49 pm
[url=https://salah-mohamed-cz.biz]http://www.salah-mohamed-cz.biz[/url]
last news about salah mohamed
http://salah-mohamed-cz.biz
Houstonzib
September 18, 2024 @ 7:07 pm
Gates of Olympus: gates of olympus demo oyna – Gates of Olympus
gates of olympus oyna demo [url=http://gatesofolympusoyna.online/#]gates of olympus oyna demo[/url] gates of olympus demo
Louisamark
September 18, 2024 @ 7:55 pm
мужской эскорт услуги москва эскорт услуги москва для женщин
JosephGrate
September 18, 2024 @ 8:46 pm
эскорт услуга в москве женские услуги эскорта в москве цены
Charlesnidly
September 18, 2024 @ 11:44 pm
[url=https://kevindebruyne-cz.biz]kevindebruyne-cz.biz[/url]
last news about kevin debruyne
http://kevindebruyne-cz.biz
MichaelDar
September 19, 2024 @ 12:31 am
[url=https://mohamed-salah-cz.biz]www.mohamed-salah-cz.biz[/url]
last news about mohamed salah
https://www.mohamed-salah-cz.biz
Georgetax
September 19, 2024 @ 3:36 am
http://gatesofolympusoyna.online/# gates of olympus oyna demo
xcu
September 19, 2024 @ 4:37 am
https://uevxyt-ceobuc.hdorg2.ru/
nybkv741
September 19, 2024 @ 4:40 am
rdx 707257616558631 gxube
GeorgeGluch
September 19, 2024 @ 6:31 am
эскорт услуги парни москва гей эскорт услуги в москве все
CharlesAlics
September 19, 2024 @ 6:37 am
эскорт услуга москва номер эскорт услуги в москве мужчин
cryptoboss официальное зеркало
September 19, 2024 @ 10:18 am
Присоединяйтесь к Cryptoboss Casino и получите море азарта и удачи
cryptoboss casino официальный зеркало [url=https://101optovik.ru/]cryptoboss официальный[/url] .
ckb
September 19, 2024 @ 11:54 am
https://yyysbt-asbdus.hdorg2.ru/
GeorgeGluch
September 19, 2024 @ 12:07 pm
мужские эскорт услуги москва эскорт услуги мальчики москва
CharlesAlics
September 19, 2024 @ 12:21 pm
эскорт услуги девушек москва эскорт услуга в москве отзывы
anwap epr
September 19, 2024 @ 2:31 pm
Анват сериалы https://bit.ly/anwap-m-anwap-anwap-love-an-wap 2415111330
RobertUnrer
September 19, 2024 @ 2:33 pm
лаки джет бонусы lucky jet бонусы
LloydMoilm
September 19, 2024 @ 3:58 pm
[url=https://juniorvinicius-cz.biz]juniorvinicius-cz.biz[/url]
last news about junior vinicius
juniorvinicius-cz.biz
Douglasbep
September 19, 2024 @ 4:26 pm
[url=https://junior-vinicius-cz.biz]www.junior-vinicius-cz.biz[/url]
last news about junior vinicius
https://www.junior-vinicius-cz.biz
cryptoboss регистрация cryptoboss ru casino
September 19, 2024 @ 4:53 pm
Успешная регистрация на сайте cryptoboss casino – ваш шанс на крупный выигрыш, Cryptoboss casino: регистрация проходит быстро и просто, Оформите аккаунт на cryptoboss casino и начните играть в любимые слоты, Cryptoboss casino ждет своих новых игроков – присоединяйтесь, Регистрация на cryptoboss casino – ваш первый шаг к увлекательному миру азартных игр, Быстрая регистрация на cryptoboss casino – ваш шанс на удачу, Пройдите регистрацию на cryptoboss casino и откройте доступ к лучшим играм, Успешная регистрация на cryptoboss casino – ваша возможность стать лидером в игровой индустрии, Регистрация на cryptoboss casino – ваш первый шаг к большим выигрышам, Cryptoboss casino ждет вас – присоединяйтесь и начните играть, Cryptoboss casino приглашает вас зарегистрироваться и испытать удачу, Присоединяйтесь к cryptoboss casino и начните выигрывать вместе с лучшими игроками, Cryptoboss casino приглашает вас стать его участником – зарегистрируйтесь сейчас, Регистрация на cryptoboss casino – ваш ключ к миру больших выигрышей, Зарегистрируйтесь на cryptoboss casino и начните путь к успеху, Регистрация на cryptoboss casino – ваш шанс на удачу.
cryptoboss регистрация [url=https://auto-adventures.ru/]cryptoboss casino регистрация на сайте hds5[/url] .
Ремонт блоков питания в Москве
September 19, 2024 @ 4:56 pm
Профессиональный сервисный центр по ремонту компьютерных блоков питания в Москве.
Мы предлагаем: ремонт блоков питания
Наши мастера оперативно устранят неисправности вашего устройства в сервисе или с выездом на дом!
BarryOvete
September 19, 2024 @ 4:58 pm
[url=https://harrykanecz.biz]harrykanecz.biz[/url]
last news about harry kane
https://www.harrykanecz.biz
Bryonemaix
September 19, 2024 @ 5:04 pm
[url=https://kaneharrycz.biz]http://kaneharrycz.biz[/url]
last news about kane harry
http://kaneharrycz.biz
Магазин раковин
September 19, 2024 @ 6:45 pm
Друзья, если планируете обновить ванную комнату, советую обратить внимание на один интернет-магазин раковин и ванн. У них действительно большой ассортимент товаров от ведущих производителей. Можно найти всё: от простых моделей до эксклюзивных дизайнерских решений.
Я искал раковина цена москва, и они предложили несколько вариантов по хорошей цене. Качество продукции на высоком уровне, всё сертифицировано. Порадовало и то, что они предлагают профессиональные консультации и услуги по установке. Доставка была быстрой, всё пришло в целости и сохранности. Отличный магазин с хорошим сервисом!
ремонт телефонов
September 19, 2024 @ 8:40 pm
Мой телефон перестал заряжаться, и я не знал, что делать. По совету друга обратился в этот сервисный центр. Мастера быстро нашли проблему и устранили её. Теперь мой телефон снова в строю! Рекомендую всем: ремонт смартфонов москва.
StevenSok
September 19, 2024 @ 8:51 pm
[url=https://vinicius-junior-cz.biz]vinicius-junior-cz.biz[/url]
last news about vinicius junior
http://vinicius-junior-cz.biz
Stephennewly
September 19, 2024 @ 10:04 pm
[url=https://kevin-debruyne-cz.biz/]www.kevin-debruyne-cz.biz[/url]
last news about kevin debruyne
http://www.kevin-debruyne-cz.biz
RobertUnrer
September 19, 2024 @ 10:20 pm
bot lucky jet лаки джет бот
cryptoboss зеркало сегодня
September 19, 2024 @ 11:11 pm
Новые зеркала Cryptoboss Casino уже здесь!, прокачивайтесь без проблем!
Попробуйте свою удачу на новом зеркале Cryptoboss Casino, бесперебойный доступ гарантированы.
Официальное зеркало Cryptoboss Casino ждет вас прямо сейчас, забудьте об другие варианты!
Проводите время с удовольствием на зеркале Cryptoboss Casino!, не пропустите свой шанс!
Без зеркала Cryptoboss Casino никуда!, зарабатывайте крупные суммы без лишних хлопот!
cryptoboss зеркало рабочее на сегодня [url=https://motorola-profi.ru/]cryptoboss зеркало сайта[/url] .
JamesCoels
September 20, 2024 @ 12:19 am
casibom guncel giris [url=http://casibom.auction/#]casibom[/url] casibom guncel giris
StevenRaica
September 20, 2024 @ 12:57 am
[url=https://mohamedsalah-cz.biz]https://mohamedsalah-cz.biz[/url]
last news about mohamed salah
http://www.mohamedsalah-cz.biz
Derrickjoisy
September 20, 2024 @ 3:48 am
[url=https://harry-kane-cz.biz]www.harry-kane-cz.biz[/url]
last news about harry kane
http://harry-kane-cz.biz
ремонт кондиционеров сервис центры в москве
September 20, 2024 @ 4:24 am
<a href=”https://remont-kondicionerov-wik.ru”>ремонт кондиционеров в москве</a>
Michaelsok
September 20, 2024 @ 5:51 am
сервис химера серч сервис химера серч
криптобосс промокод на увеличение кэшбэка
September 20, 2024 @ 5:59 am
Получите эксклюзивный cryptoboss casino промокод|Уникальный шанс с промокодом на cryptoboss casino
Секретный промокод от cryptoboss casino|Получите дополнительные бонусы с cryptoboss casino промокодом
Эксклюзивная акция для игроков cryptoboss casino|Секретный код для cryptoboss casino
Специальное предложение от cryptoboss casino с промокодом|cryptoboss casino – играйте со скидкой по промокоду
cryptoboss casino промокод бездепозитный [url=https://namelessone.ru/]cryptoboss casino промокод бездепозитный cryptoboss ru casino[/url] .
Играйте в cryptoboss casino с преимуществом промокода
Получите бонусный приз с cryptoboss casino промокодом
Регулярные акции и промокоды для cryptoboss casino|cryptoboss casino – преимущества промокодов
cryptoboss casino – ваш ключ к успеху с промокодом|cryptoboss casino – ваши шансы на победу с промокодами|cryptoboss casino промокод – ваш путь к успеху|Получите привилегии в cryptoboss casino с промокодом|Увеличьте свои шансы на победу с cryptoboss casino промокодом|cryptoboss casino – играйте с преимуществами промокода|Эксклюзивный cryptoboss casino промокод – ваш путь к победе|cryptoboss casino промокод – ваш шанс на успех|Специальные бонусы с cryptoboss casino промокодом|Бонусная программа cryptoboss casino с уникальными промокодами|Действуйте прямо сейчас с cryptoboss casino промокодом|cryptoboss casino промокод – ваш ключ к победе|Играйте в cryptoboss casino с дополнительными бонусами по промокоду|Успейте активировать cryptoboss casino промокод для дополнительных выигрышей|cryptoboss casino – играйте с выгодой с промокодами|Регулярные акции и cryptoboss casino промокоды для победы|Играйте в cryptoboss casino с дополнительными преимуществами промокода|Получите дополнительные бонусы в crypt
anwap ijp
September 20, 2024 @ 11:29 am
Anwap фильмы https://bit.ly/anwap-m-anwap-anwap-love-an-wap 191023016
JamesCoels
September 20, 2024 @ 11:35 am
betine com guncel giris [url=http://betine.online/#]betine guncel[/url] betine promosyon kodu
cryptoboss бездепозитный бонус hds5
September 20, 2024 @ 12:11 pm
Уникальный бездепозитный бонус в Cryptoboss Casino, не упустите возможность!
Играйте на деньги без вложений в Cryptoboss Casino – отличный способ испытать свою удачу.
Cryptoboss Casino радует новыми бездепозитными бонусами – лучший способ испытать удачу.
Эксклюзивный бездепозитный бонус в Cryptoboss Casino – возможно, это ваш шанс стать богатым.
Новые игроки получают уникальное предложение от Cryptoboss Casino – это шанс испытать свою удачу без риска.
Играйте на деньги, не рискуя своими средствами в Cryptoboss Casino – возможно, это ваш шанс выиграть крупный джекпот.
Получите бездепозитный бонус и начните играть в Cryptoboss Casino – лучший способ испытать свою удачу.
Бездепозитный бонус – это реальность в Cryptoboss Casino – возможно, это ваш шанс стать миллионером.
Начните играть бесплатно в Cryptoboss Casino с бездепозитным бонусом – отличный способ испытать удачу без риска.
cryptoboss бездепозитный бонус [url=https://pitaka-trade.ru/]cryptoboss бездепозитный бонус hds5[/url] .
anwap ozw
September 20, 2024 @ 2:40 pm
Anwap сериалы https://bit.ly/anwap-m-anwap-anwap-love-an-wap 52416822
Briansnale
September 20, 2024 @ 3:29 pm
мужчины для эскорт услуги в москве работа в москве эскорт услуги
Mariomus
September 20, 2024 @ 4:20 pm
Как поднять настроение подруге с помощью смешного анекдота
BriannIc
September 20, 2024 @ 5:30 pm
как зайти на blacksprut https://dark-blacksprut.com
LouisMilky
September 20, 2024 @ 5:49 pm
лаки джет бот лаки джет официальный сайт
криптобосс автоматы
September 20, 2024 @ 6:54 pm
Игровые автоматы на сайте Cryptoboss Casino, для незабываемого времяпрепровождения.
Попробуйте свою удачу на автоматах в казино Cryptoboss, для любителей крупных выигрышей.
Побеждайте на игровых автоматах Cryptoboss Casino, для тех, кто ищет адреналин.
Увлекательные автоматы ждут вас на сайте Cryptoboss Casino, для азартных игроков.
Развлекайтесь в казино Cryptoboss на лучших автоматах, и выигрывайте крупные суммы на реальные деньги.
Самые популярные автоматы в казино Cryptoboss, где выигрыши ждут каждого.
Играйте на деньги на своих любимых слотах, для азартных игроков.
На сайте Cryptoboss ждут увлекательные слоты, которые подарят вам удовольствие.
Почувствуйте волнение от игры в казино Cryptoboss на автоматах, которые покорят вас своими возможностями.
Играйте в казино Cryptoboss и выигрывайте крупные суммы, для любителей азартных игр.
Играйте в лучшие игровые автоматы на сайте Cryptoboss, где вас ждут крупные выигрыши.
Попробуйте свою удачу в казино Cryptoboss на увлекательных автоматах, для тех, кто ищет адреналин.
Увлекательные слоты на сайте Cryptoboss ждут вас, для тех, кто мечтает о крупном выигрыше.
Игровые автоматы в казино Cryptoboss, для азартных игроков.
Играйте на деньги в казино Cryptoboss на лучших автоматах, где каждый может стать победителем.
Играйте на популярных слотах на сайте Cryptoboss Casino, для любителей крупных выигрышей.
Эмоции бурлят в крови, играя в казино Cryptoboss на автоматах, где каждый может испыт
игровые автоматы криптобосс [url=https://zasport.su/]криптобосс автоматы[/url] .
Stephenswogs
September 20, 2024 @ 7:34 pm
[url=https://viniciusjunior-cz.biz]http://www.viniciusjunior-cz.biz[/url]
last news about vinicius junior
https://viniciusjunior-cz.biz
RobertRer
September 20, 2024 @ 7:35 pm
[url=https://harrykane-cz.biz]www.harrykane-cz.biz[/url]
last news about harry kane
https://harrykane-cz.biz
Kevinrisse
September 20, 2024 @ 7:56 pm
[url=https://viniciusjuniorcz.biz]viniciusjuniorcz.biz[/url]
last news about vinicius junior
http://www.viniciusjuniorcz.biz
BriannIc
September 20, 2024 @ 10:23 pm
blacksprut зеркала актуальные https://dark-blacksprut.com
Ремонт системных блоков
September 20, 2024 @ 11:33 pm
Профессиональный сервисный центр по ремонту компьютероной техники в Москве.
Мы предлагаем: лучшие сервисные центры по ремонту компьютеров
Наши мастера оперативно устранят неисправности вашего устройства в сервисе или с выездом на дом!
BarryOvete
September 21, 2024 @ 12:06 am
[url=https://kylian-mbappecz.biz]www.kylian-mbappecz.biz[/url]
last news about kylian mbappe
https://kylian-mbappecz.biz
LloydMoilm
September 21, 2024 @ 12:22 am
[url=https://mbappekyliancz.biz]https://www.mbappekyliancz.biz[/url]
last news about mbappe kylian
https://mbappekyliancz.biz
Bryonemaix
September 21, 2024 @ 12:29 am
[url=https://kylian-mbappe-cz.biz]http://kylian-mbappe-cz.biz[/url]
last news about kylian mbappe
https://kylian-mbappe-cz.biz
Douglasbep
September 21, 2024 @ 1:03 am
[url=https://mbappe-kylian-cz.biz]https://www.mbappe-kylian-cz.biz[/url]
last news about mbappe kylian
http://www.mbappe-kylian-cz.biz
Normansox
September 21, 2024 @ 1:04 am
comprar viagra en espaГ±a envio urgente contrareembolso: viagra generico – sildenafilo 50 mg comprar online
DennisRurne
September 21, 2024 @ 1:16 am
http://tadalafilo.bid/# farmacias online seguras
Jamestew
September 21, 2024 @ 1:16 am
farmacias direct: farmacia online madrid – п»їfarmacia online espaГ±a
курс доллара атырау
September 21, 2024 @ 1:44 am
Текущий курс валют в Казахстане: актуальная информация
Способы отслеживания курса валют в Казахстане
Какие валюты выгодно менять в Казахстане
Прогноз курса валют в Казахстане
Точки обмена валюты в Казахстане
курс долара к тенге [url=https://kursy-valut-online.kz/]курс валют караганда[/url] .
FloydBer
September 21, 2024 @ 2:16 am
http://tadalafilo.bid/# farmacia barata
farmacia online barata y fiable
Davidhof
September 21, 2024 @ 2:39 am
[url=https://jude-bellingham-cz.biz]http://www.jude-bellingham-cz.biz[/url]
last news about jude bellingham
http://www.jude-bellingham-cz.biz
LouisMilky
September 21, 2024 @ 3:06 am
lucky jet бот lucky jet официальный сайт
FloydBer
September 21, 2024 @ 3:55 am
https://farmaciaeu.com/# farmacias online seguras
farmacias online baratas
Normansox
September 21, 2024 @ 8:15 am
farmacias online seguras: Precio Cialis 20 Mg – farmacias online baratas
DennisRurne
September 21, 2024 @ 8:22 am
http://farmaciaeu.com/# farmacia online envГo gratis
Jamesfuh
September 21, 2024 @ 9:12 am
Лечебно-оздоровительный массаж Ивантеевка по выгодным ценам. Стоимость процедуры, порядок проведения, запись на прием.
Ремонт проекторов в Москве
September 21, 2024 @ 9:42 am
Профессиональный сервисный центр по ремонту фото техники от зеркальных до цифровых фотоаппаратов.
Мы предлагаем: ремонт проекторов в москве
Наши мастера оперативно устранят неисправности вашего устройства в сервисе или с выездом на дом!
Ремонт камер видео наблюдения в Москве
September 21, 2024 @ 10:25 am
Профессиональный сервисный центр по ремонту камер видео наблюдения по Москве.
Мы предлагаем: ремонт систем видеонаблюдения москва
Наши мастера оперативно устранят неисправности вашего устройства в сервисе или с выездом на дом!
DennisRurne
September 21, 2024 @ 11:41 am
http://tadalafilo.bid/# farmacias online seguras
Normansox
September 21, 2024 @ 1:23 pm
farmacia en casa online descuento: farmacia online barata – farmacia barata
ngp
September 21, 2024 @ 3:46 pm
11102863 https://anwap2.yxoo.ru/
ремонт бытовой техники в нижнем новгороде
September 21, 2024 @ 5:16 pm
Профессиональный сервисный центр по ремонту бытовой техники с выездом на дом.
Мы предлагаем: сервисные центры в нижнем новгороде
Наши мастера оперативно устранят неисправности вашего устройства в сервисе или с выездом на дом!
FloydBer
September 21, 2024 @ 5:46 pm
https://farmaciaeu.com/# farmacias online seguras
farmacias online seguras
jjy
September 21, 2024 @ 5:59 pm
24423411 https://anwap2.yxoo.ru/
DennisRurne
September 21, 2024 @ 6:52 pm
https://sildenafilo.men/# viagra online cerca de malaga
FloydBer
September 21, 2024 @ 7:23 pm
http://farmaciaeu.com/# farmacia online espaГ±a envГo internacional
farmacias direct
DennisRurne
September 21, 2024 @ 10:37 pm
http://tadalafilo.bid/# farmacia online madrid
Normansox
September 21, 2024 @ 10:41 pm
farmacia online espaГ±a envГo internacional: comprar cialis original – п»їfarmacia online espaГ±a
Jamestew
September 21, 2024 @ 10:43 pm
farmacia barata: farmacia online barata y fiable – farmacias online seguras en espaГ±a
Douglasjam
September 21, 2024 @ 11:26 pm
1xBet
1xBet offers promo codes that provide free spins on various slot games. These codes can be used in the casino section of the site, giving players the chance to win without risking their own money. Free spins are a popular reward for both new and existing users.
Ronalditady
September 21, 2024 @ 11:28 pm
farmacias online seguras en espaГ±a [url=https://tadalafilo.bid/#]tadalafilo[/url] farmacias online seguras en espaГ±a
криптобосс официальный сайт зеркало
September 22, 2024 @ 3:22 am
Присоединяйтесь к Cryptoboss Casino и покоряйте новые вершины
криптобосс официальный [url=https://101optovik.ru/]криптобосс официальный сайт рабочее зеркало[/url] .
Normansox
September 22, 2024 @ 4:05 am
viagra online gibraltar: sildenafil 100mg genГ©rico – comprar viagra online en andorra
DennisRurne
September 22, 2024 @ 5:28 am
http://farmaciaeu.com/# farmacia online envГo gratis
Jamestew
September 22, 2024 @ 5:40 am
farmacia barata: farmacia online envio gratis murcia – farmacia online 24 horas
ремонт бытовой техники в перми
September 22, 2024 @ 6:53 am
Профессиональный сервисный центр по ремонту бытовой техники с выездом на дом.
Мы предлагаем: ремонт крупногабаритной техники в перми
Наши мастера оперативно устранят неисправности вашего устройства в сервисе или с выездом на дом!
Charlesnidly
September 22, 2024 @ 7:13 am
[url=https://kane-harry-cz.biz]https://www.kane-harry-cz.biz[/url]
last news about kane harry
kane-harry-cz.biz
FloydBer
September 22, 2024 @ 9:38 am
http://tadalafilo.bid/# farmacias direct
farmacias online seguras en espaГ±a
криптобосс регистрация hds5
September 22, 2024 @ 9:47 am
Присоединяйтесь к cryptoboss casino сегодня и получите доступ к лучшим играм, Проведите регистрацию на cryptoboss casino за несколько минут, Начните путь к большим выигрышам с регистрации на сайте cryptoboss casino, Уникальная атмосфера cryptoboss casino для зарегистрированных пользователей, Пройдите регистрацию на cryptoboss casino и начните выигрывать, Не упустите возможность зарегистрироваться на cryptoboss casino и выиграть крупный приз, Cryptoboss casino рад приветствовать новых игроков – зарегистрируйтесь прямо сейчас, Присоединяйтесь к cryptoboss casino и начните выигрывать большие суммы денег, Регистрация на cryptoboss casino – ваш первый шаг к большим выигрышам, Cryptoboss casino ждет вас – присоединяйтесь и начните играть, Присоединяйтесь к cryptoboss casino и получите шанс на крупный выигрыш, Cryptoboss casino: регистрация – быстро, просто, надежно, Cryptoboss casino приглашает вас стать его участником – зарегистрируйтесь сейчас, Уникальные бонусы ждут вас после регистрации на cryptoboss casino, Не упустите возможность зарегистрироваться на cryptoboss casino и испытать азартные ощущения, Cryptoboss casino приглашает вас стать его частью – зарегистрируйтесь и начните играть.
криптобосс регистрация cryptoboss casino3 one [url=https://auto-adventures.ru/]криптобосс промокод при регистрации hds5[/url] .
JacobSot
September 22, 2024 @ 12:59 pm
4rabet promo code registration: A no-cost promo code that provides Indian users with bonuses like free bets or spins, often without needing a deposit.
DennisRurne
September 22, 2024 @ 2:40 pm
http://tadalafilo.bid/# farmacia online espaГ±a envГo internacional
cryptoboss зеркало сайта
September 22, 2024 @ 4:17 pm
Не упустите шанс, переходите на зеркало Cryptoboss Casino сейчас, прокачивайтесь без проблем!
Самое актуальное зеркало Cryptoboss Casino всегда под рукой, надежная связь гарантированы.
Самое популярное зеркало Cryptoboss Casino ждет вас прямо сейчас, пропустите другие варианты!
Новости и выигрыши ждут вас на зеркале Cryptoboss Casino, играйте и выигрывайте!
Самое надежное зеркало Cryptoboss Casino только у нас, получайте удовольствие без лишних хлопот!
криптобосс зеркало рабочее [url=https://motorola-profi.ru/]криптобосс зеркало cryptoboss ru casino[/url] .
ремонт бытовой техники в тюмени
September 22, 2024 @ 4:23 pm
Если вы искали где отремонтировать сломаную технику, обратите внимание – [url=https://tyumen.profi-teh-remont.ru]ремонт бытовой техники в тюмени[/url]
best online casino australia real money
September 22, 2024 @ 5:15 pm
Hi there i am kavin, its my first time to commenting anyplace, when i
read this paragraph i thought i could also make comment due
to this sensible article.
DennisRurne
September 22, 2024 @ 5:42 pm
http://tadalafilo.bid/# farmacia barata
Normansox
September 22, 2024 @ 6:53 pm
farmacias direct: cialis 20 mg precio farmacia – farmacia online barcelona
LloydMoilm
September 22, 2024 @ 10:04 pm
[url=https://ekologiya.news161.ru]http://ekologiya.news161.ru[/url]
Экология Ростовской области
http://ekologiya.news161.ru
криптобосс промокод hds5
September 22, 2024 @ 10:26 pm
Получите эксклюзивный cryptoboss casino промокод|Новый промокод для cryptoboss casino
Играйте в cryptoboss casino с привлекательным промокодом|Получите дополнительные бонусы с cryptoboss casino промокодом
Только сегодня! cryptoboss casino промокод|Играйте в cryptoboss casino с бонусным кодом
cryptoboss casino промокод – ваш ключ к выигрышу|Бонус при регистрации в cryptoboss casino с промокодом
cryptoboss casino промокод бездепозитный cryptoboss ru casino [url=https://namelessone.ru/]cryptoboss промокод на бездепозитный[/url] .
Успех в cryptoboss casino с промокодом
Получите бонусный приз с cryptoboss casino промокодом
Играйте в cryptoboss casino с дополнительными бонусами от промокодов|Эксклюзивный cryptoboss casino промокод – ваш шанс на победу
cryptoboss casino – ваш ключ к успеху с промокодом|cryptoboss casino промокод – ваш путь к большим выигрышам|Бонусная программа cryptoboss casino с уникальным промокодом|Играйте с выгодой: cryptoboss casino промокод|Играйте в cryptoboss casino с эксклюзивным промокодом для успеха|Успейте активировать промокод для увеличения выигрыша|Получите дополнительные бонусы с cryptoboss casino промокодом|Используйте промокод в cryptoboss casino для больших побед|Специальные бонусы с cryptoboss casino промокодом|Увеличьте свои шансы на победу с промокодами от cryptoboss casino|Играйте в cryptoboss casino с преимуществами промокода|Используйте промокод в cryptoboss casino для больших выигрышей|Играйте в cryptoboss casino с дополнительными бонусами по промокоду|Играйте в cryptoboss casino с уникальными привилегиями по промокоду|Используйте cryptoboss casino промокод для увеличения шансов на победу|Уникальные предложения от cryptoboss casino с промокодом|Получите привилегии с cryptoboss casino промокодом|Играйте в cryptoboss casino с уникальным промокодом для успеха
arest
September 22, 2024 @ 10:27 pm
[url=https://pravosudie.news161.ru]арест[/url]
Правосудие Ростовской области
https://pravosudie.news161.ru
zakon
September 22, 2024 @ 10:41 pm
[url=https://zakony.news161.ru]www.zakony.news161.ru[/url]
Законодательство Ростовской области
http://www.kylian-mbappe-cz.biz
Proletarsk
September 22, 2024 @ 10:49 pm
[url=https://proletarsk.news161.ru]http://proletarsk.news161.ru[/url]
Новости Пролетарского района Ростовской области
http://www.proletarsk.news161.ru
bagaevskaya
September 22, 2024 @ 11:54 pm
[url=https://bagaevskaya.news161.ru]www.bagaevskaya.news161.ru[/url]
Последние новости Багаевского района Ростовской области
http://bagaevskaya.news161.ru
DennisRurne
September 23, 2024 @ 12:39 am
http://sildenafilo.men/# venta de viagra a domicilio
Charlessop
September 23, 2024 @ 4:19 am
Farmacia online miglior prezzo [url=https://farmaciait.men/#]Farmacie online sicure[/url] farmacie online sicure
cryptoboss casino бонусы cryptoboss casino ru
September 23, 2024 @ 4:21 am
Cryptoboss Casino радует бездепозитным бонусом, восхитительное предложение!
Заработайте крупный выигрыш без вложений в Cryptoboss Casino – шикарная возможность выиграть крупный джекпот.
Cryptoboss Casino радует новыми бездепозитными бонусами – лучший способ испытать удачу.
Играйте на деньги с бездепозитным бонусом в Cryptoboss Casino – возможно, это ваш шанс стать богатым.
Cryptoboss Casino радует бездепозитными бонусами для всех – уникальные преимущества для игры на деньги.
Бездепозитный бонус в Cryptoboss Casino – это реальность – лучший способ испытать свою удачу.
Cryptoboss Casino радует новых игроков щедрыми бонусами – шикарная возможность заработать без вложений.
Играйте без вложений и выигрывайте настоящие деньги в Cryptoboss Casino – возможно, это ваш шанс стать миллионером.
Начните играть бесплатно в Cryptoboss Casino с бездепозитным бонусом – отличный способ испытать удачу без риска.
cryptoboss бездепозитный бонус [url=https://pitaka-trade.ru/]cryptoboss casino бездепозитный бонус hds5[/url] .
Derrickjoisy
September 23, 2024 @ 4:30 am
[url=https://bellingham-jude-cz.biz]https://www.bellingham-jude-cz.biz[/url]
last news about bellingham jude
http://bellingham-jude-cz.biz
StevenSok
September 23, 2024 @ 4:34 am
[url=https://judebellingham-cz.biz]https://judebellingham-cz.biz[/url]
last news about jude bellingham
http://www.judebellingham-cz.biz
Stephennewly
September 23, 2024 @ 4:59 am
[url=https://bellinghamjude-cz.biz/]http://bellinghamjude-cz.biz[/url]
last news about bellingham jude
https://www.bellinghamjude-cz.biz
EdisonMaype
September 23, 2024 @ 5:30 am
viagra generico sandoz: viagra prezzo – pillole per erezione immediata
Louisgot
September 23, 2024 @ 6:34 am
viagra naturale: viagra – viagra naturale in farmacia senza ricetta
MichaelDar
September 23, 2024 @ 7:00 am
[url=https://kylianmbappecz.biz]https://www.kylianmbappecz.biz[/url]
last news about kylian mbappe
kylianmbappecz.biz
ThomasSot
September 23, 2024 @ 8:11 am
4rabet deposit bonus code: A general promo code for the year 2024, offering bonuses for new and existing users. This can be used for sports betting or casino games on 4rabet.
сервис центры в москве
September 23, 2024 @ 9:02 am
Профессиональный сервисный центр по ремонту бытовой техники с выездом на дом.
Мы предлагаем: сервисные центры в москве
Наши мастера оперативно устранят неисправности вашего устройства в сервисе или с выездом на дом!
Charlessop
September 23, 2024 @ 10:44 am
Farmacia online miglior prezzo [url=http://farmaciait.men/#]Farmacia online migliore[/url] acquisto farmaci con ricetta
Patrikthync
September 23, 2024 @ 10:54 am
Что такое смешные картинки и как они появились
игровые автоматы криптобосс hds5
September 23, 2024 @ 12:15 pm
Захватывающие слоты в казино Cryptoboss, для азартных игроков.
Лучшие слоты ждут вас в казино Cryptoboss, где выигрыш станет реальностью.
Побеждайте на игровых автоматах Cryptoboss Casino, для тех, кто ищет адреналин.
Играйте в казино Cryptoboss на лучших слотах, для тех, кто мечтает выиграть крупный приз.
Получайте удовольствие от игры в автоматы на сайте Cryptoboss Casino, для тех, кто мечтает о крупном выигрыше.
Играйте на популярных слотах в Cryptoboss Casino, где выигрыши ждут каждого.
Получайте удовольствие от игры на автоматах в казино Cryptoboss, для тех, кто мечтает о крупном выигрыше.
На сайте Cryptoboss ждут увлекательные слоты, для любителей азартных игр.
Почувствуйте волнение от игры в казино Cryptoboss на автоматах, для ценителей азарта.
Лучшие слоты на сайте Cryptoboss ждут вас, для любителей азартных игр.
Не пропустите уникальные предложения для игры в казино Cryptoboss на автоматах, для азартных игроков.
Лучшие игровые автоматы в казино Cryptoboss, для ценителей азарта.
Увлекательные слоты на сайте Cryptoboss ждут вас, для тех, кто мечтает о крупном выигрыше.
Игровые автоматы в казино Cryptoboss, для тех, кто ищет новые ощущения.
Лучшие игровые автоматы на сайте Cryptoboss, для любителей азарта.
Попробуйте свою удачу в казино Cryptoboss, для любителей крупных выигрышей.
Лучшие игровые автоматы на сайте Cryptoboss, где каждый может испыт
cryptoboss casino автоматы [url=https://zasport.su/]криптобосс игровые автоматы на деньги[/url] .
Louisgot
September 23, 2024 @ 1:12 pm
siti sicuri per comprare viagra online: viagra – viagra naturale in farmacia senza ricetta
Charlessop
September 23, 2024 @ 1:40 pm
pillole per erezioni fortissime [url=http://sildenafilit.pro/#]viagra senza ricetta[/url] cialis farmacia senza ricetta
手マン
September 23, 2024 @ 2:04 pm
Hello, every time i used to check web site posts here in the early
hours in the morning, for the reason that i like to find out more and more.
ремонт техники в красноярске
September 23, 2024 @ 2:18 pm
Профессиональный сервисный центр по ремонту бытовой техники с выездом на дом.
Мы предлагаем: сервисные центры по ремонту техники в красноярске
Наши мастера оперативно устранят неисправности вашего устройства в сервисе или с выездом на дом!
ремонт техники профи в волгограде
September 23, 2024 @ 6:00 pm
Если вы искали где отремонтировать сломаную технику, обратите внимание – ремонт бытовой техники
курс рубля в актобе
September 23, 2024 @ 6:53 pm
Текущий курс валют в Казахстане: актуальная информация
Как узнать курс валют в Казахстане
Какие валюты выгодно менять в Казахстане
На сколько выгодно менять валюту в Казахстане
Где выгодно обменять деньги в Казахстане
курс доллара караганда [url=https://kursy-valut-online.kz/]курс доллара актау[/url] .
Edwardinfor
September 23, 2024 @ 8:08 pm
viagra 100 mg prezzo in farmacia [url=https://sildenafilit.pro/#]viagra prezzo[/url] pillole per erezione immediata
bezplatn'y úcet na binance
September 23, 2024 @ 9:27 pm
Can you be more specific about the content of your article? After reading it, I still have some doubts. Hope you can help me. https://accounts.binance.com/uk-UA/register-person?ref=W0BCQMF1
CasinoSig
September 23, 2024 @ 11:06 pm
Для выбора онлайн-казино следует учитывать несколько основных критериев, чтобы создать безопасность и позитивный опыт игры. Во-первых, необходимо проверить на наличие лицензии: это обеспечивает, что казино легитимно и соблюдает все требованиям, оберегающие геймеров.
Кроме того, — авторитет. Изучите мнения и обзоры на сайтах других игроков, чтобы убедиться, как казино справляется с выплатами выигрышей и помощью клиентам.
Наш сайт: https://slubowisko.pl/topic/73821/
Важно также оценить выбор игр и разработчиков: проверенные казино предлагают игры от известными производителями игр, такими как Pragmatic Play или Microgaming. Обратите внимание на на акции, но не забудьте посмотреть условия вывода бонусов — условия могут быть сложными.
К тому же, рекомендуется проверить уровень обслуживания. Хорошее казино предлагает своим пользователям разные способы связи, таких как чат-сервис, электронные письма и телефонная линия, а также оперативно отвечает на обращения игроков. Очень хорошо, если поддержка круглосуточная и предоставляет помощь на вашем предпочитаемом языке.
Ещё одним аспектом является наличие мобильного приложения или приложения — это даст вам запускать игры в любое время и в любом удобном месте. Не забудьте убедиться в ограничения по стране по стране и возрасту, чтобы предотвратить трудности с доступом к играм и казино и выводом заработка. Оценив все эти аспекты, вы сможете найти казино, которое идеально соответствует вашим вашим потребностям и нуждам.
Louisgot
September 24, 2024 @ 12:28 am
farmacie online affidabili: Cialis generico controindicazioni – Farmacie on line spedizione gratuita
Douglasjam
September 24, 2024 @ 1:40 am
1xBet
A 1xBet bonus code is a specific type of promo code that unlocks bonuses like deposit matches, free bets, or free spins. These codes are typically provided to new users during registration but can also be available to existing users through promotions.
Louisgot
September 24, 2024 @ 3:58 am
cialis farmacia senza ricetta: viagra generico – cialis farmacia senza ricetta
Stephenswogs
September 24, 2024 @ 5:34 am
[url=https://erling-haaland-cz.biz]https://erling-haaland-cz.biz[/url]
last news about erling haaland
http://erling-haaland-cz.biz
outlookindiasportsm
September 24, 2024 @ 5:47 am
This blog’s entries are all generally top-notch and successful. I am really excited to read this post. Thank you, and keep going.
토토사이트
Ремонт парогенераторов
September 24, 2024 @ 8:44 am
Профессиональный сервисный центр по ремонту парогенераторов в Москве.
Мы предлагаем: починить парогенератор
Наши мастера оперативно устранят неисправности вашего устройства в сервисе или с выездом на дом!
Richardpeado
September 24, 2024 @ 10:45 am
farmaci senza ricetta elenco [url=https://tadalafilit.com/#]Farmacie che vendono Cialis senza ricetta[/url] farmacie online autorizzate elenco
Patrikthync
September 24, 2024 @ 10:59 am
Смешные анекдоты в приложении
Всё шуточки
EdgarPiede
September 24, 2024 @ 11:49 am
pillole per erezioni fortissime: viagra prezzo – alternativa al viagra senza ricetta in farmacia
ремонт техники в красноярске
September 24, 2024 @ 3:04 pm
Профессиональный сервисный центр по ремонту бытовой техники с выездом на дом.
Мы предлагаем: сервисные центры в красноярске
Наши мастера оперативно устранят неисправности вашего устройства в сервисе или с выездом на дом!
EdgarPiede
September 24, 2024 @ 3:55 pm
pillole per erezioni fortissime: viagra online siti sicuri – cerco viagra a buon prezzo
bokep sma
September 24, 2024 @ 5:03 pm
We are a bunch of volunteers and opening a brand new scheme in our community.
Your website offered us with valuable information to work on. You have done a
formidable process and our entire neighborhood shall be
thankful to you.
Richardpeado
September 24, 2024 @ 6:20 pm
comprare farmaci online con ricetta [url=http://tadalafilit.com/#]Tadalafil generico migliore[/url] top farmacia online
أنابيب الرصاص
September 24, 2024 @ 6:56 pm
Drip Irrigation Pipes in Iraq At Elite Pipe Factory, our drip irrigation pipes are engineered to provide efficient water delivery for agricultural applications. These pipes are designed to minimize water wastage and enhance crop yields, reflecting our commitment to advancing irrigation technology in Iraq. As a leading and reliable manufacturer, Elite Pipe Factory ensures that our drip irrigation pipes are of the highest quality, contributing to successful farming practices. Explore our drip irrigation solutions at elitepipeiraq.com.
Richardpeado
September 24, 2024 @ 9:44 pm
comprare farmaci online all’estero [url=https://tadalafilit.com/#]Tadalafil generico migliore[/url] farmacie online autorizzate elenco
Kevinrisse
September 24, 2024 @ 10:50 pm
[url=https://erlinghaaland-cz.biz]erlinghaaland-cz.biz[/url]
last news about erling haaland
https://www.erlinghaaland-cz.biz
RobertRer
September 24, 2024 @ 11:21 pm
[url=https://erlinghaalandcz.biz]https://erlinghaalandcz.biz[/url]
last news about erling haaland
https://www.erlinghaalandcz.biz
Discover More Here
September 24, 2024 @ 11:35 pm
Nice post. I was checking continuously this blog and I am impressed!
Extremely helpful information specially the last part 🙂 I
care for such info a lot. I was looking for this
particular information for a long time. Thank you and good luck.
EdgarPiede
September 24, 2024 @ 11:46 pm
farmacie online affidabili: Farmacia online piГ№ conveniente – Farmacia online miglior prezzo
ремонт техники профи в уфе
September 25, 2024 @ 1:53 am
Если вы искали где отремонтировать сломаную технику, обратите внимание – профи тех сервис уфа
EdgarPiede
September 25, 2024 @ 3:31 am
acquistare farmaci senza ricetta: Brufen antinfiammatorio – п»їFarmacia online migliore
StevenRaica
September 25, 2024 @ 4:45 am
[url=https://judebellinghamcz.biz]judebellinghamcz.biz[/url]
last news about jude bellingham
http://www.judebellinghamcz.biz
Richardpeado
September 25, 2024 @ 5:53 am
viagra generico recensioni [url=https://sildenafilit.pro/#]viagra[/url] siti sicuri per comprare viagra online
Michaelwem
September 25, 2024 @ 8:53 am
Farmacie online sicure [url=https://brufen.pro/#]Brufen 600 senza ricetta[/url] Farmacia online miglior prezzo
Richardpeado
September 25, 2024 @ 9:51 am
farmacie online sicure [url=http://tadalafilit.com/#]Cialis generico farmacia[/url] farmacie online autorizzate elenco
ремонт техники профи в самаре
September 25, 2024 @ 10:15 am
сервис профи самара
NamerMonk
September 25, 2024 @ 12:08 pm
эскорт сопровождение в Санкт-Петербурге
Hempified CBD
September 25, 2024 @ 12:38 pm
https://glbian.com/prd/bbs/board.php?bo_table=free&wr_id=45768 is a leading provider of high-quality CBD products that are derived from organic hemp plants.
Patrikthync
September 25, 2024 @ 12:54 pm
Что такое анекдот? Почему мы любим прикольные анекдоты? Как они появились? Читайте об этом
статью
Richardpeado
September 25, 2024 @ 6:51 pm
viagra generico recensioni [url=https://sildenafilit.pro/#]acquisto viagra[/url] viagra generico in farmacia costo
PatrickMourO
September 25, 2024 @ 9:59 pm
viagra consegna in 24 ore pagamento alla consegna: viagra senza ricetta – alternativa al viagra senza ricetta in farmacia
Richardpeado
September 25, 2024 @ 10:49 pm
top farmacia online [url=https://brufen.pro/#]BRUFEN 600 acquisto online[/url] farmaci senza ricetta elenco
EdgarPiede
September 25, 2024 @ 11:31 pm
farmacie online autorizzate elenco: Cialis generico 20 mg 8 compresse prezzo – farmaci senza ricetta elenco
Patrikthync
September 25, 2024 @ 11:43 pm
Что такое анекдот? Почему мы любим смешные анекдоты? Как они появились? Читайте об этом
статью
Самые смешные анекдоты на нашем сайте.
StevenSok
September 26, 2024 @ 1:08 am
[url=https://moussamaregaar.biz]https://www.moussamaregaar.biz[/url]
last news about moussa marega
moussamaregaar.biz
Derrickjoisy
September 26, 2024 @ 1:45 am
[url=https://moussa-marega-ar.biz]www.moussa-marega-ar.biz[/url]
last news about moussa marega
https://moussa-marega-ar.biz
MichaelDar
September 26, 2024 @ 2:05 am
[url=https://moussa-maregaar.biz]https://www.moussa-maregaar.biz[/url]
last news about moussa marega
https://moussa-maregaar.biz
EdgarPiede
September 26, 2024 @ 3:25 am
Farmacia online piГ№ conveniente: Ibuprofene 600 prezzo senza ricetta – acquisto farmaci con ricetta
ремонт кондиционеров сервис центры в москве
September 26, 2024 @ 5:39 am
[url=https://remont-kondicionerov-wik.ru]сервисный центр кондиционеров[/url]
Richardpeado
September 26, 2024 @ 7:18 am
comprare farmaci online all’estero [url=http://farmaciait.men/#]Farmacie online sicure[/url] farmacie online affidabili
Michaelwem
September 26, 2024 @ 8:44 am
acquistare farmaci senza ricetta [url=https://farmaciait.men/#]Farmacie online sicure[/url] п»їFarmacia online migliore
PatrickMourO
September 26, 2024 @ 8:52 am
farmacie online affidabili: BRUFEN 600 prezzo in farmacia – farmacie online affidabili
Garlandtoimi
September 26, 2024 @ 9:21 am
Что такое смешной анекдот? История забавного анекдота, какие бывают.
Ремонт бытовой техники на выезде
September 26, 2024 @ 9:52 am
Сервисный центр предлагает отремонтировать морозильной камеры centek ремонт морозильных камер centek адреса
Фильм Джентельмены
September 26, 2024 @ 11:30 am
How old is snoop dogg. Au. Terrorism. Watergate. Frond. https://bit.ly/dzhentlmeny-films-dzhentlmeny-2
ArmandosRup
September 26, 2024 @ 11:39 am
Смешные приколы на разные темы
Поднимите себе настроение!
Richardpeado
September 26, 2024 @ 11:47 am
Farmacia online miglior prezzo [url=https://farmaciait.men/#]farmacia online migliore[/url] farmacia online
Фильм Джентельмены
September 26, 2024 @ 2:51 pm
When does summer start. United states flag. Tim cook. Static. https://bit.ly/dzhentlmeny-films-dzhentlmeny-2
EdgarPiede
September 26, 2024 @ 3:20 pm
farmacia online: Brufen 600 prezzo – acquisto farmaci con ricetta
Garlandtoimi
September 26, 2024 @ 8:39 pm
Смешные картинки http://prikoly-shutki.ru/kartinki-prikolnye Что такое мемы.
Stephenswogs
September 26, 2024 @ 10:02 pm
[url=https://maregamoussaar.biz]www.maregamoussaar.biz[/url]
last news about marega moussa
http://www.maregamoussaar.biz
JacobSot
September 27, 2024 @ 12:54 am
Бесплатный промокод на сайт ищет каждый потенциальный клиент компании. Почему? Мало кто откажется от щедрой приветственной награды, позволяющей увеличить сумму первого депозита, заработав до ?9100 бонуса (с 24 апреля бонус увеличился до 10 400). Промокоды – это маркетинговый инструмент, с помощью которого рекламные партнеры букмекера привлекают новых бетторов. Поэтому в современной практике сам букмекер редко распространяет коды, за исключением флагманских акций или раздачи купонов в рамках специальных ивентов. Не все бонусы предполагают наличие рекламного кода. Для активации большинства из них достаточно просто пополнить счет или делать ставки на спорт, получая за это баллы активности. В последующем есть возможность в рамках программы лояльности БК «Мелбет» обменивать баллы на коды для бесплатных ставок (фрибеты) или на реальные денежные средства. Раздобыть промокод Melbet Вы всегда сможете на нашем сайте. При этом Вы можете быть уверенными в том, что это именно актуальное предложение. Также рекомендуем подписаться на информационную рассылку БК, где компания при помощи SMS и почтовых уведомлений держит в курсе событий любителей ставок.
Davidpania
September 27, 2024 @ 1:31 am
https://brufen.pro/# BRUFEN 600 bustine prezzo
Brufen 600 prezzo con ricetta
Davidpania
September 27, 2024 @ 1:50 am
http://brufen.pro/# BRUFEN 600 bustine prezzo
Brufen 600 senza ricetta
сервис центры в ростове на дону
September 27, 2024 @ 1:51 am
Профессиональный сервисный центр по ремонту бытовой техники с выездом на дом.
Мы предлагаем:ремонт бытовой техники в ростове на дону
Наши мастера оперативно устранят неисправности вашего устройства в сервисе или с выездом на дом!
EdgarPiede
September 27, 2024 @ 3:22 am
farmacie online sicure: migliori farmacie online 2024 – Farmacia online miglior prezzo
фильмы
September 27, 2024 @ 3:56 am
Kleptomaniac. Tom hardy movies and tv shows. Lawyer. Hg. Lisa kudrow. https://bit.ly/hdrezka-hdrezka-hdrezka
Charlesnidly
September 27, 2024 @ 8:29 am
[url=https://haalanderlingcz.biz]http://haalanderlingcz.biz[/url]
last news about haaland erling
https://www.haalanderlingcz.biz
Gartandtoimi
September 27, 2024 @ 8:34 am
Как поднять настроение с помошью мемов. Свежие анекдоты
https://sumkin.ru/forum/member.php?u=45672
ArnoldNog
September 27, 2024 @ 9:54 am
neurontin 4000 mg: neurontin 214 – neurontin 300 mg caps
ArnoldNog
September 27, 2024 @ 9:55 am
lasix 40mg: buy furosemide – lasix 100 mg
Sergiorax
September 27, 2024 @ 10:56 am
https://furosemide.men/# furosemide 100mg
пирамида уровней дилтса
September 27, 2024 @ 11:01 am
Kean university. Guinea pig. Russian ukraine war. Biltmore. Vision. Kintsugi. The three body problem. Fae. Navel.
Daviddoomi
September 27, 2024 @ 12:22 pm
Buy compounded semaglutide online: Semaglutide pharmacy price – buy rybelsus
AlfredMef
September 27, 2024 @ 12:25 pm
buy gabapentin [url=https://gabapentin.site/#]neurontin capsules 300mg[/url] neurontin 600 mg coupon
пирамида роберта дилтса
September 27, 2024 @ 2:13 pm
Takashi murakami. Dyson. Thorazine. Black widow. Rensselaer polytechnic institute. Endearing. Alive movie.
AnthonySuinA
September 27, 2024 @ 2:52 pm
sollers авто автобус sollers
Josephcek
September 27, 2024 @ 3:01 pm
prednisone nz: prednisone 54899 – prednisone 10mg cost
Daviddoomi
September 27, 2024 @ 3:41 pm
neurontin capsules 600mg: neurontin 2018 – ordering neurontin online
Douglasjam
September 27, 2024 @ 5:55 pm
1xBet
For casino players, 1xBet provides promo codes that offer free spins, deposit bonuses, or cashback on losses. These codes can be used in the casino section of the site to enhance the gaming experience.
AnthonySuinA
September 27, 2024 @ 6:39 pm
sollers atlant фургон автобус sollers
AlfredMef
September 27, 2024 @ 6:56 pm
lasix furosemide 40 mg [url=https://furosemide.men/#]buy furosemide[/url] furosemide 100 mg
Josephcek
September 27, 2024 @ 7:39 pm
lasix 100mg: lasix for sale – lasix dosage
ArnoldNog
September 27, 2024 @ 9:42 pm
prednisone 5 mg cheapest: can you buy prednisone over the counter uk – 20 mg of prednisone
ArnoldNog
September 27, 2024 @ 9:42 pm
lasix dosage: buy furosemide – lasix for sale
RobertRer
September 27, 2024 @ 9:58 pm
[url=https://marega-moussa-ar.biz]http://marega-moussa-ar.biz[/url]
last news about marega moussa
https://www.marega-moussa-ar.biz
AlfredMef
September 27, 2024 @ 10:02 pm
order neurontin over the counter [url=https://gabapentin.site/#]32 neurontin[/url] neurontin brand name
Gartandtoimi
September 27, 2024 @ 10:17 pm
Как поднять настроение с помошью мемов. прикольные картинки
https://goodgame.ru/user/1653043
Daviddoomi
September 27, 2024 @ 10:51 pm
lasix dosage: cheap lasix – lasix 100mg
Sergiorax
September 27, 2024 @ 11:34 pm
https://rybelsus.tech/# rybelsus generic
Sergiorax
September 28, 2024 @ 1:12 am
http://ventolininhaler.pro/# ventolin generic brand
DamianSot
September 28, 2024 @ 1:17 am
Промокод при регистрации позволит получить подарки – это и бонусное пополнение счета, и бесплатные ставки. На самом деле, это зависит от стремления букмекера сделать шаг навстречу своим клиентам. Всевозможные бонусы и акции становятся магнитом, привлекают новых бетторов, но в то же время вносят разнообразие в игру тех, кто уже считает, что их нечем удивить. Чтобы новый игрок получил на счет приветственный бонус, он должен ввести промокоды бесплатно на ставку в соответствующее поле в регистрационной анкете. Эти средства будут находиться на дополнительном (бонусном) балансе – эти средства нельзя вывести, но на них можно делать ставки.
Daviddoomi
September 28, 2024 @ 2:59 am
over the counter prednisone pills: generic prednisone pills – buying prednisone mexico
top_alarm_clocks_490
September 28, 2024 @ 3:52 am
buy electronic alarm clock buy loud alarm clock
GrahamHes
September 28, 2024 @ 7:18 am
Кракен сайт – это онлайн-площадка, работающая в скрытых сетях интернета, где пользователи могут покупать и продавать различные виды наркотиков. Для доступа к Kraken onion необходимо использовать специальное программное обеспечение, позволяющее обходить блокировки и обеспечивающее анонимность пользователей.
StevenRaica
September 28, 2024 @ 9:01 am
[url=https://moussa-dembele-ar.biz]https://moussa-dembele-ar.biz[/url]
last news about moussa dembele
http://moussa-dembele-ar.biz
Josephgox
September 28, 2024 @ 9:49 am
apo prednisone: prednisone over the counter south africa – cost of prednisone 40 mg
JaredNeoks
September 28, 2024 @ 10:30 am
time clocks for quickbooks minecraft clocks
Сервисный центр профи в Москве
September 28, 2024 @ 11:08 am
Сервисный центр предлагает мастерские ремонта телевизоров modena ремонт телевизора modena на дому
Daviddoomi
September 28, 2024 @ 11:24 am
prednisone purchase canada: prednisone 50 mg prices – buy prednisone 10mg online
ремонт телевизоров
September 28, 2024 @ 11:36 am
сервисный центре предлагает тв ремонт – ремонт телевизоров в москве
Josephcek
September 28, 2024 @ 1:07 pm
ventolin uk pharmacy: buy Ventolin – generic ventolin inhaler
WilliamTer
September 28, 2024 @ 1:30 pm
Сайт о биодобавках https://биодобавки.рф подробная информация о видах добавок, их действии и пользе. Рекомендации по выбору для поддержки здоровья на основе актуальных исследований.
Daviddoomi
September 28, 2024 @ 2:56 pm
semaglutide: semaglutide – rybelsus cost
Gartandtoimi
September 28, 2024 @ 4:29 pm
Как поднять настроение с помошью мемов. Свежие анекдоты
http://pax.nichost.ru/forum/view_profile.php?UID=138230
Sergiorax
September 28, 2024 @ 5:14 pm
http://gabapentin.site/# neurontin cap 300mg price
binance us registrati
September 28, 2024 @ 5:20 pm
Your point of view caught my eye and was very interesting. Thanks. I have a question for you.
Josephgox
September 28, 2024 @ 5:26 pm
can you buy prednisone without a prescription: prednisone 20 mg tablet price – 40 mg prednisone pill
Любовь на троих
September 28, 2024 @ 5:56 pm
Синоним к слову рождается. Это в психологии.
Московские школы сос. Photographer logo. Тест на вид характера.
Выберите предмет психологии. Некритическое.
Почему я забываю прошлое.
Michaelper
September 28, 2024 @ 6:22 pm
Сайт о биодобавках https://биодобавки.рф предлагает проверенную информацию о натуральных добавках для здоровья. Узнайте, как выбрать подходящие средства для улучшения иммунитета, повышения энергии и поддержания активного образа жизни. Подробные описания и советы помогут сделать осознанный выбор.
Casino Plus Login
September 28, 2024 @ 6:50 pm
Kеep on woгking, ցreat job!.Casino Plus Login
Sergiorax
September 28, 2024 @ 6:51 pm
https://furosemide.men/# lasix
Daviddoomi
September 28, 2024 @ 10:30 pm
lasix generic: lasix tablet – lasix for sale
Kevinrisse
September 28, 2024 @ 11:09 pm
[url=https://moussadembelear.biz]https://moussadembelear.biz[/url]
last news about moussa dembele
http://www.moussadembelear.biz
Capitlip
September 28, 2024 @ 11:17 pm
Аренда яхт Пхукет https://arendayachtphuket.ru/ Забронируйте частную яхту для романтического ужина на закате, наслаждаясь потрясающими видами и вкусной кухней.
Josephgox
September 28, 2024 @ 11:24 pm
where to buy prednisone in canada: where can i buy prednisone online without a prescription – buy prednisone online paypal
Josephcek
September 28, 2024 @ 11:25 pm
rybelsus price: rybelsus – cheap Rybelsus 14 mg
Capitlip
September 28, 2024 @ 11:41 pm
Аренда яхты на Пхукете https://arendayachtphuket.ru/ Выбирайте между различными яхтами — от стильных катамаранов до роскошных моторных яхт, чтобы удовлетворить любые ваши пожелания.
Derrickjoisy
September 29, 2024 @ 12:31 am
[url=https://dembele-moussa-ar.biz]https://dembele-moussa-ar.biz[/url]
last news about dembele moussa
https://www.dembele-moussa-ar.biz
JackSot
September 29, 2024 @ 12:37 am
Букмекер Мелбет неслучайно пользуется популярностью среди игроков. Если вы желаете делать ставки на UFC, воспользуйтесь сайтом оператора Мелбет. Здесь предложена одна из лучших росписей в режиме Live и в линии. Обширная вариативность ставок – на исход, продолжительность состязания и формы досрочных побед, а также сравнительно высокие коэффициенты сделают вашу игру наиболее насыщенной. Для поддержания интересов клиентов, букмекер Мелбет предлагает разнообразные бонусные программы, которые дают определенные привилегии игрокам. Они могут выражаться в фрибетах, бонусах к депозиту или в виде кэшбэка. Чтобы открыть доступ ко всем бонусам от букмекера, используйте промокод для регистрации. Бонусы, которые предлагает букмекер действующим клиентам, можно использовать на ставках по любым спортивным направлениям. Если вы интересуетесь смешанными единоборствами, на сайте Мелбет можно найти порядка сотни предложений по заключению пари на актуальные матчи. Выбор ставок доступен как в линии, так и в Live-разделе. Чтобы воспользоваться бонусами от букмекера и перейти к заключению пари на спорт, необходимо создать личную учетную запись на сайте. Рассказываем, как это сделать.
StevenSok
September 29, 2024 @ 12:51 am
[url=https://moussa-dembelear.biz]https://moussa-dembelear.biz[/url]
last news about moussa dembelear
http://www.moussa-dembelear.biz
Daviddoomi
September 29, 2024 @ 2:07 am
gabapentin 300mg: neurontin price uk – how to get neurontin
криптобосс промокод hds5
September 29, 2024 @ 5:49 am
Используйте промокод в cryptoboss casino и получите уникальные бонусы|Уникальный шанс с промокодом на cryptoboss casino
Бонусный код для cryptoboss casino|Используйте промокод в cryptoboss casino и увеличьте свои шансы на победу
Успейте воспользоваться привилегией cryptoboss casino промокода|Уникальное предложение от cryptoboss casino с промокодом
Специальное предложение от cryptoboss casino с промокодом|cryptoboss casino – играйте со скидкой по промокоду
cryptoboss casino промокод бездепозитный [url=https://namelessone.ru/]cryptoboss casino промокод бездепозитный cryptoboss ru casino[/url] .
Играйте в cryptoboss casino с преимуществом промокода
Воспользуйтесь промокодом в cryptoboss casino для дополнительных выигрышей
Играйте в cryptoboss casino с дополнительными бонусами от промокодов|Получите уникальное предложение с cryptoboss casino промокодами
Играйте в cryptoboss casino с бонусным промокодом для победы|cryptoboss casino промокод – ваш путь к большим выигрышам|cryptoboss casino промокод – ваш путь к успеху|Играйте с выгодой: cryptoboss casino промокод|Играйте в cryptoboss casino с эксклюзивным промокодом для успеха|Успейте активировать промокод для увеличения выигрыша|Играйте в cryptoboss casino с уникальным промокодом и выиграйте больше|Уникальное предложение для игроков cryptoboss casino: промокод|Специальные бонусы с cryptoboss casino промокодом|Бонусная программа cryptoboss casino с уникальными промокодами|Играйте в cryptoboss casino с преимуществами промокода|cryptoboss casino промокод – ваш ключ к победе|Играйте в cryptoboss casino с дополнительными бонусами по промокоду|Играйте в cryptoboss casino с уникальными привилегиями по промокоду|Получите дополнительные бонусы в cryptoboss casino с промокодом|Уникальные предложения от cryptoboss casino с промокодом|Увеличьте свои шансы на победу в cryptoboss casino с промокодом|Эксклюзивный cryptoboss casino промокод – ваш путь к выигрышу
seasonvar
September 29, 2024 @ 6:58 am
Mein kampf. Valentine. Solar eclipse. Bisexual. Fox animal. Kristen stewart movies. Steak tartare. https://20242025.g-u.su/
seasonvar
September 29, 2024 @ 7:51 am
Objective. Shrek 2. Black cat. Scrubs cast. Albino. Bethlehem pa. Ether meaning. https://20242025.g-u.su/
seasonvar
September 29, 2024 @ 8:42 am
Lakers. Rock and roll hall of fame. Steal. The getty. California academy of sciences. Target field. Northwestern university. Hi. Progesterone. https://20242025.g-u.su/
Douglasjam
September 29, 2024 @ 9:02 am
1xBet
1xBet offers daily promo codes that provide bonuses such as free bets, deposit matches, or free spins. These codes are valid for the day they are issued and can be used to boost your betting experience.
Gartandtoimi
September 29, 2024 @ 9:13 am
Как поднять настроение с помошью мемов. Ржачные картинки.
https://qooh.me/vychislavyakov
Daviddoomi
September 29, 2024 @ 9:51 am
prednisone canada pharmacy: prednisone 3 tablets daily – cost of prednisone 10mg tablets
Sergiorax
September 29, 2024 @ 10:25 am
https://prednisolone.pro/# prednisone 12 tablets price
escorts agency Doncaster
September 29, 2024 @ 11:03 am
Hey great website! Does running a blog such as this require a lot of work?
I’ve very little knowledge of programming but I had been hoping to start my own blog in the near future.
Anyhow, should you have any recommendations or techniques for
new blog owners please share. I know this is off subject
however I simply needed to ask. Appreciate it!
ремонт macbook в москве
September 29, 2024 @ 11:08 am
Профессиональный сервисный центр по ремонту компьютеров и ноутбуков в Москве.
Мы предлагаем: ремонт макбук про москва
Наши мастера оперативно устранят неисправности вашего устройства в сервисе или с выездом на дом!
Sergiorax
September 29, 2024 @ 12:06 pm
https://gabapentin.site/# 800mg neurontin
vhoamicuu
September 29, 2024 @ 12:07 pm
A league of their own. Prince edward island. Notting hill. Ansel adams. Lose. Lampoon. Police. Life movie. https://2480.g-u.su/
Robertbeems
September 29, 2024 @ 12:35 pm
Сайт предоставляет информацию о биодобавках http://биодобавки.рф состав, польза и рекомендации по применению. Здесь вы найдёте обзоры эффективных добавок для улучшения здоровья, иммунитета и энергии на основе научных данных и экспертных мнений.
wskzsmriw
September 29, 2024 @ 1:01 pm
Helen mirren. Spider man into the spider verse. Minnesota twins. Ted kennedy. Euro symbol. Daddy long legs. Angler fish. Yugioh. https://2480.g-u.su/
Daviddoomi
September 29, 2024 @ 1:50 pm
gabapentin 300: neurontin 400 mg price – neurontin 100 mg
Josephgox
September 29, 2024 @ 3:00 pm
neurontin for sale: neurontin 300 mg cap – neurontin price comparison
madjxquui
September 29, 2024 @ 3:44 pm
2012. Vance. Tarpon. Tacos. Gisele. Peace sign. https://2480.g-u.su/
tqwyupngl
September 29, 2024 @ 6:00 pm
Redemption. Atlantis. Seppuku. Devils lake. Zodiac dates. Apartheid. Daniel. Alito. Freddy rodriguez. https://2480.g-u.su/
Stephenswogs
September 29, 2024 @ 7:46 pm
[url=https://marcelo-brozovic-ar.biz]https://www.marcelo-brozovic-ar.biz[/url]
last news about marcelo brozovic
marcelo-brozovic-ar.biz
ArmandokRup
September 29, 2024 @ 7:51 pm
Прикольные анекдоты. История анекдотов.
Stephennewly
September 29, 2024 @ 8:07 pm
[url=https://haaland-erling-cz.biz/]https://www.haaland-erling-cz.biz[/url]
last news about haaland erling
https://haaland-erling-cz.biz
xvsqvxllf
September 29, 2024 @ 8:26 pm
Calypso. Hilaria baldwin. Catamaran. Utah jazz. Cody rhodes. Polypropylene. Cacophony. Leave it to beaver. Aphorism. https://2480.g-u.su/
ngcihqiyn
September 29, 2024 @ 9:17 pm
Kurt vonnegut. Perseid meteor shower. Spine anatomy. Solenoid. Navy pier. Itunes. https://2480.g-u.su/
Charlesnidly
September 29, 2024 @ 9:54 pm
[url=https://marcelo-brozovicar.biz]https://marcelo-brozovicar.biz[/url]
last news about marcelo brozovic
http://www.marcelo-brozovicar.biz
nelabyksf
September 29, 2024 @ 10:07 pm
Univision. Otis redding. Eucalyptus. Taxonomy. Who is running for president in 2024. Ohare. Clown. Viagra. https://2480.g-u.su/
ThomasSot
September 29, 2024 @ 10:47 pm
Промокод – небольшая цифробуквенная комбинация, которая дает право на получение каких-то привилегий и бонусов. Система промокодов позволяет букмекерским конторам привлекать новых пользователей, поощрять их регистрацию и пополнение счета, поэтому эта схема удобна как букмекерам, так и пользователям. Вводя промокод новинка и другие бонусы для первых ставок. Обычно ввод промокода не представляет особой сложности. На сайте букмекера при регистрации будет отведено специальное поле для ввода кодовой комбинации. При выполнении всех условий компании, предоставляющей бонус, код начинает действовать сразу после ввода. Дополнительная активация не требуется. В этом случае есть свои особенности, о которых будет рассказано далее.
TimothyCrilm
September 30, 2024 @ 12:15 am
buy medicines online in india: top 10 online pharmacy in india – india online pharmacy
xzxeabstq
September 30, 2024 @ 12:21 am
Supple. Mount pleasant. Dak prescott rushing stats. Www yahoo.com. Ewan mcgregor. Bait. https://2480.g-u.su/
RogerRoR
September 30, 2024 @ 2:09 am
mexican border pharmacies shipping to usa: mexican mail order pharmacies – mexican border pharmacies shipping to usa
криптобосс промокод на увеличение выигрыша
September 30, 2024 @ 2:26 am
Используйте промокод в cryptoboss casino и получите уникальные бонусы|Специальное предложение от cryptoboss casino
Секретный промокод от cryptoboss casino|Используйте промокод в cryptoboss casino и увеличьте свои шансы на победу
Только сегодня! cryptoboss casino промокод|Секретный код для cryptoboss casino
cryptoboss casino промокод – ваш ключ к выигрышу|Играйте в cryptoboss casino с эксклюзивным промокодом
cryptoboss casino промокод hds5 [url=https://namelessone.ru/]cryptoboss промокод на бездепозитный бонус[/url] .
Получите уникальный бонус с cryptoboss casino промокодом
Используйте cryptoboss casino промокод для увеличения выигрыша
Играйте в cryptoboss casino с дополнительными бонусами от промокодов|Эксклюзивный cryptoboss casino промокод – ваш шанс на победу
cryptoboss casino – ваш ключ к успеху с промокодом|cryptoboss casino промокод – ваш путь к большим выигрышам|Специальное предложение для игроков cryptoboss casino: промокод|Уникальное предложение от cryptoboss casino с промокодом|Увеличьте свои шансы на победу с cryptoboss casino промокодом|cryptoboss casino – играйте с преимуществами промокода|Получите дополнительные бонусы с cryptoboss casino промокодом|cryptoboss casino промокод – ваш шанс на успех|cryptoboss casino – ваша удача по промокоду|cryptoboss casino промокоды – ваш путь к успеху|Играйте в cryptoboss casino с преимуществами промокода|cryptoboss casino – ваш шанс на успех с промокодом|Играйте в cryptoboss casino с дополнительными бонусами по промокоду|Успейте активировать cryptoboss casino промокод для дополнительных выигрышей|Получите дополнительные бонусы в cryptoboss casino с промокодом|cryptoboss casino – ваш ключ к успеху с промокодами|Получите привилегии с cryptoboss casino промокодом|Эксклюзивный cryptoboss casino промокод – ваш путь к выигрышу
Robertkic
September 30, 2024 @ 5:38 am
Сайт предоставляет информацию о биодобавках биодобавки.рф состав, польза и рекомендации по применению. Здесь вы найдёте обзоры эффективных добавок для улучшения здоровья, иммунитета и энергии на основе научных данных и экспертных мнений.
криптобосс промокод на дополнительные акции hds5
September 30, 2024 @ 8:00 am
Получите эксклюзивный cryptoboss casino промокод|Уникальный шанс с промокодом на cryptoboss casino
Секретный промокод от cryptoboss casino|Используйте промокод в cryptoboss casino и увеличьте свои шансы на победу
Эксклюзивная акция для игроков cryptoboss casino|Уникальное предложение от cryptoboss casino с промокодом
Специальное предложение от cryptoboss casino с промокодом|cryptoboss casino – играйте со скидкой по промокоду
криптобосс промокод на дополнительные акции [url=https://namelessone.ru/]cryptoboss промокод hds5[/url] .
Получите уникальный бонус с cryptoboss casino промокодом
Воспользуйтесь промокодом в cryptoboss casino для дополнительных выигрышей
Увеличьте свои шансы на победу с cryptoboss casino промокодами|cryptoboss casino – преимущества промокодов
Играйте в cryptoboss casino с бонусным промокодом для победы|cryptoboss casino – ваши шансы на победу с промокодами|Специальное предложение для игроков cryptoboss casino: промокод|Получите привилегии в cryptoboss casino с промокодом|Играйте в cryptoboss casino с эксклюзивным промокодом для успеха|cryptoboss casino – играйте с преимуществами промокода|Играйте в cryptoboss casino с уникальным промокодом и выиграйте больше|Используйте промокод в cryptoboss casino для больших побед|Играйте в cryptoboss casino с преимуществом промокода|cryptoboss casino промокоды – ваш путь к успеху|Действуйте прямо сейчас с cryptoboss casino промокодом|cryptoboss casino – ваш шанс на успех с промокодом|Играйте в cryptoboss casino с дополнительными бонусами по промокоду|Играйте в cryptoboss casino с уникальными привилегиями по промокоду|cryptoboss casino – играйте с выгодой с промокодами|cryptoboss casino – ваш ключ к успеху с промокодами|Играйте в cryptoboss casino с дополнительными преимуществами промокода|Эксклюзивный cryptoboss casino промокод – ваш путь к выигрышу
RogerRoR
September 30, 2024 @ 9:55 am
reputable indian online pharmacy: best online pharmacy india – india online pharmacy
ремонт техники профи в воронеже
September 30, 2024 @ 10:26 am
Если вы искали где отремонтировать сломаную технику, обратите внимание – ремонт бытовой техники
hplnsbfwf
September 30, 2024 @ 11:46 am
Cobalt. Leonard bernstein. Shelley long. Attentive. Cherokee. Whiskey. Palestine and israel. https://bit.ly/chto-bylo-dalshe-ruzil-minekayev-smotret-onlayn
zjynwalmt
September 30, 2024 @ 12:41 pm
Acute. David ogden stiers. Nicolas cage. Sorrel. Randolph scott. Jurassic park movies. Long island. Vasopressin. https://bit.ly/chto-bylo-dalshe-ruzil-minekayev-smotret-onlayn
криптобосс промокод на дополнительные акции
September 30, 2024 @ 1:25 pm
Получите эксклюзивный cryptoboss casino промокод|Уникальный шанс с промокодом на cryptoboss casino
Играйте в cryptoboss casino с привлекательным промокодом|Увеличьте свой выигрыш с cryptoboss casino промокодом
Успейте воспользоваться привилегией cryptoboss casino промокода|Играйте в cryptoboss casino с бонусным кодом
Увеличьте свои шансы на победу в cryptoboss casino промокодом|Играйте в cryptoboss casino с эксклюзивным промокодом
криптобосс промокод на увеличение выигрыша [url=https://namelessone.ru/]криптобосс промокод на дополнительные акции[/url] .
Играйте в cryptoboss casino с преимуществом промокода
Используйте cryptoboss casino промокод для увеличения выигрыша
Увеличьте свои шансы на победу с cryptoboss casino промокодами|Эксклюзивный cryptoboss casino промокод – ваш шанс на победу
Успейте воспользоваться cryptoboss casino промокодом и выиграть больше|cryptoboss casino промокод – ваш путь к большим выигрышам|Бонусная программа cryptoboss casino с уникальным промокодом|Получите привилегии в cryptoboss casino с промокодом|Играйте в cryptoboss casino с эксклюзивным промокодом для успеха|Бонусы и привилегии с cryptoboss casino промокодом|Эксклюзивный cryptoboss casino промокод – ваш путь к победе|Используйте промокод в cryptoboss casino для больших побед|Играйте в cryptoboss casino с преимуществом промокода|Бонусная программа cryptoboss casino с уникальными промокодами|Получите уникальные бонусы с cryptoboss casino промокодом|cryptoboss casino промокод – ваш ключ к победе|Играйте в cryptoboss casino с дополнительными бонусами по промокоду|Играйте в cryptoboss casino с уникальными привилегиями по промокоду|Используйте cryptoboss casino промокод для увеличения шансов на победу|Уникальные предложения от cryptoboss casino с промокодом|Играйте в cryptoboss casino с дополнительными преимуществами промокода|Играйте в cryptoboss casino с уникальным промокодом для успеха
hcfpjohrj
September 30, 2024 @ 1:35 pm
Iowa caucus. Rome. Party animals. Trees. Alaska. Costa mesa. S&p500. Wizard. At&t. https://bit.ly/chto-bylo-dalshe-ruzil-minekayev-smotret-onlayn
Gartandtoimi
September 30, 2024 @ 1:42 pm
Ржачные приколы
Как появились приколы. Узнайте, как улучшить себе настроение!
MichaelDar
September 30, 2024 @ 2:54 pm
[url=https://dembelemoussaar.biz]https://www.dembelemoussaar.biz[/url]
last news about dembele moussa
http://www.dembelemoussaar.biz
StanleyJat
September 30, 2024 @ 4:44 pm
[url=][/url]
Временная регистрация в СПб: Быстро и Легально!
Ищете, где оформить временную регистрацию в Санкт-Петербурге?
Мы гарантируем быстрое и легальное оформление без очередей и лишних документов.
Ваше спокойствие – наша забота!
Минимум усилий • Максимум удобства • Полная легальность
Свяжитесь с нами прямо сейчас!
Временная регистрация
[url=][/url]
tgnews
September 30, 2024 @ 7:21 pm
[url=https://t.me/news_161_ru]www.t.me/news_161_ru[/url]
новости ростовской области
https://t.me/news_161_ru
StevenRaica
October 1, 2024 @ 12:01 am
[url=https://brozovic-marceloar.biz]http://www.brozovic-marceloar.biz[/url]
last news about brozovic marcelo
http://www.brozovic-marceloar.biz
Ремонт кондиционеров в Москве
October 1, 2024 @ 2:37 am
Профессиональный сервисный центр по ремонту кондиционеров в Москве.
Мы предлагаем: вызвать мастера по ремонту кондиционеров
Наши мастера оперативно устранят неисправности вашего устройства в сервисе или с выездом на дом!
Jameskem
October 1, 2024 @ 3:19 am
https://mexicanpharma.icu/# mexican rx online
StevenMem
October 1, 2024 @ 3:46 am
[url=][/url]
Новые методы лечения болезни Паркинсона в клинике Neuro Implant Clinic.
Акупунктура уха – новый метод лечения болезни Паркинсона, Альцгеймера, Рассеянного Склероза.
Выездное лечение в разные Страны.
Отзывы нашего метода на официальном сайте Neuro Implant Clinic.
[url=][/url]
Ремонт моноблоков
October 1, 2024 @ 4:58 am
Профессиональный сервисный центр по ремонту моноблоков в Москве.
Мы предлагаем: замена экрана моноблока
Наши мастера оперативно устранят неисправности вашего устройства в сервисе или с выездом на дом!
Ремонт гироскутеров в Москве
October 1, 2024 @ 5:05 am
Профессиональный сервисный центр по ремонту гироскутеров в Москве.
Мы предлагаем: сервис гироскутеров в москве
Наши мастера оперативно устранят неисправности вашего устройства в сервисе или с выездом на дом!
JohnSot
October 1, 2024 @ 5:42 am
Промокод при регистрации позволит получить подарки – это и бонусное пополнение счета, и бесплатные ставки. На самом деле, это зависит от стремления букмекера сделать шаг навстречу своим клиентам. Всевозможные бонусы и акции становятся магнитом, привлекают новых бетторов, но в то же время вносят разнообразие в игру тех, кто уже считает, что их нечем удивить. Чтобы новый игрок получил на счет приветственный бонус, он должен ввести melbet промокод при регистрации в соответствующее поле в регистрационной анкете. Эти средства будут находиться на дополнительном (бонусном) балансе – эти средства нельзя вывести, но на них можно делать ставки.
Jameskem
October 1, 2024 @ 8:32 am
http://mexicanpharma.icu/# buying prescription drugs in mexico
RobertRer
October 1, 2024 @ 10:15 am
[url=https://marcelobrozovicar.biz]http://marcelobrozovicar.biz[/url]
last news about marcelo brozovic
https://marcelobrozovicar.biz
RogerRoR
October 1, 2024 @ 12:59 pm
п»їbest mexican online pharmacies: medication from mexico pharmacy – medication from mexico pharmacy
StanleyJat
October 1, 2024 @ 1:01 pm
[url=][/url]
Временная регистрация в Санкт-Петербурге: Быстро и Легально!
Ищете, где оформить временную регистрацию в СПб?
Мы гарантируем быстрое и легальное оформление без очередей и лишних документов.
Ваше спокойствие – наша забота!
Минимум усилий • Максимум удобства • Полная легальность
Свяжитесь с нами прямо сейчас!
Временная регистрация в СПб
[url=][/url]
криптобосс промокод hds5
October 1, 2024 @ 2:13 pm
Успейте воспользоваться промокодом для cryptoboss casino|Специальное предложение от cryptoboss casino
Играйте в cryptoboss casino с привлекательным промокодом|Используйте промокод в cryptoboss casino и увеличьте свои шансы на победу
Только сегодня! cryptoboss casino промокод|Играйте в cryptoboss casino с бонусным кодом
Увеличьте свои шансы на победу в cryptoboss casino промокодом|Играйте в cryptoboss casino с эксклюзивным промокодом
криптобосс промокод на дополнительные акции hds5 [url=https://namelessone.ru/]cryptoboss casino промокод бездепозитный[/url] .
Получите уникальный бонус с cryptoboss casino промокодом
Используйте cryptoboss casino промокод для увеличения выигрыша
Увеличьте свои шансы на победу с cryptoboss casino промокодами|cryptoboss casino – преимущества промокодов
Успейте воспользоваться cryptoboss casino промокодом и выиграть больше|Получите дополнительные бонусы с cryptoboss casino промокодом|Специальное предложение для игроков cryptoboss casino: промокод|Уникальное предложение от cryptoboss casino с промокодом|Играйте в cryptoboss casino с эксклюзивным промокодом для успеха|Успейте активировать промокод для увеличения выигрыша|Получите дополнительные бонусы с cryptoboss casino промокодом|Используйте промокод в cryptoboss casino для больших побед|cryptoboss casino – ваша удача по промокоду|Увеличьте свои шансы на победу с промокодами от cryptoboss casino|Действуйте прямо сейчас с cryptoboss casino промокодом|cryptoboss casino – ваш шанс на успех с промокодом|Бонусы и скидки с cryptoboss casino промокодом|Играйте в cryptoboss casino с уникальными привилегиями по промокоду|Получите дополнительные бонусы в cryptoboss casino с промокодом|Уникальные предложения от cryptoboss casino с промокодом|Увеличьте свои шансы на победу в cryptoboss casino с промокодом|Играйте в cryptoboss casino с уникальным промокодом для успеха
Jameskem
October 1, 2024 @ 2:25 pm
https://indiadrugs.pro/# indianpharmacy com
Binance Account
October 1, 2024 @ 3:34 pm
Thank you for your sharing. I am worried that I lack creative ideas. It is your article that makes me full of hope. Thank you. But, I have a question, can you help me?
ArmandokRup
October 1, 2024 @ 4:30 pm
Прикольные анекдоты. Что такое анекдот.
Gartandtoimi
October 1, 2024 @ 4:45 pm
Смешные анекдоты. Что такое анекдот.
Jameskem
October 1, 2024 @ 7:56 pm
http://indiadrugs.pro/# top 10 pharmacies in india
MikelKaria
October 1, 2024 @ 8:34 pm
[url=https://khvichakvaratskheliaar.biz]http://khvichakvaratskheliaar.biz[/url]
last news about khvicha kvaratskhelia
http://khvichakvaratskheliaar.biz
Derrickjoisy
October 1, 2024 @ 9:51 pm
[url=https://lautaro-martinezar.biz]https://lautaro-martinezar.biz[/url]
last news about lautaro martinez
https://www.lautaro-martinezar.biz
Kevinrisse
October 1, 2024 @ 10:13 pm
[url=https://lautaro-martinez-ar.biz]http://www.lautaro-martinez-ar.biz[/url]
last news about lautaro martinez
lautaro-martinez-ar.biz
StevenSok
October 1, 2024 @ 10:52 pm
[url=https://brozovicmarceloar.biz]www.brozovicmarceloar.biz[/url]
last news about brozovic marcelo
https://www.brozovicmarceloar.biz
English adult cam shows
October 1, 2024 @ 11:41 pm
Hi there! I’m at work surfing around your blog from my new apple iphone!
Just wanted to say I love reading through your blog and look forward to all your posts!
Carry on the outstanding work!
AndrewVOg
October 2, 2024 @ 12:43 am
[url=https://martinezlautaroar.biz]www.martinezlautaroar.biz[/url]
last news about martine zlautaro
https://martinezlautaroar.biz
JeremyBuiva
October 2, 2024 @ 12:46 am
[url=https://khvicha-kvaratskheliaar.biz]khvicha-kvaratskheliaar.biz[/url]
last news about khvicha kvaratskhelia
khvicha-kvaratskheliaar.biz
JacobSot
October 2, 2024 @ 1:48 am
промокод при регистрации melbet стоит ввести для повышенного бонуса на первый депозит. Большинство букмекерских контор используют такую практику привлечения новых игроков и выдают значительные бонусы. Актуальный промокод Мелбет на 2024 год – RS777. Ввести его могут исключительно новые игроки, у которых еще нет аккаунта в Melbet. Не рекомендуем идти на хитрость и заново проходить процесс регистрации, так как служба безопасности тщательно отслеживает мультиаккаунты. Максимальная сумма бонуса при использовании промокода Мелбет – 10400 рублей. Это значит, что при вводе средств на баланс после регистрации новому игроку будет зачислено еще 130% от первого депозита в качестве поощрения от букмекера.
pxzneervs
October 2, 2024 @ 2:29 am
Elton john songs. Astrology chart. Lid. Erroneous. Georgetown university. Fred claus. Smog. Great expectations. https://hd-film-online.domdrakona.su
porn blonde
October 2, 2024 @ 3:17 am
Heya i’m for the first time here. I found
this board and I in finding It truly useful
& it helped me out much. I hope to present something back
and aid others such as you helped me.
syhpmodwt
October 2, 2024 @ 3:24 am
Ruby bridges. Suriname. Peter krause. Assimilate. Engine. Dialogue. Density of water. Panda. Texas. https://hd-film-online.domdrakona.su
ремонт бытовой техники в тюмени
October 2, 2024 @ 3:40 am
Профессиональный сервисный центр по ремонту бытовой техники с выездом на дом.
Мы предлагаем: ремонт бытовой техники в тюмени
Наши мастера оперативно устранят неисправности вашего устройства в сервисе или с выездом на дом!
dzrtkedrb
October 2, 2024 @ 4:17 am
Academy. Juju. Allosaurus. Time in arizona. Buck. Wagner. Rose of sharon. https://hd-film-online.domdrakona.su
hoxovwzct
October 2, 2024 @ 6:45 am
Elvis. Gilded age. Luka dončić. Hulk. Communication. Wahoo. https://hd-film-online.domdrakona.su
Gartandtoimi
October 2, 2024 @ 12:13 pm
Как поддерживать хорошее настроение с помощью забавных приколов
xoazboukd
October 2, 2024 @ 3:15 pm
Christmas eve. Malcom x. Uncle. Joke. Dolly. Olive tree. https://123-123-movies-123-movie-movies-123.domdrakona.su
ckczftcst
October 2, 2024 @ 4:10 pm
Chris elliott. Hitachi. James harden stats. Henry. Sofia vergara movies and shows. Ad hoc meaning. https://123-123-movies-123-movie-movies-123.domdrakona.su
pefvtxrah
October 2, 2024 @ 5:03 pm
Celtics. Cash and carry. What is hamas. Usa map. Bernadette peters. Norwegian. https://123-123-movies-123-movie-movies-123.domdrakona.su
EasyCash
October 2, 2024 @ 6:16 pm
Оказался в сложной ситуации – нужно было срочно отправить деньги родственникам. В банке отказали, но я нашёл список МФО, которые дают [url=http://epicleaks.cc/Thread-Good-teen-girls?pid=1331418&mode=threaded]деньги без отказа на карту[/url] до 30 000 рублей без лишних проверок. Это стало настоящим спасением! Деньги пришли моментально, а я решил все свои проблемы. Теперь знаю, что [url=https://tinyteria.com/duis-id-elementum-turpis/]займ онлайн срочно на карту без отказа[/url] — это лучший способ быстро получить нужную сумму.
Ремонт Айпадов в Москве
October 2, 2024 @ 6:54 pm
Профессиональный сервисный центр по ремонту планшетов в том числе Apple iPad.
Мы предлагаем: ремонт планшетов на дому
Наши мастера оперативно устранят неисправности вашего устройства в сервисе или с выездом на дом!
gtwetlowb
October 2, 2024 @ 7:30 pm
Biosphere. Kristen bell movies and tv shows. Hbcu. Inferno. Doctors without borders. Slavery. Dennis rodman. https://123-123-movies-123-movie-movies-123.domdrakona.su
Robertdup
October 2, 2024 @ 10:16 pm
[url=https://martinez-lautaro-ar.biz]https://www.martinez-lautaro-ar.biz[/url]
last news about martinez lautaro
http://www.martinez-lautaro-ar.biz
Micahham
October 2, 2024 @ 11:11 pm
[url=https://khvicha-kvaratskhelia-ar.biz]http://www.khvicha-kvaratskhelia-ar.biz[/url]
last news about khvicha kvaratskhelia
khvicha-kvaratskhelia-ar.biz
Gartandtoimi
October 2, 2024 @ 11:35 pm
Как развеселить друга с помощью смешных приколов
Ремонт посудомоечных машин в Москве
October 2, 2024 @ 11:43 pm
Профессиональный сервисный центр по ремонту посудомоечных машин с выездом на дом в Москве.
Мы предлагаем: ремонт посудомоечной машины хайсен на дому
Наши мастера оперативно устранят неисправности вашего устройства в сервисе или с выездом на дом!
Jameskem
October 3, 2024 @ 12:04 am
http://mexicanpharma.icu/# pharmacies in mexico that ship to usa
Charlesnidly
October 3, 2024 @ 12:10 am
[url=https://lautaromartinezar.biz]http://lautaromartinezar.biz[/url]
last news about lautaro martinez
https://lautaromartinezar.biz
Stephentot
October 3, 2024 @ 9:30 am
http://clssansordonnance.icu/# pharmacie en ligne france livraison internationale
HeathDex
October 3, 2024 @ 9:58 am
pharmacie en ligne sans ordonnance: Acheter Cialis – pharmacie en ligne livraison europe
JosephMossy
October 3, 2024 @ 12:03 pm
[url=][/url]
Временная регистрация в Санкт-Петербурге: Быстро и Легально!
Ищете, где оформить временную регистрацию в СПБ?
Мы гарантируем быстрое и легальное оформление без очередей и лишних документов.
Ваше спокойствие – наша забота!
Минимум усилий • Максимум удобства • Полная легальность
Свяжитесь с нами прямо сейчас!
Временная регистрация в СПБ
[url=][/url]
Charlescoamp
October 3, 2024 @ 12:19 pm
pharmacie en ligne sans ordonnance [url=https://clssansordonnance.icu/#]Cialis generique prix[/url] pharmacie en ligne france pas cher
Gartandtoimi
October 3, 2024 @ 12:58 pm
Как поддерживать хорошее настроение с помощью смешных приколов
PatrickFes
October 3, 2024 @ 1:59 pm
выкуп автомобиля из лизинга выкуп авто из лизинга
AndrewVOg
October 3, 2024 @ 2:36 pm
[url=https://play-dog-house.ru]play-dog-house.ru[/url]
last news about dog house
http://play-dog-house.ru
Stephentot
October 3, 2024 @ 2:37 pm
https://vgrsansordonnance.com/# Viagra homme prix en pharmacie sans ordonnance
sweet bonanza bonus buy
October 3, 2024 @ 2:43 pm
Top Reasons to Play Sweet Bonanza Today Looking for a new slot to try? Sweet Bonanza offers exciting features, vibrant graphics, and the chance for huge multipliers. Discover why it’s a favorite among slot enthusiasts.
[url=https://sweet-bonanza-game.ru/]slot bonanza[/url]
JeremyBuiva
October 3, 2024 @ 3:38 pm
[url=https://avtomat-dog-house.ru]https://avtomat-dog-house.ru[/url]
last news about dog house
https://www.avtomat-dog-house.ru
StevenSok
October 3, 2024 @ 4:24 pm
[url=https://download-casino-bonsai.ru]https://download-casino-bonsai.ru[/url]
last news about casino bonsai
https://www.download-casino-bonsai.ru
PatrickFes
October 3, 2024 @ 5:10 pm
выкуп авто с пробегом https://vikup-avto2.ru
MikelKaria
October 3, 2024 @ 5:36 pm
[url=https://game-dog-house.ru]http://game-dog-house.ru[/url]
last news about game dog house
http://game-dog-house.ru
HeathDex
October 3, 2024 @ 6:09 pm
pharmacie en ligne france livraison internationale: pharmacie en ligne pas cher – Achat mГ©dicament en ligne fiable
MichaelDar
October 3, 2024 @ 6:46 pm
[url=https://angel-di-maria-ar.biz]https://www.angel-di-maria-ar.biz[/url]
last news about angel di maria
http://www.angel-di-maria-ar.biz
sweetbonanza
October 3, 2024 @ 6:48 pm
Candy-Themed Fun in Sweet Bonanza – A Visual Feast! Immerse yourself in the candy-filled world of Sweet Bonanza. With vibrant, colorful graphics and sweets at every turn, this slot offers a visual feast alongside exciting gameplay.
[url=https://sweet-bonanza-game.ru/]slot bonanza[/url]
WillieVok
October 3, 2024 @ 6:58 pm
Indian porn https://desiporn.one in Hindi. Only high-quality porn videos, convenient search and regular updates.
HeathDex
October 3, 2024 @ 8:52 pm
Pharmacie en ligne livraison Europe: pharmacie en ligne pas cher – п»їpharmacie en ligne france
RobertRer
October 3, 2024 @ 9:25 pm
[url=https://angel-dimaria-ar.biz]https://www.angel-dimaria-ar.biz[/url]
last news about angel dimaria
https://angel-dimaria-ar.biz
Stephennewly
October 3, 2024 @ 9:27 pm
[url=https://angel-dimariaar.biz/]http://www.angel-dimariaar.biz[/url]
last news about angel dimaria
https://www.angel-dimariaar.biz
StevenRaica
October 3, 2024 @ 9:44 pm
[url=https://kvaratskhelia-khvicha-ar.biz]https://kvaratskhelia-khvicha-ar.biz[/url]
last news about kvaratskhelia khvicha
https://kvaratskhelia-khvicha-ar.biz
Gartandtoimi
October 4, 2024 @ 12:41 am
Смешные анекдоты https://shutochki.wordpress.com/2024/09/27/smeshnye-anekdoty/
Stephentot
October 4, 2024 @ 12:53 am
https://pharmaciepascher.pro/# Achat mГ©dicament en ligne fiable
Stephenswogs
October 4, 2024 @ 1:08 am
[url=https://kvaratskheliakhvichaar.biz]http://kvaratskheliakhvichaar.biz[/url]
last news about kvaratskhelia khvicha
https://kvaratskheliakhvichaar.biz
Ремонт МФУ
October 4, 2024 @ 1:18 am
Профессиональный сервисный центр по ремонту МФУ в Москве.
Мы предлагаем: где можно починить мфу
Наши мастера оперативно устранят неисправности вашего устройства в сервисе или с выездом на дом!
HeathDex
October 4, 2024 @ 2:43 am
Pharmacie en ligne livraison Europe: pharmacie en ligne france pas cher – pharmacies en ligne certifiГ©es
Douglasjam
October 4, 2024 @ 2:59 am
1xBet Free Promo Codes https://idematapp.com/wp-content/pages/1xbet_promo_codes_free_bonus_offers.html
1xBet offers free promo codes that provide users with bonuses without requiring a deposit. These codes are a great way to try out the platform and win real money without risking your own funds.
MichaelDancy
October 4, 2024 @ 4:26 am
Porn videos https://thetittyfuck.com watch online for free. A selection of high-quality porn videos, a collection of sex videos
Charlescoamp
October 4, 2024 @ 5:19 am
Viagra sans ordonnance livraison 48h [url=https://vgrsansordonnance.com/#]Viagra sans ordonnance 24h[/url] Viagra femme ou trouver
HeathDex
October 4, 2024 @ 5:32 am
Pharmacie sans ordonnance: Cialis generique prix – п»їpharmacie en ligne france
dzennews
October 4, 2024 @ 7:43 am
[url=https://dzen.ru/news161ru]www.dzen.ru/news161ru[/url]
новости ростовской области
dzen.ru/news161ru
Gartandtoimi
October 4, 2024 @ 8:09 am
КОРОТКИЕ ШУТКИ И АНЕКДОТЫ
Как развить чувство юмора
HeathDex
October 4, 2024 @ 10:57 am
pharmacie en ligne sans ordonnance: Cialis generique achat en ligne – pharmacie en ligne avec ordonnance
JeremyBuiva
October 4, 2024 @ 4:32 pm
[url=https://apk-casino-bonsai.ru]www.apk-casino-bonsai.ru[/url]
last news about casino bonsai
http://www.apk-casino-bonsai.ru
AndrewVOg
October 4, 2024 @ 4:52 pm
[url=https://dimaria-angel-ar.biz]dimaria-angel-ar.biz[/url]
last news about dimaria angel
http://dimaria-angel-ar.biz
Nonprofit finances
October 4, 2024 @ 5:27 pm
Great post. I’m dealing with a few of these issues as well..
MikelKaria
October 4, 2024 @ 5:36 pm
[url=https://casino-bonsai-download.ru]www.casino-bonsai-download.ru[/url]
last news about casino bonsai
http://www.casino-bonsai-download.ru
Ремонт принтеров в Москве
October 4, 2024 @ 7:29 pm
Профессиональный сервисный центр по ремонту принтеров в Москве.
Мы предлагаем: мастер по ремонту принтеров москва
Наши мастера оперативно устранят неисправности вашего устройства в сервисе или с выездом на дом!
Charlescoamp
October 4, 2024 @ 9:32 pm
Pharmacie Internationale en ligne [url=http://clssansordonnance.icu/#]cialis prix[/url] acheter mГ©dicament en ligne sans ordonnance
Derrickjoisy
October 4, 2024 @ 10:30 pm
[url=https://game-doghouse.ru]game-doghouse.ru[/url]
last news about dog house
http://www.game-doghouse.ru
Charlescoamp
October 4, 2024 @ 11:05 pm
Pharmacie sans ordonnance [url=https://clssansordonnance.icu/#]Acheter Cialis[/url] Achat mГ©dicament en ligne fiable
Kevinrisse
October 4, 2024 @ 11:25 pm
[url=https://play-doghouse.ru]https://www.play-doghouse.ru[/url]
last news about play doghouse
http://www.play-doghouse.ru
claire miller
October 5, 2024 @ 12:21 am
Need assistance with your Service and Operations Management in IT assignments? Our experts are here to help. We provide comprehensive guidance on topics like ITIL framework, service level agreements, incident management, problem management, change management, and more. Analyse the organisational and people factors that influence the delivery and operationalisation of IT services. Get expert solutions, timely delivery, and plagiarism-free work. Contact us today for top-notch IT Service and Operations Management assignment help.
Douglasjam
October 5, 2024 @ 12:43 am
The Best Promo Code for 1xBet https://actuchomage.org/includes/wkl/code_promo_69.html
The best promo codes for 1xBet are those that offer significant bonuses, such as large deposit matches, free bets, or free spins. These codes can greatly enhance the betting experience by providing extra value for users.
24kbet
October 5, 2024 @ 1:46 am
我努力让所有的声音都受到尊重,并创造一个积极的环境。我希望每个人都能自由表达自己的意见,参与有意义的对话,并相互支持。让我们一起创造一个能够激励和支持彼此的地方!
另外,我非常感谢大家能够将我的博客分享给更多的人。谢谢!
24kbet
CharlestaK
October 5, 2024 @ 3:45 am
[url=][/url]
[b]Продажа мини-погрузчиков Lonking[/b]
Продажа мини-погрузчиков Lonking на территории России от официального
дистрибьютора. Новая многофункциональная техника для любых задач.
Наши машины предназначены для того, чтобы упростить вашу работу:
от строительных площадок до складских операций.
Высокая эффективность, надежность и инновационные решения — все,
что вам нужно для успешных проектов. Погрузите свой бизнес в будущее
с мини-погрузчиками Lonking!
47% российских покупателей выбрали мини-погрузчики Lonking в 2023 году
продано более 1200 единиц.
Lonking
[url=][/url]
ArmandokRup
October 5, 2024 @ 4:12 am
Лучшие анекдоты
Подними себе настроение
sweet bonanza online casino
October 5, 2024 @ 4:14 am
Play Sweet Bonanza – Spin Your Way to Big Wins! Experience the sweet thrill of Sweet Bonanza, a fun and colorful slot game. With cascading reels and high multipliers, you’re just a spin away from massive rewards!
[url=https://sweet-bonanza-game.ru/]slot bonanza[/url]
Stephentot
October 5, 2024 @ 4:48 am
http://vgrsansordonnance.com/# Viagra homme prix en pharmacie
HeathDex
October 5, 2024 @ 5:53 am
vente de mГ©dicament en ligne: pharmacie en ligne pas cher – pharmacie en ligne pas cher
Gartandtoimi
October 5, 2024 @ 7:45 am
Веселые еврейские анекдоты
Подними настроение
Mirtinvest.ru
October 5, 2024 @ 8:09 am
На [url=https://mirtinvest.ru/]mirtinvest.ru[/url] ты найдёшь более 40 проверенных МФО, которые выдают микрокредиты онлайн без отказов. Для всех от 18 лет, с минимальными требованиями и ставкой не выше 0,8% в день — это настоящий шанс решить свои финансовые вопросы быстро и безопасно. Мы работаем только с лицензированными компаниями, так что можешь доверять нам. Пора действовать — займи деньги легко и без забот!
Ремонт плоттеров
October 5, 2024 @ 8:46 am
Профессиональный сервисный центр по ремонту плоттеров в Москве.
Мы предлагаем: цены на ремонт плоттеров
Наши мастера оперативно устранят неисправности вашего устройства в сервисе или с выездом на дом!
slot bonanza
October 5, 2024 @ 9:05 am
Sweet Bonanza Free Spins Feature – How It Works Unlock the exciting free spins feature in Sweet Bonanza and enjoy higher chances of landing huge multipliers and combos. Learn how to trigger the free spins for even more sweet rewards!
[url=https://sweet-bonanza-game.ru/]slot bonanza[/url]
Charlesnidly
October 5, 2024 @ 12:26 pm
[url=https://saud-abdulhamid-ar.biz]https://www.saud-abdulhamid-ar.biz[/url]
last news about saud abdulhamid
https://saud-abdulhamid-ar.biz
Mfozaima
October 5, 2024 @ 12:28 pm
[url=https://t.me/s/mfo_2024_online]Новые займы[/url] на карту за минуту. [url=https://t.me/s/mfo_2024_online]МФО 2024[/url] предлагают кредиты без проверок и отказов.
Kennethlof
October 5, 2024 @ 1:09 pm
продвижение товаров на вайлдберриз https://prodvizhenie-wildberries.ru
Charlescoamp
October 5, 2024 @ 2:03 pm
п»їpharmacie en ligne france [url=http://pharmaciepascher.pro/#]pharmacie en ligne[/url] pharmacie en ligne livraison europe
LarryEteds
October 5, 2024 @ 2:31 pm
Федерация – это проводник в мир покупки запрещенных товаров, можно купить гашиш, купить мефедрон, купить кокаин, купить меф, купить экстази, купить альфа пвп, купить гаш в различных городах. Москва, Санкт-Петербург, Краснодар, Владивосток, Красноярск, Норильск, Екатеринбург, Мск, СПБ, Хабаровск, Новосибирск, Казань и еще 100+ городов.
HeathDex
October 5, 2024 @ 2:36 pm
Viagra gГ©nГ©rique pas cher livraison rapide: Viagra generique en pharmacie – Viagra homme sans ordonnance belgique
JeremyBuiva
October 5, 2024 @ 4:19 pm
[url=https://roberto-firmino-ar.biz]www.roberto-firmino-ar.biz[/url]
last news about roberto firmino
http://www.roberto-firmino-ar.biz
Kennethlof
October 5, 2024 @ 4:29 pm
продвижение на вайлдберриз под ключ раскрутка товаров на вайлдберриз
AndrewVOg
October 5, 2024 @ 4:47 pm
[url=https://saudabdulhamidar.biz]http://www.saudabdulhamidar.biz[/url]
last news about saud abdulhamid
http://saudabdulhamidar.biz
MikelKaria
October 5, 2024 @ 5:29 pm
[url=https://roberto-firminoar.biz]https://roberto-firminoar.biz[/url]
last news about roberto firmino
https://www.roberto-firminoar.biz
ArmandokRup
October 5, 2024 @ 5:31 pm
Свежие приколы
Stephentot
October 5, 2024 @ 5:58 pm
https://vgrsansordonnance.com/# Viagra Pfizer sans ordonnance
LarryEteds
October 5, 2024 @ 5:58 pm
Федерация – это проводник в мир покупки запрещенных товаров, можно купить гашиш, купить мефедрон, купить кокаин, купить меф, купить экстази, купить альфа пвп, купить гаш в различных городах. Москва, Санкт-Петербург, Краснодар, Владивосток, Красноярск, Норильск, Екатеринбург, Мск, СПБ, Хабаровск, Новосибирск, Казань и еще 100+ городов.
HeathDex
October 5, 2024 @ 8:57 pm
acheter mГ©dicament en ligne sans ordonnance: Cialis generique achat en ligne – Achat mГ©dicament en ligne fiable
Parrainage Binance
October 5, 2024 @ 8:58 pm
Thank you for your sharing. I am worried that I lack creative ideas. It is your article that makes me full of hope. Thank you. But, I have a question, can you help me?
StevenSok
October 5, 2024 @ 9:03 pm
[url=https://casino-bonsai.ru]https://casino-bonsai.ru[/url]
last news about casino bonsai
casino-bonsai.ru
Micahham
October 5, 2024 @ 10:10 pm
[url=https://bonsai-casino.ru]http://bonsai-casino.ru[/url]
last news about bonsai casino
http://bonsai-casino.ru
Gartandtoimi
October 5, 2024 @ 11:08 pm
Смешные еврейские анекдоты
Подними настроение
Ремонт техники в Москве
October 5, 2024 @ 11:30 pm
Сервисный центр предлагает ремонт сигвеев electrotown недорого ремонт сигвей electrotown адреса
Ремонт серверов в Москве
October 6, 2024 @ 1:59 am
Профессиональный сервисный центр по ремонту серверов в Москве.
Мы предлагаем: ремонт серверного оборудования
Наши мастера оперативно устранят неисправности вашего устройства в сервисе или с выездом на дом!
Haroldjaf
October 6, 2024 @ 2:14 am
rybelsus coupon: rybelsus cost – semaglutide cost
Franktouth
October 6, 2024 @ 4:03 am
http://rybelsus.shop/# rybelsus cost
BarryLig
October 6, 2024 @ 4:20 am
https://rybelsus.shop/# buy semaglutide online
Robertdup
October 6, 2024 @ 4:56 am
[url=https://angeldimariaar.biz]http://www.angeldimariaar.biz[/url]
last news about angel dimaria
http://www.angeldimariaar.biz
bonanza casino online
October 6, 2024 @ 7:20 am
Candy-Themed Fun in Sweet Bonanza – A Visual Feast! Immerse yourself in the candy-filled world of Sweet Bonanza. With vibrant, colorful graphics and sweets at every turn, this slot offers a visual feast alongside exciting gameplay.
[url=https://sweet-bonanza-game.ru/]slot bonanza[/url]
stake us states
October 6, 2024 @ 8:48 am
Hey! This post could not be written any better! Reading this post
reminds me of my old room mate! He always kept talking about this.
I will forward this article to him. Pretty sure he will have a good read.
Thank you for sharing!
сервис центры в уфе
October 6, 2024 @ 10:30 am
Профессиональный сервисный центр по ремонту бытовой техники с выездом на дом.
Мы предлагаем:ремонт крупногабаритной техники в уфе
Наши мастера оперативно устранят неисправности вашего устройства в сервисе или с выездом на дом!
Haroldjaf
October 6, 2024 @ 11:18 am
buy semaglutide pills: buy semaglutide pills – cheapest rybelsus pills
play sweet bonanza
October 6, 2024 @ 11:42 am
Sweet Bonanza Slot – Top 5 Reasons to Play Want to know why Sweet Bonanza is such a hit? We’ve compiled the top 5 reasons players love this game, from huge multipliers to the fun, candy-themed graphics.
[url=https://sweet-bonanza-game.ru/]slot bonanza[/url]
OscarSot
October 6, 2024 @ 12:06 pm
free bet promo codes: Online betting sites regularly offer promotions to attract and retain players. These can include welcome bonuses, free bets, cashback offers, and boosted odds on specific events.
Ремонт объективов
October 6, 2024 @ 12:08 pm
Профессиональный сервисный центр по ремонту объективов в Москве.
Мы предлагаем: ремонт объектив фотоаппарат
Наши мастера оперативно устранят неисправности вашего устройства в сервисе или с выездом на дом!
JeremyBuiva
October 6, 2024 @ 3:26 pm
[url=https://mohammedalowaisar.biz]https://mohammedalowaisar.biz[/url]
last news about mohammed alowais
http://mohammedalowaisar.biz
AndrewVOg
October 6, 2024 @ 3:32 pm
[url=https://firmino-roberto-ar.biz]http://firmino-roberto-ar.biz[/url]
last news about firmino roberto
https://firmino-roberto-ar.biz
MikelKaria
October 6, 2024 @ 5:03 pm
[url=https://alowaismohammedar.biz]www.alowaismohammedar.biz[/url]
last news about alowais mohammed
http://alowaismohammedar.biz
Gartandtoimi
October 6, 2024 @ 5:34 pm
Смешные свежие анекдоты и шутки
Заходи посмеяться
top djs
October 6, 2024 @ 7:15 pm
It’s difficult to find well-informed people about this subject, however, you seem like you
know what you’re talking about! Thanks
Read My Reviews Here
October 6, 2024 @ 7:21 pm
Improvements in budget friendly 3D CAD software have considerably impacted the design landscape, allowing a wider stable of customers to get access to high-end style tools. Affordable CAD software has actually reinvented the means professionals operate, creating it extra reliable, and cost-efficient, http://linoit.com/users/LeonardsdHardy/canvases/software%20discount%20store
Stephennewly
October 6, 2024 @ 8:51 pm
[url=https://saud-abdulhamidar.biz/]http://saud-abdulhamidar.biz[/url]
last news about saud abdulhamid
http://www.saud-abdulhamidar.biz
ArmandokRup
October 6, 2024 @ 9:56 pm
Веселые анекдоты и шутки
Подними настроение
Stephenswogs
October 7, 2024 @ 12:47 am
[url=https://robertofirminoar.biz]www.robertofirminoar.biz[/url]
last news about roberto firmino
robertofirminoar.biz
24kbet
October 7, 2024 @ 3:10 am
I think this is one of the most significant information for me. And i’m glad reading your article. But should remark on some general things, The website style is perfect, the articles is really great nice. – cockfight login
Franktouth
October 7, 2024 @ 4:08 am
https://rybelsus.shop/# buy rybelsus online
Ahmedmooge
October 7, 2024 @ 6:00 am
ремонт сколов автостекол https://zamena-avtostekol-spb.ru
RobertRer
October 7, 2024 @ 7:51 am
[url=https://abdulhamidsaud-ar.biz]https://abdulhamidsaud-ar.biz[/url]
last news about abdulhamid saud
http://www.abdulhamidsaud-ar.biz
DamianSot
October 7, 2024 @ 8:15 am
best betting sites bonuses: Offers provided by betting platforms to reward both new and existing users. These offers can include free bets, deposit bonuses, and special promotions on major sporting events.
jxvknkzsk
October 7, 2024 @ 10:26 am
Quetzalcoatl. Daenerys targaryen. Bollywood. Fennec fox. Ford motor company. House of representatives. Back muscles. Vital. Colbert. https://81.200.117.113
StevenRaica
October 7, 2024 @ 10:51 am
[url=https://abdulhamid-saud-ar.biz]www.abdulhamid-saud-ar.biz[/url]
last news about abdulhamid saud
https://www.abdulhamid-saud-ar.biz
JeremyJex
October 7, 2024 @ 11:49 am
ozempic generic [url=https://ozempic.art/#]ozempic cost[/url] Ozempic without insurance
cnihueplg
October 7, 2024 @ 12:02 pm
Lovejoy. Browns score. San carlos. How old is simone biles. Aplastic anemia. Iguazu falls. Cliff burton. Uefa euro. November zodiac. https://81.200.117.113
Raymondusefe
October 7, 2024 @ 12:42 pm
[url=][/url]
Продажа мини-погрузчиков Lonking
Продажа мини-погрузчиков Lonking на территории России от официального
дистрибьютора. Новая многофункциональная техника для любых задач.
Наши машины предназначены для того, чтобы упростить вашу работу:
от строительных площадок до складских операций.
Высокая эффективность, надежность и инновационные решения — все,
что вам нужно для успешных проектов. Погрузите свой бизнес в будущее
с мини-погрузчиками Lonking!
47% российских покупателей выбрали мини-погрузчики Lonking в 2023 году продано более 1200 единиц.
Мини-погрузчики Lonking
[url=][/url]
bvkqdohcb
October 7, 2024 @ 1:25 pm
Norah jones. Retention. University of kansas. F scott fitzgerald. Cheeseburger. Will howard. Liability insurance. Calligraphy alphabet. Generation y. https://81.200.117.113
Gartandtoimi
October 7, 2024 @ 3:42 pm
Лучшие анекдоты
Заходи посмеяться
Jamesdyelf
October 7, 2024 @ 4:49 pm
gates of olympus казино gates of olympus slot
auzintkeu
October 7, 2024 @ 5:14 pm
Redlining. Blue jay bird. Calla lily. Turkish angora. Oxnard. Lake baikal. Structure. https://81.200.117.113
mostbet_992
October 7, 2024 @ 5:19 pm
мостбет промокод рабочий при регистрации мостбет промокод AZ3202 как пользоваться
JeremyBuiva
October 7, 2024 @ 5:49 pm
[url=https://obzor-gates-of-olympus.ru]https://obzor-gates-of-olympus.ru[/url]
last news about gates of olympus
http://obzor-gates-of-olympus.ru
ArmandokRup
October 7, 2024 @ 6:27 pm
Как улучшить настроение другу? Посмотрите
смешные анекдоты и поделитесь с близкими.
AndrewVOg
October 7, 2024 @ 6:59 pm
[url=https://alowais-mohammed-ar.biz]https://alowais-mohammed-ar.biz[/url]
last news about alowais mohammed
http://alowais-mohammed-ar.biz
MikelKaria
October 7, 2024 @ 7:21 pm
[url=https://gates-of-olympus-obzor.ru]http://gates-of-olympus-obzor.ru[/url]
last news about gates of olympus
http://gates-of-olympus-obzor.ru
cheap sex webcams
October 7, 2024 @ 7:27 pm
This is a topic that is near to my heart…
Best wishes! Exactly where are your contact details though?
Jamesdyelf
October 7, 2024 @ 8:10 pm
gates of olympus на деньги игра gates of olympus
Ремонт сигвеев в Москве
October 7, 2024 @ 8:14 pm
Профессиональный сервисный центр по ремонту сигвеев в Москве.
Мы предлагаем: сигвей сервис
Наши мастера оперативно устранят неисправности вашего устройства в сервисе или с выездом на дом!
mostbet_305
October 7, 2024 @ 9:01 pm
мостбет промокод рабочий как пользоваться 2024 промокод мостбет 2024
Davidcig
October 7, 2024 @ 10:32 pm
Online casino https://specii79.online/2024/10/06/bc-game-nigeria-the-ultimate-destination-for-gamers with high bonuses, roulette and slots, instant payouts. Register on the official mirror.
займы без отказа
October 7, 2024 @ 10:50 pm
На [url=https://mirtinvest.ru/]mirtinvest.ru[/url] ты найдёшь более 40 проверенных МФО, которые выдают микрокредиты онлайн без отказов. Для всех от 18 лет, с минимальными требованиями и ставкой не выше 0,8% в день — это настоящий шанс решить свои финансовые вопросы быстро и безопасно. Мы работаем только с лицензированными компаниями, так что можешь доверять нам. Пора действовать — займи деньги легко и без забот!
ALL-CREDIT моментальный займ на карту
October 7, 2024 @ 10:52 pm
Выбор надёжной микрофинансовой организации — ключ к успешному получению займа. На нашем [url=https://all-credit.ru/zaym-25000-rubley/ ]сайте[/url] вы найдете советы, как правильно выбрать МФО для онлайн займа. Эксперты рекомендуют обращать внимание на процентные ставки, условия погашения и репутацию компании. Мы собрали актуальную информацию о надежных МФО, которые предлагают займы на выгодных условиях. Переходите на [url=https://all-credit.ru/zaymy-s-avtomaticheskim-odobreniem/ ]страницу[/url], чтобы узнать больше о том, как выбрать подходящую компанию, подать заявку и получить деньги на карту в 2024 году. Правильный выбор МФО — залог финансовой безопасности и уверенности.
Derrickjoisy
October 7, 2024 @ 10:56 pm
[url=https://mohammed-alowais-ar.biz]https://www.mohammed-alowais-ar.biz[/url]
last news about mohammed alowais
http://www.mohammed-alowais-ar.biz
PinUpboacy
October 8, 2024 @ 1:04 am
pin up kz: пинап кз – пинап кз
Douglasjam
October 8, 2024 @ 1:44 am
1x Bet Promo Codes https://idematapp.com/wp-content/pages/1xbet_promo_codes_free_bonus_offers.html
1x Bet promo codes are alphanumeric strings that unlock bonuses on the 1xBet platform, such as free bets, deposit matches, or free spins. These codes are often provided as part of welcome offers or special promotions.
Davidcig
October 8, 2024 @ 2:29 am
Online casino https://webtowar.ru/2024/10/07/your-finnish-guide-to-slotspalace-suomi-casino with high bonuses, roulette and slots, instant payouts. Register on the official mirror.
AndrewCot
October 8, 2024 @ 4:32 am
лаки джет онлайн lucky jet официальный сайт
Franktouth
October 8, 2024 @ 5:15 am
https://ozempic.art/# buy cheap ozempic
Ремонт сетевых хранилищ в Москве
October 8, 2024 @ 5:45 am
Профессиональный сервисный центр по ремонту сетевых хранилищ в Москве.
Мы предлагаем: профессиональный ремонт сетевых хранилищ
Наши мастера оперативно устранят неисправности вашего устройства в сервисе или с выездом на дом!
Baikalwheels.ru
October 8, 2024 @ 6:20 am
Наша компания Baikal-Wheels также предлагает эксклюзивные кованые диски на заказ и [url=https://baikalwheels.ru/catalog?type=kovanye_diski]кованые диски купить интернет магазин[/url] которые отличаются прочностью и легкостью, улучшающей динамические характеристики автомобиля. Приобретая продукцию в компании Baikal-Wheels, вы получаете не только проверенные изделия от лучших производителей, но и профессиональный подход с удобной доставкой прямо к двери.
Gartandtoimi
October 8, 2024 @ 6:59 am
Смешные мемы. Как они появились.
AndrewCot
October 8, 2024 @ 8:00 am
lucky jet регистрация лаки джет сигналы
Психолог сейчас
October 8, 2024 @ 10:04 am
Наш Психолог сейчас
ZairobaSyday
October 8, 2024 @ 10:36 am
Тебе срочно нужны деньги, но все отказывают? [url=https://t.me/kino_film_20221/161]Займы с плохой историей без проверок[/url] – это твой спасательный круг! Мы собрали проверенные компании, которые выдают деньги на карту быстро и без лишних вопросов. Не трать время на ненужные попытки, подпишись на канал и получи свой займ уже сегодня! Твой шанс на успех – здесь!
Vivod iz zapoya rostov_xfkl
October 8, 2024 @ 10:54 am
вывод из запоя круглосуточно ростов на дону [url=https://vyvod-iz-zapoya-rostov16.ru/]вывод из запоя круглосуточно ростов на дону[/url] .
Charlesnidly
October 8, 2024 @ 11:02 am
[url=https://firminorobertoar.biz]https://www.firminorobertoar.biz[/url]
last news about firmino roberto
https://www.firminorobertoar.biz
Vivod iz zapoya rostov_orOl
October 8, 2024 @ 11:02 am
вывод. из. запоя. анонимно. ростов. [url=vyvod-iz-zapoya-rostov15.ru]вывод. из. запоя. анонимно. ростов.[/url] .
Психолог сейчас
October 8, 2024 @ 11:41 am
Наш Психолог сейчас
Психолог сейчас
October 8, 2024 @ 1:05 pm
Не паблик Психолог сейчас
Equalizer Exchange
October 8, 2024 @ 2:11 pm
I’ve been exploring for a bit for any high-quality articles or blog posts on this sort of space .
Exploring in Yahoo I finally stumbled upon this web site.
Studying this information So i’m glad to exhibit
that I have an incredibly excellent uncanny feeling I came upon just what I needed.
I so much certainly will make certain to do not
put out of your mind this website and give it a glance on a relentless
basis.
SheldonEruse
October 8, 2024 @ 2:20 pm
Aviator онлайн Авиатор игра
JeremyBuiva
October 8, 2024 @ 2:35 pm
[url=https://zeus-vs-hades-wiki.ru]http://zeus-vs-hades-wiki.ru[/url]
last news about zeus vs hades
https://www.zeus-vs-hades-wiki.ru
AndrewVOg
October 8, 2024 @ 4:12 pm
[url=https://casino-crazy-time.ru]www.casino-crazy-time.ru[/url]
last news about casino crazy time
http://casino-crazy-time.ru
DonaldSiz
October 8, 2024 @ 4:21 pm
[url=][/url]
Продажа мини-погрузчиков Lonking
Продажа мини-погрузчиков Lonking на территории России от официального дистрибьютора.
Новая многофункциональная техника для любых задач.
Наши машины предназначены для того, чтобы упростить вашу работу: от строительных площадок
до складских операций.
Высокая эффективность, надежность и инновационные решения — все, что вам нужно
для успешных проектов. Погрузите свой бизнес в будущее с мини-погрузчиками Lonking!
47% российских покупателей выбрали мини-погрузчики Lonking в 2023 году продано более 1200 единиц.
Наши сайт: Мини-погрузчики Lonking
[url=][/url]
JamesOwepe
October 8, 2024 @ 4:22 pm
apartment in a new building from the developer https://helium-balloons-delivery.com
MLM bitcoin opportunity
October 8, 2024 @ 4:29 pm
Can you tell us more about this? I’d like to find out more details.
купить сейф
October 8, 2024 @ 4:40 pm
В магазине сейфов предлагают купить сейф купить сейф москва
www.fluencycheck.com
October 8, 2024 @ 4:54 pm
Typically, physical and energised discrepancies can arise from emotional chaos and acupuncture for anxiety, http://www.fluencycheck.com,.
SheldonEruse
October 8, 2024 @ 6:08 pm
Aviator онлайн Aviator бонусы
BarryLig
October 8, 2024 @ 6:18 pm
https://ozempic.art/# buy cheap ozempic
Vivod iz zapoya rostov_geOn
October 8, 2024 @ 7:20 pm
вывод из запоя цены ростов-на-дону [url=www.vyvod-iz-zapoya-rostov18.ru/]www.vyvod-iz-zapoya-rostov18.ru/[/url] .
Narkolog na dom krasnodar_fikl
October 8, 2024 @ 7:41 pm
вызов нарколога на дом цена [url=http://www.narkolog-na-dom-krasnodar15.ru]вызов нарколога на дом цена[/url] .
JamesOwepe
October 8, 2024 @ 7:51 pm
buy an apartment in a new building from the developer air balloons
Mirtinvest.ru онлайн займы всем без звонков
October 8, 2024 @ 8:51 pm
На [url=https://mirtinvest.ru/]mirtinvest.ru[/url] вы найдёте более 40 микрофинансовых организаций, готовых предложить микрокредиты на карту онлайн без отказов. Все компании лицензированы и работают с соблюдением максимальной ставки в 0,8% в день. Мы предлагаем простую процедуру оформления, минимальные требования к заемщикам и возможность получения денег уже с 18 лет. Наш сервис создан для тех, кто ценит своё время и ищет надежные решения для своих финансовых нужд.
Gartandtoimi
October 8, 2024 @ 9:02 pm
ржачные картинки. Как они появились.
ALL-CREDIT займ под 0% без скрытых комиссий
October 8, 2024 @ 9:55 pm
Выбор МФО — это как выбор хорошего друга: вам нужны надёжность, честность и быстрое решение ваших проблем. На нашем [url=https://all-credit.ru/zaym-10000-rubley/ ]сайте[/url] мы подобрали для вас “друзей” — топовые МФО, которые готовы выручить вас займом на карту в любой момент. Хотите деньги без лишней суеты и бюрократии? Легко! Зачем бегать по банкам и собирать кучу бумаг, если всё можно оформить онлайн? Переходите на [url=https://all-credit.ru/dengi-v-dolg-ot-mfo-bez-proverki-ki/ ]страницу[/url], где вы найдёте не просто компании, а настоящих помощников в мире финансов. Оцените выгодные условия, нажмите пару кнопок — и деньги у вас. Всё просто, как разговор с лучшим другом!
Vivod iz zapoya rostov_rtsn
October 8, 2024 @ 11:17 pm
вывод. из. запоя. на. дому. ростов. [url=www.vyvod-iz-zapoya-rostov17.ru/]вывод. из. запоя. на. дому. ростов.[/url] .
Robertspony
October 9, 2024 @ 12:15 am
pinup az [url=http://pinupaz.bid/#]pin up azerbaijan[/url] pin-up oyunu
Kevinrisse
October 9, 2024 @ 1:59 am
[url=https://alowaismohammedar.biz]https://www.alowaismohammedar.biz[/url]
last news about alowais mohammed
https://www.alowaismohammedar.biz
Sarah jos
October 9, 2024 @ 2:11 am
Keep up the great work .Face Toner!
Sarah jos
October 9, 2024 @ 2:12 am
Keep up the great work .
Sarah jos
October 9, 2024 @ 2:12 am
https://sarahjosbeauty.com/products/sarah-jo-s-beauty-chateau-non-cbd-toner
Sarah jos
October 9, 2024 @ 2:16 am
Thank you for your sharing. https://sarahjosbeauty.com/products/sarah-jo-s-beauty-chateau-non-cbd-toner
Micahham
October 9, 2024 @ 2:50 am
[url=https://gates-of-olympus-info.ru]http://www.gates-of-olympus-info.ru[/url]
last news about gates of olympus
http://www.gates-of-olympus-info.ru
3KAUTO
October 9, 2024 @ 3:00 am
Today, while I was at work, my cousin stole my iphone and
tested to see if it can survive a forty foot drop, just so she can be
a youtube sensation. My apple ipad is now destroyed and she
has 83 views. I know this is totally off topic but I had to share it
with someone!
Douglasjam
October 9, 2024 @ 3:16 am
1xBet Sign Up Bonus https://actuchomage.org/includes/wkl/code_promo_69.html
The 1xBet sign-up bonus is a special offer available to new users when they register on the platform. This bonus usually includes a deposit match, giving new users extra funds to start betting on sports or casino games.
Gabrieldak
October 9, 2024 @ 6:42 am
пин ап казино вход http://pinupaz.bid/# pin up azerbaijan
пин ап 634
Forrestjed
October 9, 2024 @ 7:42 am
пин ап зеркало: пин ап официальный сайт – pin up казино
Gartandtoimi
October 9, 2024 @ 9:30 am
Прикольные анекдоты.
StevenSok
October 9, 2024 @ 10:56 am
[url=https://gates-of-olympus-wiki.ru]https://gates-of-olympus-wiki.ru[/url]
last news about gates of olympus
http://www.gates-of-olympus-wiki.ru
DonniejaF
October 9, 2024 @ 11:48 am
[url=https://casino-gates-of-olympus.ru]https://casino-gates-of-olympus.ru[/url]
last news about casino gates-of-olympus
casino-gates-of-olympus.ru
IvoryKAB
October 9, 2024 @ 11:55 am
Надежные МФО с гарантированным одобрением [url=https://maps.google.is/url?q=https://all-credit.ru/zaim-bez-otkaza-na-kartu/]микрозаймы на карту без отказа [/url] до 30 000 рублей для всех желающих без проверок.
Robertspony
October 9, 2024 @ 12:29 pm
pin up guncel giris [url=http://pinupturkey.pro/#]pin up aviator[/url] pin up giris
Gabrieldak
October 9, 2024 @ 12:39 pm
пин ап http://pinupturkey.pro/# pin up giris
пин ап казино вход
Сейфы второго класса взломостойкости
October 9, 2024 @ 1:44 pm
В магазине сейфов предлагают взломостойкие сейфы 2 класса купить сейф 2
VirgilFramb
October 9, 2024 @ 2:15 pm
Casino and slot https://bigimg.ru/2024/10/07/neon54-greece-casino-online-fun-real-rewards machines online. Play slot machines with bonus, casino for money. Registration and login to the working mirror
Forrestjed
October 9, 2024 @ 2:28 pm
pin up casino: pin up azerbaijan – pinup az
seo18.su
October 9, 2024 @ 2:58 pm
В digital агентстве ПАРОВОЗ мы занимаемся профессионально на [url=https://seo18.su/servicetype/seo-prodvizhenie-sajta-v-google/]seo сайта заказать[/url] и создании качественных веб-проектов. С более чем десятью годами опыта, мы гарантируем эффективное продвижение сайтов в ТОП поисковых систем, используя современные технологии и инструменты. Наша команда постоянно улучшает свои умения, что позволяет нам достигать неизменных результатов для клиентов.
Сегодня [url=https://seo18.su/]продвижение сайтов цены seo fortuna[/url] – это ключ к достижению целей вашего бизнеса. В условиях высокой конкуренции важно оставаться в ТОПе в Гугл и Yandex, чтобы увеличивать пользователей. Мы приглашаем на пробный период, где вы проверите наш профессионализм и проверите действенность наших методов.
KevinAlobe
October 9, 2024 @ 3:12 pm
Free online https://thetranny.com porn videos and movies. Only girls with dicks. On our site there are trans girls for every taste!
JeremyBuiva
October 9, 2024 @ 3:57 pm
[url=https://sugar-rush-casino-play.ru]https://www.sugar-rush-casino-play.ru[/url]
last news about sugar rush casino
http://www.sugar-rush-casino-play.ru
Leroytwime
October 9, 2024 @ 4:21 pm
Профессиональный сервисный центр ремонт телефонов москва дешево ближайший сервис по ремонту телефонов
AndrewVOg
October 9, 2024 @ 4:26 pm
[url=https://midas-golden-touch-obzor.ru]http://midas-golden-touch-obzor.ru[/url]
last news about midas golden touch
http://www.midas-golden-touch-obzor.ru
Реферальный бонус на binance
October 9, 2024 @ 4:40 pm
I don’t think the title of your article matches the content lol. Just kidding, mainly because I had some doubts after reading the article. https://accounts.binance.com/pt-BR/register-person?ref=YY80CKRN
MichaelDar
October 9, 2024 @ 5:13 pm
[url=https://mohammed-alowaisar.biz]https://mohammed-alowaisar.biz[/url]
last news about mohammed alowais
mohammed-alowaisar.biz
MikelKaria
October 9, 2024 @ 5:17 pm
[url=https://midas-golden-touch-wiki.ru]http://www.midas-golden-touch-wiki.ru[/url]
last news about midas golden touch
http://www.midas-golden-touch-wiki.ru
VirgilFramb
October 9, 2024 @ 6:08 pm
Casino and slot https://posutochno-1.online/2024/10/06/elevate-your-winnings-with-1win-benin machines online. Play slot machines with bonus, casino for money. Registration and login to the working mirror
Gabrieldak
October 9, 2024 @ 7:55 pm
пинап казино http://pinupaz.bid/# pin up 306
пинап казино
Stephennewly
October 9, 2024 @ 9:16 pm
[url=https://crazy-time-obzor.ru/]http://crazy-time-obzor.ru[/url]
last news about crazy time obzor
crazy-time-obzor.ru
StevenRaica
October 9, 2024 @ 9:40 pm
[url=https://casino-zeus-vs-hades.ru]https://casino-zeus-vs-hades.ru[/url]
last news about casino zeus vs hades
http://www.casino-zeus-vs-hades.ru
itacat.ru
October 9, 2024 @ 10:22 pm
Займы — главный финансовый тренд 2024 года [url=https://itacat.ru/]срочные онлайн займы на карту[/url] . Оформление стало проще, чем когда-либо: достаточно заполнить онлайн-заявку, и деньги уже на вашей карте [url=https://itacat.ru/]взять займ срочно без отказа на карту[/url] . Это доступно каждому, независимо от уровня дохода или кредитной истории [url=https://itacat.ru/]кредиты без отказа срочно[/url] . Моментальное одобрение и быстрый перевод средств на карту делают займы идеальным решением для экстренных ситуаций [url=https://itacat.ru/]мфо которые дают деньги всем без отказа[/url] . Более того, многие компании предлагают первый займ бесплатно! Теперь не нужно ждать или проходить бесконечные проверки — займ доступен всем от 18 лет, быстро и без отказов!
Forrestjed
October 9, 2024 @ 10:22 pm
pin-up oyunu: pinup azerbaycan – pin up casino
кино веном 2
October 9, 2024 @ 11:07 pm
веном фильм смотреть онлайн веном 2 часть веном смотреть онлайн
Scottcaurl
October 9, 2024 @ 11:12 pm
Porn video https://zeenite.com watch online from pages for free in FullHD quality. Fresh archive of selected porn and tender sex scenes from all over the world!
смотреть смотреть фильм веном
October 10, 2024 @ 12:40 am
фильм веном 2 смотреть веном смотреть веном 2 смотреть онлайн ок
Gartandtoimi
October 10, 2024 @ 1:57 am
Прикольные картинки.
веном фильм смотреть онлайн
October 10, 2024 @ 2:04 am
смотреть фильм веном 2 веном 2 скачать веном 2 смотреть онлайн ок
Robertdup
October 10, 2024 @ 2:40 am
[url=https://play-crazy-time.ru]play-crazy-time.ru[/url]
last news about play crazy time
https://play-crazy-time.ru
сео продвижение медицинских сайтов
October 10, 2024 @ 3:04 am
Тут делают продвижение стратегия с роботами комплексное продвижение медицинских сайтов
seo services consultants
October 10, 2024 @ 3:05 am
Hi, this weekend is nice in favor of me, since this point in time i am reading this enormous
informative article here at my house.
сервис центры в волгограде
October 10, 2024 @ 3:23 am
Профессиональный сервисный центр по ремонту бытовой техники с выездом на дом.
Мы предлагаем: сервисные центры по ремонту техники в волгограде
Наши мастера оперативно устранят неисправности вашего устройства в сервисе или с выездом на дом!
Gabrieldak
October 10, 2024 @ 3:48 am
пин ап http://pinupru.site/# pin up
пин ап казино
создание медицинских сайтов под ключ
October 10, 2024 @ 5:11 am
Тут делают продвижение разработка сайтов для медицинских центров разработка медицинских сайтов
Forrestjed
October 10, 2024 @ 7:01 am
пин ап вход: pin up зеркало – пин ап вход
Ремонт планшетов
October 10, 2024 @ 8:29 am
Профессиональный сервисный центр по ремонту планшетов в Москве.
Мы предлагаем: ремонт стекла планшета
Наши мастера оперативно устранят неисправности вашего устройства в сервисе или с выездом на дом!
Gartandtoimi
October 10, 2024 @ 9:56 am
Смешные картинки.
Микрозаймы
October 10, 2024 @ 9:57 am
[url=https://mikro-zaim-online.ru/]займы без проверок и отказа онлайн[/url]
Gabrieldak
October 10, 2024 @ 10:58 am
пинап кз http://pinupru.site/# pin up
пин ап
Robertspony
October 10, 2024 @ 10:58 am
pinup az [url=http://pinupaz.bid/#]pinup azerbaycan[/url] pin up azerbaijan
Derrickjoisy
October 10, 2024 @ 11:35 am
[url=https://sugar-rush-casino-info.ru]www.sugar-rush-casino-info.ru[/url]
last news about sugar rush casino
http://www.sugar-rush-casino-info.ru
Stephenswogs
October 10, 2024 @ 12:00 pm
[url=https://play-zeus-vs-hades.ru]www.play-zeus-vs-hades.ru[/url]
last news about zeus vs hades
https://www.play-zeus-vs-hades.ru
AngelUTETA
October 10, 2024 @ 2:19 pm
bs2best at
Forrestjed
October 10, 2024 @ 2:56 pm
pin up guncel giris: pin-up casino – pin up bet
GeorgeZosse
October 10, 2024 @ 3:50 pm
Вызов сантехника https://santekhnik-moskva.blogspot.com/p/v-moskve.html на дом в Москве и Московской области в удобное для вас время. Сантехнические работы любой сложности, от ремонта унитаза, устранение засора, до замены труб.
Vivod iz zapoya krasnodar_tlot
October 10, 2024 @ 4:57 pm
вывод из запоя [url=http://www.vyvod-iz-zapoya-krasnodar15.ru]http://www.vyvod-iz-zapoya-krasnodar15.ru[/url] .
AndrewVOg
October 10, 2024 @ 5:26 pm
[url=https://midas-golden-touch-info.ru]www.midas-golden-touch-info.ru[/url]
last news about midas golden touch
midas-golden-touch-info.ru
Займы
October 10, 2024 @ 5:32 pm
[url=https://mikro-zaim-online.ru/]займы под 0 процентов для всех[/url]
instagram story viewer_bnMr
October 10, 2024 @ 5:35 pm
stories anonymously without [url=http://www.anonimstories.com ]stories anonymously without[/url] .
MikelKaria
October 10, 2024 @ 5:40 pm
[url=https://sugar-rush-casino-slots.ru]http://sugar-rush-casino-slots.ru[/url]
last news about sugar rush casino
sugar-rush-casino-slots.ru
Santoneine
October 10, 2024 @ 5:52 pm
https://pinupturkey.pro/# pin up bet
ArmandokRup
October 10, 2024 @ 6:01 pm
Свежие видео приколы.
Robertspony
October 10, 2024 @ 6:07 pm
pin up azerbaijan [url=https://pinupaz.bid/#]pinup az[/url] pin up azerbaijan
Gabrieldak
October 10, 2024 @ 6:22 pm
пин ап казино https://pinupkz.tech/# пин ап казино онлайн
пин ап казахстан
Narkolog na dom krasnodar_cbka
October 10, 2024 @ 7:17 pm
нарколог на дом [url=https://narkolog-na-dom-krasnodar17.ru/]нарколог на дом[/url] .
Narkolog na dom krasnodar_pmml
October 10, 2024 @ 7:30 pm
вызвать нарколога на дом [url=https://narkolog-na-dom-krasnodar16.ru]https://narkolog-na-dom-krasnodar16.ru[/url] .
Gartandtoimi
October 10, 2024 @ 7:39 pm
Смешные пошлые анекдоты.
взломостойкие сейфы
October 10, 2024 @ 8:19 pm
В магазине сейфов предлагают сейфы взломостойкие сейф взломостойкий цена
Charlesnidly
October 10, 2024 @ 8:55 pm
[url=https://crazy-time-casino.ru]http://crazy-time-casino.ru[/url]
last news about crazy time casino
http://www.crazy-time-casino.ru
ClementAcing
October 10, 2024 @ 9:24 pm
В DPM+ мы знаем, насколько важно качественное [url=https://dpmplus.ru/]архитектурное проектирование[/url] для достижения успеха в девелопменте. Наша программа предоставляет комплексный подход к обучению, начиная с разработки концепции и заканчивая управлением проектами. Присоединяйтесь к нам, чтобы стать настоящим экспертом и поднять свою карьеру на новый уровень!
Forrestjed
October 10, 2024 @ 10:28 pm
pin up: pin up azerbaijan – pin up azerbaijan
Robertspony
October 11, 2024 @ 1:43 am
pin-up bonanza [url=http://pinupturkey.pro/#]pin up casino[/url] pin up casino guncel giris
Gabrieldak
October 11, 2024 @ 2:08 am
pin up казино http://pinupturkey.pro/# pin up
пин ап казино вход
seo медицинских сайтов
October 11, 2024 @ 3:53 am
Тут делают продвижение медицинское seo сео продвижение медицинского сайта
создание медицинского сайта под ключ
October 11, 2024 @ 5:10 am
Тут делают продвижение разработка сайт медицинского центра создание сайтов для клиники
inspirational quotes
October 11, 2024 @ 6:46 am
Attractive portion of content. I just stumbled upon your site and in accession capital to say that I acquire actually loved account your weblog posts.
Anyway I will be subscribing in your feeds and even I achievement you get
admission to persistently quickly.
Быстрые займы за 10 минут
October 11, 2024 @ 8:07 am
[url=https://all-credit.ru/zaim-bez-otkaza-na-kartu/]Займы без отказов[/url] – получите деньги до 30 тысяч рублей за 5 минут. Простое оформление по паспорту, даже с плохой кредитной историей, лучше, чем брать кредит в банке.
Micahham
October 11, 2024 @ 9:52 am
[url=https://play-midas-golden-touch.ru]http://play-midas-golden-touch.ru[/url]
last news about play midas golden
http://play-midas-golden-touch.ru
MichaelHoash
October 11, 2024 @ 10:02 am
neurontin 600 mg cost [url=https://gabapentin.auction/#]cheapest gabapentin[/url] buying neurontin without a prescription
KelvinDer
October 11, 2024 @ 11:43 am
[url=][/url]
Интернет-магазин плитки и керамики «ИнфоПлитка»
Интернет-магазин керамической плитки и керамогранита «Infoplitka» (infoplitka.ru)
предлагает широкий ассортимент высококачественной плитки и керамогранита от ведущих производителей.
Мы стремимся предложить нашим клиентам только лучшее, поэтому в ассортименте представлены товары,
отвечающие самым высоким стандартам качества и дизайна.
Мы понимаем, что выбор керамической плитки и керамогранита – это важный этап в создании
комфортного и стильного интерьера. Поэтому наша команда профессионалов готова помочь вам в
подборе идеального варианта для вашего проекта. Независимо от того, нужны ли вам плитка
для ванной комнаты, кухни, гостиной, или же керамогранит для облицовки пола, вы всегда найдете
у нас разнообразные и актуальные коллекции, соответствующие последним тенденциям в области
дизайна интерьеров.
Официальный сайт «ИнфоПлитка»
[url=][/url]
займ без отказа
October 11, 2024 @ 1:30 pm
[url=https=https://все-займы-тут.рф/]Микрозаймы онлайн на карту без отказа[/url] — это отличный вариант для тех, кто ищет мгновенные решения. Без проверок и сложных условий, деньги поступают моментально.
MatthewAdact
October 11, 2024 @ 2:04 pm
buy cheap neurontin: neurontin 4 mg – brand neurontin 100 mg canada
SamuelAlaws
October 11, 2024 @ 4:01 pm
http://semaglutide.win/# rybelsus cost
JeremyBuiva
October 11, 2024 @ 5:02 pm
[url=https://slot-casino-info.ru]www.slot-casino-info.ru[/url]
last news about slot casino
http://slot-casino-info.ru
Gartandtoimi
October 11, 2024 @ 5:33 pm
Веселые приколы https://teletype.in/@anekdoty/prikoly-nastroenie.
MichaelHoash
October 11, 2024 @ 5:35 pm
rybelsus cost [url=http://semaglutide.win/#]cheap Rybelsus 14 mg[/url] Buy semaglutide pills
AndrewVOg
October 11, 2024 @ 5:38 pm
[url=https://slots-casino-info.ru]slots-casino-info.ru[/url]
last news about slots casino
slots-casino-info.ru
MikelKaria
October 11, 2024 @ 5:54 pm
[url=https://dog-house-casino-wiki.ru]dog-house-casino-wiki.ru[/url]
last news about dog house
http://www.dog-house-casino-wiki.ru
Free multiplayer games
October 11, 2024 @ 7:42 pm
Have you ever considered creating an ebook or guest authoring on other sites?
I have a blog centered on the same ideas you discuss
and would really like to have you share some stories/information. I
know my readers would enjoy your work. If you’re even remotely interested,
feel free to shoot me an e-mail.
купить угги
October 11, 2024 @ 9:21 pm
Покупай [url=https://uggs-russia.com/]угги в Москве[/url] и наслаждайся зимними прогулками в тепле.
разработка сайт медицинского центра
October 11, 2024 @ 10:28 pm
Тут делают продвижение разработка сайт медицинского центра разработка медицинского сайта
MatthewAdact
October 11, 2024 @ 10:30 pm
how to get zithromax over the counter: zithromax best price – zithromax online australia
RobertRer
October 11, 2024 @ 11:04 pm
[url=https://crazy-time-wiki.ru]https://www.crazy-time-wiki.ru[/url]
last news about crazy time
https://www.crazy-time-wiki.ru
StevenSok
October 11, 2024 @ 11:07 pm
[url=https://sugar-rush-casino-wiki.ru]https://www.sugar-rush-casino-wiki.ru[/url]
last news about sugar rush casino
https://www.sugar-rush-casino-wiki.ru
MichaelHoash
October 12, 2024 @ 2:04 am
stromectol online canada [url=http://stromectol.agency/#]stromectol price[/url] ivermectin gel
сервис центры в воронеже
October 12, 2024 @ 2:13 am
Профессиональный сервисный центр по ремонту бытовой техники с выездом на дом.
Мы предлагаем: сервис центры бытовой техники воронеж
Наши мастера оперативно устранят неисправности вашего устройства в сервисе или с выездом на дом!
Ремонт телефонов
October 12, 2024 @ 2:23 am
Профессиональный сервисный центр сервисный центр мобильных телефонов сервисный центр ремонт телефонов
Gartandtoimi
October 12, 2024 @ 2:38 am
Веселые анекдоты https://telegra.ph/Smeshnye-anekdoty-10-09.
Stevenprath
October 12, 2024 @ 5:09 am
https://zithromax.company/# zithromax online australia
generic zithromax online paypal
MatthewAdact
October 12, 2024 @ 6:13 am
neurontin 100 mg caps: gabapentin price – buy neurontin online uk
ArmandokRup
October 12, 2024 @ 7:45 am
Свежие еврейские анекдоты https://sites.google.com/view/shutki-anekdoty/%D0%B0%D0%BD%D0%B5%D0%BA%D0%B4%D0%BE%D1%82%D1%8B-%D0%BF%D1%80%D0%BE-%D0%B5%D0%B2%D1%80%D0%B5%D0%B5%D0%B2.
MichaelHoash
October 12, 2024 @ 9:29 am
ivermectin price canada [url=https://stromectol.agency/#]stromectol best price[/url] ivermectin tablet price
ixxx_ahkt
October 12, 2024 @ 10:22 am
free porn sax [url=https://ixxxsexvideo.com/]ixxxsexvideo.com[/url] .
Kevinrisse
October 12, 2024 @ 10:35 am
[url=https://sugar-rush-casino-obzor.ru]sugar-rush-casino-obzor.ru[/url]
last news about sugar rush casino
http://sugar-rush-casino-obzor.ru
Vivod iz zapoya krasnodar_wrka
October 12, 2024 @ 11:24 am
вывод из запоя на дому краснодар круглосуточно [url=https://vyvod-iz-zapoya-krasnodar16.ru/]вывод из запоя на дому краснодар круглосуточно[/url] .
Stevenprath
October 12, 2024 @ 1:20 pm
https://semaglutide.win/# buy rybelsus
buy zithromax without presc
MatthewAdact
October 12, 2024 @ 2:16 pm
buy amoxicillin canada: where to buy amoxicillin over the counter – amoxicillin 500mg pill
SamuelAlaws
October 12, 2024 @ 2:23 pm
https://gabapentin.auction/# neurontin online
Gartandtoimi
October 12, 2024 @ 2:43 pm
Свежие картинки с надписями https://prikoly-tut.blogspot.com/2024/10/smeshnye-kartinki.html.
Vivod iz zapoya ekaterinbyrg_lhmr
October 12, 2024 @ 3:24 pm
срочный вывод из запоя [url=http://www.vyvod-iz-zapoya-ekaterinburg14.ru]срочный вывод из запоя[/url] .
DonaldViash
October 12, 2024 @ 3:26 pm
bs2site.at
MichaelHoash
October 12, 2024 @ 4:23 pm
stromectol online canada [url=https://stromectol.agency/#]order stromectol[/url] ivermectin medication
Ремонт электросамокатов в Москве
October 12, 2024 @ 4:48 pm
Профессиональный сервисный центр по ремонту электросамокатов в Москве.
Мы предлагаем: тюнинг электросамокатов в москве
Наши мастера оперативно устранят неисправности вашего устройства в сервисе или с выездом на дом!
Stevenprath
October 12, 2024 @ 8:04 pm
http://stromectol.agency/# stromectol
buy azithromycin zithromax
MatthewAdact
October 12, 2024 @ 9:11 pm
Semaglutide pharmacy price: buy rybelsus – Rybelsus 7mg
ArmandokRup
October 12, 2024 @ 10:16 pm
Веселые анекдоты про евреев https://sites.google.com/view/shutki-anekdoty/%D0%B0%D0%BD%D0%B5%D0%BA%D0%B4%D0%BE%D1%82%D1%8B-%D0%BF%D1%80%D0%BE-%D0%B5%D0%B2%D1%80%D0%B5%D0%B5%D0%B2.
MichaelHoash
October 12, 2024 @ 10:59 pm
Buy compounded semaglutide online [url=https://semaglutide.win/#]buy semaglutide online[/url] Semaglutide pharmacy price
Stephennewly
October 12, 2024 @ 11:35 pm
[url=https://slot-midas-golden-touch.ru/]http://slot-midas-golden-touch.ru[/url]
last news about slot midas golden touch
https://www.slot-midas-golden-touch.ru
SamuelAlaws
October 13, 2024 @ 12:10 am
https://stromectol.agency/# price of stromectol
Gartandtoimi
October 13, 2024 @ 12:23 am
Смешные фотоприколы https://prikoly-tut.blogspot.com/2024/10/smeshnye-kartinki.html.
MichaelDar
October 13, 2024 @ 12:52 am
[url=https://slots-casino-obzor.ru]https://www.slots-casino-obzor.ru[/url]
last news about slots casino
http://www.slots-casino-obzor.ru
Vivod iz zapoya krasnodar_mtmi
October 13, 2024 @ 1:45 am
вывод из запоя круглосуточно [url=https://www.vyvod-iz-zapoya-krasnodar17.ru]https://www.vyvod-iz-zapoya-krasnodar17.ru[/url] .
Vivod iz zapoya ekaterinbyrg_gnPr
October 13, 2024 @ 2:08 am
вывод из запоя на дому [url=http://www.vyvod-iz-zapoya-ekaterinburg15.ru]вывод из запоя на дому[/url] .
RogelioImpok
October 13, 2024 @ 2:20 am
[url=][/url]
Найти специалиста по независимой экспертизе и оценке!
Сайт-агрегатор компаний и услуг в сфере независимой экспертизы и оценки.
Мы создали этот проект, чтобы помочь вам найти надежных и опытных профессионалов
для решения ваших задач.
Главная цель — сделать процесс поиска специалистов по независимой экспертизе
и оценке максимально простым и эффективным. Мы стремимся предоставить вам
доступ к компаниям, которые гарантируют высокое качество услуг.
С нами вы сможете быстро найти нужного эксперта и сравнить различные предложения.
На нашем сайте собраны карточки компаний, каждая из которых содержит подробную
информацию о предоставляемых услугах. Посетители могут фильтровать предложения
по различным критериям:
Локация
Направление экспертизы
Стоимость услуг
Отзывы клиентов
Наш сайт Специалисты по независимой экспертизе и оценке.
[url=][/url]
MatthewAdact
October 13, 2024 @ 4:18 am
rybelsus cost: Semaglutide pharmacy price – Buy semaglutide pills
MichaelHoash
October 13, 2024 @ 5:35 am
can i buy amoxicillin over the counter in australia [url=https://amoxil.llc/#]buy amoxil[/url] where to buy amoxicillin 500mg
Gartandtoimi
October 13, 2024 @ 6:40 am
Веселые приколы за день https://prikolnyekartinki.mystrikingly.com/blog/0ade1e0d9a9.
ArmandokRup
October 13, 2024 @ 8:50 am
Веселые анекдоты https://telegra.ph/Smeshnye-anekdoty-10-09.
Derrickjoisy
October 13, 2024 @ 9:30 am
[url=https://slot-casino-wiki.ru]slot-casino-wiki.ru[/url]
last news about slot casino
http://www.slot-casino-wiki.ru
Stevenprath
October 13, 2024 @ 9:46 am
https://semaglutide.win/# rybelsus cost
can i buy zithromax over the counter
SamuelAlaws
October 13, 2024 @ 9:54 am
http://gabapentin.auction/# neurontin brand name 800mg
uggs
October 13, 2024 @ 10:52 am
Мечтаешь о тепле и стиле? Выбирай [url=https://uggs-russia.com/]женские угги[/url] с доставкой по России.
MatthewAdact
October 13, 2024 @ 11:13 am
neurontin 300: gabapentin best price – prescription drug neurontin
MichaelHoash
October 13, 2024 @ 12:10 pm
minocycline mr [url=https://stromectol.agency/#]stromectol for sale[/url] ivermectin 2%
AndrewVOg
October 13, 2024 @ 3:12 pm
[url=https://big-bamboo-casino-slots.ru]http://big-bamboo-casino-slots.ru[/url]
last news about big bamboo casino
http://big-bamboo-casino-slots.ru
MikelKaria
October 13, 2024 @ 3:13 pm
[url=https://dog-house-casino-play.ru]https://dog-house-casino-play.ru[/url]
last news about dog house
dog-house-casino-play.ru
Stevenprath
October 13, 2024 @ 4:56 pm
https://stromectol.agency/# minocycline 50mg pills
zithromax 250 mg
Gartandtoimi
October 13, 2024 @ 5:01 pm
Свежие пошлые анекдоты https://ru.pinterest.com/vseshutochki/%D0%BF%D0%BE%D1%88%D0%BB%D1%8B%D0%B5-%D0%B0%D0%BD%D0%B5%D0%BA%D0%B4%D0%BE%D1%82%D1%8B/.
rzufw312
October 13, 2024 @ 6:15 pm
t70285 read more here c7655028
Going Here
October 13, 2024 @ 6:40 pm
Keep on working, great job!
MatthewAdact
October 13, 2024 @ 6:44 pm
stromectol for head lice: stromectol for sale – ivermectin new zealand
Stevenloach
October 13, 2024 @ 6:46 pm
bs2best.at
See Details
October 13, 2024 @ 6:46 pm
I do not even know how I ended up here, but I thought this post was good.
I don’t know who you are but certainly you are going to a famous blogger if you aren’t already 😉 Cheers!
binance kaydi
October 13, 2024 @ 7:12 pm
Your point of view caught my eye and was very interesting. Thanks. I have a question for you.
MichaelHoash
October 13, 2024 @ 7:16 pm
buy cheap zithromax online [url=https://zithromax.company/#]buy zithromax z-pak online[/url] zithromax 500mg price in india
Robertdup
October 13, 2024 @ 8:32 pm
[url=https://slots-casino-wiki.ru]https://slots-casino-wiki.ru[/url]
last news about slots casino
slots-casino-wiki.ru/a>
index
October 13, 2024 @ 9:05 pm
hey there and thank you for your info – I have certainly picked up
anything new from right here. I did however expertise a few technical points using this web site, as I experienced to reload
the web site many times previous to I could get it to load correctly.
I had been wondering if your web hosting is OK?
Not that I’m complaining, but sluggish loading instances times will often affect your placement in google and can damage
your quality score if ads and marketing with Adwords.
Well I’m adding this RSS to my e-mail and can look out for much more of your respective fascinating content.
Ensure that you update this again very soon.
JeremyBuiva
October 13, 2024 @ 9:07 pm
[url=https://big-bamboo-casino-wiki.ru]https://www.big-bamboo-casino-wiki.ru[/url]
last news about big bamboo casino
https://www.big-bamboo-casino-wiki.ru
Ремонт Apple iMac в Москве
October 13, 2024 @ 10:29 pm
Профессиональный сервисный центр по ремонту моноблоков iMac в Москве.
Мы предлагаем: ремонт аймак москва
Наши мастера оперативно устранят неисправности вашего устройства в сервисе или с выездом на дом!
ArmandokRup
October 13, 2024 @ 11:45 pm
Смешные анекдоты https://vk.com/vseshutochki.
Stevenprath
October 14, 2024 @ 1:17 am
https://gabapentin.auction/# neurontin 30 mg
where can i purchase zithromax online
MichaelHoash
October 14, 2024 @ 2:35 am
zithromax 500 mg lowest price pharmacy online [url=https://zithromax.company/#]order zithromax[/url] zithromax pill
Vivod iz zapoya ekaterinbyrg_foMn
October 14, 2024 @ 6:45 am
вывод из запоя в екатеринбурге [url=www.vyvod-iz-zapoya-ekaterinburg16.ru]вывод из запоя в екатеринбурге[/url] .
ekokoja kypit_mbSi
October 14, 2024 @ 7:15 am
экокожа купить [url=www.ekokozha-kupit11.ru]экокожа купить[/url] .
MelvinMop
October 14, 2024 @ 8:42 am
Gay Boys Porn https://gay0day.com HD is the best gay porn tube to watch high definition videos of horny gay boys jerking, sucking their mates and fucking on webcam
euroshop18.ru
October 14, 2024 @ 8:50 am
Не стоит тратить время на поиски [url=https://euroshop18.ru/katalog/kuxonnyie-vyityazhki/akpo-wk-4-mio-eco-ii-60-sm-belyj.html]полновстраиваемая вытяжка 60 см[/url] в других магазинах. На euroshop18.ru вы найдёте всё необходимое по выгодной цене. Покупать здесь — это разумное вложение в комфорт вашего дома.
MichaelHoash
October 14, 2024 @ 9:27 am
zithromax online paypal [url=http://zithromax.company/#]zithromax best price[/url] where can i buy zithromax medicine
Займ
October 14, 2024 @ 9:33 am
Ищете быстрые решения? Оформите [url=https://all-credit/]срочно займ на карту[/url] за 15 минут! Процедура полностью онлайн, деньги моментально на карте.
MatthewAdact
October 14, 2024 @ 9:46 am
stromectol price us: stromectol price – what is minocycline 100 mg used for
Derrickjoisy
October 14, 2024 @ 10:28 am
[url=https://lucky-jet-casino-slots.ru]http://lucky-jet-casino-slots.ru[/url]
last news about lucky jet casino
http://www.lucky-jet-casino-slots.ru
euroshop
October 14, 2024 @ 11:44 am
Улыбнитесь новым возможностям с [url=https://euroshop18.ru/katalog/kuxonnyie-vyityazhki/naklonnye-vytyazhki.html]наклонная вытяжка купить[/url] на euroshop18.ru! Пусть каждый день на кухне будет полон радости и вдохновения благодаря нашей качественной технике.
Stephenswogs
October 14, 2024 @ 1:05 pm
[url=https://doghouse-casino-play.ru]www.doghouse-casino-play.ru[/url]
last news about doghouse casino
https://www.doghouse-casino-play.ru
Conradtix
October 14, 2024 @ 3:02 pm
bs2site
Stevenprath
October 14, 2024 @ 3:16 pm
http://zithromax.company/# zithromax online usa no prescription
buy zithromax canada
AndrewVOg
October 14, 2024 @ 4:36 pm
[url=https://sweet-bonanza-casino-play.ru]sweet-bonanza-casino-play.ru[/url]
last news about sweet bonanza casino
http://sweet-bonanza-casino-play.ru
купить сейф третьего класса
October 14, 2024 @ 4:42 pm
магазин сейфов предлагает сейф 3 класса сейф 3 класс
Займы
October 14, 2024 @ 4:42 pm
Выбирайте лучший способ получить деньги без процентов! На нашем сайте [url=https://all-credit/]займы на карту без процентов[/url] доступны с 18 лет, не теряйте возможность оформить займ под 0%.
ArmandokRup
October 14, 2024 @ 4:56 pm
Смешные приколы https://teletype.in/@anekdoty/prikoly-nastroenie.
rabota v Kazahstane_qtka
October 14, 2024 @ 5:11 pm
свежие вакансии [url=www.umicum.kz/]свежие вакансии[/url] .
rabota v Almati_zxMr
October 14, 2024 @ 5:15 pm
работа в алматы график 1/3 [url=https://www.trudvsem.kz]работа в алматы график 1/3[/url] .
MikelKaria
October 14, 2024 @ 5:56 pm
[url=https://lucky-jet-casino-info.ru]lucky-jet-casino-info.ru[/url]
last news about lucky jet casino
http://lucky-jet-casino-info.ru
Gartandtoimi
October 14, 2024 @ 6:24 pm
Смешные приколы https://prikolyshutki.wordpress.com/2024/10/11/prikoly-umor/.
rabota v Astane_hjoi
October 14, 2024 @ 6:46 pm
ежедневная работа в астане [url=www.sitsen.kz/]ежедневная работа в астане[/url] .
StevenSok
October 14, 2024 @ 7:19 pm
[url=https://doghouse-casino-wiki.ru]www.doghouse-casino-wiki.ru[/url]
last news about doghouse casino
doghouse-casino-wiki.ru
one88
October 14, 2024 @ 7:33 pm
Incredible! This blog looks exactly like my old one!
It’s on a totally different subject but it has pretty much the same page
layout and design. Outstanding choice of colors!
создание медицинского сайта под ключ
October 14, 2024 @ 8:04 pm
Тут делают продвижение seo для медицинского центра разработка сайтов медицинских центров
RobertRer
October 14, 2024 @ 9:09 pm
[url=https://big-bamboo-casino-obzor.ru]http://big-bamboo-casino-obzor.ru[/url]
last news about big bamboo casino
https://www.big-bamboo-casino-obzor.ru
seo медицинских сайтов
October 14, 2024 @ 9:42 pm
Тут делают продвижение стратегия с роботами сео продвижение медицинского сайта
bycesoir
October 14, 2024 @ 10:13 pm
[url=https://bycesoir.com/]Займ на карту срочно без отказа онлайн[/url]
Ремонт автомагнитол
October 14, 2024 @ 11:37 pm
Профессиональный сервисный центр по ремонту автомагнитол в Москве.
Мы предлагаем: сервис центр ремонта магнитол
Наши мастера оперативно устранят неисправности вашего устройства в сервисе или с выездом на дом!
Stevenloach
October 15, 2024 @ 2:34 am
bs2best
StevenRaica
October 15, 2024 @ 6:45 am
[url=https://big-bamboo-casino-play.ru]http://www.big-bamboo-casino-play.ru[/url]
last news about big bamboo
big-bamboo-casino-play.ru
ArmandokRup
October 15, 2024 @ 7:01 am
Веселые приколы https://teletype.in/@anekdoty/prikoly-nastroenie.
Gartandtoimi
October 15, 2024 @ 7:43 am
Свежие приколы http://anekdotshutka.ru.
IgnacioRar
October 15, 2024 @ 9:01 am
cs2 gambling sites cs2 gambling sites
RogerBeame
October 15, 2024 @ 9:12 am
csgo gambling sites csgo gambling sites
mfzyo900
October 15, 2024 @ 9:35 am
b178759
find out more d2922455
Charlesnidly
October 15, 2024 @ 12:35 pm
[url=https://big-bamboo-casino-info.ru]big-bamboo-casino-info.ru[/url]
last news about big bamboo casino
https://big-bamboo-casino-info.ru
GregoryZooke
October 15, 2024 @ 12:41 pm
top erection pills http://indianpharmdelivery.com/# best online pharmacy india
JeremyBuiva
October 15, 2024 @ 4:27 pm
[url=https://sweet-bonanza-casino-wiki.ru]www.sweet-bonanza-casino-wiki.ru[/url]
last news about sweet bonanza casino
https://sweet-bonanza-casino-wiki.ru
Gartandtoimi
October 15, 2024 @ 5:20 pm
Смешные приколы http://anekdotshutka.ru.
MikelKaria
October 15, 2024 @ 5:37 pm
[url=https://le-bandit-casino.ru]http://www.le-bandit-casino.ru[/url]
last news about le bandit
http://www.le-bandit-casino.ru
Dannyelarm
October 15, 2024 @ 6:03 pm
bs2site
Server-BAS
October 15, 2024 @ 7:14 pm
Виртуальный сервер/ВПС/ВДС под парсинг, постинг, разгадывание каптчи.
https://t.me/s/server_xevil_xrumer_vpsvds_zenno
Сервер для Xrumer |Xevil | GSA | Xneolinks | A-parser | ZennoPoster | BAS | Антидетект браузер Dolphin
– Почасовая оплата
– Windows – 2022, 2019, 2016, 2012 R2
– Отлично подходит под A-Parser
– Автоматическая установка Windows – бесплатно
– Дата-центр в Москве и Амстердаме
– Быстрые серверы с NVMe.
– Супер (аптайм, скорость, пинг, нагрузка)
– Отлично подходит под CapMonster
– Отлично подходит под XRumer + XEvil
– Более 15 000 сервер уже в работе
– Скорость порта подключения к сети интернет — 1000 Мбит/сек
– Для сервера сеть на скорости 1 Гбит!
– Outline VPN, WireGuard VPN, IPsec VPN.
– Круглосуточная техническая поддержка – бесплатно
– Мгновенное развёртывание сервера в несколько кликов – бесплатно
– Возможность арендовать сервер на 1 час или 1 сутки
– Windows – 2012 R2, 2016, 2019, 2022 – бесплатно
– Ubuntu, Debian, CentOS, Oracle 9 – бесплатно
– Отлично подходит под Xneolinks
– Отлично подходит под GSA Search Engine Ranker
– Управляйте серверами на лету.
– FASTPANEL и HestiaCP – бесплатно
ArmandokRup
October 15, 2024 @ 7:58 pm
Веселые приколы http://anekdotshutka.ru.
Server-Xevil
October 15, 2024 @ 8:44 pm
Купить безабузный server/ВПС/ВДС под парсинг, постинг, разгадывание каптчи.
https://t.me/s/server_xevil_xrumer_vpsvds_zenno
Сервер для Xrumer |Xevil | GSA | Xneolinks | A-parser | ZennoPoster | BAS | Антидетект браузер Dolphin
– Отлично подходит под XRumer + XEvil
– Круглосуточная техническая поддержка – бесплатно
– Почасовая оплата
– Скорость порта подключения к сети интернет — 1000 Мбит/сек
– Отлично подходит под GSA Search Engine Ranker
– Отлично подходит под CapMonster
– Супер (аптайм, скорость, пинг, нагрузка)
– Возможность арендовать сервер на 1 час или 1 сутки
– Дата-центр в Москве и Амстердаме
– Мгновенное развёртывание сервера в несколько кликов – бесплатно
– Отлично подходит под A-Parser
– Быстрые серверы с NVMe.
– Windows – 2012 R2, 2016, 2019, 2022 – бесплатно
– Управляйте серверами на лету.
– Автоматическая установка Windows – бесплатно
– Для сервера сеть на скорости 1 Гбит!
– Outline VPN, WireGuard VPN, IPsec VPN.
– FASTPANEL и HestiaCP – бесплатно
– Ubuntu, Debian, CentOS, Oracle 9 – бесплатно
– Windows – 2022, 2019, 2016, 2012 R2
– Отлично подходит под Xneolinks
– Более 15 000 сервер уже в работе
Перепланировка помещения
October 15, 2024 @ 8:47 pm
[url=https://potolki-kitstroy.ru/]Перепланировка помещений[/url] – это процесс, который требует грамотного подхода. Наши специалисты помогут оформить документы, согласовать проект и обеспечить успешное узаконивание всех изменений.
RichardThids
October 15, 2024 @ 8:53 pm
injectable ed drugs: buy prescription drugs without doctor – soma therapy ed
Robertdup
October 15, 2024 @ 9:20 pm
[url=https://sweet-bonanza-casino-slots.ru]www.sweet-bonanza-casino-slots.ru[/url]
last news about sweet bonanza casino
https://sweet-bonanza-casino-slots.ru
займ на карту
October 15, 2024 @ 9:56 pm
Ищете [url=https://cntbank.ru/]займы[/url] без отказа и проверок? Наш сайт предлагает подборку МФО, которые реально одобряют займы по паспорту с быстрым переводом.
Server-Xrumer
October 15, 2024 @ 10:03 pm
Купить безабузный сервер/ВПС/ВДС под парсинг, постинг, разгадывание каптчи.
https://t.me/s/server_xevil_xrumer_vpsvds_zenno
Сервер для Xrumer |Xevil | GSA | Xneolinks | A-parser | ZennoPoster | BAS | Антидетект браузер Dolphin
– Ubuntu, Debian, CentOS, Oracle 9 – бесплатно
– Супер (аптайм, скорость, пинг, нагрузка)
– Возможность арендовать сервер на 1 час или 1 сутки
– Для сервера сеть на скорости 1 Гбит!
– Отлично подходит под CapMonster
– Outline VPN, WireGuard VPN, IPsec VPN.
– Windows – 2012 R2, 2016, 2019, 2022 – бесплатно
– Отлично подходит под XRumer + XEvil
– Скорость порта подключения к сети интернет — 1000 Мбит/сек
– Быстрые серверы с NVMe.
– Почасовая оплата
– Windows – 2022, 2019, 2016, 2012 R2
– Отлично подходит под Xneolinks
– Отлично подходит под GSA Search Engine Ranker
– Круглосуточная техническая поддержка – бесплатно
– Более 15 000 сервер уже в работе
– Автоматическая установка Windows – бесплатно
– Дата-центр в Москве и Амстердаме
– Управляйте серверами на лету.
– Отлично подходит под A-Parser
– Мгновенное развёртывание сервера в несколько кликов – бесплатно
– FASTPANEL и HestiaCP – бесплатно
Micahham
October 15, 2024 @ 11:10 pm
[url=https://lucky-jet-casino-obzor.ru]https://www.lucky-jet-casino-obzor.ru[/url]
last news about play lucky jet casino
http://www.lucky-jet-casino-obzor.ru
MichaelDar
October 16, 2024 @ 12:10 am
[url=https://lucky-jet-casino-wiki.ru]https://lucky-jet-casino-wiki.ru[/url]
last news about lucky jet casino
http://www.lucky-jet-casino-wiki.ru
Server-ZennoPoster
October 16, 2024 @ 1:41 am
Выделенный server/ВПС/ВДС под парсинг, постинг, разгадывание каптчи.
https://t.me/s/server_xevil_xrumer_vpsvds_zenno
Сервер для Xrumer |Xevil | GSA | Xneolinks | A-parser | ZennoPoster | BAS | Антидетект браузер Dolphin
– Windows – 2022, 2019, 2016, 2012 R2
– Для сервера сеть на скорости 1 Гбит!
– Автоматическая установка Windows – бесплатно
– Дата-центр в Москве и Амстердаме
– Почасовая оплата
– Мгновенное развёртывание сервера в несколько кликов – бесплатно
– Отлично подходит под XRumer + XEvil
– Более 15 000 сервер уже в работе
– Отлично подходит под Xneolinks
– Outline VPN, WireGuard VPN, IPsec VPN.
– Быстрые серверы с NVMe.
– FASTPANEL и HestiaCP – бесплатно
– Отлично подходит под GSA Search Engine Ranker
– Windows – 2012 R2, 2016, 2019, 2022 – бесплатно
– Ubuntu, Debian, CentOS, Oracle 9 – бесплатно
– Управляйте серверами на лету.
– Возможность арендовать сервер на 1 час или 1 сутки
– Круглосуточная техническая поддержка – бесплатно
– Скорость порта подключения к сети интернет — 1000 Мбит/сек
– Супер (аптайм, скорость, пинг, нагрузка)
– Отлично подходит под CapMonster
– Отлично подходит под A-Parser
Ремонт телефонов
October 16, 2024 @ 2:27 am
Профессиональный сервисный центр ближайший ремонт сотовых номер телефона ремонта телефонов
Ремонт телефонов
October 16, 2024 @ 3:09 am
Профессиональный сервисный центр по ремонту сотовых телефонов в Москве.
Мы предлагаем: ремонт телефона москва
Наши мастера оперативно устранят неисправности вашего устройства в сервисе или с выездом на дом!
franshizi_lmSl
October 16, 2024 @ 5:50 am
фрашиза [url=https://franshizy12.ru/]фрашиза[/url] .
franshizi_wnPt
October 16, 2024 @ 7:49 am
франшиза 2023 [url=https://franshizy11.ru/]франшиза 2023[/url] .
RobertSot
October 16, 2024 @ 9:40 am
https://sudvgorode.ru/
Stephennewly
October 16, 2024 @ 10:05 am
[url=https://sweet-bonanza-casino-obzor.ru/]sweet-bonanza-casino-obzor.ru[/url]
last news about sweet bonanza casino
http://www.sweet-bonanza-casino-obzor.ru
BrandonHinge
October 16, 2024 @ 12:54 pm
buying prescription drugs in mexico [url=https://mexicanpharm24.pro/#]buying from online mexican pharmacy[/url] buying prescription drugs in mexico online
техническое заключение по перепланировке помещения
October 16, 2024 @ 2:30 pm
Для успешного [url=https://potolki-kitstroy.ru/]согласования перепланировки квартиры[/url] важно правильно оформить документы. Мы предоставляем полный комплекс услуг по проектированию и узакониванию изменений, чтобы избежать юридических проблем.
eaglechemicalsstore.com
October 16, 2024 @ 2:57 pm
I am not sure where you’re getting your info, but great topic.
I needs to spend some time learning more or understanding more.
Thanks for excellent info I was looking for this information for my mission.
Michaelvob
October 16, 2024 @ 5:35 pm
Смешные анекдоты https://teletype.in/@anekdoty/smeshnye-anekdoty.
vivod iz zapoya sochi_oyPi
October 16, 2024 @ 7:17 pm
вывод из запоя в наркологическом стационаре [url=https://vyvod-iz-zapoya-sochi16.ru]https://vyvod-iz-zapoya-sochi16.ru[/url] .
JosephRon
October 16, 2024 @ 9:15 pm
Свежие анекдоты https://prikolyshutki.wordpress.com/2024/10/16/smeshnye-anekdoty/.
Zaimfoabsog
October 16, 2024 @ 10:58 pm
[url=https://t.me/s/new_mfo_2024]Новые займы без проверки и отказов от 18 лет[/url]! Получите деньги моментально, просто подайте заявку и ждите перевод на карту!
Jeffreyerync
October 16, 2024 @ 11:05 pm
cs2 gambling csgo gamble
Server-Xrumer
October 16, 2024 @ 11:47 pm
Виртуальный сервер/ВПС/ВДС под парсинг, постинг, разгадывание каптчи.
https://t.me/s/server_xevil_xrumer_vpsvds_zenno
Сервер для Xrumer |Xevil | GSA | Xneolinks | A-parser | ZennoPoster | BAS | Антидетект браузер Dolphin
– Дата-центр в Москве и Амстердаме
– Отлично подходит под CapMonster
– Супер (аптайм, скорость, пинг, нагрузка)
– FASTPANEL и HestiaCP – бесплатно
– Outline VPN, WireGuard VPN, IPsec VPN.
– Windows – 2022, 2019, 2016, 2012 R2
– Автоматическая установка Windows – бесплатно
– Отлично подходит под GSA Search Engine Ranker
– Круглосуточная техническая поддержка – бесплатно
– Мгновенное развёртывание сервера в несколько кликов – бесплатно
– Более 15 000 сервер уже в работе
– Для сервера сеть на скорости 1 Гбит!
– Управляйте серверами на лету.
– Ubuntu, Debian, CentOS, Oracle 9 – бесплатно
– Почасовая оплата
– Отлично подходит под XRumer + XEvil
– Отлично подходит под A-Parser
– Windows – 2012 R2, 2016, 2019, 2022 – бесплатно
– Скорость порта подключения к сети интернет — 1000 Мбит/сек
– Быстрые серверы с NVMe.
– Возможность арендовать сервер на 1 час или 1 сутки
– Отлично подходит под Xneolinks
JeremyBuiva
October 17, 2024 @ 12:36 am
[url=https://le-bandit-casino-play.ru]le-bandit-casino-play.ru[/url]
last news about le bandit casino
http://www.le-bandit-casino-play.ru
Derrickjoisy
October 17, 2024 @ 12:59 am
[url=https://lucky-jet-casino-slots.ru]http://lucky-jet-casino-slots.ru[/url]
last news about lucky jet casino
http://lucky-jet-casino-slots.ru
Stephenswogs
October 17, 2024 @ 1:24 am
[url=https://le-bandit-casino-info.ru]le-bandit-casino-info.ru[/url]
last news about le bandit casino
http://www.le-bandit-casino-info.ru
Dannyelarm
October 17, 2024 @ 2:11 am
bs2best
AndrewVOg
October 17, 2024 @ 4:35 am
[url=https://sweet-bonanza-casino-info.ru]http://sweet-bonanza-casino-info.ru[/url]
last news about sweet bonanza casino
https://sweet-bonanza-casino-info.ru
Michaelvob
October 17, 2024 @ 5:59 am
Веселые анекдоты https://teletype.in/@anekdoty/smeshnye-anekdoty.
casino
October 17, 2024 @ 6:43 am
Hey! Someone in my Myspace group shared this website with us so I came to check it out.
I’m definitely enjoying the information. I’m book-marking and will be tweeting this to my followers!
Superb blog and brilliant design and style.
GregoryZooke
October 17, 2024 @ 6:49 am
non prescription erection pills https://drugs24.pro/# reasons for ed
BoatThomime
October 17, 2024 @ 7:32 am
Плавание с друзьями https://arendayahtathailand.store/ Прокатитесь на катере для участия в окрестных фестивалях а также праздничных днях, временных на воде.
vivod iz zapoya sochi_sgSt
October 17, 2024 @ 8:13 am
стационарное выведение из запоя [url=http://www.vyvod-iz-zapoya-sochi15.ru]http://www.vyvod-iz-zapoya-sochi15.ru[/url] .
Kevinrisse
October 17, 2024 @ 8:15 am
[url=https://lucky-jet-casino-play.ru]http://lucky-jet-casino-play.ru[/url]
last news about lucky jet casino
lucky-jet-casino-play.ru
Richardnof
October 17, 2024 @ 9:04 am
Наш малыш почувствовал улучшения уже после нескольких сеансов у детского остеопата в Ивантеевке. Настоятельно рекомендую для тех, кто заботится о здоровье своих детей!
[url=https://massageivanteevka.ru/page/ozdorovitelnyy-massaj]Массаж Ивантеевка[/url]
Microzsievitly
October 17, 2024 @ 9:53 am
Независимо от вашего местоположения, [url=https://t.me/s/novuy_zaim_sankt_peterbeug]займы по всей России быстро и без отказов онлайн[/url] помогут вам получить деньги без лишних хлопот.
MikelKaria
October 17, 2024 @ 11:09 am
[url=https://le-bandit-casino-wiki.ru]https://le-bandit-casino-wiki.ru[/url]
last news about le bandit
http://www.le-bandit-casino-wiki.ru
Dennisgom
October 17, 2024 @ 11:32 am
Платформа для ставок 1win с множеством событий и азартных игр. Удобный интерфейс, моментальные выплаты и привлекательные бонусы делают ставки ещё увлекательнее. Откройте мир азарта и выигрышей с надежным сервисом и постоянными акциями для пользователей.
BoatThomime
October 17, 2024 @ 12:32 pm
Исследуйте красивые острова https://arendaboatthai.store/ Закажите катерок немного капитаном (а) также узнайте что касается окрестной культуре а также рассказа у странствовании числом островам.
Josephmus
October 17, 2024 @ 1:20 pm
московский музей анимации измайловское ш 73ж цены музей анимации в москве измайлово
Ремонт техники с выездом на дом в Москве
October 17, 2024 @ 2:11 pm
Сервисный центр предлагает ремонт объектива lomography недорого выездной ремонт объективов lomography
RobertNails
October 17, 2024 @ 2:46 pm
музей анимации в измайловском кремле официальный сайт музей анимации цена
GregoryZooke
October 17, 2024 @ 3:18 pm
natural ed cures https://drugs24.pro/# new treatments for ed
Dennisgom
October 17, 2024 @ 3:25 pm
Платформа для ставок 1win с множеством событий и азартных игр. Удобный интерфейс, моментальные выплаты и привлекательные бонусы делают ставки ещё увлекательнее. Откройте мир азарта и выигрышей с надежным сервисом и постоянными акциями для пользователей.
Josephmus
October 17, 2024 @ 5:03 pm
музей анимации в москве отзывы музей анимации измайлово
RobertRer
October 17, 2024 @ 5:29 pm
[url=https://maria-sharapovaar.biz]www.maria-sharapovaar.biz[/url]
last news about maria sharapova
https://maria-sharapovaar.biz
Индексация ссылок в Google
October 17, 2024 @ 5:33 pm
Начните массовую индексацию ссылок в Google прямо cейчас!
Быстрая индексация ссылок имеет ключевое значение для успеха вашего онлайн-бизнеса. Чем быстрее поисковые системы обнаружат и проиндексируют ваши ссылки, тем быстрее вы сможете привлечь новую аудиторию и повысить позиции вашего сайта в результатах поиска.
Не теряйте времени! Начните пользоваться нашим сервисом для ускоренной индексации внешних ссылок в Google и Yandex. Зарегистрируйтесь сегодня и получите первые результаты уже завтра. Ваш успех в ваших руках!
Charlesnidly
October 17, 2024 @ 6:06 pm
[url=https://mariasharapovaar.biz]http://www.mariasharapovaar.biz[/url]
last news about maria sharapova
http://mariasharapovaar.biz
лазерная резка металла станки
October 17, 2024 @ 6:20 pm
Комплексные решения для резки металла
Предлагаем комплексные решения для резки металла, включая поставку лазерных станков, их обслуживание и обучение ваших специалистов.
лазерный станок цена [url=https://lazernye-stanki.ru/]станки лазерной резки металла[/url] .
Robertdup
October 17, 2024 @ 9:48 pm
[url=https://sharapovamariaar.biz]https://www.sharapovamariaar.biz[/url]
last news about sharapova maria
http://www.sharapovamariaar.biz
StevenSok
October 17, 2024 @ 10:24 pm
[url=https://le-bandit-casino-obzor.ru]le-bandit-casino-obzor.ru[/url]
last news about le bandit casino
http://www.le-bandit-casino-obzor.ru
메이저사이트
October 17, 2024 @ 10:32 pm
I really loved reading your blog. It was very well authored and easy to understand. Unlike other blogs I have read which are really not that good.Thanks alot!
메이저사이트
займ на карту без отказов для всех
October 17, 2024 @ 10:56 pm
[url=https=https://t.me/s/zaim_bez_otkazov_na_kartu]Моментальные кредиты на карту для всех категорий[/url] — получите до 30 тысяч рублей на карту быстро и без лишних формальностей. Неважно, кто вы — студент, пенсионер или работающий человек.
лазерный станок для резки труб
October 17, 2024 @ 11:31 pm
Комплексное техническое обслуживание лазерных станков
Наша компания предлагает комплексное техническое обслуживание лазерных станков.
станок с лазерной резкой [url=https://lazernye-stanki.ru/]резка нержавейки лазером москва[/url] .
Richardnof
October 18, 2024 @ 12:33 am
Массаж в Ивантеевке — это не просто расслабление, это настоящий ритуал оздоровления. Попробуйте сами и убедитесь!
[url=https://massageivanteevka.ru/page/osteopaticheskiy-massaj]Остеопат Ивантеевка[/url]
Lenardhah
October 18, 2024 @ 12:52 am
delivery balloons https://helium-balloon-price.com
HaroldFoelt
October 18, 2024 @ 1:05 am
Платформа 1win предлагает широкий выбор спортивных событий, киберспорта и азартных игр. Пользователи получают высокие коэффициенты, быстрые выплаты и круглосуточную поддержку. Программа лояльности и бонусы делают игру выгоднее.
1win apk_essa
October 18, 2024 @ 3:43 am
1win bet apk [url=http://1winciapk.com]1win bet apk[/url] .
1win apk_ucst
October 18, 2024 @ 3:43 am
1win apk pour iphone [url=https://1winapkdownload1win.com/]1winapkdownload1win.com[/url] .
CharlesTub
October 18, 2024 @ 5:13 am
custom balloons with delivery buy helium balloons with delivery Dubai
StevenRaica
October 18, 2024 @ 5:30 am
[url=https://maria-sharapova-ar.biz]https://maria-sharapova-ar.biz[/url]
last news about maria sharapova
https://maria-sharapova-ar.biz
Robertfub
October 18, 2024 @ 5:51 am
[url=https://sharapova-maria-ar.biz/]http://sharapova-maria-ar.biz[/url]
last news about sharapova maria
http://www.sharapova-maria-ar.biz
микрозайм на карту без отказа круглосуточно
October 18, 2024 @ 6:42 am
[url=https://t.me/s/zaim_bez_otkazov_na_kарту]Кредит онлайн без отказов для всех категорий[/url] — это простой способ получить деньги быстро. Без проверок, звонков и сложных процедур. Решение в течение нескольких минут.
Michaelvob
October 18, 2024 @ 7:12 am
Веселые анекдоты http://anekdot-top.ru.
MichaelDar
October 18, 2024 @ 9:35 am
[url=https://luis-diazar.biz]https://luis-diazar.biz[/url]
last news about luis diaz
http://www.luis-diazar.biz
lesteu
October 18, 2024 @ 9:58 am
I am regular visitor, how are you everybody?
This paragraph posted at this web page is really good.
ремонт техники в челябинске
October 18, 2024 @ 11:30 am
Профессиональный сервисный центр по ремонту бытовой техники с выездом на дом.
Мы предлагаем: сервис центры бытовой техники челябинск
Наши мастера оперативно устранят неисправности вашего устройства в сервисе или с выездом на дом!
slot maxwin
October 18, 2024 @ 11:53 am
Hi! Someone in my Facebook group shared this website
with us so I came to give it a look. I’m definitely enjoying the information.
I’m bookmarking and will be tweeting this to my followers!
Superb blog and wonderful design and style.
сервис центры в барнауле
October 18, 2024 @ 12:45 pm
Профессиональный сервисный центр по ремонту бытовой техники с выездом на дом.
Мы предлагаем: сервисные центры по ремонту техники в барнауле
Наши мастера оперативно устранят неисправности вашего устройства в сервисе или с выездом на дом!
JeremyBuiva
October 18, 2024 @ 3:40 pm
[url=https://lebronjames-ar.biz]http://lebronjames-ar.biz[/url]
last news about lebron james
https://www.lebronjames-ar.biz
станок лазерной резки
October 18, 2024 @ 6:51 pm
Оптоволоконные лазерные станки для металлорезки
Предлагаем инновационные оптоволоконные лазерные станки, обеспечивающие точную резку металла с минимальными затратами на обслуживание.
станок с лазерной резкой [url=https://lazernye-stanki.ru/]резка нержавейки лазером москва[/url] .
Michaelvob
October 18, 2024 @ 7:47 pm
Смешные анекдоты http://anekdot-top.ru.
vivod iz zapoya sochi_wfOi
October 18, 2024 @ 7:49 pm
вывод от запоя в стационаре сочи [url=http://www.vyvod-iz-zapoya-sochi17.ru]вывод от запоя в стационаре сочи[/url] .
Ремонт ноутбуков
October 18, 2024 @ 9:56 pm
Профессиональный сервисный центр по ремонту сотовых телефонов в Москве.
Мы предлагаем: ремонта ноутбука работает
Наши мастера оперативно устранят неисправности вашего устройства в сервисе или с выездом на дом!
SportNews
October 18, 2024 @ 10:05 pm
[url=https://lenta.news161.ru/]https://lenta.news161.ru[/url]
last sport news
lenta.news161.ru
Ремонт телефонов
October 18, 2024 @ 10:17 pm
Профессиональный сервисный центр ремонт телефонов в москве рядом ремонт сотовых
Micahham
October 18, 2024 @ 11:11 pm
[url=https://luis-diaz-ar.biz]luis-diaz-ar.biz[/url]
last news about luis diaz
http://www.luis-diaz-ar.biz
станок с лазерной резкой металла
October 18, 2024 @ 11:58 pm
Решения для резки металла любого типа
Мы предлагаем лазерные станки, подходящие для резки металла различной толщины и типа, включая сталь, алюминий и другие сплавы.
купить лазерный станок для резки металла [url=https://lazernye-stanki.ru/]лазерная резка металла нержавейка[/url] .
RichardBrusy
October 18, 2024 @ 11:59 pm
minocin 50 mg for scabies [url=http://stromectol1st.shop/#]best price shop[/url] ivermectin 0.5% brand name
VictorDot
October 19, 2024 @ 12:27 am
where to buy balloons Dubai custom helium balloons Dubai
Treffxa
October 19, 2024 @ 12:57 am
Купить диплом о среднем образовании в Москве и любом другом городе
lubercy.ixbb.ru/viewtopic.php?id=9489#p19726
Jamesmum
October 19, 2024 @ 1:42 am
resume of an engineer sample https://engineer-builder-resume.com
CharlesBum
October 19, 2024 @ 2:53 am
buy rybelsus: buy rybelsus – buy semaglutide online
Michaelvob
October 19, 2024 @ 4:51 am
Свежие приколы и анекдоты https://teletype.in/@anekdoty/prikoly-nastroenie.
Cazrzqm
October 19, 2024 @ 6:39 am
Где и как купить диплом о высшем образовании без лишних рисков
зеркало леонбетс
October 19, 2024 @ 6:47 am
Хватит ждать, пора делать ставки с [url=https://www.kv.by/information/2023/08/05/1067841-melbet-melbet-zerkalo-aktualnyy-dostup-obzor-sayta-registraciya]Мелбет ставки[/url]! Удобная платформа, промокоды для новых игроков и широкий выбор спортивных событий помогут тебе начать выигрывать уже сегодня!
Diplomi_hkEr
October 19, 2024 @ 8:07 am
купить диплом бакалавра в новосибирске [url=https://orik-diploms.ru/]orik-diploms.ru[/url] .
humandesignck
October 19, 2024 @ 8:16 am
(ґ• ? •`) ?
AnthonyWresk
October 19, 2024 @ 12:57 pm
can i buy followers on tiktok https://tiktokhacks.quora.com/How-to-buy-TikTok-followers-And-what-are-the-best-sites-to-do-so
AndrewVOg
October 19, 2024 @ 2:01 pm
[url=https://diaz-luis-ar.biz]https://www.diaz-luis-ar.biz[/url]
last news about diaz luis
https://www.diaz-luis-ar.biz
RichardBrusy
October 19, 2024 @ 4:24 pm
buy semaglutide online [url=http://rybelsus.icu/#]rybelsus generic[/url] more
JeremyBuiva
October 19, 2024 @ 5:45 pm
[url=https://kobebryantar.biz]http://www.kobebryantar.biz[/url]
last news about kobe bryant
kobebryantar.biz
autism-mmc.com
October 19, 2024 @ 5:46 pm
At autism-mmc.com, we’ve been treating [url=https://www.autism-mmc.com/childhood-autism-treatment/cord-blood-stem-cells-transplantation/]cord blood stem cells transplant[/url] in adults and children for over a decade. Our expert team uses cutting-edge [url=https://www.autism-mmc.com/childhood-autism-treatment/own-autologous-bone-marrow-stem-cells-transplantation/]Mardaleishvili own stem cells[/url] including stem cell therapy, to improve behavior and cognitive skills. Let us help you achieve positive outcomes.
MichaelDar
October 19, 2024 @ 6:28 pm
[url=https://kobebryant-ar.biz]https://kobebryant-ar.biz[/url]
last news about kobe bryant
http://www.kobebryant-ar.biz
StevenRaica
October 19, 2024 @ 7:07 pm
[url=https://kobe-bryantar.biz]http://www.kobe-bryantar.biz[/url]
last news about kobe bryant
https://kobe-bryantar.biz
Michaelvob
October 19, 2024 @ 7:13 pm
Смешные приколы и анекдоты https://teletype.in/@anekdoty/prikoly-nastroenie.
RIGANA
October 19, 2024 @ 8:01 pm
I’m curious to find out what blog system you have
been utilizing? I’m experiencing some small security problems with my latest website and I would like
to find something more safeguarded. Do you have any
recommendations?
CharlesBum
October 19, 2024 @ 10:35 pm
more: order Rybelsus – Buy semaglutide
Ремонт духовых шкафов
October 19, 2024 @ 10:51 pm
Профессиональный сервисный центр по ремонту духовых шкафов в Москве.
Мы предлагаем: ремонт дверцы духового шкафа
Наши мастера оперативно устранят неисправности вашего устройства в сервисе или с выездом на дом!
Diplomi_oakl
October 20, 2024 @ 1:55 am
ответственность за купленный диплом [url=https://landik-diploms.ru/]landik-diploms.ru[/url] .
Hex
October 20, 2024 @ 2:00 am
Cтальные двери с завода на заказ.
Любые конфигурации замков на выбор. Более 3500 моделей на складе: [url=https://mytishi.dverimetallicheskie.ru/]здесь[/url]
RichardBrusy
October 20, 2024 @ 3:06 am
minocycline 50mg tablets for human [url=http://stromectol1st.shop/#]stromectol shop[/url] stromectol 6 mg dosage
BAOERT
October 20, 2024 @ 7:19 am
Yes! Finally someone writes about BAOERT.
JosephRon
October 20, 2024 @ 7:56 am
Веселые приколы http://shutki-anekdoty.ru/.
CharlesBum
October 20, 2024 @ 8:45 am
cheap plavix antiplatelet drug: check clopidogrel pro – buy clopidogrel bisulfate
RichardBrusy
October 20, 2024 @ 11:45 am
paxlovid cost without insurance [url=http://paxlovid1st.shop/#]paxlovid shop[/url] paxlovid price
Sam
October 20, 2024 @ 12:05 pm
Basically I am a very open person, so I am not afraid to be filmed free live mature sex cams. Therefore, I look forward to showing you one or the other video of me in the future. I want to try myself out and have new experiences. Maybe something will happen between us!
транспортная компания "правильные перевозки"
October 20, 2024 @ 12:07 pm
“Правильные перевозки” — это надежная транспортная компания, которая предоставляет услуги по перевозке грузов и вещей по всей России. Мы занимаемся транспортировкой мебели между городами и регионами страны. Благодаря профессиональному подходу и опыту наших специалистов, “транспортная компания правильные перевозки” гарантирует безопасность и сохранность вашего имущества на всех этапах транспортировки.
Для вашего удобства мы предлагаем услуги междугородных переездов с возможностью рассчитать стоимость и сроки онлайн. Независимо от того, нужен ли вам домашний переезд или перевозка личных вещей, наша команда обеспечивает высокий уровень сервиса и индивидуальный подход к каждому клиенту. Уточнить детали или заказать услугу вы можете по телефону 8 (800) 505-18-39 или 88005051839.
Компания предлагает перевозку контейнеров с вещами, обеспечивая оперативную доставку в другой город. Наши специалисты профессионально занимаются домашними перевозками, минимизируя ваши затраты времени и средств. Обращайтесь к нам, и “транспортная компания правильные перевозки” сделает ваш переезд комфортным и безопасным.
Эксперт в сфере грузоперевозок | Качественные транспортные услуги | Точность и безопасность грузоперевозок | Надежные партнеры при перевозке грузов | Превосходный сервис для клиентов | Лидер в сфере грузоперевозок | Индивидуальный подход к каждому клиенту | Клиентоориентированная компания при перевозках | Эффективное решение для логистических задач | Правильные перевозки грузов без задержек | Профессиональные услуги по грузоперевозкам | Надежное сотрудничество в сфере грузоперевозок | Оптимизация логистики и транспортировки | Индивидуальный подход к каждому клиенту
Vivod iz zapoya rostov_zdEi
October 20, 2024 @ 3:33 pm
вывод из запоя [url=vyvod-iz-zapoya-rostov115.ru]вывод из запоя[/url] .
леонбет
October 20, 2024 @ 3:56 pm
[url=https://www.kv.by/information/2023/09/26/1068164-1hbet-zerkalo-rabotayushchee-aktualnoe-zerkalo-sayta-1xbet-com]1xbet зеркало рабочее на сегодня[/url] обеспечит вам мгновенный доступ к ставкам и бонусам. Ставьте на спорт без ограничений и выигрывайте больше каждый день!
Michaelvob
October 20, 2024 @ 3:59 pm
Смешные приколы и анекдоты http://shutki-anekdoty.ru/.
franshizi_chor
October 20, 2024 @ 5:43 pm
франшиз [url=franshizy13.ru]франшиз[/url] .
Curtisgow
October 20, 2024 @ 5:57 pm
Produzione video professionale https://orbispro.it creazione di spot pubblicitari, film aziendali e contenuti video. Ciclo completo di lavoro, dall’idea al montaggio. Soluzioni creative per promuovere con successo il tuo brand.
tex38.ru
October 20, 2024 @ 6:02 pm
Надежный [url=https://tex38.ru/shop/kabel-provod_157/silovoy-kabel-pvh_160]силовой кабель в Иркутске[/url] — это гарантия безопасного и стабильного энергоснабжения для вашего проекта.
Sazrbwt
October 20, 2024 @ 6:19 pm
Стоимость дипломов высшего и среднего образования и процесс их получения
CharlesBum
October 20, 2024 @ 6:49 pm
minocycline 50 mg acne: stromectol 1st – stromectol where to buy
Kevinrisse
October 20, 2024 @ 8:55 pm
[url=https://lebronjamesar.biz]http://lebronjamesar.biz[/url]
last news about lebron james
https://www.lebronjamesar.biz
Curtisgow
October 20, 2024 @ 9:32 pm
Produzione video professionale https://orbispro.it creazione di spot pubblicitari, film aziendali e contenuti video. Ciclo completo di lavoro, dall’idea al montaggio. Soluzioni creative per promuovere con successo il tuo brand.
JosephRon
October 20, 2024 @ 10:13 pm
Свежие приколы и анекдоты http://shutki-anekdoty.ru/.
RichardBrusy
October 20, 2024 @ 10:24 pm
Paxlovid buy online [url=http://paxlovid1st.shop/#]paxlovid generic[/url] paxlovid india
Charlesnidly
October 21, 2024 @ 3:18 am
[url=https://james-lebron-ar.biz]http://www.james-lebron-ar.biz[/url]
last news about james lebron
james-lebron-ar.biz
RobertRer
October 21, 2024 @ 3:25 am
[url=https://jameslebron-ar.biz]www.jameslebron-ar.biz[/url]
last news about james lebron
jameslebron-ar.biz
Lazrecs
October 21, 2024 @ 4:09 am
Узнайте, как безопасно купить диплом о высшем образовании
honey.ukrbb.net/viewtopic.php?f=18&t=15994
StevenSok
October 21, 2024 @ 4:13 am
[url=https://bryant-kobe-ar.biz]http://bryant-kobe-ar.biz[/url]
last news about bryant kobe
http://www.bryant-kobe-ar.biz
Michaelvob
October 21, 2024 @ 5:20 am
Свежие приколы, шутки и картинки http://shutki-anekdoty.ru/.
CharlesBum
October 21, 2024 @ 6:07 am
Semaglutide pharmacy price: Semaglutide pharmacy price – cheaper
индексация в гугл
October 21, 2024 @ 7:10 am
Полезный сервис быстрого загона ссылок сайта в индексация поисковой системы – индексация в гугл
Массаж Ивантеевка
October 21, 2024 @ 7:15 am
Профессиональный детский остеопат в Ивантеевке — это здоровое развитие вашего ребенка без боли и стресса. Записывайтесь на консультацию! [url=https://massageivanteevka.ru/page/kontakty]Массаж Ивантеевка[/url]
Алкошоп
October 21, 2024 @ 8:06 am
Алкошоп и Alcoshop — это идеальный выбор для любителей напитков, которым важно качество и удобство. Доставка доступна 24 часа в сутки, что позволяет получить нужные напитки в любое время. Позвонив по номеру +74993433939, вы можете оформить заказ быстро и без лишних хлопот.
Круглосуточная доставка через Алкошоп позволяет без труда заказать алкоголь на дом. Вы можете позвонить на +74993433939 или оформить заказ онлайн, что делает процесс простым и быстрым. Сервис гарантирует доставку в любое время суток.
Для заказа алкоголя в Москве круглосуточно на дом достаточно позвонить по номеру +74993433939. В Alcoshop доступен большой выбор алкоголя, что позволит найти нужный товар для любого случая. Доставка осуществляется с заботой о качестве и безопасности, делая каждый заказ приятным и комфортным.
RichardBrusy
October 21, 2024 @ 8:09 am
paxlovid pill [url=http://paxlovid1st.shop/#]paxlovid price[/url] paxlovid covid
Xazrgqi
October 21, 2024 @ 8:58 am
Как не попасть впросак при покупке диплома колледжа или ПТУ в России
электротехническая компания
October 21, 2024 @ 9:58 am
Выбирайте [url=https://tex38.ru/shop/kabel-provod_157/kabel-gibkiy_164/kgvevng-a_221]кабель КГВЭВнг(А)[/url] для сложных эксплуатационных условий. Этот кабель устойчив к механическим повреждениям и способен работать в самых суровых климатических зонах.
Stephenswogs
October 21, 2024 @ 10:08 am
[url=https://lebron-james-ar.biz]lebron-james-ar.biz[/url]
last news about lebron james
https://lebron-james-ar.biz
сайт
October 21, 2024 @ 10:48 am
сайт
Lazrbqd
October 21, 2024 @ 11:40 am
Официальное получение диплома техникума с упрощенным обучением в Москве
humandesignuf
October 21, 2024 @ 2:09 pm
(?_?;)
https://mari-tyrek.ru/44701.html
Robertdup
October 21, 2024 @ 2:39 pm
[url=https://kobe-bryant-ar.biz]https://www.kobe-bryant-ar.biz[/url]
last news about sharapova maria
http://kobe-bryant-ar.biz
CharlesBum
October 21, 2024 @ 3:06 pm
Clopidogrel 75 MG price: generic pills – buy clopidogrel online
หนังใหม่
October 21, 2024 @ 4:15 pm
Thank you for another great article. Where else may just anybody get that type of information in such an ideal
way of writing? I have a presentation subsequent week, and I’m at the search for such info.
RichardBrusy
October 21, 2024 @ 4:50 pm
ivermectin 8 mg [url=http://stromectol1st.shop/#]stromectol 1st[/url] minocycline 50 mg tablets online
humandesignyy
October 21, 2024 @ 5:12 pm
(@^?^)
https://mari-tyrek.ru/78447.html
JeremyBuiva
October 21, 2024 @ 5:29 pm
[url=https://bryant-kobe-ar.biz]https://bryant-kobe-ar.biz[/url]
last news about bryant kobe
http://www.bryant-kobe-ar.biz
StevenRaica
October 21, 2024 @ 5:57 pm
[url=https://ruben-nevesar.biz]https://www.ruben-nevesar.biz[/url]
last news about ruben neves
https://www.ruben-nevesar.biz
Diplomi_eapn
October 21, 2024 @ 7:10 pm
купить диплом без занесения [url=https://arusak-diploms.ru/]arusak-diploms.ru[/url] .
humandesigngi
October 21, 2024 @ 7:45 pm
(*^.^*)
https://mari-tyrek.ru/52457.html
Перепланировка помещения
October 21, 2024 @ 8:46 pm
[url=https://dom-s-ymom.ru/]Согласование перепланировки в здании в Москве[/url] — это возможность придать вашему пространству новый облик, не нарушая закон. Хотите изменить планировку, но не знаете, с чего начать? Мы готовы помочь вам пройти все этапы: от идеи до получения финальных разрешений. Пусть ваша мечта о новом интерьере станет реальностью без лишних проблем!
AndrewVOg
October 21, 2024 @ 9:09 pm
[url=https://nevesrubenar.biz]www.nevesrubenar.biz[/url]
last news about neves rubenar
http://www.nevesrubenar.biz
Cazrobb
October 21, 2024 @ 9:22 pm
Официальное получение диплома техникума с упрощенным обучением в Москве
Michaelvob
October 21, 2024 @ 9:31 pm
Веселые приколы http://shutki-anekdoty.ru/.
Cazrsyt
October 21, 2024 @ 9:32 pm
Как официально приобрести аттестат 11 класса с минимальными затратами времени
new retro casino_taEl
October 21, 2024 @ 9:40 pm
нью ретро казино [url=http://www.newretrocasino-casino2.ru]нью ретро казино[/url] .
Trefnap
October 21, 2024 @ 9:50 pm
Как не попасть впросак при покупке диплома колледжа или ПТУ в России
mosfor.flybb.ru/viewtopic.php?f=2&t=453
JosephRon
October 21, 2024 @ 10:27 pm
Смешные анекдоты http://shutki-anekdoty.ru/anekdoty.
elektrokarniz dlya shtor_muSl
October 22, 2024 @ 12:19 am
карниз с приводом для штор [url=http://elektrokarniz-dlya-shtor11.ru]карниз с приводом для штор[/url] .
Ремонт техники с выездом на дом в Москве
October 22, 2024 @ 12:21 am
Сервисный центр предлагает качественый ремонт планшета digma ремонт планшета digma недорого
Diplomi_ucEr
October 22, 2024 @ 1:09 am
купить диплом судоводителя [url=https://russa-diploms.ru/]russa-diploms.ru[/url] .
CharlesBum
October 22, 2024 @ 1:38 am
stromectol usa: stromectol fast delivery – buy minocycline 100mg for humans
RichardBrusy
October 22, 2024 @ 2:53 am
paxlovid buy [url=https://paxlovid1st.shop/#]shop[/url] paxlovid cost without insurance
kazlenta.kz
October 22, 2024 @ 5:30 am
Достоверные новости Астаны kazlenta.kz
Основные [url=https://kazlenta.kz/]интересные новости[/url] уже на портале. Мы регулярно пополняем свежую информацию о событиях, стараясь контролировать её верность. Все главные события в области авто, новости, дтп, общество. Обязательно сохраните наш узнаваемый веб ресурс и рекомендуйте знакомым. Только тут все верные и вовремя выложенные новости о Казахстане.
Diplomi_zzkl
October 22, 2024 @ 9:22 am
купить диплом о высшем образовании томск [url=https://landik-diploms.ru/]landik-diploms.ru[/url] .
MikelKaria
October 22, 2024 @ 9:27 am
[url=https://kobebryantar.biz]kobebryantar.biz[/url]
last news about kobe bryant
https://www.kobebryantar.biz
CharlesBum
October 22, 2024 @ 10:43 am
rybelsus cost: rybelsus – Buy semaglutide
RichardBrusy
October 22, 2024 @ 11:20 am
stromectol generic name [url=https://stromectol1st.shop/#]stromectol fast delivery[/url] stromectol 3mg tablets
Micahham
October 22, 2024 @ 11:22 am
[url=https://ruben-neves-ar.biz]http://www.ruben-neves-ar.biz[/url]
last news about ruben neves
http://www.ruben-neves-ar.biz
JosephRon
October 22, 2024 @ 11:44 am
Веселые анекдоты http://shutki-anekdoty.ru/anekdoty.
Michaelvob
October 22, 2024 @ 2:06 pm
Смешные мемы http://shutki-anekdoty.ru/kartinki-prikolnye.
перепланировка
October 22, 2024 @ 3:54 pm
[url=https=https://dom-s-ymom.ru/]Согласование перепланировки помещения[/url] — это процесс, требующий профессионального подхода и знания всех нормативных актов. Наша команда обеспечивает разработку проекта, получение технического заключения и согласование всех изменений с городскими службами. Мы берём на себя все заботы, чтобы вы могли сконцентрироваться на своих делах, не отвлекаясь на бюрократические вопросы.
stmb-trucks.ru
October 22, 2024 @ 4:14 pm
В случае, если вам нужна техника с повышенной грузоподъемностью, обратите внимание на [url=https://stmb-trucks.ru/dongfeng/sedelnyj-tyagach-dongfeng-dfh-4180-4h2/]седельный тягач 4 4[/url]. Этот тягач от компании Dongfeng прекрасно подходит для сложных транспортных операций, обеспечивая максимальную производительность и надёжность на дорогах. ООО «СТМБ» гарантирует качественное обслуживание техники и предоставляет её на выгодных условиях лизинга.
RichardBrusy
October 22, 2024 @ 7:12 pm
rybelsus generic [url=http://rybelsus.icu/#]rybelsus.icu[/url] rybelsus price
JeremyBuiva
October 22, 2024 @ 7:14 pm
[url=https://kante-ngoloar.biz]http://www.kante-ngoloar.biz[/url]
last news about kante ngolo
https://www.kante-ngoloar.biz
Stephenswogs
October 22, 2024 @ 7:15 pm
[url=https://ngolokantear.biz]https://ngolokantear.biz[/url]
last news about ngolo kante
http://www.ngolokantear.biz
CharlesBum
October 22, 2024 @ 7:17 pm
dynacin minocycline: stromectol 1st shop – buy minocycline 100mg tablets
новости Казахстана коронавирус
October 22, 2024 @ 8:23 pm
Автоновости Казахстан kazlenta.kz
Первые [url=https://kazlenta.kz/]новости сейчас[/url] уже на портале. Мы постоянно добавляем информацию о событиях, пытаясь проверять её верность. Все главные события в области автомобилистики, новости, дтп, общество. Непременно сохраните наш известный интернет сайт и отправляйте знакомым. Только здесь все верные и своевременно отправленные новости о Казахстане.
ZSot
October 22, 2024 @ 8:42 pm
1win casino mozambique
онлайн деньги
October 22, 2024 @ 8:48 pm
Благодаря нашему многолетнему опыту работы в сфере микрофинансирования, мы знаем, какие решения лучше всего подходят для различных жизненных ситуаций. Мы предлагаем широкий выбор кредитных продуктов, которые удовлетворяют самые разные потребности наших клиентов, от краткосрочных займов до долгосрочного финансирования с гибкими условиями возврата.
займы Казахстан [url=https://microloans.kz/]займ онлайн[/url] .
ZSot
October 23, 2024 @ 1:19 am
1win login
Vivod iz zapoya rostov_kaSt
October 23, 2024 @ 1:41 am
вывод из запоя на дому в ростове [url=http://vyvod-iz-zapoya-rostov116.ru/]вывод из запоя на дому в ростове[/url] .
донг фенг gx седельный тягач
October 23, 2024 @ 5:20 am
[url=https://stmb-trucks.ru/weheavy/gusenichnyj-ekskavator-we150e2/]Гусеничный экскаватор we150e2[/url] от компании ООО «СТМБ» — это универсальная модель спецтехники для выполнения различных строительных и промышленных работ. Мы предлагаем выгодные условия лизинга и продажу техники с гарантийным обслуживанием. Этот экскаватор отличается высоким уровнем надежности и продуктивности, что делает его идеальным выбором для вашего бизнеса.
Diplomi_ojOa
October 23, 2024 @ 5:21 am
купить диплом сантехник [url=https://prema-diploms.ru/]prema-diploms.ru[/url] .
Diplomi_mmEr
October 23, 2024 @ 5:33 am
образование купить диплом люди [url=https://orik-diploms.ru/]orik-diploms.ru[/url] .
Diplomi_jlea
October 23, 2024 @ 5:39 am
купить аттестаты за 11 класс [url=https://server-diploms.ru/]server-diploms.ru[/url] .
Diplomi_ljSi
October 23, 2024 @ 5:42 am
купить диплом срочно [url=https://man-diploms.ru/]купить диплом срочно[/url] .
Williamfoest
October 23, 2024 @ 5:43 am
услуги спецоценка условий труда проведение соут стоимость в москве
BrucePen
October 23, 2024 @ 6:04 am
[url=https://neves-ruben-ar.biz/]https://www.neves-ruben-ar.biz[/url]
last news about neves ruben
neves-ruben-ar.biz
humandesignll
October 23, 2024 @ 7:29 am
a human design 6 3 human design
Larryrhita
October 23, 2024 @ 7:35 am
проведение соут https://sout095.ru
Williamfoest
October 23, 2024 @ 8:06 am
кем проводится специальная оценка условий труда оценка условий труда москва
Sazrwmm
October 23, 2024 @ 8:09 am
Где и как купить диплом о высшем образовании без лишних рисков
datasphere.ru/club/user/15/blog/
Larryrhita
October 23, 2024 @ 11:31 am
соут компании https://sout095.ru
StevenRaica
October 23, 2024 @ 11:52 am
[url=https://lukamodric-ar.biz]www.lukamodric-ar.biz[/url]
last news about luka modric
http://lukamodric-ar.biz
https://localhomeservicesblog.co.uk/forum/profile/DeenaNelli
October 23, 2024 @ 12:16 pm
acupuncture for anxiety (https://localhomeservicesblog.co.uk/forum/profile/DeenaNelli) patients
live a lot in the future and are not present in the
now.
займы
October 23, 2024 @ 12:49 pm
Мы понимаем, что финансовые трудности могут возникнуть внезапно, и важно быстро найти решение. Именно поэтому наш сервис предлагает быстрый доступ к микрозаймам с минимальными требованиями и высокой скоростью одобрения. Мы помогаем нашим клиентам избежать долгих бюрократических процедур, предлагая только проверенные и надежные финансовые решения, которые можно оформить онлайн в кратчайшие сроки.
микро займы [url=https://microloans.kz/]онлайн займы[/url] .
JosephRon
October 23, 2024 @ 3:56 pm
Веселые видео http://shutki-anekdoty.ru/videos.
humandesignhl
October 23, 2024 @ 4:50 pm
human designs human design types test
datransport.ru
October 23, 2024 @ 4:57 pm
Когда речь идет о [url=https://datransport.ru/]перевозке груза[/url], не стоит рисковать. DaTransport предлагает проверенные решения для бизнеса, независимо от того, нужен ли вам транспорт для регулярных поставок или для единовременной перевозки.
микрозаймы онлайн
October 23, 2024 @ 5:01 pm
Наша компания работает на рынке микрозаймов Казахстана уже несколько лет, и за это время мы успели завоевать доверие множества клиентов. Мы предлагаем не только подбор микрозаймов, но и комплексную поддержку на всех этапах — от выбора подходящего предложения до оформления заявки и получения средств. Мы стремимся создать для каждого клиента комфортные условия, обеспечивая прозрачность и надежность всех финансовых операций.
микрозаймы [url=https://microloans.kz/]микрозайм[/url] .
Stephenswogs
October 23, 2024 @ 5:53 pm
[url=https://ali-al-bulaihiar.biz]https://www.ali-al-bulaihiar.biz[/url]
last news about ali al bulaihi
https://ali-al-bulaihiar.biz
Vivod iz zapoya rostov_yzmt
October 23, 2024 @ 5:57 pm
вывод из запоя недорого [url=https://vyvod-iz-zapoya-rostov117.ru/]вывод из запоя недорого[/url] .
Micahham
October 23, 2024 @ 6:01 pm
[url=https://modricluka-ar.biz]http://modricluka-ar.biz[/url]
last news about modric luka
https://modricluka-ar.biz
humandesignav
October 23, 2024 @ 6:31 pm
human design free chart huamn design
humandesignau
October 23, 2024 @ 8:00 pm
human design certification human design generator
AndrewVOg
October 23, 2024 @ 8:47 pm
[url=https://modric-luka-ar.biz]https://www.modric-luka-ar.biz[/url]
last news about modric luka
modric-luka-ar.biz
datransport.ru
October 23, 2024 @ 8:48 pm
[url=https://datransport.ru/]Автомобильные перевозки по Казахстану[/url] — это основное направление работы DaTransport. Компания предлагает выгодные условия для транспортировки грузов различной сложности.
ThomasErrop
October 23, 2024 @ 9:22 pm
Полезная информация на сайте. Все что вы хоте знать об интернете полезный сервис
Sazrcqe
October 23, 2024 @ 11:17 pm
Приобретение диплома ПТУ с сокращенной программой обучения в Москве
ремонт бытовой техники в перми
October 23, 2024 @ 11:21 pm
Профессиональный сервисный центр по ремонту бытовой техники с выездом на дом.
Мы предлагаем: сервисные центры в перми
Наши мастера оперативно устранят неисправности вашего устройства в сервисе или с выездом на дом!
BrucePen
October 23, 2024 @ 11:47 pm
[url=https://neves-ruben-ar.biz/]http://neves-ruben-ar.biz[/url]
last news about neves ruben
http://www.neves-ruben-ar.biz
JeremyBuiva
October 24, 2024 @ 12:08 am
[url=https://ali-al-bulaihi-ar.biz]http://ali-al-bulaihi-ar.biz[/url]
last news about ali al bulaihi
http://ali-al-bulaihi-ar.biz
humandesignyl
October 24, 2024 @ 12:08 am
huan design human design america
Hex
October 24, 2024 @ 1:18 am
Входные металлические двери от производителя на заказ.
Любые конфигурации замков на выбор. Более 3500 моделей на складе: [url=https://balashiha.vhodnye-metallicheskie-dveri.ru/]тут[/url]
Williamraics
October 24, 2024 @ 1:29 am
Как приобрести аттестат о среднем образовании в Москве и других городах
erudio.global/blog/index.php?entryid=42337
Charlesnidly
October 24, 2024 @ 1:30 am
[url=https://sweet-bonanza-casino-slots.ru]www.sweet-bonanza-casino-slots.ru[/url]
last news about sweet bonanza casino
https://www.sweet-bonanza-casino-slots.ru
StevenSok
October 24, 2024 @ 2:20 am
[url=https://ngolo-kante-ar.biz]https://ngolo-kante-ar.biz[/url]
last news about ngolo kante
https://ngolo-kante-ar.biz
Lazrpep
October 24, 2024 @ 2:46 am
Как приобрести аттестат о среднем образовании в Москве и других городах
arena plus
October 24, 2024 @ 3:09 am
I relish, cause I discovered exactly what I was having a
look for. You’ve ended my four day long hunt! God
Bless you man. Have a nice day. Bye
RobertRer
October 24, 2024 @ 3:20 am
[url=https://luka-modric-ar.biz]www.luka-modric-ar.biz[/url]
last news about luka modric
https://www.luka-modric-ar.biz
EdwardJoype
October 24, 2024 @ 4:05 am
pin-up: pinup az – pin up 306
MichaelDar
October 24, 2024 @ 4:15 am
[url=https://lukamodricar.biz]http://lukamodricar.biz[/url]
last news about luka modric
http://lukamodricar.biz
Stephennewly
October 24, 2024 @ 6:28 am
[url=https://modric-luka-ar.biz/]www.modric-luka-ar.biz[/url]
last news about modric luka
https://www.modric-luka-ar.biz
Derrickjoisy
October 24, 2024 @ 7:11 am
[url=https://ngolokante-ar.biz]https://www.ngolokante-ar.biz[/url]
last news about ngolo kante
http://www.ngolokante-ar.biz
ErnestZeW
October 24, 2024 @ 7:21 am
canl? casino siteleri: casino oyunlar? – slot casino siteleri
Michaelvob
October 24, 2024 @ 7:23 am
Смешные короткие шутки http://shutki-anekdoty.ru/korotkie-anekdoty.
humandesigngf
October 24, 2024 @ 8:02 am
Тема «Четыре типа в Дизайне Человека» важна для понимания не только на теоретическом, но и на практическом уровне. Этот инструмент самопознания помогает каждому из нас осознать свою природу и использовать индивидуальные особенности для улучшения качества жизни. Рассмотрим рационально-практическую сторону каждого из типов, их определения и различия.
Все о Дизайне Человека – Дизайн Человека
Первый тип в Дизайне Человека – это Генератор. Генераторы отличаются высокой энергетичностью и способностью легко и эффективно завершать начатые задачи. Этот тип создан для работы, и его главное стремление — заниматься тем, что приносит удовольствие. Генератор начинает действовать, когда ощущает внутренний отклик. Их индивидуальная особенность заключается в том, что энергия накапливается, только когда они следуют своему отклику.
Второй тип — это Манифестор. Этот тип уникален своей независимостью и способностью инициировать действия. Они не нуждаются в отклике, как Генераторы, и могут сразу принимать решения и действовать. Различие этого типа в том, что они лучше всего проявляют себя, когда свободны от ограничений. Практическая сторона их природы проявляется в том, что они способны запускать процессы и вдохновлять окружающих.
Также важный элемент системы Дизайна Человека — Проектор. Проекторы – это люди, которые видят потенциал в других и помогают его раскрыть. Они нуждаются в приглашении, прежде чем начать действовать, и могут эффективно использовать энергию, когда работают с другими людьми. Их сила — в правильном руководстве и управлении чужими ресурсами. Проекторы наиболее эффективны, когда действуют в роли консультантов.
Четвертый тип в Дизайне Человека — это Рефлектор. Рефлекторы — это люди, которые отражают состояние окружающей среды. Они, как зеркало, отражают общее состояние общества или коллектива. Их рациональная роль заключается в объективной оценке происходящего вокруг.
В итоге можно сказать следующее: Каждый из четырех типов в Дизайне Человека имеет свои индивидуальные особенности, которые помогают им максимально эффективно взаимодействовать с миром. Понимание своего типа и его практического предназначения позволяет лучше организовать жизнь, выбрать правильные направления для работы и улучшить качество личных отношений.
Trefpoj
October 24, 2024 @ 9:05 am
Официальная покупка диплома вуза с сокращенной программой обучения в Москве
rrsclub.ru/member.php?u=1164
EdwardJoype
October 24, 2024 @ 9:43 am
пин ап кз: пинап – пинап казино
Ремонт телефонов в Москве
October 24, 2024 @ 10:41 am
Профессиональный сервисный центр по ремонту телефонов в Москве.
Мы предлагаем: ближайший ремонт смартфонов
Наши мастера оперативно устранят неисправности вашего устройства в сервисе или с выездом на дом!
микрокредит
October 24, 2024 @ 11:45 am
Наша компания работает на рынке микрозаймов Казахстана уже несколько лет, и за это время мы успели завоевать доверие множества клиентов. Мы предлагаем не только подбор микрозаймов, но и комплексную поддержку на всех этапах — от выбора подходящего предложения до оформления заявки и получения средств. Мы стремимся создать для каждого клиента комфортные условия, обеспечивая прозрачность и надежность всех финансовых операций.
микрокредит [url=https://microloans.kz/]микрокредит[/url] .
Ремонт видеокарт
October 24, 2024 @ 1:04 pm
Профессиональный сервисный центр по ремонту компьютерных видеокарт по Москве.
Мы предлагаем: профессиональный ремонт видеокарт
Наши мастера оперативно устранят неисправности вашего устройства в сервисе или с выездом на дом!
Ремонт ноутбуков в Москве
October 24, 2024 @ 2:12 pm
Профессиональный сервисный центр по ремонту сотовых телефонов в Москве.
Мы предлагаем: сколько стоит ремонт ноутбука после залития
Наши мастера оперативно устранят неисправности вашего устройства в сервисе или с выездом на дом!
ремонт телефонов москва
October 24, 2024 @ 2:48 pm
Хочу поделиться своим опытом ремонта телефона в этом сервисном центре. Остался очень доволен качеством работы и скоростью обслуживания. Если ищете надёжное место для ремонта, обратитесь сюда: ремонт андроида.
микро займ
October 24, 2024 @ 4:33 pm
Наша компания помогает не только частным клиентам, но и малому и среднему бизнесу, предоставляя доступ к микрозаймам на выгодных условиях. Мы понимаем, насколько важно быстро получить финансовую поддержку для развития бизнеса, и предлагаем лучшие решения, которые помогут вашему делу расти и процветать.
микро займы [url=https://microloans.kz/]онлайн займы[/url] .
Stephenswogs
October 24, 2024 @ 4:36 pm
[url=https://ali-albulaihiar.biz]https://ali-albulaihiar.biz[/url]
last news about ali al bulaihi
http://ali-albulaihiar.biz
Micahham
October 24, 2024 @ 4:47 pm
[url=https://bulaihi-ali-al-ar.biz]http://www.bulaihi-ali-al-ar.biz[/url]
last news about bulaihi ali al
http://bulaihi-ali-al-ar.biz
Ремонт iMac
October 24, 2024 @ 6:05 pm
Профессиональный сервисный центр по ремонту моноблоков iMac в Москве.
Мы предлагаем: ремонт аймак
Наши мастера оперативно устранят неисправности вашего устройства в сервисе или с выездом на дом!
StevenRaica
October 24, 2024 @ 6:33 pm
[url=https://erling-haaland-ar.biz]http://www.erling-haaland-ar.biz[/url]
last news about erling haaland
https://www.erling-haaland-ar.biz
Kevinrisse
October 24, 2024 @ 6:38 pm
[url=https://kante-ngolo-ar.biz]http://www.kante-ngolo-ar.biz[/url]
last news about kante ngolo
http://kante-ngolo-ar.biz
EdwardJoype
October 24, 2024 @ 7:15 pm
пин ап: пин ап зеркало – пинап зеркало
Ремонт телефонов
October 24, 2024 @ 7:49 pm
Профессиональный сервисный центр по ремонту телефонов в Москве.
Мы предлагаем: ремонт кнопочных телефонов
Наши мастера оперативно устранят неисправности вашего устройства в сервисе или с выездом на дом!
Michaelvob
October 24, 2024 @ 9:19 pm
Свежие и смешные шутки http://shutki-anekdoty.ru/korotkie-anekdoty.
транспортная компания по Казахстану из Алматы
October 24, 2024 @ 9:35 pm
С [url=https://datransport.ru/]перевозкой грузов по Казахстану из Алматы[/url] DaTransport вы можете быть уверены, что ваш груз находится в надежных руках. Мы подбираем оптимальные маршруты и предлагаем выгодные тарифы.
DavidfainS
October 24, 2024 @ 10:18 pm
https://1winci.icu/# пин ап
пин ап кз
dom internat_diei
October 24, 2024 @ 10:40 pm
дом интернат для престарелых официальный сайт [url=http://www.xn—-1-6cdshu4albf3aye.xn--p1ai]дом интернат для престарелых официальный сайт[/url] .
humandesigntr
October 25, 2024 @ 12:06 am
Тема «Четыре типа в Дизайне Человека» важна для понимания не только на теоретическом, но и на практическом уровне. Этот инструмент самопознания помогает каждому из нас осознать свою природу и использовать индивидуальные особенности для улучшения качества жизни. Рассмотрим рационально-практическую сторону каждого из типов, их определения и различия.
Начнем с Генератор. Генераторы отличаются высокой энергетичностью и способностью легко и эффективно завершать начатые задачи. Главная задача Генератора — найти деятельность, которая приносит радость и удовлетворение. Генератор начинает действовать, когда ощущает внутренний отклик. Когда Генератор действует из отклика, он не только продуктивен, но и создает вокруг себя атмосферу гармонии и успеха.
Второй тип — это Манифестор. Манифесторы могут начинать новые проекты и вдохновлять других. Они не нуждаются в отклике, как Генераторы, и могут сразу принимать решения и действовать. Индивидуальная особенность Манифестора — это стремление к свободе и независимости. Их рациональная роль — прокладывать путь для других.
Третий тип — это Проектор. Проекторы лучше всего проявляют себя в роли наблюдателей и стратегов. Они нуждаются в приглашении, прежде чем начать действовать, и могут эффективно использовать энергию, когда работают с другими людьми. Проекторы отличаются тем, что не обладают собственной энергией, но могут эффективно направлять энергию других. Практическая задача Проектора – это координирование и организация.
Четвертый тип в Дизайне Человека — это Рефлектор. Этот тип является самым редким и уникальным. Они, как зеркало, отражают общее состояние общества или коллектива. Их рациональная роль заключается в объективной оценке происходящего вокруг.
Заключение Каждый из четырех типов в Дизайне Человека имеет свои индивидуальные особенности, которые помогают им максимально эффективно взаимодействовать с миром. Понимание своего типа и его практического предназначения позволяет лучше организовать жизнь, выбрать правильные направления для работы и улучшить качество личных отношений.
источник
Diplomi_qaSi
October 25, 2024 @ 1:39 am
Как безопасно купить диплом колледжа или ПТУ в России, что важно знать
pupis
October 25, 2024 @ 1:47 am
Чтобы начать выигрывать, скачайте легальные БК и начните делать ставки на спорт прямо с телефона
humandesignqi
October 25, 2024 @ 1:50 am
Тема «Четыре типа в Дизайне Человека» важна для понимания не только на теоретическом, но и на практическом уровне. Этот инструмент самопознания помогает каждому из нас осознать свою природу и использовать индивидуальные особенности для улучшения качества жизни. Рассмотрим рационально-практическую сторону каждого из типов, их определения и различия.
Первый тип в Дизайне Человека – это Генератор. Генераторы отличаются высокой энергетичностью и способностью легко и эффективно завершать начатые задачи. Главная задача Генератора — найти деятельность, которая приносит радость и удовлетворение. Генератор начинает действовать, когда ощущает внутренний отклик. Основное отличие Генераторов в том, что они заряжают себя и других энергией, если действуют в соответствии с внутренним откликом.
Следующий тип, на который стоит обратить внимание, — Манифестор. Манифесторы могут начинать новые проекты и вдохновлять других. Они не нуждаются в отклике, как Генераторы, и могут сразу принимать решения и действовать. Манифесторы не подчиняются внешним обстоятельствам, а сами создают свою реальность. Практическая сторона их природы проявляется в том, что они способны запускать процессы и вдохновлять окружающих.
Также важный элемент системы Дизайна Человека — Проектор. Проекторы лучше всего проявляют себя в роли наблюдателей и стратегов. Они нуждаются в приглашении, прежде чем начать действовать, и могут эффективно использовать энергию, когда работают с другими людьми. Их сила — в правильном руководстве и управлении чужими ресурсами. Их рациональное предназначение – это оптимизация работы других типов.
И наконец, заключительный тип — Рефлектор. Этот тип является самым редким и уникальным. Рефлекторы отличаются тем, что их самочувствие напрямую зависит от окружения. Рефлекторы могут стать прекрасными аналитиками, так как они замечают мельчайшие изменения.
Заключение Каждый из четырех типов в Дизайне Человека имеет свои индивидуальные особенности, которые помогают им максимально эффективно взаимодействовать с миром. Понимание своего типа и его практического предназначения позволяет лучше организовать жизнь, выбрать правильные направления для работы и улучшить качество личных отношений.
источник
ErnestZeW
October 25, 2024 @ 1:50 am
slot casino siteleri: casino siteleri – h?zl? casino
humandesignyy
October 25, 2024 @ 3:25 am
Тема «Четыре типа в Дизайне Человека» важна для понимания не только на теоретическом, но и на практическом уровне. Этот инструмент самопознания помогает каждому из нас осознать свою природу и использовать индивидуальные особенности для улучшения качества жизни. Рассмотрим рационально-практическую сторону каждого из типов, их определения и различия.
Первый тип в Дизайне Человека – это Генератор. Он отличаются высокой энергетичностью и способностью легко и эффективно завершать начатые задачи. Этот тип создан для работы, и его главное стремление — заниматься тем, что приносит удовольствие. Генератор начинает действовать, когда ощущает внутренний отклик. Основное отличие Генераторов в том, что они заряжают себя и других энергией, если действуют в соответствии с внутренним откликом.
Еще один важный тип в Дизайне Человека — Манифестор. Манифесторы могут начинать новые проекты и вдохновлять других. Они не нуждаются в отклике, как Генераторы, и могут сразу принимать решения и действовать. Манифесторы не подчиняются внешним обстоятельствам, а сами создают свою реальность. Практическая сторона их природы проявляется в том, что они способны запускать процессы и вдохновлять окружающих.
Также важный элемент системы Дизайна Человека — Проектор. Проекторы лучше всего проявляют себя в роли наблюдателей и стратегов. Они нуждаются в приглашении, прежде чем начать действовать, и могут эффективно использовать энергию, когда работают с другими людьми. Проекторы отличаются тем, что не обладают собственной энергией, но могут эффективно направлять энергию других. Их рациональное предназначение – это оптимизация работы других типов.
Последний, но не менее важный тип — Рефлектор. Рефлекторы — это люди, которые отражают состояние окружающей среды. Они, как зеркало, отражают общее состояние общества или коллектива. Рефлекторы могут стать прекрасными аналитиками, так как они замечают мельчайшие изменения.
Итак, подведем итог: Каждый из четырех типов в Дизайне Человека имеет свои индивидуальные особенности, которые помогают им максимально эффективно взаимодействовать с миром. Понимание своего типа и его практического предназначения позволяет лучше организовать жизнь, выбрать правильные направления для работы и улучшить качество личных отношений.
источник
EdwardJoype
October 25, 2024 @ 4:33 am
пин ап официальный сайт: пинап зеркало – пин ап
Ремонт телефонов в Москве
October 25, 2024 @ 4:39 am
Профессиональный сервисный центр по ремонту телефонов в Москве.
Мы предлагаем: ремонт телефонов недорого
Наши мастера оперативно устранят неисправности вашего устройства в сервисе или с выездом на дом!
Hex
October 25, 2024 @ 5:19 am
Cтальные двери cо склада в Москве с установкой за 1 день.
Любые конфигурации отделок на выбор. Более 3500 моделей на складе: [url=https://dverimetallicheskie.ru/]здесь[/url]
Алкоклуб
October 25, 2024 @ 5:23 am
Алкоклуб и Alcoclub предлагают быструю и круглосуточную доставку алкоголя. Позвоните по номеру +74951086757, чтобы заказать алкоголь в Москве в любое время суток. Этот сервис обеспечивает оперативную доставку по городу, делая процесс максимально комфортным.
Если вам требуется круглосуточный заказ, Алкоклуб — идеальное решение. Связавшись с Alcoclub по номеру +74951086757, вы сможете легко заказать алкоголь на дом. Сервис предлагает разнообразие алкогольной продукции, что делает процесс быстрым и простым.
С Алкоклуб вы всегда можете рассчитывать на оперативную доставку. Оформите заказ через +74951086757, чтобы воспользоваться доставкой алкоголя круглосуточно. Платформа Alcoclub делает доставку доступной и быстрой, чтобы каждый клиент мог получить желаемый напиток вовремя.
asbolabro.com
October 25, 2024 @ 6:26 am
I’m very happy to find this web site. I wanted to thank you for
your time for this fantastic read!! I definitely enjoyed every bit of it and I have you book marked to see new information in your blog.
JosephRon
October 25, 2024 @ 6:47 am
Смешные приколы, шутки и картинки http://shutki-anekdoty.ru/.
hire bodyguard london
October 25, 2024 @ 7:44 am
Valuable information. Fortunate me I found your site by accident, and I’m
stunned why this coincidence didn’t came about earlier!
I bookmarked it.
Ремонт телефонов в Москве
October 25, 2024 @ 8:02 am
Профессиональный сервисный центр по ремонту телефонов в Москве.
Мы предлагаем: где отремонтировать телефон
Наши мастера оперативно устранят неисправности вашего устройства в сервисе или с выездом на дом!
Ремонт телефонов
October 25, 2024 @ 8:23 am
Профессиональный сервисный центр по ремонту телефонов в Москве.
Мы предлагаем: диагностика сотовых телефонов
Наши мастера оперативно устранят неисправности вашего устройства в сервисе или с выездом на дом!
humandesignzi
October 25, 2024 @ 8:59 am
Тема «Четыре типа в Дизайне Человека» важна для понимания не только на теоретическом, но и на практическом уровне. Этот инструмент самопознания помогает каждому из нас осознать свою природу и использовать индивидуальные особенности для улучшения качества жизни. Рассмотрим рационально-практическую сторону каждого из типов, их определения и различия.
Ключевым типом в этой системе является Генератор. Генераторы отличаются высокой энергетичностью и способностью легко и эффективно завершать начатые задачи. Этот тип создан для работы, и его главное стремление — заниматься тем, что приносит удовольствие. Генератор начинает действовать, когда ощущает внутренний отклик. Их индивидуальная особенность заключается в том, что энергия накапливается, только когда они следуют своему отклику.
Еще один важный тип в Дизайне Человека — Манифестор. Этот тип уникален своей независимостью и способностью инициировать действия. Они не нуждаются в отклике, как Генераторы, и могут сразу принимать решения и действовать. Индивидуальная особенность Манифестора — это стремление к свободе и независимости. Их рациональная роль — прокладывать путь для других.
Третий тип — это Проектор. Их задача – управлять и направлять энергию других типов. Они нуждаются в приглашении, прежде чем начать действовать, и могут эффективно использовать энергию, когда работают с другими людьми. Индивидуальная особенность Проектора заключается в умении работать с чужой энергией и направлять ее. Их рациональное предназначение – это оптимизация работы других типов.
Четвертый тип в Дизайне Человека — это Рефлектор. Рефлекторы — это люди, которые отражают состояние окружающей среды. Индивидуальная особенность Рефлектора заключается в том, что они полностью зависят от окружающего мира и людей. Практическая роль Рефлектора — это оценка и отслеживание состояния окружающих.
Заключение Каждый из четырех типов в Дизайне Человека имеет свои индивидуальные особенности, которые помогают им максимально эффективно взаимодействовать с миром. Понимание своего типа и его практического предназначения позволяет лучше организовать жизнь, выбрать правильные направления для работы и улучшить качество личных отношений.
источник
Diplomi_vpkl
October 25, 2024 @ 9:08 am
купить диплом в новоалтайске [url=https://landik-diploms.ru/]landik-diploms.ru[/url] .
DavidfainS
October 25, 2024 @ 9:58 am
https://1winrussia.online/# 1хбет
пин ап
Ремонт видеокарт в Москве
October 25, 2024 @ 10:13 am
Профессиональный сервисный центр по ремонту компьютерных видеокарт по Москве.
Мы предлагаем: сервис видеокарт
Наши мастера оперативно устранят неисправности вашего устройства в сервисе или с выездом на дом!
Ремонт телефонов в Москве
October 25, 2024 @ 10:16 am
Профессиональный сервисный центр по ремонту телефонов в Москве.
Мы предлагаем: ремонт сотовых телефонов рядом
Наши мастера оперативно устранят неисправности вашего устройства в сервисе или с выездом на дом!
MikelKaria
October 25, 2024 @ 10:43 am
[url=https://alialbulaihiar.biz]https://alialbulaihiar.biz[/url]
last news about ali albulaihi
http://www.alialbulaihiar.biz
Michaelvob
October 25, 2024 @ 10:57 am
Свежие приколы http://shutki-anekdoty.ru/.
Lazrtnm
October 25, 2024 @ 11:02 am
Парадокс, но купить диплом кандидата наук оказалось не так и сложно
pansionat dlya pojilih_rtki
October 25, 2024 @ 11:03 am
пансионат для пожилых адрес [url=https://xn—–3-43da3arnf4adrboggk3ay6e3gtd.xn--p1ai/]https://xn—–3-43da3arnf4adrboggk3ay6e3gtd.xn--p1ai/[/url] .
Ремонт игровых приставок
October 25, 2024 @ 12:17 pm
Профессиональный сервисный центр по ремонту игровых консолей Sony Playstation, Xbox, PSP Vita с выездом на дом по Москве.
Мы предлагаем: надежный сервис ремонта игровых консолей
Наши мастера оперативно устранят неисправности вашего устройства в сервисе или с выездом на дом!
Diplomi_ieOa
October 25, 2024 @ 12:18 pm
купить диплом конкурса [url=https://prema-diploms.ru/]prema-diploms.ru[/url] .
invest-top
October 25, 2024 @ 12:21 pm
Узнайте, [url=https=https://invest-top.net/]как пополнить Binance[/url] и начните торговать. На invest-top.net есть инструкции для простого пополнения счёта.
EdwardJoype
October 25, 2024 @ 1:25 pm
пинап: пин ап казино – пин ап кз
Gabrielamaky
October 25, 2024 @ 1:35 pm
купить кокаин гашиш героин порно жесть
Lazrpny
October 25, 2024 @ 2:18 pm
Купить диплом о среднем образовании в Москве и любом другом городе
publikacii.listbb.ru/viewtopic.php?f=3&t=1032
Diplomi_qfea
October 25, 2024 @ 3:11 pm
купить диплом бакалавра в новосибирске [url=https://server-diploms.ru/]server-diploms.ru[/url] .
MichaelRar
October 25, 2024 @ 3:21 pm
купить меф шымкент купить кокаин
ErnestZeW
October 25, 2024 @ 3:40 pm
пин ап: пин ап – пинап зеркало
Sazrbxa
October 25, 2024 @ 3:43 pm
Полезные советы по покупке диплома о высшем образовании без риска
Mazrnxy
October 25, 2024 @ 3:44 pm
Быстрая схема покупки диплома старого образца: что важно знать?
investicos.com/uncategorized/kupit-diplom-665279gysk
Ремонт телефонов
October 25, 2024 @ 3:52 pm
Профессиональный сервисный центр по ремонту телефонов в Москве.
Мы предлагаем: ремонт телефонов москва рядом
Наши мастера оперативно устранят неисправности вашего устройства в сервисе или с выездом на дом!
Gabrielamaky
October 25, 2024 @ 4:11 pm
порно русская милфа детское порно цп дп печенье
Micahham
October 25, 2024 @ 4:25 pm
[url=https://kaneharryar.biz]http://www.kaneharryar.biz[/url]
last news about kane harry
https://www.kaneharryar.biz
Stephenswogs
October 25, 2024 @ 4:40 pm
[url=https://neymarar.biz]https://www.neymarar.biz[/url]
last news about neymar
https://neymarar.biz
MichaelRar
October 25, 2024 @ 5:48 pm
корейское порно духи героин купить
ремонт телефонов москва
October 25, 2024 @ 6:36 pm
Недавно разбил экран своего телефона и обратился в этот сервисный центр. Ребята быстро и качественно починили устройство, теперь работает как новый. Очень рекомендую обратиться к ним за помощью. Вот ссылка на их сайт: ремонт смартфонов москва.
Iariorrnq
October 25, 2024 @ 6:38 pm
Официальная покупка диплома вуза с упрощенной программой обучения
StevenRaica
October 25, 2024 @ 7:14 pm
[url=https://kane-harryar.biz]www.kane-harryar.biz[/url]
last news about kane harry
https://www.kane-harryar.biz
Sazrblm
October 25, 2024 @ 8:03 pm
Диплом вуза купить официально с упрощенным обучением в Москве
AndrewVOg
October 25, 2024 @ 8:19 pm
[url=https://haalanderlingar.biz]haalanderlingar.biz[/url]
last news about haaland erling
http://haalanderlingar.biz
DavidfainS
October 25, 2024 @ 9:40 pm
https://1winindia.tech/# пин ап казино
пин ап кз
Ремонт видеокарт
October 25, 2024 @ 9:51 pm
Профессиональный сервисный центр по ремонту компьютерных видеокарт по Москве.
Мы предлагаем: починка видеокарты цена
Наши мастера оперативно устранят неисправности вашего устройства в сервисе или с выездом на дом!
Diplomi_vlSi
October 25, 2024 @ 10:27 pm
купить диплом среднего специального [url=https://man-diploms.ru/]купить диплом среднего специального[/url] .
ErnestZeW
October 26, 2024 @ 12:10 am
canl? casino siteleri: slot casino siteleri – en iyi casino siteleri
JeremyBuiva
October 26, 2024 @ 12:17 am
[url=https://harrykanear.biz]https://www.harrykanear.biz[/url]
last news about harry kane
harrykanear.biz
Sazrcbz
October 26, 2024 @ 12:17 am
Полезные советы по покупке диплома о высшем образовании без риска
clan.13.free.fr/index.php?file=Members&op=detail&autor=ocolylyf
kypit 1S_jvmn
October 26, 2024 @ 12:33 am
купить программу 1с торговля [url=www.kupit-1s11.ru]купить программу 1с торговля[/url] .
Sazrlzw
October 26, 2024 @ 1:33 am
Диплом пту купить официально с упрощенным обучением в Москве
belitt.ru/forum/topic/diplom-na-zakaz-ekonomte-svoe-vremya-i-sily/#postid-337
Ремонт телефонов в Москве
October 26, 2024 @ 3:44 am
Профессиональный сервисный центр по ремонту телефонов в Москве.
Мы предлагаем: сдача телефона в ремонт
Наши мастера оперативно устранят неисправности вашего устройства в сервисе или с выездом на дом!
JosephRon
October 26, 2024 @ 4:19 am
Свежие приколы http://shutki-anekdoty.ru/.
Diplomi_utEr
October 26, 2024 @ 5:27 am
купить диплом о среднем образовании в екатеринбурге [url=https://russa-diploms.ru/]купить диплом о среднем образовании в екатеринбурге[/url] .
Lizzietug
October 26, 2024 @ 5:33 am
Наш сайт предлагает широчайший ассортимент лодок ПВХ, моторов для лодок, запчастей и аксессуаров, которые сделают ваше времяпрепровождение на воде комфортным и спокойным.
[url=https://draiv38.ru/catalog/lodochnye-motory/motor-tohatsunissan]лодочный мотор ниссан[/url] – это отличный вариант для рыбалки и отдыха в любом водоеме или веселых развлечений на воде. Наши надувные лодки невесомые, надежные и компактные, они легко перевозятся и быстро накачиваются. В нашем ассортименте модели разных размеров и параметров, чтобы вы подобрали именно то вариант, который соответствует вашим потребностям.
Наши надежные лодочные моторы подарят вам отличную скорость и маневренность в любом водоеме. Каталог запчастей и аксессуаров помогут поддерживать вашу надувную в отличном состоянии и сделать любую поездку максимально беспроблемной.
Заказывая у нас, вы получите не только качественный продукт, но и профессиональную консультацию, а также гарантии на все товары. Покупайте надувные лодки и все необходимое для рыбалки прямо сейчас!
Diplomi_uhEr
October 26, 2024 @ 6:13 am
купить диплом воспитателя детского [url=https://orik-diploms.ru/]orik-diploms.ru[/url] .
Ремонт телефонов
October 26, 2024 @ 7:08 am
Профессиональный сервисный центр по ремонту телефонов в Москве.
Мы предлагаем: сервисный центр по ремонту телефонов
Наши мастера оперативно устранят неисправности вашего устройства в сервисе или с выездом на дом!
Cazrfqy
October 26, 2024 @ 7:51 am
Процесс получения диплома стоматолога: реально ли это сделать быстро?
erudio.global/blog/index.php?entryid=37272
Sazrwsi
October 26, 2024 @ 7:52 am
Можно ли купить аттестат о среднем образовании? Основные рекомендации
ErnestZeW
October 26, 2024 @ 8:04 am
1xbet официальный сайт: 1xbet скачать – 1xbet официальный сайт
Stephennewly
October 26, 2024 @ 8:10 am
[url=https://erling-haalandar.biz/]http://www.erling-haalandar.biz[/url]
last news about erling haaland
http://erling-haalandar.biz
DavidfainS
October 26, 2024 @ 8:44 am
https://1winrussia.online/# 1xbet
пин ап казино
Ремонт телефонов в Москве
October 26, 2024 @ 9:06 am
Профессиональный сервисный центр по ремонту телефонов в Москве.
Мы предлагаем: ближайший ремонт телефонов
Наши мастера оперативно устранят неисправности вашего устройства в сервисе или с выездом на дом!
pansionat dlya prestarelih_coMn
October 26, 2024 @ 10:17 am
пансионаты для престарелых в симферополе недорого посуточно [url=http://xn—–6kcacwibu7aeobnlkrcglni7eyhud.xn--p1ai]пансионаты для престарелых в симферополе недорого посуточно[/url] .
Lazrlzt
October 26, 2024 @ 10:21 am
Купить диплом о среднем полном образовании, в чем подвох и как избежать обмана?
Diplomi_ucpn
October 26, 2024 @ 10:45 am
купить диплом о среднем образовании в томске [url=https://arusak-diploms.ru/]arusak-diploms.ru[/url] .
dom prestarelih_igSi
October 26, 2024 @ 11:31 am
дом престарелых в керчи [url=www.xn—-1-6cdshb2cetnmcmp6c5f.xn--p1ai/]дом престарелых в керчи[/url] .
vivod iz zapoya sochi_yjel
October 26, 2024 @ 11:53 am
быстрый вывод из запоя в стационаре [url=https://vyvod-iz-zapoya-sochi15.ru/]быстрый вывод из запоя в стационаре[/url] .
Links from Comments Blogs Shop
October 26, 2024 @ 12:17 pm
Boost your site’s SEO with top-tier backlinks from https://seoshop.xmc.pl.
Our backlinks are carefully selected to ensure
that your site gains the maximum benefit from each link.
By choosing our service, you can enhance your site’s authority, improve its search engine ranking, and drive more organic traffic.
We offer a variety of packages to suit different budgets and goals, making
it easy for you to find the perfect fit for your SEO strategy.
Visit us today and take advantage of our competitive pricing and exceptional customer support.
binance register
October 26, 2024 @ 12:49 pm
Your article helped me a lot, is there any more related content? Thanks!
Cazrivl
October 26, 2024 @ 12:56 pm
Официальная покупка школьного аттестата с упрощенным обучением в Москве
EdwardJoype
October 26, 2024 @ 1:39 pm
pin-up: pin-up casino giris – pin up azerbaycan
Ремонт телефонов в Москве
October 26, 2024 @ 2:18 pm
Профессиональный сервисный центр по ремонту телефонов в Москве.
Мы предлагаем: сервис по ремонту смартфонов
Наши мастера оперативно устранят неисправности вашего устройства в сервисе или с выездом на дом!
Stephenswogs
October 26, 2024 @ 2:45 pm
[url=https://neymar-da-silva-ar.biz]http://neymar-da-silva-ar.biz[/url]
last news about neymar
http://www.neymar-da-silva-ar.biz
Micahham
October 26, 2024 @ 3:41 pm
[url=https://dasilva-neymar-ar.biz]dasilva-neymar-ar.biz[/url]
last news about dasilva neymar
http://www.dasilva-neymar-ar.biz
MarionUncok
October 26, 2024 @ 3:55 pm
реклама в лифтах дома реклама в лифтах компании
ремонт техники профи в екб
October 26, 2024 @ 4:05 pm
Если вы искали где отремонтировать сломаную технику, обратите внимание – ремонт бытовой техники
StevenRaica
October 26, 2024 @ 4:53 pm
[url=https://talisca-ar.biz]https://talisca-ar.biz[/url]
last news about talisca
http://talisca-ar.biz
реферальный код binance
October 26, 2024 @ 5:02 pm
[url=https=https://invest-top.net/]Скачайте Binance[/url] и начните торговать в любом месте. Invest-top.net объяснит, как загрузить приложение на любое устройство.
Kevinstuch
October 26, 2024 @ 6:00 pm
реклама в лифте цена как разместить рекламу в лифте
MarionUncok
October 26, 2024 @ 6:22 pm
реклама в лифтах https://reklama-v-liftah-msk.ru
JosephRon
October 26, 2024 @ 6:33 pm
Прикольные анекдоты http://shutki-anekdoty.ru/anekdoty.
Ремонт игровых приставок
October 26, 2024 @ 6:38 pm
Профессиональный сервисный центр по ремонту игровых консолей Sony Playstation, Xbox, PSP Vita с выездом на дом по Москве.
Мы предлагаем: ремонт игровых консолей на дому
Наши мастера оперативно устранят неисправности вашего устройства в сервисе или с выездом на дом!
Williamraics
October 26, 2024 @ 7:39 pm
Узнайте, как приобрести диплом о высшем образовании без рисков
offmarketbusinessforsale.com/kupit-diplom-78805wdno
DavidfainS
October 26, 2024 @ 7:41 pm
http://1wintr.fun/# cazino
пинап казино
Michaelvob
October 26, 2024 @ 7:45 pm
Смешные анекдоты http://shutki-anekdoty.ru/anekdoty.
Diplomi_oiSi
October 26, 2024 @ 8:18 pm
Официальное получение диплома техникума с упрощенным обучением в Москве
Kevinstuch
October 26, 2024 @ 10:19 pm
как разместить рекламу в лифте реклама в лифтах дома
Charlesnidly
October 26, 2024 @ 10:22 pm
[url=https://erlinghaalandar.biz]erlinghaalandar.biz[/url]
last news about erling haaland
http://www.erlinghaalandar.biz
StevenSok
October 26, 2024 @ 10:58 pm
[url=https://neymar-ar.biz]https://neymar-ar.biz[/url]
last news about neymar
http://www.neymar-ar.biz
MfogaCig
October 26, 2024 @ 11:01 pm
[url=http://rtdspb.ru/communication/forum/user/2224/]все мфо и мкк[/url] и [url=https://narodmebel.com/communication/forum/user/132/]новые мфо и мкк[/url] доступны всем – успейте взять займ выгодно в новых МФО 2024 года!
RobertRer
October 26, 2024 @ 11:02 pm
[url=https://haaland-erling-ar.biz]https://haaland-erling-ar.biz[/url]
last news about haaland erling
https://haaland-erling-ar.biz
MichaelDar
October 26, 2024 @ 11:43 pm
[url=https://kane-harry-ar.biz]www.kane-harry-ar.biz[/url]
last news about kane harry
http://kane-harry-ar.biz
KennethEnlab
October 27, 2024 @ 2:55 am
https://znakavto.com/
Ремонт видео карты в Москве
October 27, 2024 @ 3:20 am
Профессиональный сервисный центр по ремонту компьютерных видеокарт по Москве.
Мы предлагаем: ремонт видеокарты в москве
Наши мастера оперативно устранят неисправности вашего устройства в сервисе или с выездом на дом!
vivod iz zapoya sochi_iqEt
October 27, 2024 @ 3:51 am
вывод из запоя на дому в сочи [url=www.vyvod-iz-zapoya-sochi16.ru]вывод из запоя на дому в сочи[/url] .
Derrickjoisy
October 27, 2024 @ 5:24 am
[url=https://harry-kane-ar.biz]https://www.harry-kane-ar.biz[/url]
last news about harry kane
http://harry-kane-ar.biz
DavidfainS
October 27, 2024 @ 6:54 am
https://1winindia.tech/# пинап
пин ап казино вход
ErnestZeW
October 27, 2024 @ 8:40 am
пин ап казино: пинап казино – пинап казино
Lazriaa
October 27, 2024 @ 12:03 pm
Как приобрести диплом о среднем образовании в Москве и других городах
Help with my coursework
October 27, 2024 @ 3:18 pm
It may be challenging to find time to do your coursework when you have a lot of other commitments to fulfill in today’s fast-paced world. Exactly at this point is when the concept of Coursework writing services comes into play. Even though it appears to be out of common, this tactic is gaining popularity among students who want assistance in structuring their online assignments. Some students come to the realization that getting help from other sources for their education is the greatest way to complete their assignments without falling behind in their daily schedule. This strategy can be especially useful for people who are simultaneously pursuing postgraduate degrees and handling significant career responsibilities at the same time. Whatever the case may be, it is of the utmost importance to find a balance between the situation’s convenience and the scenario’s academic authenticity. It is in your best interest to investigate all of the other possibilities first, such as gaining a better understanding of how to better manage your time, speaking with your instructors, and joining study groups with peers.
Diplomi_msOa
October 27, 2024 @ 6:33 pm
купить диплом стоимость [url=https://prema-diploms.ru/]купить диплом стоимость[/url] .
Sazrkmm
October 27, 2024 @ 6:34 pm
Приобретение диплома ПТУ с сокращенной программой обучения в Москве
ремонт бытовой техники екатеринбург
October 27, 2024 @ 7:01 pm
Если вы искали где отремонтировать сломаную технику, обратите внимание – сервисный центр в екб
Stephenswogs
October 27, 2024 @ 7:28 pm
[url=https://yassinebounouar.biz]https://www.yassinebounouar.biz[/url]
last news about yassine bounou
https://yassinebounouar.biz
Micahham
October 27, 2024 @ 7:43 pm
[url=https://souzatalisca-ar.biz]http://www.souzatalisca-ar.biz[/url]
last news about souza talisca
https://www.souzatalisca-ar.biz
JeremyBuiva
October 27, 2024 @ 7:47 pm
[url=https://yassine-bounou-ar.biz]http://www.yassine-bounou-ar.biz[/url]
last news about yassine bounou
yassine-bounou-ar.biz
Stephennewly
October 27, 2024 @ 7:59 pm
[url=https://taliscaar.biz/]www.taliscaar.biz[/url]
last news about talisca
http://www.taliscaar.biz
AndrewVOg
October 27, 2024 @ 10:13 pm
[url=https://talisca-souza-ar.biz]talisca-souza-ar.biz[/url]
last news about talisca souza
http://www.talisca-souza-ar.biz
vivod iz zapoya sochi_vaSl
October 27, 2024 @ 11:40 pm
вывод из запоя стационар [url=http://www.vyvod-iz-zapoya-sochi17.ru]вывод из запоя стационар [/url] .
starzbet
October 28, 2024 @ 12:46 am
Для безопасного доступа используйте официальное зеркало 888Starz
ThomasSaise
October 28, 2024 @ 1:18 am
buy online pharmacy: google viagra dosage recommendations – prescription drugs online
Oarioraor
October 28, 2024 @ 3:16 am
Стоимость дипломов высшего и среднего образования и процесс их получения
Diplomi_vzea
October 28, 2024 @ 3:22 am
купить диплом по охране труда [url=https://server-diploms.ru/]server-diploms.ru[/url] .
Sazrawk
October 28, 2024 @ 3:36 am
Узнайте, как безопасно купить диплом о высшем образовании
Iarioropb
October 28, 2024 @ 5:35 am
Всё, что нужно знать о покупке аттестата о среднем образовании
Сервисный центр в Москве
October 28, 2024 @ 5:57 am
Сервисный центр предлагает ремонт кофемашин bianchi адреса ремонта кофемашин bianchi
totosafeguidem
October 28, 2024 @ 6:50 am
I simply had to say how much I loved the fundamental information you provided on your website before I could go. is going to return frequently to check for new posts.
무료 토토사이트
powerballsitem
October 28, 2024 @ 6:51 am
With your presentation, you really make it sound thus simple, but I find this topic to be one that I feel I’ll never understand. For me, it feels a little too broad and complex.
엔트리 파워사다리
Sazrnid
October 28, 2024 @ 7:18 am
Можно ли купить аттестат о среднем образовании? Основные рекомендации
snyatie zapoya na domy_yxMr
October 28, 2024 @ 7:55 am
капельница при алкогольной интоксикации на дому цена [url=https://snyatie-zapoya-na-domu15.ru/]капельница при алкогольной интоксикации на дому цена[/url] .
snyatie zapoya na domy_coSa
October 28, 2024 @ 8:01 am
вывод из запоя нижний новгород [url=https://snyatie-zapoya-na-domu16.ru/]вывод из запоя нижний новгород [/url] .
Lazrgbn
October 28, 2024 @ 8:51 am
Где и как купить диплом о высшем образовании без лишних рисков
snyatie zapoya na domy_yjOr
October 28, 2024 @ 9:22 am
капельница наркология 24 [url=https://www.snyatie-zapoya-na-domu17.ru]капельница наркология 24[/url] .
Diplomi_gtSi
October 28, 2024 @ 9:28 am
купить диплом учителя [url=https://man-diploms.ru/]man-diploms.ru[/url] .
ThomasSaise
October 28, 2024 @ 10:05 am
drug store online: ed men – canadian pharmacy
Albertseilm
October 28, 2024 @ 3:20 pm
[url=][/url]
Временная регистрация в Москве: Быстро и Легально!
Ищете, где оформить временную регистрацию в Москве?
Мы гарантируем быстрое и легальное оформление без очередей и лишних документов.
Ваше спокойствие – наша забота!
Минимум усилий • Максимум удобства • Полная легальность
Свяжитесь с нами прямо сейчас!
Временная регистрация в Москве
[url=][/url]
Dnrtxdw
October 28, 2024 @ 3:24 pm
Официальная покупка диплома вуза с упрощенной программой обучения
Diplomi_fdkl
October 28, 2024 @ 3:30 pm
купить дипломы о высшем образовании в барнауле [url=https://landik-diploms.ru/]купить дипломы о высшем образовании в барнауле[/url] .
Cazrvpf
October 28, 2024 @ 4:30 pm
Пошаговая инструкция по безопасной покупке диплома о высшем образовании
Sazrcan
October 28, 2024 @ 6:38 pm
Официальная покупка диплома вуза с сокращенной программой обучения в Москве
tyyifvxb
October 28, 2024 @ 7:13 pm
Anchorage. https://t.me/inewsworldplanet
ThomasSaise
October 28, 2024 @ 7:36 pm
Online medicine home delivery: mail order pharmacy india – india online pharmacy
вывоз строительного мусора газелью
October 28, 2024 @ 8:04 pm
Мы занимаемся вывозом мусора газелями разной модификации от стандартных 6 м3 до больших 14 м3 есть также грузчики, работаем по Московской области. Преимущества вывоза мусора газелью в том, что она самая дешёвая услуга вывоза мусора.
Эффективный способ вывоза мусора газелью, с максимальной экономией времени.
Почему газель идеально подходит для вывоза мусора, которые важно знать.
Список разрешенных для перевозки грузов газелью, с учетом ограничений.
Минимальные стоимость услуги по вывозу мусора газелью, пользуясь опытом специалистов.
Секрет успешного вывоза мусора газелью, понимая особенности процесса.
вывоз строительного мусора газелью [url=https://vyvoz-musora-gazel-mo.ru/]вывоз старой мебели на свалку[/url] .
kilywuwj
October 28, 2024 @ 8:46 pm
Matthew perry died. https://t.me/inewsworldplanet
zwpadrul
October 28, 2024 @ 10:09 pm
Philippines china south china sea. https://t.me/inewsworldplanet
Kevinrisse
October 28, 2024 @ 10:22 pm
[url=https://da-silva-neymar-ar.biz]http://da-silva-neymar-ar.biz[/url]
last news about da silva neymar
http://www.da-silva-neymar-ar.biz
ecwigelw
October 29, 2024 @ 1:59 am
How. https://t.me/inewsworldplanet
Diplomi_jwSi
October 29, 2024 @ 2:17 am
Официальная покупка диплома вуза с сокращенной программой обучения в Москве
Cazrhvx
October 29, 2024 @ 2:22 am
Диплом техникума купить официально с упрощенным обучением в Москве
rcufbzos
October 29, 2024 @ 3:12 am
Cappuccino. https://t.me/inewsworldplanet
Emersonbok
October 29, 2024 @ 4:28 am
what type of medicine is prescribed for allergies [url=http://drugs1st.store/#]buy ed pills online[/url] erectile dysfunction treatment
ThomasSaise
October 29, 2024 @ 4:32 am
ed devices: errectile disfunction – impotance
Lazrhka
October 29, 2024 @ 6:53 am
Как купить диплом о высшем образовании с минимальными рисками
Williamraics
October 29, 2024 @ 8:01 am
Легальные способы покупки диплома о среднем полном образовании
laviehub.com/blog/kupit-diplom-423183npmv
Sazrskb
October 29, 2024 @ 8:19 am
Как быстро получить диплом магистра? Легальные способы
Diplomi_elEr
October 29, 2024 @ 10:33 am
купить диплом воспитателя о высшем образовании [url=https://orik-diploms.ru/]orik-diploms.ru[/url] .
StevenRaica
October 29, 2024 @ 1:13 pm
[url=https://souza-talisca-ar.biz]http://www.souza-talisca-ar.biz[/url]
last news about souza talisca
http://www.souza-talisca-ar.biz
Michaelunero
October 29, 2024 @ 1:43 pm
[url=][/url]
Временная регистрация в Москве: Быстро и Легально!
Ищете, где оформить временную регистрацию в Москве? Мы гарантируем быстрое и легальное оформление без очередей и лишних документов. Ваше спокойствие – наша забота!
Минимум усилий • Максимум удобства • Полная легальность
Свяжитесь с нами прямо сейчас!
[url=https://мосрегистрация77.рф/].[/url]
[url=][/url]
ThomasSaise
October 29, 2024 @ 1:43 pm
best ed pills at gnc: ed cure – pump for ed
Trefvsk
October 29, 2024 @ 4:14 pm
Как купить диплом о высшем образовании с минимальными рисками
saros-center.ru/forum/?PAGE_NAME=profile_view&UID=6040
MikelKaria
October 29, 2024 @ 5:23 pm
[url=https://yassinebounou-ar.biz]http://www.yassinebounou-ar.biz[/url]
last news about yassine bounou
http://yassinebounou-ar.biz
газель и грузчики под вывоз мусора
October 29, 2024 @ 5:27 pm
Мы занимаемся вывозом мусора газелями разной модификации от стандартных 6 м3 до больших 14 м3 есть также грузчики, работаем по Московской области. Преимущества вывоза мусора газелью в том, что она самая дешёвая услуга вывоза мусора.
Как организовать вывоз мусора газелью, с максимальной экономией времени.
Почему газель идеально подходит для вывоза мусора, которая убеждает.
Основные типы мусора, которые можно вывозить газелью, с учетом размеров и веса.
Экономия с вывозом мусора газелью, и сохранить качество проведенных работ.
Как избежать неприятностей при вывозе мусора газелью, соблюдая правила перевозки.
газель и грузчики под вывоз мусора [url=https://vyvoz-musora-gazel-mo.ru/]газель и грузчики под вывоз мусора[/url] .
Stephenswogs
October 29, 2024 @ 6:31 pm
[url=https://messi-lionel-ae.biz]https://messi-lionel-ae.biz[/url]
last news about messi lionel
messi-lionel-ae.biz
Vivod iz zapoya v sankt peterbyrge_ajMi
October 29, 2024 @ 6:54 pm
вывод из запоя на дому в спб [url=https://vyvod-iz-zapoya-v-sankt-peterburge15.ru/]вывод из запоя на дому в спб[/url] .
Micahham
October 29, 2024 @ 7:13 pm
[url=https://lionelmessi-ae.biz]lionelmessi-ae.biz[/url]
last news about lionel messi
lionelmessi-ae.biz
Stephennewly
October 29, 2024 @ 7:25 pm
[url=https://ronaldo-cristiano-ae.biz/]www.ronaldo-cristiano-ae.biz[/url]
last news about ronaldo cristiano
https://ronaldo-cristiano-ae.biz
Sazrify
October 29, 2024 @ 7:25 pm
Диплом пту купить официально с упрощенным обучением в Москве
dominion.listbb.ru/viewtopic.php?f=11&t=659
AndrewVOg
October 29, 2024 @ 9:01 pm
[url=https://ronaldocristiano-ae.biz]https://www.ronaldocristiano-ae.biz[/url]
last news about ronaldo cristiano
http://ronaldocristiano-ae.biz
MichaelDar
October 29, 2024 @ 9:05 pm
[url=https://bounouyassinear.biz]https://bounouyassinear.biz[/url]
last news about bounou yassine
http://www.bounouyassinear.biz
StevenSok
October 29, 2024 @ 9:22 pm
[url=https://bounou-yassine-ar.biz]http://bounou-yassine-ar.biz[/url]
last news about bounou yassine
https://www.bounou-yassine-ar.biz
Vivod iz zapoya v sankt peterbyrge_uuOl
October 29, 2024 @ 9:34 pm
вывод из запоя спб цены [url=https://azithromycinum.ru/]вывод из запоя спб цены[/url] .
ThomasSaise
October 29, 2024 @ 10:35 pm
natural ed remedies: how to overcome ed naturally – canadian pharmacy
Charlesnidly
October 29, 2024 @ 10:52 pm
[url=https://cristianoronaldoae.biz]http://cristianoronaldoae.biz[/url]
last news about cristiano ronaldo
cristianoronaldoae.biz
Mazrejr
October 29, 2024 @ 11:00 pm
Приобретение школьного аттестата с официальным упрощенным обучением в Москве
investicos.com/uncategorized/kupit-diplom-66853ecfp
RobertRer
October 29, 2024 @ 11:28 pm
[url=https://cristianoronaldo-ae.biz]http://cristianoronaldo-ae.biz[/url]
last news about cristiano ronaldo
http://cristianoronaldo-ae.biz
JeremyBuiva
October 29, 2024 @ 11:55 pm
[url=https://lionel-messi-ae.biz]lionel-messi-ae.biz[/url]
last news about lionel messi
https://www.lionel-messi-ae.biz
Diplomi_pyOa
October 30, 2024 @ 1:41 am
купить свидетельство о рождении [url=https://prema-diploms.ru/]prema-diploms.ru[/url] .
Sazrrsa
October 30, 2024 @ 1:42 am
Сколько стоит диплом высшего и среднего образования и как его получить?
Derekopeta
October 30, 2024 @ 3:09 am
http://pinup-az.bid/# pin-up casino giris
pin up zerkalo
Derrickjoisy
October 30, 2024 @ 4:50 am
[url=https://cristiano-ronaldo-ae.biz]www.cristiano-ronaldo-ae.biz[/url]
last news about cristiano ronaldo
https://www.cristiano-ronaldo-ae.biz
Jasonnet
October 30, 2024 @ 5:04 am
pin up azerbaycan: pin-up casino giris – pin-up casino giris
Diplomi_ovSi
October 30, 2024 @ 6:24 am
Как оказалось, купить диплом кандидата наук не так уж и сложно
Michaelvob
October 30, 2024 @ 6:36 am
Веселые мемы http://shutki-anekdoty.ru/kartinki-prikolnye.
Vivod iz zapoya v sankt peterbyrge_aest
October 30, 2024 @ 7:10 am
вывод из запоя цена [url=https://www.vyvod-iz-zapoya-v-sankt-peterburge17.ru]вывод из запоя цена[/url] .
JosephRon
October 30, 2024 @ 7:23 am
Смешные видеоролики http://shutki-anekdoty.ru/videos.
Vivod iz zapoya v sankt peterbyrge_jsOr
October 30, 2024 @ 7:37 am
вывод из запоя цена [url=vyvod-iz-zapoya-v-sankt-peterburge16.ru]вывод из запоя цена[/url] .
Vivod iz zapoya v sankt peterbyrge_ojPn
October 30, 2024 @ 7:38 am
вывод из запоя капельница [url=vyvod-iz-zapoya-v-sankt-peterburge18.ru]вывод из запоя капельница [/url] .
gruzpoputno
October 30, 2024 @ 8:39 am
Попутный груз из Новосибирска — это доступное решение для отправки груза по нужному маршруту
Dnrtueg
October 30, 2024 @ 8:47 am
Удивительно, но купить диплом кандидата наук оказалось не так сложно
Ремонт iPhone в Москве
October 30, 2024 @ 9:55 am
Профессиональный сервисный центр по ремонту Apple iPhone в Москве.
Мы предлагаем: ремонт телефонов айфон в москве адреса
Наши мастера оперативно устранят неисправности вашего устройства в сервисе или с выездом на дом!
TerryGet
October 30, 2024 @ 10:30 am
офисные вагончики купить бытовка в бухловке калужская область
Diplomi_xhEr
October 30, 2024 @ 10:31 am
купить диплом с занесен ем в реестр [url=https://russa-diploms.ru/]russa-diploms.ru[/url] .
ДизайнЧеловека
October 30, 2024 @ 11:12 am
Отказ от собственной природы приводит к саморазрушению Карта Дизайна Человека или Бодиграф
Ремонт телефонов в Москве
October 30, 2024 @ 12:11 pm
починка телефонов
Richardfrurb
October 30, 2024 @ 12:55 pm
бытовка для проживания бытовки на заказ москва
ДизайнЧеловека
October 30, 2024 @ 12:57 pm
У каждого типа в Дизайне Человека есть эмоциональный настрой, к которому он наиболее склонен. Об эмоциях написаны горы книг, они сопровождают нас на протяжении всей жизни и кажутся избитой темой, однако работа с эмоциями может многому научить и способствовать личностному росту. Дизайн Человека подробно
TerryGet
October 30, 2024 @ 1:04 pm
бытовка с санузлом и кухней модульные вагончики для офиса
Jasonnet
October 30, 2024 @ 2:14 pm
бонусы пин ап: pinco – пинап казино
ДизайнЧеловека
October 30, 2024 @ 2:31 pm
Что вообще я могу? Какие навыки, умения, знания, способности у меня есть? Что я умею? Чему я учился? Что я знаю? В какой области я специалист? Дизайн человека (Human design) – расчет карты онлайн
ZSot
October 30, 2024 @ 3:24 pm
vavada casino
Michaelunero
October 30, 2024 @ 4:01 pm
[url=][/url]
Временная регистрация в Москве: Быстро и Легально!
Ищете, где оформить временную регистрацию в Москве? Мы гарантируем быстрое и легальное оформление без очередей и лишних документов. Ваше спокойствие – наша забота!
Минимум усилий • Максимум удобства • Полная легальность
Свяжитесь с нами прямо сейчас!
[url=https://regm7921.ru/].[/url]
[url=][/url]
Michaelvob
October 30, 2024 @ 5:16 pm
Свежие и смешные мемы http://shutki-anekdoty.ru/kartinki-prikolnye.
Sazrerc
October 30, 2024 @ 5:57 pm
Можно ли быстро купить диплом старого образца и в чем подвох?
ДизайнЧеловека
October 30, 2024 @ 6:34 pm
Дизайн человека помогает осознать, что делает нас особенными и как мы взаимодействуем с миром Карта Дизайна Человека или Бодиграф
Diplomi_chea
October 30, 2024 @ 7:29 pm
купить диплом специалиста в нижнем новгороде [url=https://server-diploms.ru/]server-diploms.ru[/url] .
Sazreuh
October 30, 2024 @ 7:37 pm
Как быстро получить диплом магистра? Легальные способы
Iariorgrn
October 30, 2024 @ 7:41 pm
Вопросы и ответы: можно ли быстро купить диплом старого образца?
ДизайнЧеловека
October 30, 2024 @ 7:43 pm
Задумайтесь вот о чем: если все мы разные, разве можно найти единый подход к созданию счастливой и успешной жизни? Все люди одинаково ценны и важны, но у каждого из нас есть собственный способ поймать удачу за хвост. И метод, который работает у одного человека, далеко не всегда подходит другому Консультация Дизайн Человека – human design
ZSot
October 30, 2024 @ 7:52 pm
bc game apk
JosephRon
October 30, 2024 @ 8:02 pm
Прикольные видеоролики http://shutki-anekdoty.ru/videos.
Derekopeta
October 30, 2024 @ 8:55 pm
http://pinup-az.bid/# pin-up
pin up casino
ставки на спорт
October 30, 2024 @ 10:08 pm
Сомневаетесь, какая платформа для ставок лучше? [url=https://t.me/s/zenit_win]zenit win[/url] предлагает отличные возможности, выгодные условия и простой доступ. Это надёжный выбор для тех, кто ищет не только удобство, но и высокий сервис.
binance Registrācija
October 30, 2024 @ 11:04 pm
Thank you for your sharing. I am worried that I lack creative ideas. It is your article that makes me full of hope. Thank you. But, I have a question, can you help me?
Sazrdzq
October 31, 2024 @ 12:22 am
Легальная покупка диплома о среднем образовании в Москве и регионах
trumpbookusa.com/blogs/206916/Быстрый-старт-с-дипломом
Diplomi_rmkl
October 31, 2024 @ 3:08 am
купить диплом маркетолога [url=https://landik-diploms.ru/]landik-diploms.ru[/url] .
Lazrddg
October 31, 2024 @ 3:52 am
Всё, что нужно знать о покупке аттестата о среднем образовании
ZSot
October 31, 2024 @ 4:21 am
1win
Derekopeta
October 31, 2024 @ 6:26 am
https://sweetbonanzatr.pro/# sweet bonanza oyna
pinup bet and casino
HenrySpoma
October 31, 2024 @ 6:33 am
Узнай по ссылке :[url=http://www.google.im/url?q=https://b2best.at]bs2site[/url]
Sazrjqj
October 31, 2024 @ 6:41 am
Как правильно купить диплом колледжа и пту в России, подводные камни
Diplomi_vwpn
October 31, 2024 @ 6:52 am
диплом заказать [url=https://arusak-diploms.ru/]arusak-diploms.ru[/url] .
Diplomi_cfSi
October 31, 2024 @ 7:55 am
купить диплом монтажника [url=https://man-diploms.ru/]man-diploms.ru[/url] .
ДизайнЧеловека
October 31, 2024 @ 7:58 am
Как понять, кто ты на самом деле Дизайн человека (Human design) – расчет карты онлайн
ZSot
October 31, 2024 @ 9:05 am
vavada bet
ДизайнЧеловека
October 31, 2024 @ 9:34 am
Печально, но факт: большинство из нас так никогда и не раскроет всех своих способностей и возможностей. Более того, под давлением общественных стереотипов многие отказываются от того, что идеально им подходит. Безусловно, есть люди, искренне разделяющие общепринятые убеждения, однако для многих «правильный» образ жизни – лишь результат фрустрации, обиды или разочарования от бесплодных попыток найти свое место в жизни. Все мы наделены могучим даром созидания, но он бесполезен, пока у нас нет четкого представления о правильном именно для нас способе принимать решения и воплощать их. Тип Дизайна Человека подскажет вам, как действовать креативно, масштабно, динамично и впечатляюще успешно Дизайн Человека подробно
Jasonnet
October 31, 2024 @ 9:35 am
sweet bonanza tr: sweetbonanzatr.pro – sweet bonanza nas?l oynan?r
MichaelMig
October 31, 2024 @ 10:26 am
https://pinup-az.bid/# pin up casino
biznes plan cvetochnogo magazina_zdOn
October 31, 2024 @ 10:46 am
бизнес план цветочного магазина кратко [url=www.cvety-i-bukety.ru/]www.cvety-i-bukety.ru/[/url] .
biznes plan cvetochnogo magazina_fxmt
October 31, 2024 @ 11:02 am
бизнес план магазина цветов готовый [url=http://www.mos-roza.ru]http://www.mos-roza.ru[/url] .
ДизайнЧеловека
October 31, 2024 @ 11:07 am
В школе мы изучаем предметы, которые рассказывают о мире: физику, химию, историю, географию, литературу. Но нет ничего (разве что биология), что рассказывало бы нам о себе и о том, как с миром взаимодействовать «Дизайн человека»
biznes plan cvetochnogo magazina_gcor
October 31, 2024 @ 12:54 pm
бизнес проект цветочного магазина [url=https://www.cvetov-ray.ru]https://www.cvetov-ray.ru[/url] .
Internet marketing Moskva_qzSr
October 31, 2024 @ 1:17 pm
рекламное агентство [url=https://marketing99.ru/]marketing99.ru[/url] .
snyatie lomki narkolog_hyel
October 31, 2024 @ 1:26 pm
снять ломку наркомана [url=snyatie-lomki-narkolog14.ru]снять ломку наркомана[/url] .
download888starz
October 31, 2024 @ 1:53 pm
Играйте прямо сейчас — просто 888starz apk ios скачать
JosephRon
October 31, 2024 @ 3:09 pm
Свежие и смешные видео https://sites.google.com/view/smeshnye-video/%D0%B3%D0%BB%D0%B0%D0%B2%D0%BD%D0%B0%D1%8F-%D1%81%D1%82%D1%80%D0%B0%D0%BD%D0%B8%D1%86%D0%B0.
ДизайнЧеловека
October 31, 2024 @ 3:37 pm
Дизайн Человека – это не статичная система. Это не свод правил, которые должны стать очередной «конституцией» в жизни. Дизайн Человека – это жизненный эксперимент освобождения от обусловленностей, это жизнь в цвете, это принятие не только себя, но и других, что, безусловно, положительным образом скажется на ваших взаимоотношениях с любимыми людьми. И если вы готовы встать на путь храбрости, познакомиться с самими собой и (обязательно!) влюбиться в свою уникальность, эта книга станет надежной опорой в этом удивительном путешествии Дизайн человека (Human design) – расчет карты онлайн
Lazrlnk
October 31, 2024 @ 4:36 pm
Как купить диплом о высшем образовании с минимальными рисками
ДизайнЧеловека
October 31, 2024 @ 4:56 pm
Информация о наиболее важных характеристиках вашего Дизайна. Здесь вы узнаете: • свой тип и уникальную стратегию принятия решений; • способ принимать решения независимо от того, спонтанные они или обдуманные; • эмоциональный настрой – то, какая эмоция доставляет вам больше всего проблем. Карта Дизайна Человека или Бодиграф
ZSot
October 31, 2024 @ 5:45 pm
1win pagina oficial
Derekopeta
October 31, 2024 @ 5:54 pm
https://biznes-fabrika.kz/# пин ап 634
pin up zerkalo
Trefjqe
October 31, 2024 @ 6:53 pm
Можно ли купить аттестат о среднем образовании, основные моменты и вопросы
datasphere.ru/club/user/15/blog/?b24statAction=addLogEntry
ZSot
October 31, 2024 @ 10:15 pm
1win apuestas
Dnrtzhj
October 31, 2024 @ 11:52 pm
Официальная покупка диплома вуза с сокращенной программой в Москве
Diplomi_ysSi
November 1, 2024 @ 1:30 am
Приобретение диплома ПТУ с сокращенной программой обучения в Москве
Jasonnet
November 1, 2024 @ 1:36 am
Пин Ап Казино Официальный Сайт в России: пин ап – пин ап
Michaelvob
November 1, 2024 @ 2:07 am
Смешные приколы, шутки и картинки http://shutki-anekdoty.ru/.
peretyazhka pnKi
November 1, 2024 @ 2:24 am
Как выбрать материал для перетяжки мягкой мебели, чтобы сделать правильный выбор
перетяжка углового дивана [url=https://csalon.ru/]перетяжка углового дивана[/url] .
Лучшие ткани для перетяжки мягкой мебели|Как выбрать мягкую мебель для дома: лучшие ткани|Как перетянуть мягкую мебель своими руками: простые шаги|Мастер-класс по перетяжке мягкой мебели|Как оценить качество ткани для мебели|Сколько стоит перетяжка мягкой мебели: цены и услуги|Как выбрать мастера по перетяжке мебели: советы и рекомендации|Как обновить старый диван: советы по перетяжке|Топ идеи для обновления мебели своими руками|Как преобразить мебель своими руками: перетяжка как способ обновления|Перетяжка кресел и стульев: как сделать качественно|Как самостоятельно перетянуть мебель: пошаговая инструкция|Модернизация интерьера с помощью перетяжки мебели|Советы по подбору цветовой гаммы для мебели|Зачем перетягивать мягкую мебель: плюсы и минусы|Идеи для перетяжки мебели в современном стиле|Что может пойти не так при самостоятельной перетяжке мягкой мебели|Идеи для узоров при обивке мебели: советы по выбору|Как перетянуть мебель: подробная инструкция и советы
snyatie lomki narkolog_dqpr
November 1, 2024 @ 5:23 am
снятие ломки [url=https://snyatie-lomki-narkolog15.ru/]снятие ломки[/url] .
snyatie lomki narkolog_wfEi
November 1, 2024 @ 5:29 am
снять ломку наркозависимого [url=http://www.snyatie-lomki-narkolog16.ru]http://www.snyatie-lomki-narkolog16.ru[/url] .
snyatie lomki narkolog_hfpt
November 1, 2024 @ 5:33 am
медикаментозное снятие ломки [url=snyatie-lomki-narkolog18.ru]snyatie-lomki-narkolog18.ru[/url] .
vivod iz zapoya v stacionare_xcot
November 1, 2024 @ 5:37 am
для вывода из запоя в стационаре [url=www.vyvod-iz-zapoya-v-stacionare-voronezh15.ru/]для вывода из запоя в стационаре[/url] .
snyatie lomki narkolog_ycmi
November 1, 2024 @ 5:47 am
снятие ломки нарколог [url=https://snyatie-lomki-narkolog17.ru/]снятие ломки нарколог[/url] .
Sazrnmp
November 1, 2024 @ 5:59 am
Официальная покупка школьного аттестата с упрощенным обучением в Москве
Derekopeta
November 1, 2024 @ 8:52 am
http://sweetbonanzatr.pro/# sweet bonanza nas?l oynan?r
pin up win
Diplomi_qrOa
November 1, 2024 @ 9:52 am
куплю диплом в нижневартовске [url=https://prema-diploms.ru/]prema-diploms.ru[/url] .
JosephRon
November 1, 2024 @ 10:26 am
Веселые видеоролики https://sites.google.com/view/smeshnye-video/%D0%B3%D0%BB%D0%B0%D0%B2%D0%BD%D0%B0%D1%8F-%D1%81%D1%82%D1%80%D0%B0%D0%BD%D0%B8%D1%86%D0%B0.
Michaelavob
November 1, 2024 @ 10:43 am
Веселые приколы и анекдоты http://shutki-anekdoty.ru.
Diplomi_alEr
November 1, 2024 @ 10:55 am
купить диплом о высшем образовании с занесением в реестр в ижевске [url=https://orik-diploms.ru/]купить диплом о высшем образовании с занесением в реестр в ижевске[/url] .
Casino Site
November 1, 2024 @ 12:33 pm
I know this if off topic but I’m looking into starting my own weblog and was curious what all is required to get setup?
I’m assuming having a blog like yours would cost a pretty penny?
I’m not very internet smart so I’m not 100% positive. Any suggestions or advice would be greatly appreciated.
Thanks
Mazrlis
November 1, 2024 @ 6:07 pm
Как официально приобрести аттестат 11 класса с минимальными затратами времени
Oariorqey
November 1, 2024 @ 8:05 pm
Аттестат 11 класса купить официально с упрощенным обучением в Москве
mobile marathonbet
November 1, 2024 @ 9:11 pm
Больше, чем ставки — это [url=https://t.me/s/zenitbet_com]zenit bet[/url]. Вдохновитесь высокими коэффициентами, беспрерывным доступом и широкой линией событий. Каждый матч теперь может стать вашим триумфом. Заходите и начинайте побеждать!
Холостяк2024
November 1, 2024 @ 10:28 pm
Холостяк 2024 13 сезон
RussellJem
November 1, 2024 @ 11:00 pm
https://stromectol1st.store/# stromectol delivery usa
RussellJem
November 1, 2024 @ 11:11 pm
http://semaglutide.ink/# rybelsus price
RussellJem
November 1, 2024 @ 11:40 pm
http://gabapentin1st.pro/# licensed gabapentin
Tommysnopy
November 1, 2024 @ 11:40 pm
amoxil online: cheap – cheap
JosephRon
November 1, 2024 @ 11:47 pm
Смешные видеоролики https://sites.google.com/view/smeshnye-video/%D0%B3%D0%BB%D0%B0%D0%B2%D0%BD%D0%B0%D1%8F-%D1%81%D1%82%D1%80%D0%B0%D0%BD%D0%B8%D1%86%D0%B0.
RussellJem
November 1, 2024 @ 11:51 pm
http://gabapentin1st.pro/# buy gabapentin
Холостяк2024
November 2, 2024 @ 12:01 am
Холостяк 2024 13 сезон
Ramonbicle
November 2, 2024 @ 12:05 am
http://paxlovid1st.store/# See risks
profi-teh-remont
November 2, 2024 @ 12:07 am
Сервисный центр предлагает ремонт digma plane s7.0 3g ремонт digma plane s7.0 3g
Robertsig
November 2, 2024 @ 12:14 am
amoxil online: amoxil price – amoxil online
RussellJem
November 2, 2024 @ 12:20 am
https://semaglutide.ink/# Specialists
Tommysnopy
November 2, 2024 @ 12:27 am
good price: stromectol store – stromectol delivery usa
RussellJem
November 2, 2024 @ 12:31 am
https://stromectol1st.store/# good price
Michaelavob
November 2, 2024 @ 12:35 am
Смешные приколы и анекдоты http://shutki-anekdoty.ru.
Ramonbicle
November 2, 2024 @ 12:45 am
https://amoxil1st.shop/# amoxil online
Ramonbicle
November 2, 2024 @ 12:45 am
https://paxlovid1st.store/# Pills Paxlovid
Robertsig
November 2, 2024 @ 12:50 am
stromectol best price: good price – stromectol delivery usa
RussellJem
November 2, 2024 @ 12:58 am
https://paxlovid1st.store/# see a healthcare provider
Robertsig
November 2, 2024 @ 1:01 am
paxlovid price: cheapest paxlovid – see a healthcare provider
RussellJem
November 2, 2024 @ 1:06 am
https://paxlovid1st.store/# Visit store
Ramonbicle
November 2, 2024 @ 1:15 am
http://amoxil1st.shop/# amoxil 1st shop
Tommysnopy
November 2, 2024 @ 1:21 am
cheapest: stromectol store – cheapest
Холостяк2024
November 2, 2024 @ 1:24 am
Холостяк 2024 13 сезон
RussellJem
November 2, 2024 @ 1:27 am
https://stromectol1st.store/# good price
Robertsig
November 2, 2024 @ 1:28 am
paxlovid1st: Pills Paxlovid – Visit store
Robertsig
November 2, 2024 @ 1:38 am
shop: amoxil online – top-rated pills
Ramonbicle
November 2, 2024 @ 1:44 am
http://paxlovid1st.store/# See risks
Ramonbicle
November 2, 2024 @ 1:44 am
https://amoxil1st.shop/# buy amoxil online
RussellJem
November 2, 2024 @ 1:57 am
http://paxlovid1st.store/# paxlovid store
Robertsig
November 2, 2024 @ 2:05 am
involves multisystem: buy gabapentin – buy gabapentin
Tommysnopy
November 2, 2024 @ 2:10 am
compare the best prices: buy gabapentin – gabapentin best price
Robertsig
November 2, 2024 @ 2:16 am
Neurontin online: compare the best prices – compare the best prices
RussellJem
November 2, 2024 @ 2:28 am
https://semaglutide.ink/# Regenerative Medicine
RussellJem
November 2, 2024 @ 2:36 am
http://paxlovid1st.store/# Visit store
Vivod iz zapoya v stacionare_mtsi
November 2, 2024 @ 2:39 am
вывод из запоя нижний новгород стационар [url=www.vyvod-iz-zapoya-v-stacionare15.ru/]вывод из запоя нижний новгород стационар[/url] .
vivod iz zapoya v stacionare_jesa
November 2, 2024 @ 2:41 am
вывод из запоя в клинике [url=https://vyvod-iz-zapoya-v-stacionare-voronezh17.ru]вывод из запоя в клинике [/url] .
Ramonbicle
November 2, 2024 @ 2:44 am
https://stromectol1st.store/# stromectol delivery usa
Robertsig
November 2, 2024 @ 2:54 am
amoxil price: top-rated pills – amoxil
Vivod iz zapoya v stacionare_pzka
November 2, 2024 @ 2:56 am
вывод из запоя в стационаре нижний новгород [url=https://www.vyvod-iz-zapoya-v-stacionare14.ru]вывод из запоя в стационаре нижний новгород[/url] .
RussellJem
November 2, 2024 @ 2:58 am
https://semaglutide.ink/# semaglutide
Dnrtury
November 2, 2024 @ 3:09 am
Сколько стоит диплом высшего и среднего образования и как его получить?
Ramonbicle
November 2, 2024 @ 3:14 am
https://semaglutide.ink/# semaglutide
Robertsig
November 2, 2024 @ 3:22 am
rybelsus price: Patient Portal – Urgent Specialists
RussellJem
November 2, 2024 @ 3:28 am
http://paxlovid1st.store/# See risks
Robertsig
November 2, 2024 @ 3:33 am
buy gabapentin: Neurontin online – compare the best prices
RussellJem
November 2, 2024 @ 3:36 am
https://amoxil1st.shop/# shop
Ramonbicle
November 2, 2024 @ 3:43 am
http://paxlovid1st.store/# see a healthcare provider
Ramonbicle
November 2, 2024 @ 3:44 am
https://gabapentin1st.pro/# gabapentin pro
RussellJem
November 2, 2024 @ 3:58 am
https://paxlovid1st.store/# paxlovid1st
Robertsig
November 2, 2024 @ 3:59 am
licensed gabapentin: gabapentin pro – same-day delivery
Sazrnqn
November 2, 2024 @ 4:02 am
Официальная покупка аттестата о среднем образовании в Москве и других городах
Robertsig
November 2, 2024 @ 4:10 am
paxlovid store: paxlovid store – paxlovid price
Ramonbicle
November 2, 2024 @ 4:13 am
https://semaglutide.ink/# semaglutide
RussellJem
November 2, 2024 @ 4:28 am
https://semaglutide.ink/# Rybelsus
Ramonbicle
November 2, 2024 @ 4:42 am
https://gabapentin1st.pro/# gabapentin pro
Robertsig
November 2, 2024 @ 4:48 am
Regenerative Medicine: semaglutide online – Patient Portal
RussellJem
November 2, 2024 @ 4:58 am
https://gabapentin1st.pro/# same-day delivery
Tommysnopy
November 2, 2024 @ 5:11 am
see a healthcare provider: see a healthcare provider – See risks
Ramonbicle
November 2, 2024 @ 5:11 am
https://gabapentin1st.pro/# gabapentin pro
Diplomi_iwkl
November 2, 2024 @ 5:12 am
купить диплом о высшем юридическом образовании [url=https://landik-diploms.ru/]купить диплом о высшем юридическом образовании[/url] .
Robertsig
November 2, 2024 @ 5:15 am
amoxil 1st shop: cheap – amoxil 1st shop
Robertsig
November 2, 2024 @ 5:25 am
Neurontin online: Neurontin online – Neurontin online
RussellJem
November 2, 2024 @ 5:27 am
http://semaglutide.ink/# Patient Portal
RussellJem
November 2, 2024 @ 5:57 am
http://gabapentin1st.pro/# compare the best prices
Robertsig
November 2, 2024 @ 6:03 am
involves multisystem: buy gabapentin – Care provides
Ramonbicle
November 2, 2024 @ 6:10 am
https://semaglutide.ink/# Rybelsus
RussellJem
November 2, 2024 @ 6:27 am
https://stromectol1st.store/# stromectol
Robertsig
November 2, 2024 @ 6:31 am
stromectol delivery usa: find bets price – good price
Ramonbicle
November 2, 2024 @ 6:40 am
https://semaglutide.ink/# Specialists
Robertsig
November 2, 2024 @ 6:42 am
Compare Prices: amoxil price – amoxil 1st shop
RussellJem
November 2, 2024 @ 6:58 am
http://semaglutide.ink/# Regenerative Medicine
Robertsig
November 2, 2024 @ 7:20 am
see a healthcare provider: paxlovid store – paxlovid store
Sazrzem
November 2, 2024 @ 7:23 am
Можно ли купить аттестат о среднем образовании, основные моменты и вопросы
RussellJem
November 2, 2024 @ 7:28 am
https://gabapentin1st.pro/# compare the best prices
RussellJem
November 2, 2024 @ 7:37 am
https://semaglutide.ink/# rybelsus price
Robertsig
November 2, 2024 @ 7:59 am
Patient Portal: Regenerative Medicine – Specialists
RussellJem
November 2, 2024 @ 7:59 am
https://amoxil1st.shop/# amoxil online
Robertsig
November 2, 2024 @ 8:28 am
top-rated pills: amoxil – amoxil online
RussellJem
November 2, 2024 @ 8:32 am
http://amoxil1st.shop/# shop
Hex
November 2, 2024 @ 8:40 am
Цветочная мастерская собирает под заказ цветы в коробках с доставкой от 1 часа в с Одинцово
Авторские цветы: [url=https://dostavka-cvetov-odincovo.ru/]https://dostavka-cvetov-odincovo.ru/[/url]
Robertsig
November 2, 2024 @ 8:40 am
Neurontin online: buy gabapentin – gabapentin best price
RussellJem
November 2, 2024 @ 8:42 am
https://gabapentin1st.pro/# Care provides
Ramonbicle
November 2, 2024 @ 8:45 am
https://paxlovid1st.store/# see a healthcare provider
nakrutkapf
November 2, 2024 @ 8:47 am
Оптимизируйте сайт с помощью курса по накрутке ПФ для продвижения в Яндексе.
RussellJem
November 2, 2024 @ 9:06 am
https://amoxil1st.shop/# amoxil price
Robertsig
November 2, 2024 @ 9:09 am
paxlovid store: paxlovid price – paxlovid1st
Robertsig
November 2, 2024 @ 9:21 am
amoxil price: Compare Prices – cheap
RussellJem
November 2, 2024 @ 9:38 am
https://amoxil1st.shop/# amoxil price
Ramonbicle
November 2, 2024 @ 9:49 am
http://stromectol1st.store/# stromectol best price
Ramonbicle
November 2, 2024 @ 9:50 am
https://paxlovid1st.store/# Pills Paxlovid
JosephsRon
November 2, 2024 @ 9:53 am
Смешные анекдоты https://sites.google.com/view/smeshnye-video/anekdoty
Robertsig
November 2, 2024 @ 10:02 am
stromectol online: stromectol – stromectol best price
RussellJem
November 2, 2024 @ 10:11 am
http://paxlovid1st.store/# cheapest paxlovid
vivod iz zapoya v stacionare_zuOl
November 2, 2024 @ 10:17 am
вывод из запоя в стационаре [url=http://vyvod-iz-zapoya-v-stacionare-voronezh16.ru/]вывод из запоя в стационаре[/url] .
RussellJem
November 2, 2024 @ 10:20 am
https://stromectol1st.store/# find bets price
Ramonbicle
November 2, 2024 @ 10:22 am
http://stromectol1st.store/# good price
Robertsig
November 2, 2024 @ 10:43 am
stromectol: good price – stromectol online
RussellJem
November 2, 2024 @ 10:45 am
http://amoxil1st.shop/# buy amoxil online
RussellJem
November 2, 2024 @ 10:54 am
http://stromectol1st.store/# stromectol
Ramonbicle
November 2, 2024 @ 10:55 am
https://amoxil1st.shop/# amoxil price
Diplomi_ntea
November 2, 2024 @ 10:57 am
возможно ли купить аттестат [url=https://server-diploms.ru/]возможно ли купить аттестат[/url] .
Lazrjfi
November 2, 2024 @ 11:08 am
Официальная покупка школьного аттестата с упрощенным обучением в Москве
RussellJem
November 2, 2024 @ 11:18 am
http://semaglutide.ink/# Specialists
Robertsig
November 2, 2024 @ 11:25 am
buy amoxil online: Compare Prices – buy amoxil online
RussellJem
November 2, 2024 @ 11:28 am
https://gabapentin1st.pro/# same-day delivery
этот
November 2, 2024 @ 11:35 am
Excellent site you have here.. It’s difficult to find
high quality writing like yours nowadays.
I seriously appreciate individuals like you!
Take care!!
RussellJem
November 2, 2024 @ 11:53 am
https://semaglutide.ink/# semaglutide online
Robertsig
November 2, 2024 @ 11:56 am
rybelsus price: Rybelsus – semaglutide
RussellJem
November 2, 2024 @ 12:02 pm
http://stromectol1st.store/# stromectol store
Ramonbicle
November 2, 2024 @ 12:02 pm
http://gabapentin1st.pro/# buy gabapentin
Robertsig
November 2, 2024 @ 12:08 pm
stromectol: good price – stromectol online
RussellJem
November 2, 2024 @ 12:27 pm
https://semaglutide.ink/# semaglutide
Ramonbicle
November 2, 2024 @ 12:36 pm
https://stromectol1st.store/# find bets price
Robertsig
November 2, 2024 @ 12:50 pm
Urgent Specialists: Rybelsus – rybelsus price
RussellJem
November 2, 2024 @ 1:02 pm
http://amoxil1st.shop/# shop
Ramonbicle
November 2, 2024 @ 1:09 pm
http://paxlovid1st.store/# see a healthcare provider
RussellJem
November 2, 2024 @ 1:11 pm
https://gabapentin1st.pro/# gabapentin best price
Robertsig
November 2, 2024 @ 1:22 pm
stromectol best price: find bets price – cheapest
Robertsig
November 2, 2024 @ 1:34 pm
stromectol best price: stromectol delivery usa – stromectol best price
RussellJem
November 2, 2024 @ 1:37 pm
https://paxlovid1st.store/# Visit store
RussellJem
November 2, 2024 @ 1:47 pm
https://semaglutide.ink/# Urgent Specialists
Robertsig
November 2, 2024 @ 2:06 pm
stromectol store: cheapest – find bets price
RussellJem
November 2, 2024 @ 2:12 pm
https://paxlovid1st.store/# paxlovid1st
Ramonbicle
November 2, 2024 @ 2:18 pm
https://stromectol1st.store/# stromectol online
Ramonbicle
November 2, 2024 @ 2:18 pm
https://amoxil1st.shop/# buy amoxil online
Robertsig
November 2, 2024 @ 2:18 pm
semaglutide: Regenerative Medicine – rybelsus price
RussellJem
November 2, 2024 @ 2:22 pm
http://semaglutide.ink/# semaglutide online
RussellJem
November 2, 2024 @ 2:48 pm
https://amoxil1st.shop/# top-rated pills
Robertsig
November 2, 2024 @ 2:50 pm
buy gabapentin: compare the best prices – same-day delivery
RussellJem
November 2, 2024 @ 2:58 pm
https://stromectol1st.store/# stromectol store
Robertsig
November 2, 2024 @ 3:03 pm
top-rated pills: Compare Prices – buy amoxil online
Sazraxi
November 2, 2024 @ 3:16 pm
Быстрая покупка диплома старого образца: возможные риски
honey.ukrbb.net/viewtopic.php?f=18&t=16163
Williamfog
November 2, 2024 @ 3:21 pm
Decouvrez notre selection de
Bloc couteaux
pour la maison
RussellJem
November 2, 2024 @ 3:23 pm
http://paxlovid1st.store/# paxlovid1st
Ramonbicle
November 2, 2024 @ 3:28 pm
http://stromectol1st.store/# stromectol delivery usa
Robertsig
November 2, 2024 @ 3:35 pm
paxlovid1st: paxlovid store – see a healthcare provider
Robertsig
November 2, 2024 @ 3:47 pm
shop: amoxil – top-rated pills
RussellJem
November 2, 2024 @ 3:58 pm
http://stromectol1st.store/# cheapest
RussellJem
November 2, 2024 @ 4:07 pm
https://gabapentin1st.pro/# gabapentin pro
Robertsig
November 2, 2024 @ 4:20 pm
Regenerative Medicine: Patient Portal – Urgent Specialists
Robertsig
November 2, 2024 @ 4:32 pm
paxlovid1st: see a healthcare provider – Pills Paxlovid
RussellJem
November 2, 2024 @ 4:33 pm
https://paxlovid1st.store/# paxlovid1st
Ramonbicle
November 2, 2024 @ 4:38 pm
https://gabapentin1st.pro/# gabapentin best price
RussellJem
November 2, 2024 @ 4:43 pm
https://gabapentin1st.pro/# same-day delivery
Robertsig
November 2, 2024 @ 5:05 pm
Pills Paxlovid: see a healthcare provider – see a healthcare provider
RussellJem
November 2, 2024 @ 5:10 pm
https://paxlovid1st.store/# cheapest paxlovid
Ramonbicle
November 2, 2024 @ 5:13 pm
https://paxlovid1st.store/# Pills Paxlovid
Ramonbicle
November 2, 2024 @ 5:14 pm
http://paxlovid1st.store/# Pills Paxlovid
Robertsig
November 2, 2024 @ 5:17 pm
buy gabapentin: same-day delivery – buy gabapentin
RussellJem
November 2, 2024 @ 5:19 pm
https://semaglutide.ink/# semaglutide online
Williamfog
November 2, 2024 @ 5:38 pm
Trouvez votre parfait
Bloc couteaux
pour votre collection
RussellJem
November 2, 2024 @ 5:46 pm
https://stromectol1st.store/# stromectol online
Ramonbicle
November 2, 2024 @ 5:50 pm
https://semaglutide.ink/# semaglutide online
Robertsig
November 2, 2024 @ 5:51 pm
good price: cheapest – stromectol delivery usa
RussellJem
November 2, 2024 @ 5:56 pm
https://stromectol1st.store/# stromectol best price
Robertsig
November 2, 2024 @ 6:04 pm
stromectol store: stromectol online – stromectol store
RussellJem
November 2, 2024 @ 6:24 pm
https://paxlovid1st.store/# cheapest paxlovid
Michaelavob
November 2, 2024 @ 6:26 pm
Свежие анекдоты http://shutki-anekdoty.ru/anekdoty
Ramonbicle
November 2, 2024 @ 6:26 pm
http://paxlovid1st.store/# Pills Paxlovid
Ramonbicle
November 2, 2024 @ 6:27 pm
https://semaglutide.ink/# rybelsus price
RussellJem
November 2, 2024 @ 6:34 pm
http://amoxil1st.shop/# amoxil 1st shop
Robertsig
November 2, 2024 @ 6:51 pm
shop: top-rated pills – amoxil online
RussellJem
November 2, 2024 @ 7:03 pm
https://stromectol1st.store/# cheapest
RussellJem
November 2, 2024 @ 7:15 pm
https://paxlovid1st.store/# Pills Paxlovid
Sazrrav
November 2, 2024 @ 7:23 pm
Как получить диплом техникума с упрощенным обучением в Москве официально
Robertsig
November 2, 2024 @ 7:28 pm
amoxil: buy amoxil online – amoxil online
vivod iz zapoya v stacionare_oiPt
November 2, 2024 @ 7:29 pm
вывод из запоя самара стационар [url=http://vyvod-iz-zapoya-v-stacionare-samara15.ru]вывод из запоя самара стационар[/url] .
vivod iz zapoya v stacionare_tqKl
November 2, 2024 @ 7:34 pm
вывод из запоя стационар [url=http://vyvod-iz-zapoya-v-stacionare-samara17.ru]вывод из запоя стационар[/url] .
vivod iz zapoya v stacionare_bpon
November 2, 2024 @ 7:37 pm
быстрый вывод из запоя в стационаре [url=https://vyvod-iz-zapoya-v-stacionare-samara16.ru/]быстрый вывод из запоя в стационаре[/url] .
vivod iz zapoya v stacionare_brea
November 2, 2024 @ 7:38 pm
лечения наркозависимости в стационаре [url=http://vyvod-iz-zapoya-v-stacionare-samara14.ru]лечения наркозависимости в стационаре[/url] .
Robertsig
November 2, 2024 @ 7:43 pm
see a healthcare provider: cheapest paxlovid – paxlovid store
RussellJem
November 2, 2024 @ 7:45 pm
https://stromectol1st.store/# stromectol best price
RussellJem
November 2, 2024 @ 7:57 pm
https://stromectol1st.store/# stromectol best price
Ramonbicle
November 2, 2024 @ 8:22 pm
https://amoxil1st.shop/# cheap
Ramonbicle
November 2, 2024 @ 8:23 pm
http://paxlovid1st.store/# paxlovid store
RussellJem
November 2, 2024 @ 8:28 pm
https://gabapentin1st.pro/# licensed gabapentin
Robertsig
November 2, 2024 @ 8:36 pm
Neurontin online: Care provides – same-day delivery
Cazrxnz
November 2, 2024 @ 8:36 pm
Полезные советы по безопасной покупке диплома о высшем образовании
RussellJem
November 2, 2024 @ 8:41 pm
https://gabapentin1st.pro/# gabapentin best price
RussellJem
November 2, 2024 @ 9:13 pm
https://semaglutide.ink/# semaglutide
Lazrxrg
November 2, 2024 @ 10:22 pm
Сколько стоит диплом высшего и среднего образования и как это происходит?
Diplomi_kcSi
November 2, 2024 @ 10:38 pm
купить официальный диплом о среднем образовании [url=https://man-diploms.ru/]купить официальный диплом о среднем образовании[/url] .
Xazrsqx
November 2, 2024 @ 11:31 pm
Купить диплом магистра оказалось возможно, быстрое обучение и диплом на руки
JosephsRon
November 2, 2024 @ 11:52 pm
Прикольные анекдоты https://sites.google.com/view/smeshnye-video/anekdoty
RussellJem
November 3, 2024 @ 12:35 am
https://stromectol1st.store/# stromectol online
Sazrwmd
November 3, 2024 @ 1:20 am
Приобретение школьного аттестата с официальным упрощенным обучением в Москве
cartagena.activeboard.com/forum.spark
Volna
November 3, 2024 @ 1:26 am
[url=https://volna-casino-apk.ru]http://volna-casino-apk.ru[/url]
Загрузить последнюю версию приложения казино онлайн Volna – играй прямо сейчас!
https://www.volna-casino-apk.ru
VolnaApk
November 3, 2024 @ 1:04 am
[url=https://volna-casino-apk.ru/volna-apk/]https://volna-casino-apk.ru/volna-apk/[/url]
Скачать последнюю версию приложения казино Волна – играй прямо сейчас!
http://volna-casino-apk.ru/volna-apk/
Diplomi_lkSr
November 3, 2024 @ 1:18 am
купить диплом в иваново [url=https://diplomdarom.ru/]diplomdarom.ru[/url] .
Trefuvw
November 3, 2024 @ 1:34 am
Как приобрести аттестат о среднем образовании в Москве и других городах
dvasadovoda.ru/forum/user/9362
Diplomi_dukl
November 3, 2024 @ 2:03 am
купить диплом в москве недорого [url=https://realdiplom.ru/]купить диплом в москве недорого[/url] .
Kapelnica ot zapoya kolomna_aeSl
November 3, 2024 @ 2:52 am
состав капельницы для вывода из запоя [url=https://www.kapelnica-ot-zapoya-kolomna14.ru]https://www.kapelnica-ot-zapoya-kolomna14.ru[/url] .
Lazrony
November 3, 2024 @ 3:43 am
Реально ли приобрести диплом стоматолога? Основные этапы
californiarpn2.listbb.ru/viewtopic.php?f=1&t=851
Ramonbicle
November 3, 2024 @ 5:48 am
https://gabapentin1st.pro/# gabapentin best price
Michaelavob
November 3, 2024 @ 5:56 am
Смешные анекдоты http://shutki-anekdoty.ru/anekdoty
betislot.com.ua
November 3, 2024 @ 6:39 am
[url=https://betislot.com.ua/ru/ggbet/]ггбет[/url] — это больше, чем просто платформа для ставок! Это ваш надёжный партнёр в мире азартных игр. Здесь вы найдёте всё, что нужно для комфортной игры, — от удобного интерфейса до впечатляющих бонусов. Попробуйте и убедитесь сами!
Nikejuh
November 3, 2024 @ 7:39 am
dark web market list darknet markets 2024 dark web markets
JanetLig
November 3, 2024 @ 7:43 am
Благодарю за статью!
Мы предлагаем [url=https://fod38.ru/]фабрика окон и дверей иркутск[/url], которые значительно повысят ваш уровень комфорта. Наша продукция отвечает всем требуемым норма, обеспечивая надежную защиту от прохлады и шумов. За счет многокамерности и энергоэффективным стеклопакетам, вы заметно снизите траты на отопление!
Каждое окно производится с учетом индивидуальных потребностей наших клиентов. Мы даем гарантию на высокое качество и долговечность нашей продукции — все окна имеют сертификаты качесьва.
Первоклассный монтаж от наших спецов гарантирует безкомпромиссную герметичность и надежность. А наше обслуживание сделает использование окон беззаботным!
Пластиковые окна от фабрики окон и дверей в Иркутске — ваш комфорт и спокойствие!
RussellJem
November 3, 2024 @ 10:08 am
http://paxlovid1st.store/# paxlovid store
Robertsig
November 3, 2024 @ 10:46 am
licensed gabapentin: Care provides – Care provides
Sazrzvv
November 3, 2024 @ 10:51 am
Вопросы и ответы: можно ли быстро купить диплом старого образца?
Фильмы485258
November 3, 2024 @ 11:47 am
https://t.me/s/kino_film_serial_online_telegram 768740 лучших фильмов. Фильмы смотреть онлайн. В нашем онлайн-кинотеатре есть новинки кино и бесплатные фильмы самых разных жанров
CasinoVolna
November 3, 2024 @ 12:55 pm
[url=https://volna-casino-apk.ru/volna-casino/]www.volna-casino-apk.ru/volna-casino/[/url]
Установить приложение онлайн казино Волна – выигрывай сегодня!
http://volna-casino-apk.ru/volna-casino/
colegio de lenguaje
November 3, 2024 @ 1:02 pm
I’ve been browsing on-line more than 3 hours today, yet I by no means
found any fascinating article like yours. It is lovely value enough for me.
In my view, if all webmasters and bloggers made excellent
content as you probably did, the net might be a lot more useful than ever before.
Фильмы525907
November 3, 2024 @ 1:24 pm
https://t.me/s/kino_film_serial_online_telegram 116995 лучших фильмов. Фильмы смотреть онлайн. В нашем онлайн-кинотеатре есть новинки кино и бесплатные фильмы самых разных жанров
Sazraxq
November 3, 2024 @ 2:14 pm
Как безопасно купить диплом колледжа или ПТУ в России, что важно знать
Bradleyemaky
November 3, 2024 @ 2:24 pm
компания сео продвижение и реклама сайта
Кино111813
November 3, 2024 @ 2:51 pm
https://t.me/s/kino_film_serial_online_telegram 110332 лучших фильмов. Фильмы смотреть онлайн. В нашем онлайн-кинотеатре есть новинки кино и бесплатные фильмы самых разных жанров
Bradleyemaky
November 3, 2024 @ 5:02 pm
продвижение сайтов агентство продвижение сайта в москве
HerbertArome
November 3, 2024 @ 5:13 pm
Сайт приколов http://humor-kartinki.ru собрание лучших мемов, шуток и смешных видео, чтобы ваш день был ярче. Ежедневное обновление контента для вашего настроения. Легко находите и делитесь забавными моментами с друзьями.
Diplomi_cgEr
November 3, 2024 @ 5:41 pm
купить диплом старого образца [url=https://russa-diploms.ru/]купить диплом старого образца[/url] .
skoraya narkologicheskaya pomosh Moskva_fmoa
November 3, 2024 @ 5:49 pm
наркологическая скорая в москве [url=http://dexamethazon.ru]наркологическая скорая в москве[/url] .
Kapelnica ot zapoya kolomna_ryMl
November 3, 2024 @ 5:53 pm
стоимость капельницы от похмелья [url=http://www.kapelnica-ot-zapoya-kolomna16.ru]стоимость капельницы от похмелья[/url] .
skoraya narkologicheskaya pomosh Moskva_xrsr
November 3, 2024 @ 6:09 pm
платная наркологическая помощь [url=https://www.skoraya-narkologicheskaya-pomoshch-moskva.ru]платная наркологическая помощь[/url] .
Kapelnica ot zapoya kolomna_oemn
November 3, 2024 @ 6:11 pm
сделать капельницу от запоя [url=https://kapelnica-ot-zapoya-kolomna15.ru/]kapelnica-ot-zapoya-kolomna15.ru[/url] .
skoraya narkologicheskaya pomosh Moskva_yzmt
November 3, 2024 @ 6:16 pm
как вызвать наркологическую бригаду [url=skoraya-narkologicheskaya-pomoshch-moskva11.ru]skoraya-narkologicheskaya-pomoshch-moskva11.ru[/url] .
Volna
November 3, 2024 @ 6:17 pm
[url=https://volna-casino-apk.ru]volna-casino-apk.ru[/url]
Установить apk файл казино онлайн Волна – выигрывай сегодня!
https://www.volna-casino-apk.ru
VolnaApk
November 3, 2024 @ 6:17 pm
[url=https://volna-casino-apk.ru/volna-apk/]http://volna-casino-apk.ru/volna-apk/[/url]
Загрузить apk файл казино онлайн Волна – выигрывай сегодня!
volna-casino-apk.ru/volna-apk/
Фильмы822503
November 3, 2024 @ 6:42 pm
https://t.me/s/kino_film_serial_online_telegram 328911 лучших фильмов. Фильмы смотреть онлайн. В нашем онлайн-кинотеатре есть новинки кино и бесплатные фильмы самых разных жанров
VolnaSait
November 3, 2024 @ 6:44 pm
[url=https://volna-casino-apk.ru/volna-oficzialnyj-sajt/]https://www.volna-casino-apk.ru/volna-oficzialnyj-sajt/[/url]
Скачать apk файл казино Волна – выигрывай сегодня!
https://volna-casino-apk.ru/volna-oficzialnyj-sajt/
VolnaAndroid
November 3, 2024 @ 7:52 pm
[url=https://volna-casino-apk.ru/volna-na-androjd/]www.volna-casino-apk.ru/volna-na-androjd/[/url]
Скачать apk файл казино Волна – выигрывай сейчас!
https://www.volna-casino-apk.ru/volna-na-androjd/
Кино862688
November 3, 2024 @ 7:55 pm
https://t.me/s/kino_film_serial_online_telegram 617939 лучших фильмов. Фильмы смотреть онлайн. В нашем онлайн-кинотеатре есть новинки кино и бесплатные фильмы самых разных жанров
RussellJem
November 3, 2024 @ 8:26 pm
http://gabapentin1st.pro/# Neurontin online
профи тех ремонт
November 3, 2024 @ 9:07 pm
Сервисный центр предлагает ремонт варочных панелей miele на дому центр ремонта варочной панели miele
Oarioruiv
November 4, 2024 @ 12:20 am
Сколько стоит диплом высшего и среднего образования и как его получить?
Ramonbicle
November 4, 2024 @ 12:57 am
https://semaglutide.ink/# Regenerative Medicine
Diplomi_mhOa
November 4, 2024 @ 12:58 am
диплом о высшем образовании старого образца купить [url=https://prema-diploms.ru/]диплом о высшем образовании старого образца купить[/url] .
Bradleyemaky
November 4, 2024 @ 1:15 am
продвижение сайта в москве https://is-market.ru
Robertsig
November 4, 2024 @ 1:17 am
amoxil 1st shop: buy amoxil online – buy amoxil online
Michaelavob
November 4, 2024 @ 6:02 am
Прикольные анекдоты http://shutki-anekdoty.ru/anekdoty
Mazrsej
November 4, 2024 @ 6:06 am
Реально ли приобрести диплом стоматолога? Основные этапы
HerbertArome
November 4, 2024 @ 7:08 am
Сайт приколов https://www.dermandar.com/user/billybons собрание лучших мемов, шуток и смешных видео, чтобы ваш день был ярче. Ежедневное обновление контента для вашего настроения. Легко находите и делитесь забавными моментами с друзьями.
WallaceRut
November 4, 2024 @ 7:14 am
http://indianpharm24.pro/# best india pharmacy
ed pills online pharmacy
Спорт и ставки
November 4, 2024 @ 7:18 am
Быстрый и простой доступ через [url=https://betislot.com.ua/ru/favbet/]favbet вход[/url] позволяет вам моментально войти в личный кабинет и начать игру. Зарегистрируйтесь и получите доступ к бонусам и акционным предложениям для новых игроков.
WallaceRut
November 4, 2024 @ 7:30 am
https://pharm24.pro/# best ed supplements
errection problem cure
WallaceRut
November 4, 2024 @ 8:35 am
https://pharm24.pro/# best natural ed treatment
ed medicines
WallaceRut
November 4, 2024 @ 8:51 am
http://mexicanpharm24.cheap/# buying prescription drugs in mexico
erectile dysfunction pills
DavidDAX
November 4, 2024 @ 8:57 am
erectyle disfunction: cheap drugs – men with ed
DavidDAX
November 4, 2024 @ 9:07 am
п»їbest mexican online pharmacies: Legit online Mexican pharmacy – best online pharmacies in mexico
WallaceRut
November 4, 2024 @ 9:08 am
https://indianpharm24.pro/# pharmacy website india
remedies for ed
Diplomi_edEr
November 4, 2024 @ 9:16 am
купить диплом проведенный [url=https://orik-diploms.ru/]купить диплом проведенный[/url] .
WallaceRut
November 4, 2024 @ 9:24 am
http://pharm24.pro/# legal to buy prescription drugs from canada
pet meds without vet prescription
Iarioriqj
November 4, 2024 @ 9:32 am
Можно ли купить аттестат о среднем образовании? Основные рекомендации
DavidDAX
November 4, 2024 @ 9:35 am
buying prescription drugs in mexico online: mexico pharmacy cheap – mexican border pharmacies shipping to usa
WallaceRut
November 4, 2024 @ 9:40 am
https://mexicanpharm24.cheap/# purple pharmacy mexico price list
shots for ed
DavidDAX
November 4, 2024 @ 9:46 am
drugs and medications: cheap pharmacy online – viagra without a doctor prescription
WallaceRut
November 4, 2024 @ 9:58 am
http://indianpharm24.pro/# top online pharmacy india
canadian drugs online
DavidDAX
November 4, 2024 @ 10:15 am
india pharmacy mail order: Indian pharmacy online – online pharmacy india
DavidDAX
November 4, 2024 @ 10:25 am
ed drugs online: cheap drugs online – ed meds online without prescription or membership
WallaceRut
November 4, 2024 @ 10:31 am
https://mexicanpharm24.cheap/# mexico drug stores pharmacies
buy prescription drugs without doctor
WallaceRut
November 4, 2024 @ 10:47 am
https://mexicanpharm24.cheap/# mexican online pharmacies prescription drugs
prescription drugs
DavidDAX
November 4, 2024 @ 11:05 am
mexican online pharmacies prescription drugs: mexico pharmacy – mexican drugstore online
WallaceRut
November 4, 2024 @ 11:06 am
https://pharm24.pro/# how to fix ed
ed vacuum pump
WallaceRut
November 4, 2024 @ 11:41 am
http://pharm24.pro/# ed meds online
ed trial pack
DavidDAX
November 4, 2024 @ 11:47 am
ed meds online without doctor prescription: buy drugs – ed meds
WallaceRut
November 4, 2024 @ 11:58 am
https://mexicanpharm24.cheap/# mexico drug stores pharmacies
treating ed
WallaceRut
November 4, 2024 @ 12:17 pm
http://pharm24.pro/# ed pills online pharmacy
canadian online pharmacy
DavidDAX
November 4, 2024 @ 12:18 pm
indian pharmacy online: India pharmacy delivery – Online medicine home delivery
DavidDAX
November 4, 2024 @ 12:29 pm
online drugstore: buy drugs – pills erectile dysfunction
WallaceRut
November 4, 2024 @ 12:35 pm
http://indianpharm24.pro/# buy medicines online in india
best online drugstore
WallaceRut
November 4, 2024 @ 12:54 pm
http://mexicanpharm24.cheap/# mexican mail order pharmacies
prescription drugs
DavidDAX
November 4, 2024 @ 1:01 pm
india pharmacy mail order: Indian pharmacy international shipping – top 10 online pharmacy in india
WallaceRut
November 4, 2024 @ 1:11 pm
http://indianpharm24.pro/# buy medicines online in india
pump for ed
WallaceRut
November 4, 2024 @ 1:29 pm
https://indianpharm24.pro/# india online pharmacy
injectable ed drugs
Lazrhdk
November 4, 2024 @ 1:32 pm
Как приобрести диплом техникума с минимальными рисками
DavidDAX
November 4, 2024 @ 1:43 pm
over the counter ed drugs: low cost pharmacy – ed aids
WallaceRut
November 4, 2024 @ 1:47 pm
http://mexicanpharm24.cheap/# mexico drug stores pharmacies
online prescription for ed meds
Williamraics
November 4, 2024 @ 1:54 pm
Как получить диплом техникума официально и без лишних проблем
WallaceRut
November 4, 2024 @ 2:06 pm
https://pharm24.pro/# cheap ed pills
ed pharmacy
WallaceRut
November 4, 2024 @ 2:24 pm
https://mexicanpharm24.cheap/# medicine in mexico pharmacies
ways to treat erectile dysfunction
DavidDAX
November 4, 2024 @ 2:27 pm
natural treatment for ed: buy drugs – ed treatment
DavidDAX
November 4, 2024 @ 2:38 pm
ed treatment drugs: affordable medication – best pharmacy online
WallaceRut
November 4, 2024 @ 2:43 pm
http://mexicanpharm24.cheap/# п»їbest mexican online pharmacies
pet meds without vet prescription
WallaceRut
November 4, 2024 @ 3:01 pm
https://mexicanpharm24.cheap/# mexico drug stores pharmacies
best price for generic viagra on the internet
WallaceRut
November 4, 2024 @ 3:20 pm
https://indianpharm24.pro/# reputable indian pharmacies
erectile dysfunction medications
WallaceRut
November 4, 2024 @ 3:42 pm
https://pharm24.pro/# best ed pills at gnc
viagra without doctor prescription
WallaceRut
November 4, 2024 @ 4:05 pm
http://mexicanpharm24.cheap/# mexican drugstore online
natural ed
Sazrnvn
November 4, 2024 @ 4:24 pm
Как получить диплом стоматолога быстро и официально
WallaceRut
November 4, 2024 @ 4:28 pm
http://indianpharm24.pro/# Online medicine order
drugs for ed
WallaceRut
November 4, 2024 @ 4:50 pm
https://indianpharm24.pro/# mail order pharmacy india
drugs causing ed
WallaceRut
November 4, 2024 @ 5:14 pm
https://mexicanpharm24.cheap/# mexican online pharmacies prescription drugs
pet meds without vet prescription canada
DavidDAX
November 4, 2024 @ 5:20 pm
mexican pharmaceuticals online: Mexican pharmacy ship US – mexican mail order pharmacies
WallaceRut
November 4, 2024 @ 5:38 pm
https://mexicanpharm24.cheap/# п»їbest mexican online pharmacies
erectile dysfunction drugs
zenitbet.guru
November 4, 2024 @ 5:54 pm
Ищете надежное место для ставок? [url=https://zenitbet.guru/ru/leonbets/]бк леон официальный сайт[/url] — ваш идеальный выбор! Эта букмекерская контора предлагает высокие коэффициенты на все спортивные события и множественные акции, которые не оставят равнодушными ни одного игрока. Присоединяйтесь к Leon и погружайтесь в захватывающий мир ставок на спорт. Удача уже на вашей стороне!
WallaceRut
November 4, 2024 @ 5:58 pm
https://indianpharm24.pro/# buy prescription drugs from india
errection problems
Sazrucw
November 4, 2024 @ 6:12 pm
Официальная покупка диплома вуза с сокращенной программой обучения в Москве
DavidDAX
November 4, 2024 @ 6:13 pm
medicines for ed: cheap meds – best male enhancement pills
WallaceRut
November 4, 2024 @ 6:19 pm
https://mexicanpharm24.cheap/# mexico drug stores pharmacies
ed meds
WallaceRut
November 4, 2024 @ 6:39 pm
http://pharm24.pro/# best ed pill
what are ed drugs
DavidDAX
November 4, 2024 @ 7:02 pm
discount prescription drugs: cheaper medications – shots for ed
WallaceRut
November 4, 2024 @ 7:22 pm
https://indianpharm24.pro/# mail order pharmacy india
ed drugs
WallaceRut
November 4, 2024 @ 7:44 pm
https://indianpharm24.pro/# indian pharmacy online
pharmacy drugs
WallaceRut
November 4, 2024 @ 8:05 pm
http://pharm24.pro/# ed meds online canada
ed meds online without doctor prescription
WallaceRut
November 4, 2024 @ 8:28 pm
http://indianpharm24.pro/# best india pharmacy
erectile dysfunction cure
DavidDAX
November 4, 2024 @ 8:31 pm
mexico drug stores pharmacies: Mexican pharmacy ship US – mexican border pharmacies shipping to usa
skoraya narkologicheskaya pomosh Moskva_srMr
November 4, 2024 @ 8:39 pm
неотложная наркологическая помощь [url=https://www.skoraya-narkologicheskaya-pomoshch-moskva12.ru]неотложная наркологическая помощь[/url] .
WallaceRut
November 4, 2024 @ 8:48 pm
http://indianpharm24.pro/# india pharmacy
buy cheap prescription drugs online
vivod iz zapoya himki_niKi
November 4, 2024 @ 8:50 pm
вывод из запоя цены химки [url=http://vyvod-iz-zapoya-himki13.ru]http://vyvod-iz-zapoya-himki13.ru[/url] .
skoraya narkologicheskaya pomosh Moskva_hyki
November 4, 2024 @ 8:55 pm
как вызвать наркологическую бригаду [url=skoraya-narkologicheskaya-pomoshch-moskva13.ru]skoraya-narkologicheskaya-pomoshch-moskva13.ru[/url] .
vivod iz zapoya himki_gxel
November 4, 2024 @ 8:57 pm
вывод из запоя на дому химки [url=https://vyvod-iz-zapoya-himki11.ru/]вывод из запоя на дому химки[/url] .
vivod iz zapoya himki_ippl
November 4, 2024 @ 8:58 pm
запой на дому химки [url=www.vyvod-iz-zapoya-himki12.ru]запой на дому химки[/url] .
Volnadownload
November 4, 2024 @ 9:29 pm
[url=https://volna-casino-apk.ru/volna-skachat/]https://www.volna-casino-apk.ru/volna-skachat/[/url]
Скачать приложение казино онлайн Volna – играй прямо сейчас!
http://www.volna-casino-apk.ru/volna-skachat/
Diplomi_fxpn
November 5, 2024 @ 12:49 am
купить диплом вышка [url=https://arusak-diploms.ru/]arusak-diploms.ru[/url] .
Diplomi_pnkl
November 5, 2024 @ 12:51 am
купить высшее образование [url=https://landik-diploms.ru/]купить высшее образование[/url] .
Diplomi_kgea
November 5, 2024 @ 1:49 am
купить диплом актера [url=https://server-diploms.ru/]купить диплом актера[/url] .
EugeneVer
November 5, 2024 @ 2:47 am
Аренда виртуальных телефонных номеров для 75 городов России и 38 стран мира. Звонки онлайн на городские и мобильные (сотовые) номера, бесплатный прием [url=https://novostikubani.ru/interesnoe/kak-poluchit-virtualnyj-nomer-dlya-avito-i-obezopasit-svoyu-registracziyu]купить виртуальный номер Авито[/url]
DevonTum
November 5, 2024 @ 3:02 am
http://indianpharm24.pro/# top 10 pharmacies in india
ed problems treatment
Sazrolm
November 5, 2024 @ 3:27 am
Легальная покупка школьного аттестата с упрощенной программой обучения
nsibirsk.ru/forum/obyavleniya/topic-28354.html
WilliamCew
November 5, 2024 @ 5:54 am
home remedies for ed https://mexicanpharm24.cheap/# buying prescription drugs in mexico
DennisTon
November 5, 2024 @ 6:20 am
https://dostavim-kzn.ru/
Trefjqx
November 5, 2024 @ 6:41 am
Полезные советы по безопасной покупке диплома о высшем образовании
kai87la.flybb.ru/viewtopic.php?f=5&t=710
HerbertArome
November 5, 2024 @ 6:58 am
Сайт смешных историй http://shutki-anekdoty.ru/istorii собрание лучших веселых усторий. Ежедневное обновление контента для вашего настроения. Легко находите и делитесь забавными моментами с друзьями.
Sazrpff
November 5, 2024 @ 7:14 am
Как получить диплом о среднем образовании в Москве и других городах
HerbertArome
November 5, 2024 @ 7:17 am
Смешные приколы на сайте https://subscribe.ru/author/31612079 Самые смешные приколы. делитесь с друзьями. Легко находите и делитесь забавными моментами с друзьями.
DennisTon
November 5, 2024 @ 7:41 am
Купить металлическую профильную трубу в розницу. Стальные профильные трубы по ГОСТ и ТУ, продажа профтрубы по низкой цене за метр с доставкой [url=http://meffert-yar.ru/]стальная профильная труба купить[/url]
Dnrtegv
November 5, 2024 @ 8:13 am
Как избежать рисков при покупке диплома колледжа или ПТУ в России
леон вход
November 5, 2024 @ 8:33 am
БК Зенит предлагает своим пользователям возможность использовать рабочие зеркала для удобного доступа к сайту. На [url=https://zenitbet.guru/ru/mirror-zenitbet/]бк зенит рабочее зеркало[/url] вы сможете продолжить делать ставки на любимые спортивные события. Платформа гарантирует стабильный доступ и высокое качество услуг, что делает ее одним из лучших выборов на рынке. Убедитесь, что у вас есть актуальная ссылка для безпроблемной игры.
DanielSmops
November 5, 2024 @ 9:19 am
Представленный круглосуточный заказ эвакуатора позволит перевезти автомобиль любого типа https://kaluga.aif.ru/dontknows/kak_evakuatoram_udaetsya_bystro_priehat_k_mestu_avarii
Lazrfil
November 5, 2024 @ 10:04 am
Стоимость дипломов высшего и среднего образования и процесс их получения
gruzoperevozki54
November 5, 2024 @ 10:48 am
Надежные грузоперевозки Telegram обеспечат вам оптимальные маршруты по СФО.
Diplomi_waSi
November 5, 2024 @ 11:36 am
купить диплом перманентный макияж [url=https://man-diploms.ru/]man-diploms.ru[/url] .
Jameskib
November 5, 2024 @ 12:08 pm
Независимо от того, являетесь ли вы опытным пользователем или только начинаете познавать блокчейн, MetaMask помогает вам подключиться к децентрализованной сети: новому интернету https://www.pgyer.com/apk/apk/io.metamask
çağlayan escort
November 5, 2024 @ 1:07 pm
merhaba en genç bayanların olduğu kağıthane esort sitesi https://escortlarburda.com
DevonTum
November 5, 2024 @ 1:08 pm
http://pharm24.pro/# treatment for erectile dysfunction
viagra without a doctor prescription
Diplomi_ksSr
November 5, 2024 @ 1:13 pm
можно купить диплом техникума [url=https://diplomdarom.ru/]diplomdarom.ru[/url] .
Sazrrtt
November 5, 2024 @ 1:29 pm
Аттестат школы купить официально с упрощенным обучением в Москве
DennisTon
November 5, 2024 @ 2:51 pm
Труба стальная профильная – длинное пустотелое тело различного сечения, произведенное из углеродистой или низколегированной стали, используемое в качестве элемента металлоконструкции труба профильная москва продажа
VolnaAndroid
November 5, 2024 @ 3:07 pm
[url=https://volna-casino-apk.ru/volna-na-androjd/]http://www.volna-casino-apk.ru/volna-na-androjd/[/url]
Установить последнюю версию приложения казино Волна – играй прямо сейчас!
https://www.volna-casino-apk.ru/volna-na-androjd/
kapelnica ot zapoya luberci_qcKl
November 5, 2024 @ 3:24 pm
запой люберцы [url=https://kapelnica-ot-zapoya-lyubercy11.ru/]запой люберцы[/url] .
kapelnica ot zapoya luberci_giea
November 5, 2024 @ 3:34 pm
вывод из запоя люберцы [url=www.kapelnica-ot-zapoya-lyubercy12.ru]вывод из запоя люберцы[/url] .
vivod iz zapoya moskva_lxor
November 5, 2024 @ 3:41 pm
срочная помощь вывод из запоя москва [url=www.nazalnyj.ru]www.nazalnyj.ru[/url] .
WilliamCew
November 5, 2024 @ 3:42 pm
prescription drugs https://indianpharm24.pro/# pharmacy website india
kapelnica ot zapoya luberci_icKl
November 5, 2024 @ 3:44 pm
капельница от запоев на дому в люберцах [url=https://kapelnica-ot-zapoya-lyubercy13.ru]https://kapelnica-ot-zapoya-lyubercy13.ru[/url] .
vivod iz zapoya moskva_fwEt
November 5, 2024 @ 3:48 pm
медикаментозный вывод из запоя москва [url=https://vyvod-iz-zapoya-moskva11.ru/]https://vyvod-iz-zapoya-moskva11.ru/[/url] .
ZSot
November 5, 2024 @ 7:18 pm
1 win casino
situs bokep
November 5, 2024 @ 8:19 pm
Howdy! This is kind of off topic but I need some
help from an established blog. Is it very difficult to set up your
own blog? I’m not very techincal but I can figure things
out pretty quick. I’m thinking about creating my own but I’m not sure where to start.
Do you have any tips or suggestions? Many thanks
Mfoshkaabund
November 5, 2024 @ 8:50 pm
100 000 рублей под низкий процент в новых МФО? Это реально! Мы подобрали для вас самые надежные малоизвестные МФО, где займ одобрят каждому. Получите консультацию и оформите заявку правильно [url=https://t.me/s/mfo_2024_online]по ссылке[/url].
HerbertArome
November 5, 2024 @ 9:58 pm
Сайт смешных историй http://shutki-anekdoty.ru/istorii собрание лучших веселых усторий. Ежедневное обновление контента для вашего настроения. Легко находите и делитесь забавными моментами с друзьями.
profi-teh-remont
November 5, 2024 @ 10:25 pm
Сервисный центр предлагает замена usb разъема hp pavilion dv6 ремонт блока питания hp pavilion dv6
DevonTum
November 5, 2024 @ 10:41 pm
https://mexicanpharm24.cheap/# mexico drug stores pharmacies
pain medications without a prescription
Oarioryjf
November 5, 2024 @ 10:48 pm
Полезные советы по безопасной покупке диплома о высшем образовании
Cazrccn
November 5, 2024 @ 11:47 pm
Купить диплом дефектолога
kyc-diplom.com/diplomy-po-professii/kupit-diplom-defektologa.html
HerbertArome
November 5, 2024 @ 11:55 pm
Смешные приколы на сайте https://subscribe.ru/author/31612079 Самые смешные приколы. делитесь с друзьями. Легко находите и делитесь забавными моментами с друзьями.
ZSot
November 6, 2024 @ 12:22 am
neon54 casino login
Diplomi_fbkl
November 6, 2024 @ 12:27 am
купить диплом сантехника [url=https://realdiplom.ru/]realdiplom.ru[/url] .
WilliamCew
November 6, 2024 @ 1:16 am
ed medications list https://pharm24.pro/# erectile dysfunction medicines
Jameskib
November 6, 2024 @ 1:46 am
Надежный поставщик недорогой строительной арматуры. Качественная арматурная сталь а500с по выгодной цене с доставкой цена на арматуру
Volnadownload
November 6, 2024 @ 1:59 am
[url=https://volna-casino-apk.ru/volna-skachat/]https://volna-casino-apk.ru/volna-skachat/[/url]
Установить последнюю версию приложения онлайн казино Волна – играй прямо сейчас!
http://www.volna-casino-apk.ru/volna-skachat/
Diplomi_hsSi
November 6, 2024 @ 2:32 am
Как приобрести аттестат о среднем образовании в Москве и других городах
AndrewHyclE
November 6, 2024 @ 2:34 am
Купить онлайн металл в нашем интернет магазине достаточно просто. Для этого вам необходимо перейти в интересующий вас раздел из нашего каталога металлопрокат в москве
VolnaSait
November 6, 2024 @ 4:52 am
[url=https://volna-casino-apk.ru/volna-oficzialnyj-sajt/]http://volna-casino-apk.ru/volna-oficzialnyj-sajt/[/url]
Установить приложение онлайн казино Volna – побеждай сейчас!
https://volna-casino-apk.ru/volna-oficzialnyj-sajt/
Sazrthr
November 6, 2024 @ 5:43 am
Купить диплом о среднем полном образовании, в чем подвох и как избежать обмана?
vivod iz zapoya moskva_qlPt
November 6, 2024 @ 6:40 am
нарколог на дом вывод из запоя москва [url=https://vyvod-iz-zapoya-moskva12.ru/]нарколог на дом вывод из запоя москва[/url] .
vivod iz zapoya v stacionare balashiha_qiKa
November 6, 2024 @ 6:43 am
вывод из запоя в клинике балашихи [url=https://vyvod-iz-zapoya-v-stacionare-balashiha12.ru/]вывод из запоя в клинике балашихи[/url] .
vivod iz zapoya moskva_chOi
November 6, 2024 @ 6:44 am
вывод из запоя на дому москва цены [url=www.vyvod-iz-zapoya-moskva13.ru/]вывод из запоя на дому москва цены[/url] .
vivod iz zapoya v stacionare balashiha_hvml
November 6, 2024 @ 6:52 am
вывод из запоя балашиха [url=www.vyvod-iz-zapoya-v-stacionare-balashiha11.ru]вывод из запоя балашиха [/url] .
vivod iz zapoya v stacionare balashiha_inOl
November 6, 2024 @ 6:52 am
вывод из запоя клиника балашиха [url=http://vyvod-iz-zapoya-v-stacionare-balashiha13.ru]вывод из запоя клиника балашиха [/url] .
JorgeFuG
November 6, 2024 @ 7:32 am
buying prescription drugs in mexico: Mexican pharmacy ship US – medicine in mexico pharmacies
Dnrtbbm
November 6, 2024 @ 8:03 am
Быстрая схема покупки диплома старого образца: что важно знать?
HerbertArome
November 6, 2024 @ 8:11 am
Короткие цитаты о смысле жизни. Цитаты известных людей.
HerbertArome
November 6, 2024 @ 8:15 am
Смешные приколы и юмор. Делитесь с друзьями. Всегда есть над чем посмеяться.
DevonTum
November 6, 2024 @ 8:36 am
https://indianpharm24.pro/# world pharmacy india
best ed pills at gnc
Andrewinoms
November 6, 2024 @ 9:14 am
Арматура — это металлический прут, используемый для армирования железобетонных конструкций. Стержни арматуры изготавливаются из углеродистой купить арматуру в москве
ZSot
November 6, 2024 @ 9:33 am
1win bet mocambique
Richardsmits
November 6, 2024 @ 9:53 am
Профильные трубы – категория металлопроката, которая включает в себя изделия с сечением, отличным от круглого. Ассортимент профильных труб профильная труба москва цена
bebek escort
November 6, 2024 @ 10:49 am
en gözde kızların olduğu beşiktaş escort sitesine hoşgeldiniz https://ilanvitrin.com
ZSot
November 6, 2024 @ 2:12 pm
2 win
Hex
November 6, 2024 @ 4:57 pm
Цветочная мастерская собирает под заказ цветочные композиции с доставкой от 30 мин в с Одинцово
300 вариантов исполнения: [url=https://dostavka-cvetov-odincovo.ru/]здесь[/url]
Mazrsst
November 6, 2024 @ 5:22 pm
Диплом пту купить официально с упрощенным обучением в Москве
JorgeFuG
November 6, 2024 @ 5:36 pm
indianpharmacy com: Pharmacies in India that ship to USA – top online pharmacy india
DevonTum
November 6, 2024 @ 6:17 pm
http://pharm24.pro/# ed cure
reasons for ed
ZSot
November 6, 2024 @ 10:39 pm
neon54 casino login
Lazrxsp
November 6, 2024 @ 11:05 pm
Как купить диплом о высшем образовании с минимальными рисками
HerbertArome
November 6, 2024 @ 11:09 pm
Короткие цитаты о смысле жизни. Цитаты известных людей.
PhillipNeali
November 7, 2024 @ 12:18 am
Цены на травматические пистолеты различаются в зависимости от производителя, мощности оружия и других показателей Травмат пм купить
StephenBlory
November 7, 2024 @ 12:25 am
Онлайн магазин игр. Купить лицензионные ключи для компьютерных игр, ключи и аккаунты Steam, Origin, Uplay, Minecraft магазин игр
HerbertArome
November 7, 2024 @ 12:40 am
Смешные приколы и юмор. Делитесь с друзьями. Всегда есть над чем посмеяться.
kapelnica ot zapoya podolsk_ufkn
November 7, 2024 @ 1:00 am
выведение из запоя в подольске [url=https://www.kapelnica-ot-zapoya-podolsk11.ru]выведение из запоя в подольске[/url] .
Xazrywy
November 7, 2024 @ 1:19 am
Приобретение школьного аттестата с официальным упрощенным обучением в Москве
kapelnica ot zapoya podolsk_seor
November 7, 2024 @ 1:21 am
капельница от запоя на дому [url=http://www.kapelnica-ot-zapoya-podolsk12.ru]капельница от запоя на дому[/url] .
vivod iz zapoya korolev_vqOn
November 7, 2024 @ 1:25 am
вывод из запоя цены на дому королев [url=vyvod-iz-zapoya-korolev11.ru]вывод из запоя цены на дому королев[/url] .
vivod iz zapoya korolev_jjet
November 7, 2024 @ 1:30 am
вывод из запоя лечение королев [url=http://vyvod-iz-zapoya-korolev12.ru]http://vyvod-iz-zapoya-korolev12.ru[/url] .
kapelnica ot zapoya podolsk_huor
November 7, 2024 @ 1:35 am
капельница при алкогольной интоксикации на дому цена [url=www.kapelnica-ot-zapoya-podolsk13.ru/]капельница при алкогольной интоксикации на дому цена[/url] .
Williamraics
November 7, 2024 @ 3:12 am
Официальное получение диплома техникума с упрощенным обучением в Москве
ZSot
November 7, 2024 @ 3:28 am
vavada site
JorgeFuG
November 7, 2024 @ 5:11 am
Online medicine home delivery: indian pharmacy purchase online – reputable indian online pharmacy
DevonTum
November 7, 2024 @ 5:18 am
http://pharm24.pro/# ed dysfunction
over the counter ed drugs
Iariorpoa
November 7, 2024 @ 6:23 am
Диплом вуза купить официально с упрощенным обучением в Москве
WilliamCew
November 7, 2024 @ 6:49 am
medication online http://pharm24.pro/# ed medicines
HerbertArome
November 7, 2024 @ 7:29 am
Смешные https://smeh.ru и юмор. Делитесь с друзьями. Всегда есть над чем посмеяться.
Sazrbqc
November 7, 2024 @ 8:39 am
Как быстро и легально купить аттестат 11 класса в Москве
Dnrtxwt
November 7, 2024 @ 9:19 am
Диплом пту купить официально с упрощенным обучением в Москве
fatih escort
November 7, 2024 @ 9:30 am
hayatının en iyi vaktini geçirebileceğin tek site fatih escort sitesi https://fatihescortbul.com
Rickeypoepe
November 7, 2024 @ 9:48 am
Эффективные методы и средства для уничтожения жука короеда в доме. Где заказать профессиональные услуги обработки деревьев на участке от насекомых вредителей в Москве? https://dzen.ru/a/Zrm0OQRatQiCoOgh
Diplomi_tbPl
November 7, 2024 @ 10:11 am
купить диплом техникума в екатеринбурге [url=https://rudik-diploms365.ru/]купить диплом техникума в екатеринбурге[/url] .
Georgesewly
November 7, 2024 @ 10:45 am
buy flat Montenegro property to buy
ThomasGrido
November 7, 2024 @ 10:56 am
backlinks seo
Backlinks act as a essential part of link generation, necessary for improving a site’s ranking power and search engine position. Alongside comprehensive SEO techniques—like targeting and targeting relevant keywords—external links act a vital function in establishing a effective online presence. Our approach includes top-notch external links with tactical SEO strategy to boost your web property’s efficiency.
3-Tier Backlink System
Our three-step link-building service is intended for superior SEO impact. This strategy integrates high-quality page redirects from sites with PageRank of 8 through 10, greatly raising your webpage’s authority. Also, we employ mapping tools that map your link network, helping SEO tools to accurately identify your web structure. This is a key factor in optimizing your indexing potential and search ranking.
Varied Techniques for Website Promotion and Link Classes
We supply paid consultations to assist you on complex optimization strategies, discussing the use of various link categories. Our professionals assist in building an effective network of inbound links and adjusting the distribution of anchor and non-anchor text links. We also provide recommendations on the optimal ratio of follow and nofollow links to create a organic backlink setup that appeals to search engine algorithms.
Comprehensive Support for Blog Networks
For those interested in PBN strategies, we provide detailed knowledge on creating stable PBN websites without relying on abandoned domains, a risk-free and more effective technique to building backlinks.
Backlink Pyramids and the Strength of Layered Links
Our methods include constructing backlink pyramids—arrangements like Tier 1, Tier 2, and Tier 3 links—to increase your webpage’s ranking power. This pyramid setup enhances primary backlinks and amplifies their influence on your site ranking.
Leveraging Social Media for Digital Visibility
Social channels are effective tools not only for user engagement but also for improving SEO. With good indexing potential by online search engines, social networks can strongly support your online presence by boosting backlink range and engagement metrics.
Outreach and Consultant Advice
Our digital strategies offer advice on networking, guiding you learn how to create key backlinks and nurture link partnerships with high-authority domains. We are able to address any inquiries, delivering tailored tips and strategies suited for your specific SEO goals.
With our unique and varied optimization solutions, you get beyond more authoritative links but further the optimization insights required to create a durable, high-ranking digital platform.
vivod iz zapoya voronej_ccel
November 7, 2024 @ 11:32 am
прокапать от алкоголя на дому воронеж [url=https://vyvod-iz-zapoya-voronezh12.ru/]прокапать от алкоголя на дому воронеж [/url] .
vivod iz zapoya korolev_jfsl
November 7, 2024 @ 11:38 am
алкоголизм вывод из запоя королев [url=www.vyvod-iz-zapoya-korolev14.ru/]www.vyvod-iz-zapoya-korolev14.ru/[/url] .
vivod iz zapoya korolev_bvpt
November 7, 2024 @ 11:38 am
вывод из запоя лечение королев [url=https://vyvod-iz-zapoya-korolev13.ru]https://vyvod-iz-zapoya-korolev13.ru[/url] .
vivod iz zapoya voronej_lpei
November 7, 2024 @ 11:40 am
клиника от запоя [url=http://vyvod-iz-zapoya-voronezh11.ru]клиника от запоя[/url] .
vivod iz zapoya voronej_nxOn
November 7, 2024 @ 11:40 am
вывод из запоя воронеж [url=https://vyvod-iz-zapoya-voronezh13.ru/]вывод из запоя воронеж[/url] .
ThomasGrido
November 7, 2024 @ 11:58 am
Backlinks act as a foundational element of backlink creation, vital for improving a site’s trust and search visibility. Paired with all-encompassing SEO strategies—involving choosing and targeting pertinent search words—inbound links work a vital position in setting up a effective internet presence. Our method merges high-quality external links with calculated optimization strategy to boost your webpage’s performance.
Triple-Step Backlink System
Our three-step backlink strategy is created for optimal SEO results. This service integrates premium forwarding links from domains with PageRank levels of 8 to 10, greatly raising your site’s trust level. Additionally, we include site analyzers that organize your link architecture, assisting web crawlers to accurately identify your website’s layout. This is a important factor in improving your search visibility and indexing.
Multiple Strategies for Online Promotion and Link Classes
We deliver professional guidance to guide you through in-depth SEO strategies, covering the implementation of various link types. Our specialists guide you in an well-structured strategy of inbound links and adjusting the ratio of anchor and non-anchor text links. We further give insights on the right balance of dofollow and nofollow links to design a search-engine-friendly backlink profile that is recognized by search engine algorithms.
Thorough Assistance for Blog Networks
For customers needing in networked sites, we offer comprehensive insights on building sustainable PBN sites avoiding old domain names, a risk-free and more beneficial approach to enhancing link-building efforts.
Link Pyramids and the Effectiveness of Tiered Links
Our approaches include building link pyramids—arrangements like pyramid layers—to boost your webpage’s ranking power. This layered strategy empowers top-tier links and boosts their power on your site ranking.
Leveraging Social Platforms for Digital Visibility
Social media are valuable resources in driving online exposure while also boosting SEO. With strong search engine visibility by search engines, these platforms can strongly support your online presence by increasing link diversity and user interaction rates.
External Link Building and Specialist Support
Our SEO consulting provide guidance on outreach methods, guiding you get insights into how to build important backlinks and maintain connections with high-authority websites. We are here to answer any questions, sharing bespoke tips and tactics focused on your unique SEO targets.
With our distinctive and varied SEO packages, you get not only reliable external links but even more the planning knowledge necessary to develop a long-term, well-ranked digital platform.
ZSot
November 7, 2024 @ 12:09 pm
neon54 ????????
DevonTum
November 7, 2024 @ 2:14 pm
http://indianpharm24.pro/# buy prescription drugs from india
errectile disfunction
Treffof
November 7, 2024 @ 2:18 pm
Сколько стоит получить диплом высшего и среднего образования легально?
repetitor.ekafe.ru/faq.php
JorgeFuG
November 7, 2024 @ 2:47 pm
pharmacies in mexico that ship to usa: mexico pharmacy – pharmacies in mexico that ship to usa
Кино632234
November 7, 2024 @ 3:53 pm
https://t.me/s/kino_film_serial_online_telegram 782218 лучших фильмов. Фильмы смотреть онлайн. В нашем онлайн-кинотеатре есть новинки кино и бесплатные фильмы самых разных жанров
WilliamCew
November 7, 2024 @ 4:02 pm
top ed pills https://pharm24.pro/# amoxicillin without a doctor’s prescription
Sazrsfr
November 7, 2024 @ 4:21 pm
Пошаговая инструкция по официальной покупке диплома о высшем образовании
fishing.ukrbb.net/posting.php?mode=post&f=21&sid=129d1d15cedc79a729501c95b7c072cc
GarryElone
November 7, 2024 @ 4:42 pm
Лучшие порно видео Гей порно Бонсай скачать бесплатно без регистрации и смс. Смотреть порно онлайн в высоком качестве.
Кино325838
November 7, 2024 @ 5:13 pm
https://t.me/s/kino_film_serial_online_telegram 268589 лучших фильмов. Фильмы смотреть онлайн. В нашем онлайн-кинотеатре есть новинки кино и бесплатные фильмы самых разных жанров
Фильмы560560
November 7, 2024 @ 6:27 pm
https://t.me/s/kino_film_serial_online_telegram 746805 лучших фильмов. Фильмы смотреть онлайн. В нашем онлайн-кинотеатре есть новинки кино и бесплатные фильмы самых разных жанров
GarryElone
November 7, 2024 @ 7:04 pm
Лучшие порно видео Гей порно Бонсай скачать бесплатно без регистрации и смс. Смотреть порно онлайн в высоком качестве.
Lazrimm
November 7, 2024 @ 7:21 pm
Аттестат школы купить официально с упрощенным обучением в Москве
iqtorg.ru/forum/topic/add/forum2
Sazrqou
November 7, 2024 @ 7:26 pm
Как купить диплом о высшем образовании с минимальными рисками
oknaalum.ru
November 7, 2024 @ 7:36 pm
Добро пожаловать в OknaAlum! Мы с радостью поможем вам выбрать качественные алюминиевые окна, которые подарят вашему дому уют и тепло. Наши окна не только прекрасно выглядят, но и надёжно защищают от холода и шума. Загляните на сайт [url=https://oknaalum.ru/]oknaalum.ru[/url], чтобы узнать больше о наших продуктах и выбрать вариант, который идеально подойдёт для вашего дома. Мы всегда рядом, чтобы помочь вам с выбором!
Diplomi_rdSi
November 7, 2024 @ 9:48 pm
Полезные советы по безопасной покупке диплома о высшем образовании
Кино644774
November 7, 2024 @ 10:01 pm
https://t.me/s/kino_film_serial_online_telegram 267406 лучших фильмов. Фильмы смотреть онлайн. В нашем онлайн-кинотеатре есть новинки кино и бесплатные фильмы самых разных жанров
Фильм873156
November 7, 2024 @ 11:05 pm
https://t.me/s/kino_film_serial_online_telegram 20749 лучших фильмов. Фильмы смотреть онлайн. В нашем онлайн-кинотеатре есть новинки кино и бесплатные фильмы самых разных жанров
HerbertArome
November 7, 2024 @ 11:33 pm
Смешные https://smeh.ru и юмор. Делитесь с друзьями. Всегда есть над чем посмеяться.
Enregistrement
November 7, 2024 @ 11:34 pm
Can you be more specific about the content of your article? After reading it, I still have some doubts. Hope you can help me.
lgo dewa
November 7, 2024 @ 11:49 pm
This is the right site for anyone who wishes to find
out about this topic. You realize so much its almost hard to
argue with you (not that I personally will need to…HaHa).
You definitely put a brand new spin on a subject that’s been written about for many years.
Wonderful stuff, just excellent!
RobertDor
November 8, 2024 @ 1:17 am
Бюстгальтеры с октрытой спиной с бесплатной доставкой в интернет-магазине, актуальные цены, в наличии большой ассортимент моделей размер бюстгальтера
Brandonsmava
November 8, 2024 @ 1:25 am
online shopping pharmacy india: Order medicine from India to USA – best india pharmacy
JorgeFuG
November 8, 2024 @ 1:34 am
buying prescription drugs in mexico: mexico pharmacy – mexico drug stores pharmacies
Derekhok
November 8, 2024 @ 2:23 am
apartment real estate Montenegro
Diplomi_moOt
November 8, 2024 @ 2:47 am
купить диплом в анапе [url=https://2orik-diploms.ru/]2orik-diploms.ru[/url] .
Lewiswousy
November 8, 2024 @ 4:30 am
стоимость укладки кафельной плитки квадратный укладка кафельной плитки своими руками
Derekhok
November 8, 2024 @ 4:53 am
budva estate Montenegro property to buy
Diplomi_gkKn
November 8, 2024 @ 5:00 am
купил диплом устроился на работу [url=https://prem-diplom77.ru/]prem-diplom77.ru[/url] .
Derekfet
November 8, 2024 @ 5:08 am
deneme bonusu veren siteler 2024: deneme bonusu veren yeni siteler – deneme bonusu veren yeni siteler
LarryPab
November 8, 2024 @ 5:11 am
ultrabet [url=https://ultrabet-tr.online/#]ultrabet[/url] ultrabet guncel
Eanrsyp
November 8, 2024 @ 5:19 am
Быстрая схема покупки диплома старого образца: что важно знать?
LarryPab
November 8, 2024 @ 5:27 am
slot oyunlar? puf noktalar? [url=https://slot-tr.online/#]slot oyunlar?[/url] az parayla cok kazandiran slot oyunlar?
Diplomi_eami
November 8, 2024 @ 5:28 am
купить диплом о высшем образовании образцы [url=https://servera-diplomy199.ru/]servera-diplomy199.ru[/url] .
LarryPab
November 8, 2024 @ 5:43 am
ultrabet giris [url=https://ultrabet-tr.online/#]ultrabet yeni giris 1125[/url] ultrabet tr online
LarryPab
November 8, 2024 @ 5:59 am
Casino Siteleri [url=http://casinositeleri.win/#]Canl? Casino Siteleri[/url] Casino Siteleri
Hassanagize
November 8, 2024 @ 6:20 am
http://slot-tr.online/# slot oyunlar? puf noktalar?
deneme bonusu veren siteler yerliarama.org
Derekfet
November 8, 2024 @ 6:36 am
en cok kazand?ran slot oyunlar?: en kazancl? slot oyunlar? – az parayla cok kazandiran slot oyunlar?
oknaalum
November 8, 2024 @ 6:37 am
Если вы хотите превратить балкон или лоджию в уютное пространство, OknaAlum — ваш лучший помощник! Мы предлагаем остекление, которое защитит от непогоды и добавит вашему дому комфорта. На нашем сайте [url=https://oknaalum.ru/]oknaalum.ru[/url] вы можете узнать больше о вариантах остекления, которые сделают ваш балкон местом для отдыха и уюта. С OknaAlum ваш дом станет ещё более уютным!
LarryPab
November 8, 2024 @ 6:46 am
Deneme Bonusu Veren Siteler [url=https://casinositeleri.win/#]casino siteleri win[/url] Canl? Casino Siteleri
Jeremyvof
November 8, 2024 @ 6:55 am
As AI is on the rise in simple applications, ChatGPT has become that through which much work—from idea generation to fast questioning—is done. You can even access ChatGPT without login through some publicly accessible platforms ChatGPT without login
Hassanagize
November 8, 2024 @ 6:58 am
https://matadorbet.bid/# matadorbet giris
denemebonusuverensiteler.top
Hassanagize
November 8, 2024 @ 6:58 am
http://casinositeleri.win/# guvenilir casino siteleri
deneme bonusu veren siteler yerliarama.org
Diplomi_qkMn
November 8, 2024 @ 6:58 am
диплом о высшем образовании купить гознак [url=https://prema365-diploms.ru/]диплом о высшем образовании купить гознак[/url] .
Oariorwwr
November 8, 2024 @ 7:06 am
Полезные советы по безопасной покупке диплома о высшем образовании
DownloadMonro
November 8, 2024 @ 7:32 am
[url=https://monro-casino-apk.ru/monro-skachat]monro-casino-apk.ru/monro-skachat[/url]
Загрузить последнюю версию приложения онлайн казино Монро – выигрывай сегодня!
http://monro-casino-apk.ru/monro-skachat
Hassanagize
November 8, 2024 @ 7:35 am
https://slot-tr.online/# slot tr online
deneme bonusu veren siteler
Wesleymendy
November 8, 2024 @ 8:08 am
Преимущества покупки алкоголя в Ароматном Мире · широкий ассортимент алкогольных и безалкогольных напитков высокого качества; · индивидуальная система скидок доставка алкоголя на дом москва
vivod iz zapoya chelyabinsk_bhpr
November 8, 2024 @ 8:47 am
помощь на дому вывод из запоя [url=http://vyvod-iz-zapoya-chelyabinsk11.ru/]помощь на дому вывод из запоя[/url] .
Lazrxdy
November 8, 2024 @ 8:55 am
Диплом пту купить официально с упрощенным обучением в Москве
vivod iz zapoya chelyabinsk_ueOt
November 8, 2024 @ 9:02 am
вывод из запоя в челябинске [url=vyvod-iz-zapoya-chelyabinsk12.ru]vyvod-iz-zapoya-chelyabinsk12.ru[/url] .
HowardGat
November 8, 2024 @ 9:03 am
Люки Шагма – идеальное сочетание функциональности и эстетики для вашего интерьера. Инновационные решения от SHAGMA обеспечивают надежный доступ к коммуникациям, сохраняя безупречный внешний вид помещения. Наши люки под плитку не только решают проблему провисания, но и гармонично вписываются в любой дизайн. Выбирая SHAGMA, вы выбираете качество, долговечность и индивидуальный подход https://akak7.ru/innovacionnye-revizionnye-lyuki-shagma-vybor-professionalov.html
vivod iz zapoya ekaterinbyrg_niml
November 8, 2024 @ 9:04 am
вывод из запоя с кодированием на дому [url=https://vyvod-iz-zapoya-ekaterinburg22.ru]https://vyvod-iz-zapoya-ekaterinburg22.ru[/url] .
vivod iz zapoya ekaterinbyrg_hzer
November 8, 2024 @ 9:06 am
помощь алкоголикам на дому [url=https://www.vyvod-iz-zapoya-ekaterinburg21.ru]https://www.vyvod-iz-zapoya-ekaterinburg21.ru[/url] .
vivod iz zapoya chelyabinsk_dvel
November 8, 2024 @ 9:13 am
помощь на дому вывод из запоя [url=https://vyvod-iz-zapoya-chelyabinsk13.ru]помощь на дому вывод из запоя[/url] .
Dnrtykd
November 8, 2024 @ 11:05 am
Покупка диплома о среднем полном образовании: как избежать мошенничества?
RobertOpire
November 8, 2024 @ 12:16 pm
Как получить, переоформить и продлить лицензию на газовое оружие. Кто может приобрести газовое оружие. Необходимые документы для лицензии на приобретение где купить патроны для травмата
Lewiswousy
November 8, 2024 @ 12:36 pm
укладка кафельной плитки кухне укладка кафельной плитки в ванной
Cazryhs
November 8, 2024 @ 2:39 pm
Диплом вуза купить официально с упрощенным обучением в Москве
click ua
November 8, 2024 @ 2:44 pm
Today, while I was at work, my cousin stole my iphone and tested to see if it can survive a
thirty foot drop, just so she can be a youtube sensation. My iPad is now destroyed and she has 83 views.
I know this is completely off topic but I had to share it with someone!
QINQDBX
November 8, 2024 @ 3:03 pm
EOEHRBH RXPFTCT TPIAVIL RTBZSGB
https://9gm.ru/article?NFXQYJ
RodneyJew
November 8, 2024 @ 3:33 pm
Размытость симптоматики большинства заболеваний нервной системы требует особого подхода к диагностике. Опытные специалисты центра неврологии больницы Ассута в Израиле разрабатывают индивидуальные планы диагностических мероприятий для пациентов https://moscow.service-med.com/neurology/
EYIIVUZ
November 8, 2024 @ 4:25 pm
JHGQOUH VBPPVRU QXEQOVD KCMHGMN
https://9gm.ru/article?YPTIRR
CasinoSol
November 8, 2024 @ 5:34 pm
[url=https://sol-casino-apk.ru/sol-casino/]https://sol-casino-apk.ru/sol-casino/[/url]
Скачать приложение казино онлайн Сол – играй прямо сейчас!
https://sol-casino-apk.ru/sol-casino/
VANNKDE
November 8, 2024 @ 5:40 pm
KDNRFLD MKXNUKK NVLNBBQ DRONUOL
https://9gm.ru/article?WHNAAQ
Hassanagize
November 8, 2024 @ 6:47 pm
https://denemebonusuverensiteler.top/# deneme bonusu veren siteler
denemebonusuverensiteler.top
MonroApk
November 8, 2024 @ 7:46 pm
[url=https://monro-casino-apk.ru/monro-apk]http://monro-casino-apk.ru/monro-apk[/url]
Скачать последнюю версию приложения казино онлайн Монро – выигрывай сейчас!
https://www.monro-casino-apk.ru/monro-apk
SolAndroid
November 8, 2024 @ 7:58 pm
[url=https://sol-casino-apk.ru/sol-na-androjd/]https://www.sol-casino-apk.ru/sol-na-androjd/[/url]
Загрузить приложение казино Сол – побеждай сегодня!
https://www.sol-casino-apk.ru/sol-na-androjd/
Sazrdfg
November 8, 2024 @ 8:25 pm
Как избежать рисков при покупке диплома колледжа или ПТУ в России
сейф для оружия
November 8, 2024 @ 9:01 pm
Тут можно преобрести сейфы оружейные купить стоимость оружейного сейфа
IVOCNUS
November 8, 2024 @ 9:16 pm
ZNSNAWP NCZNLVK KOOSWYN BTECVUA
https://9gm.ru/article?AHZZAK
SolSait
November 8, 2024 @ 10:05 pm
[url=https://sol-casino-apk.ru/sol-oficzialnyj-sajt/]sol-casino-apk.ru/sol-oficzialnyj-sajt/[/url]
Установить apk файл онлайн казино Sol – побеждай сегодня!
https://www.sol-casino-apk.ru/sol-oficzialnyj-sajt/
YUSUNNZ
November 8, 2024 @ 10:18 pm
UTQRLNP ZGPGSSK RFADAIN CTOXHAX
https://9gm.ru/article?JYGPPP
SolCasinoApk
November 8, 2024 @ 10:31 pm
[url=https://sol-casino-apk.ru]sol-casino-apk.ru[/url]
Установить последнюю версию приложения казино Sol – играй сегодня!
https://www.sol-casino-apk.ru
MonroCasino
November 8, 2024 @ 10:36 pm
[url=https://monro-casino-apk.ru/monro-casino]www.monro-casino-apk.ru/monro-casino[/url]
Скачать приложение казино Monro – побеждай сегодня!
http://www.monro-casino-apk.ru/monro-casino
Soldownload
November 8, 2024 @ 10:50 pm
[url=https://sol-casino-apk.ru/sol-skachat/]https://sol-casino-apk.ru/sol-skachat/[/url]
Загрузить apk файл казино онлайн Sol – играй сегодня!
http://sol-casino-apk.ru/sol-skachat/
Sazrpnm
November 8, 2024 @ 11:36 pm
Можно ли купить аттестат о среднем образовании? Основные рекомендации
sultangazi escort
November 8, 2024 @ 11:39 pm
hayatını birleştireceğin kadınların tek adresi syltangazi escort https://sultangaziescortbul.com
MonroZercalo
November 9, 2024 @ 12:05 am
[url=https://monro-casino-apk.ru/monro-oficzialnyj-sajt]www.monro-casino-apk.ru/monro-oficzialnyj-sajt[/url]
Скачать apk файл казино онлайн Монро – играй сейчас!
http://monro-casino-apk.ru/monro-oficzialnyj-sajt
купить сейф для дома
November 9, 2024 @ 12:34 am
Тут можно сейф огнестойкий для дома купить домашние сейфы купить
candy ai
November 9, 2024 @ 1:09 am
May I simply say what a relief to uncover someone who genuinely understands what they’re discussing on the net.
You definitely know how to bring an issue to light and make it important.
More and more people really need to check this out and understand this side
of your story. I was surprised that you are not more popular given that you surely have the gift.
narkolog na dom ekaterinbyrg_fyKa
November 9, 2024 @ 1:48 am
нарколог на дом в екатеринбурге [url=http://narkolog-na-dom-ekaterinburg11.ru]нарколог на дом в екатеринбурге[/url] .
narkolog na dom ekaterinbyrg_srOl
November 9, 2024 @ 2:10 am
вызов нарколога на дом екатеринбург круглосуточно [url=https://narkolog-na-dom-ekaterinburg12.ru]вызов нарколога на дом екатеринбург круглосуточно [/url] .
elektrokarniz dlya shtor_gkOn
November 9, 2024 @ 2:13 am
карниз для штор электрический [url=https://elektrokarniz-dlya-shtor15.ru/]https://elektrokarniz-dlya-shtor15.ru/[/url] .
DavidSporm
November 9, 2024 @ 2:22 am
Бесплатные виртуальные номера страны Белоруссия с кодом +375 для приема СМС, получения активации сервисов и аренды мобильного телефона купить американский номер
onexslotsfun
November 9, 2024 @ 2:38 am
Нужен доступ к игре? Скачайте 1xslots apk и начните выигрывать прямо на Android.
ScottGed
November 9, 2024 @ 2:51 am
В настоящее время одной из ключевых современных информационных угроз для общества в сфере медиабезопасности становится вовлечение детей и подростков в деструктивные Интернет-сообщества порно онлайн сливы школьниц
Jasontiz
November 9, 2024 @ 3:31 am
срочный займ Sravnim.kz
HermanMAP
November 9, 2024 @ 3:35 am
соут специальная оценка условий труда https://sout095.ru
StephenBlory
November 9, 2024 @ 4:03 am
Ни одно застолье невозможно представить себе без освежающих безалкогольных напитков. Сейчас вам не придется тратить свое время и силы на то, чтобы ездить в магазины и покупать соки, минеральную воду доставка алкоголя москва круглосуточно alcodostavkaplanet msk ru
огнестойкие сейфы
November 9, 2024 @ 4:50 am
Тут можно преобрести сейф противопожарный купить несгораемый сейф
Jasontiz
November 9, 2024 @ 6:44 am
Микрокредит Казахстан Sravnim.kz
HermanMAP
November 9, 2024 @ 6:44 am
процедура оценки условий труда стоимость оценки условий труда одного рабочего места
Mazriko
November 9, 2024 @ 6:48 am
Всё, что нужно знать о покупке аттестата о среднем образовании
CurtisTut
November 9, 2024 @ 9:48 am
соут в новой организации специалист соут
Bruceaciff
November 9, 2024 @ 10:22 am
Ведь есть же десятки сервисов для номеров из других стран. А беларусские форумы огорожены, им подавай только их собственный картофель типа +375 купить временный номер для Телеграм
SolApk
November 9, 2024 @ 10:45 am
[url=https://sol-casino-apk.ru/sol-apk/]sol-casino-apk.ru/sol-apk/[/url]
Загрузить apk файл казино Sol – выигрывай сегодня!
https://www.sol-casino-apk.ru/sol-apk/
ManuelGag
November 9, 2024 @ 11:35 am
Melbet – начинай играть на реальные деньги на официальном сайте онлайн казино Мелбет и выигрывай крупные суммы в популярных игровых автоматах комета casino r7casinoplay com
muah ai
November 9, 2024 @ 11:40 am
I have been surfing online more than 3 hours today, but I by no means found any fascinating article
like yours. It is beautiful worth sufficient for me. In my opinion, if all website owners and bloggers made excellent content as you probably did, the internet will likely be much more useful than ever before.
발로란트 대리
November 9, 2024 @ 11:45 am
First of all I want to say great blog! I had a quick question which I’d like to ask if you don’t
mind. I was interested to know how you center yourself and clear your thoughts prior to writing.
I’ve had a tough time clearing my mind in getting my thoughts
out there. I truly do take pleasure in writing but it just seems like
the first 10 to 15 minutes are wasted just trying to figure out how to begin. Any ideas or
tips? Thanks!
JohnnyMer
November 9, 2024 @ 12:35 pm
Веб агентство в Москве выполняет разработку сайтов на платформе Битрикс под ключ. Дополнительные услуги – продвижение в поисковых системах Яндекс и Гугл, техническая поддержка, обслуживание, аудит, тестирование, наполнение контентом, контекстная реклама и маркетинг создание и разработка сайтов на bitrix
www.williamseal.com
November 9, 2024 @ 1:03 pm
These are in fact fantastic ideas in concerning blogging.
You have touched some pleasant factors here.
Any way keep up wrinting.
Lazrevi
November 9, 2024 @ 1:26 pm
Как приобрести аттестат о среднем образовании в Москве и других городах
Mazrulb
November 9, 2024 @ 2:20 pm
Полезные советы по безопасной покупке диплома о высшем образовании
iskysstvennaya koja dlya mebeli kypit_crea
November 9, 2024 @ 3:22 pm
искусственная кожа цена [url=http://iskusstvennaya-kozha-dlya-mebeli-kupit.ru]искусственная кожа цена[/url] .
stoimost soglasovaniya pereplanirovki kvartiri_hemn
November 9, 2024 @ 3:23 pm
проект перепланировки квартиры цена [url=stoimost-soglasovaniya-pereplanirovki-kvartiry.ru]проект перепланировки квартиры цена[/url] .
купить сейф для охотничьего ружья
November 9, 2024 @ 3:28 pm
Тут можно преобрести сейфы для оружия купить оружейный сейф для ружья
Diplomi_buSi
November 9, 2024 @ 5:00 pm
Стоимость дипломов высшего и среднего образования и как избежать подделок
yodayo ai
November 9, 2024 @ 5:07 pm
There’s certainly a great deal to find out about this issue.
I really like all of the points you made.
JasonDeaky
November 9, 2024 @ 5:33 pm
сколько стоит прокат сноуборда Красная Поляна https://prokat-lyzh-krasnaya-polyana.ru
Denniscrype
November 9, 2024 @ 6:04 pm
прокат лыж красная стоимость проката лыж
MelvinEthic
November 9, 2024 @ 6:12 pm
айфон 11 128 цена https://store-iphone43.ru
발로란트 대리
November 9, 2024 @ 6:23 pm
If some one desires expert view on the topic of blogging then i suggest him/her to pay
a visit this weblog, Keep up the pleasant work.
Denniscrype
November 9, 2024 @ 8:28 pm
прокат в адлере https://prokat-lyzh-krasnaya-polyana.ru
MelvinEthic
November 9, 2024 @ 8:41 pm
купить айфон 15 про макс цена айфона рублей
Dnrtklc
November 9, 2024 @ 9:54 pm
Приобретение школьного аттестата с официальным упрощенным обучением в Москве
Iariorxxc
November 9, 2024 @ 10:52 pm
Диплом техникума купить официально с упрощенным обучением в Москве
servis nakrytki podpischikov_kest
November 9, 2024 @ 11:02 pm
сервис для накрутки подписчиков инстаграм бесплатно [url=www.artmaxboost11.com]сервис для накрутки подписчиков инстаграм бесплатно[/url] .
Sazrzkf
November 10, 2024 @ 12:25 am
Как приобрести аттестат о среднем образовании в Москве и других городах
Sazrlxk
November 10, 2024 @ 12:29 am
Официальное получение диплома техникума с упрощенным обучением в Москве
Aaronveide
November 10, 2024 @ 1:26 am
Создание лендингов под ключ в студии в Москве включает проектирование, дизайн и программирование, обеспечивающее эффективность и удобство использования. Также предоставляются услуги: оптимизация для Яндекс и Google, настройка рекламных кампаний, поддержка, обслуживание и ведение маркетинга создание лендинга
dougtitcomb.com
November 10, 2024 @ 1:32 am
I am really enjoying the theme/design of your website.
Do you ever run into any browser compatibility issues?
A small number of my blog visitors have complained about my site not working correctly in Explorer but looks great in Firefox.
Do you have any advice to help fix this issue?
DownloadMonro
November 10, 2024 @ 2:07 am
[url=https://monro-casino-apk.ru/monro-skachat]www.monro-casino-apk.ru/monro-skachat[/url]
Скачать приложение онлайн казино Monro – выигрывай прямо сейчас!
http://www.monro-casino-apk.ru/monro-skachat
Geraldtraub
November 10, 2024 @ 2:19 am
Разработка лендингов в студии в Москве осуществляется с учетом современных маркетинговых стратегий и требований рынка. К основным услугам добавляются: аудит и анализ конверсии, продвижение в поисковых системах, контентное наполнение, настройка CRM и профессиональная техническая поддержка https://webstudiya-landing.ru/
Online Quranic Courses
November 10, 2024 @ 2:23 am
I blog often and I genuinely appreciate your content.
Your article has truly peaked my interest. I am going to book mark your blog and keep checking for new details about once per week.
I opted in for your Feed too.
PhillipHoOny
November 10, 2024 @ 2:24 am
Разработка сайтов на платформе 1С-Битрикс в студии в Москве включает полный цикл: от проектирования до запуска и дальнейшего сопровождения. Также предлагаются услуги: оптимизация сайта для поисковых систем, интеграция с хостингом, контентное наполнение, анализ данных и маркетинговые стратегии https://Webstudiya-Moscow.ru/
Trefitg
November 10, 2024 @ 2:44 am
Как правильно приобрести диплом колледжа или ПТУ в России, важные моменты
mosmol.flybb.ru/viewtopic.php?f=2&t=624
Soldownload
November 10, 2024 @ 3:56 am
[url=https://sol-casino-apk.ru/sol-skachat/]http://sol-casino-apk.ru/sol-skachat/[/url]
Установить приложение казино онлайн Sol – побеждай сейчас!
http://www.sol-casino-apk.ru/sol-skachat/
does mazda give free oil changes
November 10, 2024 @ 4:38 am
Let’s spread the love! Tag a friend who would appreciate this post as much as you did.
Diplomi_thpi
November 10, 2024 @ 4:40 am
купить диплом вуза в самаре [url=https://many-diplom77.ru/]many-diplom77.ru[/url] .
Diplomi_ykKn
November 10, 2024 @ 4:41 am
купить диплом вуза в казани [url=https://1russa-diploms.ru/]1russa-diploms.ru[/url] .
MichaelJeony
November 10, 2024 @ 5:43 am
Полный цикл маркетинговых услуг: от настройки рекламы, SEO-продвижения, создания сайтов до управления репутацией в интернете и PR https://makeallperfect.ru/
발로란트 대리
November 10, 2024 @ 6:25 am
Good day! Do you use Twitter? I’d like to follow you if
that would be okay. I’m definitely enjoying your blog and
look forward to new updates.
Sazrdta
November 10, 2024 @ 6:56 am
Легальные способы покупки диплома о среднем полном образовании
kai87la.flybb.ru/viewtopic.php?f=5&t=725
Cazruxp
November 10, 2024 @ 7:08 am
Полезные советы по безопасной покупке диплома о высшем образовании
Sazrtuw
November 10, 2024 @ 7:09 am
Официальная покупка аттестата о среднем образовании в Москве и других городах
купить сейф для охотничьего ружья
November 10, 2024 @ 7:20 am
Тут можно преобрести оружейные сейфы купить купить оружейный сейф
Hair Botox for Lifeless Hair
November 10, 2024 @ 7:34 am
Hola! I’ve been reading your blog for a while now and finally got the bravery to go
ahead and give you a shout out from Humble Tx! Just
wanted to say keep up the great work!
PhillipInola
November 10, 2024 @ 7:36 am
Хочешь добавить остроты в свою интимную жизнь и не знаешь, с чего начать? У нас есть то, что тебе нужно! В нашем магазине можно купить секс игрушки по самым разным вкусам: от классических вибраторов и фаллоимитаторов до инновационных игрушек для взрослых, которые подарят новые ощущения. Мы предлагаем заказать секс игрушки онлайн — это просто и удобно, ведь интим игрушки станут идеальным дополнением для разнообразных сексуальных приключений. У нас только самые качественные секс товары, созданные, чтобы доставлять максимум удовольствия. Хотите попробовать что-то новое? Стимуляторы, БДСМ аксессуары и игрушки для секса уже ждут вас. Дешево? Конечно, у нас всегда есть предложения, чтобы сделать стоимость покупки более приятной. Будь то эротические игрушки для себя или для партнера — мы знаем, как сделать вашу ночную жизнь по-настоящему яркой https://topsexshop3.co.ua/
CharlieRap
November 10, 2024 @ 7:43 am
Апартаменты Napa Gem в пешей доступности от одного из самых красивых пляжей Кипра, а также бурного веселья на шумном летнем курорте Айя-Напа недвижимость на кипре объявления
MonroCasino
November 10, 2024 @ 8:03 am
[url=https://monro-casino-apk.ru/monro-casino]https://www.monro-casino-apk.ru/monro-casino[/url]
Установить последнюю версию приложения казино Monro – играй сейчас!
monro-casino-apk.ru/monro-casino
Eddiebrase
November 10, 2024 @ 8:12 am
Продажа недвижимости на Кипре: 10 674 предложения с ценами и фото. Tranio поможет подобрать и купить жилье на Кипре стоимостью от 54 000€ кипр квартира купить
Hair Loss Treatment Innovations
November 10, 2024 @ 9:04 am
Hi, the whole thing is going fine here and ofcourse every one is sharing data, that’s in fact good,
keep up writing.
FrankPebra
November 10, 2024 @ 9:35 am
Лучшие онлайн казино Скачать казино
SolAndroid
November 10, 2024 @ 9:36 am
[url=https://sol-casino-apk.ru/sol-na-androjd/]sol-casino-apk.ru/sol-na-androjd/[/url]
Скачать apk файл онлайн казино Sol – побеждай прямо сейчас!
http://www.sol-casino-apk.ru/sol-na-androjd/
EddieCyhox
November 10, 2024 @ 9:40 am
В нейрохирургических отделениях израильских клиник сегодня производятся самые революционные операции на спинном и головном мозге, которые максимально результативны и безопасны https://methodstreatment.wixsite.com/methods
FrankPebra
November 10, 2024 @ 12:04 pm
Скачать казино Казино на деньги
luckyjetraketa
November 10, 2024 @ 12:12 pm
Для начала игры используйте промокод лаки джет и начните с бонусом.
발로란트 대리
November 10, 2024 @ 12:15 pm
I needed to thank you for this great read!! I certainly loved every bit of it.
I’ve got you bookmarked to look at new stuff you post…
MonroCasinoApk
November 10, 2024 @ 1:14 pm
[url=https://monro-casino-apk.ru]monro-casino-apk.ru[/url]
Загрузить последнюю версию приложения казино онлайн Monro – побеждай сейчас!
http://www.monro-casino-apk.ru
SolApk
November 10, 2024 @ 1:44 pm
[url=https://sol-casino-apk.ru/sol-apk/]www.sol-casino-apk.ru/sol-apk/[/url]
Загрузить приложение казино Сол – выигрывай сегодня!
https://sol-casino-apk.ru/sol-apk/
kinogo.new
November 10, 2024 @ 3:29 pm
Ожидаете новинок? [url=https://kinogo.new/xfsearch/genre/семейный/]Семейные фильмы 2025[/url] обещают быть насыщенными яркими сюжетами и трогательными моментами. Наш сайт предоставляет возможность первыми увидеть новейшие фильмы для семейного просмотра, которые порадуют и научат важным ценностям. Проводите время вместе, наслаждаясь качественным кинематографом.
Diplomi_gxMn
November 10, 2024 @ 6:32 pm
купить диплом горного инженера [url=https://prema365-diploms.ru/]prema365-diploms.ru[/url] .
발로란트 대리
November 10, 2024 @ 6:39 pm
Howdy! I could have sworn I’ve been to this blog before but after
looking at some of the posts I realized it’s new to me.
Regardless, I’m definitely delighted I stumbled upon it
and I’ll be book-marking it and checking back
regularly!
Andrewunsup
November 10, 2024 @ 6:59 pm
работ укладке кафельной плитки https://ukladka-keramogranita-spb.ru
RobertViano
November 10, 2024 @ 7:36 pm
аренда горных лыж красная поляна сколько стоит прокат лыж красная поляна
MonroAndroid
November 10, 2024 @ 8:44 pm
[url=https://monro-casino-apk.ru/monro-na-androjd]http://www.monro-casino-apk.ru/monro-na-androjd[/url]
Скачать последнюю версию приложения казино онлайн Monro – выигрывай сегодня!
https://monro-casino-apk.ru/monro-na-androjd
servis nakrytki podpischikov_rqKr
November 10, 2024 @ 9:33 pm
сервис накрутки подписчиков в телеграм [url=https://nakrutkamedia11.com]сервис накрутки подписчиков в телеграм[/url] .
Sazrniq
November 10, 2024 @ 9:42 pm
Пошаговая инструкция по официальной покупке диплома о высшем образовании
Uazrayu
November 10, 2024 @ 9:43 pm
Вопросы и ответы: можно ли быстро купить диплом старого образца?
hali.bg/attestat-kupit-kemerovo-9-klass.html
prodvijenie saitov v moskve_tzsl
November 10, 2024 @ 10:04 pm
сео продвижение сайта москва [url=prodvizhenie-sajtov-v-moskve214.ru]сео продвижение сайта москва[/url] .
prodvijenie saitov v moskve_srPt
November 10, 2024 @ 10:13 pm
заказать создание и продвижение сайта [url=https://prodvizhenie-sajtov-v-moskve215.ru]заказать создание и продвижение сайта[/url] .
prodvijenie saitov v moskve_ujel
November 10, 2024 @ 10:15 pm
продвижение сайта купить москва [url=http://www.prodvizhenie-sajtov-v-moskve216.ru]http://www.prodvizhenie-sajtov-v-moskve216.ru[/url] .
franshizi_fdOa
November 10, 2024 @ 10:17 pm
топ франшиз [url=https://franshizy17.ru/]топ франшиз[/url] .
Sheffied Escorts agency
November 10, 2024 @ 10:51 pm
I’m amazed, I must say. Seldom do I encounter a blog that’s
both equally educative and entertaining, and let me tell you, you have hit the nail on the head.
The issue is something that too few men and women are speaking intelligently about.
I’m very happy that I stumbled across this in my hunt for something regarding this.
Diplomi_koSi
November 10, 2024 @ 11:08 pm
Удивительно, но купить диплом кандидата наук оказалось не так сложно
Waynealarm
November 10, 2024 @ 11:44 pm
В мире, где каждый элемент окружения играет важную роль, особое внимание стоит уделить тому, как мы оформляем свои жилища и рабочие места. Современные технологии позволяют преобразить любое пространство, добавив в него уникальные акценты и характер http://aramvill.com/bbs/board.php?bo_table=p_photo&wr_id=99410
Andrewunsup
November 11, 2024 @ 12:10 am
укладка кафельной плитки спб https://ukladka-keramogranita-spb.ru
promokod_getkurspay_random[a..z]tmt
November 11, 2024 @ 12:29 am
Getcourse промокод [url=platezhnyj-modul-getkurs-promokod.ru]platezhnyj-modul-getkurs-promokod.ru[/url] .
발로란트 대리
November 11, 2024 @ 12:47 am
I savour, cause I found just what I used to be looking for.
You’ve ended my 4 day long hunt! God Bless you man. Have a nice day.
Bye
Thomasgak
November 11, 2024 @ 1:11 am
Мы печатаем развороты фотокниг на высококачественной профессиональной фотобумаге Fujicolor Crystal Archive Digital тип DP II (матовая или тиснёная) на современной машине Noritsu QSS 3704HD фотокнига заказать москва недорого
Patrickrer
November 11, 2024 @ 1:20 am
Фотосфера – это долгожданный момент творчества, вдохновение, радость, уникальные дизайны и невероятно красивые решения. Это творческая лаборатория, где люди на основе снимков и изображений создают шедевры для себя, родных и друзей фотоателье москвы
RobertDooma
November 11, 2024 @ 1:53 am
Откройте мир вождения с нашей автошколой, обучающей безопасным навыкам управления автомобилем. Мы предлагаем высококачественное обучение вождению, которое поможет вам уверенно управлять автомобилем в любых условиях автошкола в москве цены на обучение
MonroZercalo
November 11, 2024 @ 3:40 am
[url=https://monro-casino-apk.ru/monro-oficzialnyj-sajt]https://monro-casino-apk.ru/monro-oficzialnyj-sajt[/url]
Загрузить последнюю версию приложения казино Монро – играй прямо сейчас!
http://www.monro-casino-apk.ru/monro-oficzialnyj-sajt
Lazroow
November 11, 2024 @ 5:56 am
Как получить диплом стоматолога быстро и официально
смотреть комедии
November 11, 2024 @ 5:57 am
Платформа [url=https://kinogo.new/]смотреть kinogo[/url] собрала в себе всё необходимое для качественного и комфортного просмотра. Вам доступны лучшие фильмы и сериалы онлайн, без регистрации и в хорошем качестве. Быстрая загрузка и разнообразие жанров обеспечат вам приятный досуг и позволят найти что-то интересное для каждого настроения.
발로란트 대리
November 11, 2024 @ 6:28 am
This text is invaluable. When can I find out more?
MonroApk
November 11, 2024 @ 7:22 am
[url=https://monro-casino-apk.ru/monro-apk]www.monro-casino-apk.ru/monro-apk[/url]
Установить приложение казино Monro – побеждай прямо сейчас!
https://www.monro-casino-apk.ru/monro-apk
YHCNXHH
November 11, 2024 @ 7:58 am
Roman. Solidarity. Black codes. Corn starch. Boss baby. Steamboat springs. Adjective. Twitter.. Lisa leslie. Jason statham. Florence and the machine. Deductive reasoning. Xi. Guarantee. Middletown. Kansas city chiefs games. Market. Nicholas sparks. Japan earthquake. Flatiron building. Wembanyama. Snl cast. Anna faris. Natural disasters. Chuck berry. What is zionism. Eu countries. Kate winslet. Edgar allan poe. Cast of the fall of the house of usher. What is ovulation. Foundation. Marquee. Irene ryan. Stephen curry. Arch. Colleen hoover books. Famous quotes. Mark mcgwire. Percy jackson and the olympians. Catch 22. Noose. Medieval times. Electric guitar. Endocrine system. Keanu reeves movies. Banco de america. Calligraphy alphabet. Martini. Isha ambani. Panic attack. Shays rebellion. Commonwealth. Sophia smith. Mood. Portland. Abraham. Portugal national football team vs slovenia national football team timeline. Edward teach. Pea. https://2025.uj74.ru/WHBIY.html
Dionysus. Biomass. Recidivism. 2023. Godzilla. Marilu henner. Tangled. Confederate flag. Micheal jackson. Coral island. Supercharger. Birth stones. Oxycodone. Jimmy carter. Stockholm syndrome. Water bug. Route 66. Fallen angels. Solemn. Youtuve. Taylor swift parents. Fifth amendment. Nathan lane. Seattle seahawks. Leukocytosis. Verge. Penny hardaway. Mariners schedule. Jeremiah johnson. Extortion. Flea market. Titanium dioxide. Russell wilson stats. Sovereign. Az. Dixie chicks. Poland flag. Garnet. Hacker. Stephen curry stats. Pen. Cartier. Tophat. Penguins. Nacl. Jimmy choo. Kinetic energy. Zionist. Satyr. FГєtbol club barcelona. Bluegill. Tenure definition. Y’. Tom hardy movies and tv shows. Morbid.
Diplomi_mzKn
November 11, 2024 @ 8:05 am
диплом с занесением в реестр купить [url=https://prem-diplom77.ru/]диплом с занесением в реестр купить[/url] .
Xazrycs
November 11, 2024 @ 8:10 am
Купить диплом о среднем образовании в Москве и любом другом городе
Eanrnqt
November 11, 2024 @ 8:15 am
Где и как купить диплом о высшем образовании без лишних рисков
RobertBeity
November 11, 2024 @ 8:41 am
Для успешного использования платформы необходимо иметь доступ к личному кабинету. Этот процесс включает несколько простых шагов, которые помогут вам зайти в аккаунт и использовать все его возможности. Независимо от устройства, важен правильный подход, чтобы избежать возможных ошибок https://wiki.soarer.ru/index.php?title=%20%D0%9F%D0%BE%D1%87%D0%B5%D0%BC%D1%83%20%D0%B2%D0%B0%D0%B6%D0%BD%D0%BE%20%D0%B8%D1%81%D0%BF%D0%BE%D0%BB%D1%8C%D0%B7%D0%BE%D0%B2%D0%B0%D1%82%D1%8C%20%D0%B7%D0%B5%D1%80%D0%BA%D0%B0%D0%BB%D0%BE%201win
Lloydwaf
November 11, 2024 @ 9:10 am
https://gabapharm.com/# Buy gabapentin for humans
CBEDWYF
November 11, 2024 @ 9:22 am
Viggo mortensen movies. Geminis. Dc comics. Richter scale. Manitoba. Av. What is temu. Fuel. Ardent. Ks state. India. Summer. Antifreeze. Emp. Ali wong. Peppermint. Dehydration. Giorgia meloni. Rosh hashanah. Salt lake city. Addias. Eldorado. What happens when you die. Bowling. Starwars. Chicharrones. Retarded. Chicago cubs games. Believe. Yellowstone. Grand junction. Mammoth. Trevor noah. Bias. Viggo mortensen. Stevie ray vaughan. Indubitably. Anna karenina. David foster. Poison hemlock. Stanley cup finals. Antagonist. Milwaukee bucks standings. Forgo. Empire. Bill maher. Art garfunkel. Butterflies. Devotion. Clandestine. Tokyo skytree. https://2025.uj74.ru/MQICL.html
Arkansas. Levis. Judy garland. Hammerhead shark. Timothy olyphant. Montgomery. Succeed. Russian ukraine. Masochist meaning. La brea tar pits. Question ai. Proclamation of 1763. Maya moore. Ender’s game. Gene kelly. Circuit. Pompeii. Fox animal. Copyright symbol. Robbie robertson. Mediastinum. Metronome. Logos. Monkeys. Spice girls. Mount rainier. Muskrat. Sheepshead fish. Salman khan. Gastrocnemius. Fender. Europa. Antibiotic. Indiana jones 1. Old movie. Earwig. Tear. Diwali. Banana. Transylvania. Baked alaska. Magna carta. Clip. Anya taylor joy. Coast guard. Los angeles lakers. Magic mountain.
Lazroaa
November 11, 2024 @ 9:54 am
Процесс получения диплома стоматолога: реально ли это сделать быстро?
situs toto slot
November 11, 2024 @ 10:10 am
My brother recommended I might like this website. He was once totally right.
This post actually made my day. You can not imagine just how so much time I had
spent for this info! Thanks!
Sazrmxd
November 11, 2024 @ 10:23 am
Как избежать рисков при покупке диплома колледжа или ПТУ в России
EnriqueDAUCT
November 11, 2024 @ 10:25 am
Мы – это интернациональная команда с большим опытом работы в сфере микрофинансирования, кредитования и банковского дела. Наши сотрудники ежедневно вносят свой вклад в микрофинансирование, помогая клиентам и партнерам выбрать лучшее решение для развития бизнеса микрозаймы для бизнеса
PORDXRH
November 11, 2024 @ 10:41 am
Claude. Kamala harris vice president. Roman polanski. Kristen bell movies and tv shows. Rebecca ferguson movies and tv shows. Obsequious. Rng. Herpes simplex. John belushi. Before. Fantasia. Miss piggy. The little things 2021 cast. Griselda blanco sons. Keith urban. Ruth bader ginsburg. Diplomacy. Raven symone. Cleft palate. Joker cast. Mitochondria function. Estradiol. Barbie film. Alternative. Amanita muscaria. Private practice. Claudette colvin. Essay. The meg. Christian. Milan italy. St. patrick’s day. Brutalist architecture. Puritan pride. Salvia plant. Quordle today. Epstein barr virus. Coca-cola. Limerick. Great expectations. Murad. Pitt. Asbestos. Truly. Appendix location. Gangnam style. Troy aikman. Salman khan. Biltmore house. Clown. Retribution definition. Sympathetic nervous system. Monster movie. Characteristics. Ape. Portugal. Magma. Spectrum. Wembanyama stats. Mickey rooney. Zionist. Spinach. https://2025.uj74.ru/LFMPB.html
Amine. Bangalore. Santana. Casablanca. Rocky mountains. Eeyore. New year’s eve. Brooklyn museum. Rupaul. Eclectic. Euthanasia. Rebuke. Marion cotillard. Gerrymandering. Hsv2. Whippets. Mark twain. Tommy john surgery. John david washington. N/a. Ambergris. Glycogen. Jesus christ superstar. Pigeon. Tokyo tower. Whiplash. West wing. Patrick mahomes parents. Tony parker. Nb. Linear. Bnp. Virtue. The color purple movie. Ray lewis. Palestine and israel. Guns. Womens world cup. Monique. The guardian. Nope. Bruce almighty. Management. Pawn. Buffalo wings. Pokemon games. S&p 500. University of southern california. Curiosity. Penn state university.
EugeneBub
November 11, 2024 @ 10:47 am
Мы предлагаем разнообразные курсы, чтобы соответствовать вашим потребностям в обучении вождению. Начните свою карьеру на дороге прямо сейчас вместе с нами сдать на права москва
ErnestBex
November 11, 2024 @ 10:48 am
Предложения для открытия бизнеса ИП с нуля — редкость. Для получения займа предприниматель уже должен работать на рынке. Обычно не меньше 6 месяцев онлайн кредит на развитие бизнеса
travmatpag
November 11, 2024 @ 11:13 am
Желаете безопасно купить травматическое оружие?
[img]https://cdn4.cdn-telegram.org/file/Hykr5BYwK6nwxMhh8Sd4720mHsTOOTaHi8rFb4p9krR2TW–oKm5zOax34TpTZjiJKB6w2_rrZuDjd3OJ7XKvCaRzxZxt1gKS68pAUlWcZ3xruXcgDbR4e7qeup0cdyWAKkteOwYQys2_u0x37Nfac4wP6d8nux-HKM7bepDzgJXQwSvevfOb6RgNuMOvld2OSvZh9xZBIIVLKeTxvs-AyF-1ttHAdZKCSxmvX3rt3nVA41MT5Ktm1ZZoxH9ocEc_JJhktNVI47S1uSBQqejoTnHhgPlR4aZnygwGk6XAz3JgXl36YjfPYo_nX6RGCGzHUpYm47sNxp4_hMuFidyVA.jpg[/img]
Сейчас проверенный канал в Телеграме «Поле чудес» предлагает своим пользователям заказать травматическое оружие, а познакомиться с предложениями возможно непосредственно на канале или же при помощи опытного сотрудника, который подробно расскажет про каждый вариант.
Это ТГ-канал сумел завоевать прекрасную репутацию, предлагая пистолеты высокого качества, благодаря которым можно защитить себя и родных. Клиенты могут бесплатно получить лицензию, благодаря которой использовать и носить травматическое оружие можно официально, без нарушения действующего законодательства. Магазин гарантирует полную конфиденциальность, поэтому пользователи смогут не беспокоиться о анонимности. Если вы хотите [url=https://telegra.ph/KUPIT-TRAVMATICHESKOE-BOEVOE-ORUZHIE-11-08]купить травматический пистолет без лицензии[/url] – то тут Вы сможете отыскать то, что искали!
Уникальные возможности канала
[img]https://cdn4.cdn-telegram.org/file/EX4LuJpQVUcXMWuaNyeur9Ba1f_2QUA-F1JRpOEd1r1fVmifLPw4dxUMVZgoWUlGdZ6rLyLO3PedlyJSxHyXyDKAZDEF4e8OdXcqKyfCATmgrDuebgDIB82EeY7K6YT3Iyf1mR5f8eNx-1ApoSxgX2OMsnt04Q4TuLAia3_n-kYAvgYU0LCBxJzFifds_9R3DhBNPesTIi3as4-e_rqj3FhdI3CP1-Sp-7GHxv-6MpDegOp1Dn46oHUI11sb7rgXDaQLb8fBLNrEgoNHKlY_RwP3wpr-dwFCmykA00kyNUAZ4iVH_V-9UoucchwCAhN_0TDW1gTS80POhOwWZPGFaQ.jpg[/img]
Этот ТГ-канал предлагает оружие по достаточно привлекательным ценам, а очень многие покупатели выбирают этот онлайн-магазин за счет следующих преимуществ:
• Прекрасная репутация.
• Доставка в день заказа.
• Большой комплект.
• Привлекательные цены.
Любой покупатель совместно с оружием получает магазин с патронами, кейс для хранения и набор для чистки, благодаря которому оружие прослужит длительные годы.
Преимущественные особенности магазина
[img]https://cdn4.cdn-telegram.org/file/LphN2SanQCudo4TjhATEuMLOxRPhByXa1pY56rVw-15jhHG6ciSGTTp2dr5ta3NjeC98_pP6wFukwyNqkgwDAdWBEwLiUkVZr2fa_1EYDi-hOjGpIFMfkdbunQFeIpadHlmpWNmsELsRynX_nmDIwWjlUH9-E0ICVxrqR-qLQmWwLXjTel-JNrYnseX9D7SV4Jb0vsFhovfWcoFs8u4Sg57MhTpP7hRjQhZuSU5R3JepeSbUktIt_tMozLpXerjdvPhyWy1geP4AN2yMnPRS0okh9bEqfIGaNmm39lCDwg2G9QC_hjHTmxJeMv7YLNhTTLRD3DAWFsDeJGsa1z9Ezw.jpg[/img]
Данный ТГ-канал на протяжении длительного времени специализируется на продаже надежного оружия, поэтому в каталоге клиентов ждут варианты, благодаря которым можно не беспокоиться о своей безопасности и близких. Заказать травмат возможно просто и легко, связавшись со специалистом в Телеграме, после чего заказ будет оформлен очень быстро. В том случае, если вы хотите [url=https://telegra.ph/KUPIT-TRAVMATICHESKOE-BOEVOE-ORUZHIE-11-08]оружие купить боевой пистолет[/url] – то здесь Вы всегда отыщите то, что искали!
Заказать травматическое оружие можно с оперативной доставкой во все регионы нашей страны, получив его в день оформления, помимо всего прочего, доставка осуществляется абсолютно бесплатно в указанное место или же курьером. Именно здесь трудится сплоченная команда профессионалов, которые знают почти все про травматическое оружие и всегда могут поделиться своим мнением с заказчиками!
Dennisinamy
November 11, 2024 @ 11:59 am
Buy gabapentin for humans: buy gabapentin india – buy Gabapentin GabaPharm
Mazrbif
November 11, 2024 @ 12:33 pm
Купить диплом магистра оказалось возможно, быстрое обучение и диплом на руки
Dnrtxua
November 11, 2024 @ 12:46 pm
Возможно ли купить диплом стоматолога, и как это происходит
MultiversX web wallet
November 11, 2024 @ 1:19 pm
Excellent beat ! I would like to apprentice at the same time as you amend
your website, how can i subscribe for a blog web site? The account
aided me a appropriate deal. I were tiny bit familiar of this your broadcast provided vibrant
clear concept
kesehatan
November 11, 2024 @ 1:36 pm
Hello! Quick question that’s entirely off topic. Do you know how to make
your site mobile friendly? My website looks weird when viewing from my iphone.
I’m trying to find a theme or plugin that might be able to correct this issue.
If you have any recommendations, please share. Many thanks!
Cazriku
November 11, 2024 @ 1:38 pm
Всё, что нужно знать о покупке аттестата о среднем образовании
PAXCUGZ
November 11, 2024 @ 2:18 pm
When is christmas. The fugitive. Avatar. Italian. Maple tree. Hades. Shutter island. Iowa. Heterotroph. Blueberry. James gandolfini. Temperatura. 26th amendment. Autotroph. There. Toronto raptors. James cameron. Geocaching. Bloom’s taxonomy. Slovakia. Hugh dancy. Lark. Birth flowers. Indiana jones 5 cast. Suspension. Ruminate. Trellis. Anunnaki. Texas state. Pratt and whitney. New rochelle. Avril lavigne. Septic shock. Boston red sox. Joe burrow stats. Rohit sharma. Justin beiber. Model y. Valkyrie. Min. Pathology. Gnats. Crown. Bullet train. Something about mary. Tahiti. Rough. When is the superbowl. https://2025.uj74.ru/NMGSC.html
Leeds. Ferris buellers day off. Veal. Trisomy 18. Qr scanner. 4×4. Free bird. Jerry west. Wordle. Raging bull. Nicholas cage. Hieronymus bosch. How old is kamala harris. Middlebury college. Ken griffey jr. Jason bateman. Static. Mama’s family. Vip. Muscle. Masochist. Pica. Flat. Incredibles. Planets. Insulin. Pirate bays. Ofc meaning. Battle of saratoga. Creole. Gen z. Mad. Tom cruise. Happy halloween. Jimmy carter age. Condescending. Forest. Hofstra university. Plaintiff. Wordle game. Siberian husky. Meat. Chichen itza. Isosceles triangle. Cellular respiration. Grateful. 13. Camouflage. Louisiana purchase. Slash. Point reyes. Donald trump age.
intimdnr.one
November 11, 2024 @ 2:46 pm
Ищете [url=https://intimdnr.one/]заказ проститутки цена[/url]? На нашем сайте представлены только реальные анкеты с актуальными ценами. Выбирайте девушек, которые готовы воплотить ваши желания в жизнь. Простой и безопасный процесс заказа подарит вам незабываемые моменты. Полная конфиденциальность гарантирована.
эскорт элит москва
November 11, 2024 @ 2:53 pm
[url=https://telegra.ph/EHSKORT-MOSKVA–RED-ROOM-11-08]проститутки москва дубровка[/url]
login panutantoto panutantoto login
November 11, 2024 @ 3:03 pm
Hey there! Do you use Twitter? I’d like to follow you
if that would be okay. I’m absolutely enjoying
your blog and look forward to new posts.
IXNXIRA
November 11, 2024 @ 3:19 pm
Bruce willis. Poignant meaning. The gentlemen. Phillies vs braves. Television show. Sic. Taurus horoscope. Bioluminescence. Pasadena. Kevin bacon. African cup of nations. Hope floats. Sinus bradycardia. Christina aguilera. Scarlett johansson movies. Whirlpool. Avengers. Frank. How did hitler die. Somnolence. Pus. Toe. Desantis. Tricep. Harrisburg pa. Veep. Carly rae jepsen. Crystal palace. Relationship. Humerus. Lust. Max 401k contribution 2024. Seven movie. Polyp. Mosquito. Thanksgiving. Jacksonville florida. Midwife. Nt. Washer. Huffington post. Generous. Adept. Assimilation. Guy pearce. Brasil. Colorado avalanche. Cyber monday. https://2025.uj74.ru/WPJDA.html
Antwerp. Imbd. Snow leopard. Disney movies. Odessa tx. Artificial intelligence. Memoir. Peter gallagher. Rachel mcadams. TelГ©fono inteligente. Madden. Colorado school of mines. Masochist meaning. Linda blair. Louboutin. Ricky gervais. Superbowl sunday. Hillbilly elegy movie cast. Constituents. Baltimore. Goofy. Nazi. Salami. Thermal energy. Janet jackson. Smith college. Wrigley field. E. Vasopressin. Ivanka trump. Shamrock. Amendment. Jlo. Immediate. Beta fish. Clonazepam. Crystals. Mamas family. Maureen o’hara. Bobby flay and. Epstein island. King baldwin. Ferry. Route 66. Scurvy. Romania. Trapezoid. Circle of fifths. Transmission. Tent.
Curtisspeve
November 11, 2024 @ 4:46 pm
buy rybelsus online usa [url=http://rybpharm.com/#]rybpharm[/url] buy rybelsus canada
Sazryyb
November 11, 2024 @ 4:57 pm
Официальная покупка диплома ПТУ с упрощенной программой обучения
Vivod iz zapoya rostov_hrea
November 11, 2024 @ 6:12 pm
быстрый вывод из запоя ростов [url=http://ideya.forums.party/viewtopic.php?id=653/]http://ideya.forums.party/viewtopic.php?id=653/[/url] .
Sazrsvn
November 11, 2024 @ 6:20 pm
Как официально купить диплом вуза с упрощенным обучением в Москве
BOKEP INDONESIA
November 11, 2024 @ 6:43 pm
With havin so much content and articles do you ever
run into any problems of plagorism or copyright violation? My website has a lot of
exclusive content I’ve either created myself or outsourced
but it seems a lot of it is popping it up all over the internet without my agreement.
Do you know any techniques to help prevent content from being stolen? I’d genuinely appreciate it.
franshizi_ekka
November 11, 2024 @ 6:53 pm
по франшизе [url=http://franshizy21.ru]по франшизе[/url] .
franshizi_dhMn
November 11, 2024 @ 6:53 pm
франшизы купить [url=www.franshizy19.ru]франшизы купить[/url] .
franshizi_rbel
November 11, 2024 @ 6:53 pm
выгодные франшизы [url=http://franshizy22.ru/]выгодные франшизы[/url] .
franshizi_fvSr
November 11, 2024 @ 6:53 pm
какие есть франшизы [url=franshizy18.ru]какие есть франшизы[/url] .
Lazrxdg
November 11, 2024 @ 7:15 pm
Узнайте стоимость диплома высшего и среднего образования и процесс получения
pdapal.com/read-blog/4776
seo
November 11, 2024 @ 8:43 pm
Attractive component of content. I just stumbled upon your blog and in accession capital to say that I acquire actually
loved account your weblog posts. Any way I will be subscribing on your
feeds and even I success you get entry to consistently fast.
огнестойкие сейфы
November 11, 2024 @ 9:28 pm
Тут можно преобрести сейф противопожарный несгораемый сейф цена
DownloadJet
November 11, 2024 @ 9:59 pm
[url=https://jet-casino-apk.ru/jet-skachat]jet-casino-apk.ru/jet-skachat[/url]
Загрузить последнюю версию приложения казино онлайн Джет – играй сегодня!
https://www.jet-casino-apk.ru/jet-skachat
купить сейф для охотничьего ружья
November 11, 2024 @ 11:48 pm
Тут можно преобрести сейф под ружье купить купить оружейный шкаф
Mazrtzq
November 12, 2024 @ 1:28 am
Быстрое обучение и получение диплома магистра – возможно ли это?
Freddiegex
November 12, 2024 @ 1:37 am
Использование разнообразных источников финансирования помогает компаниям достигать устойчивого развития, минимизируя риски и максимизируя эффективность инвестиций в проекты и инновации http://www.alltab.co.kr/bbs/board.php?bo_table=free&wr_id=1745541
SolSait
November 12, 2024 @ 2:05 am
[url=https://sol-casino-apk.ru/sol-oficzialnyj-sajt/]http://sol-casino-apk.ru/sol-oficzialnyj-sajt/[/url]
Скачать apk файл казино Sol – побеждай прямо сейчас!
sol-casino-apk.ru/sol-oficzialnyj-sajt/
MonroCasinoApk
November 12, 2024 @ 2:06 am
[url=https://monro-casino-apk.ru]http://www.monro-casino-apk.ru[/url]
Скачать apk файл казино онлайн Monro – играй сейчас!
http://www.monro-casino-apk.ru
Richardhop
November 12, 2024 @ 2:29 am
Подскажите, где в Москве можно купить качественную строительную технику, такую как башенные краны или мачтовые подъемники? Интересуют надежные компании с хорошей репутацией https://ogorodland.ru/forum/topic/vygoda-lizinga-mashin-udobstvo-i-finansovaya-gibkost/
Diplomi_deSi
November 12, 2024 @ 2:49 am
Купить диплом о среднем полном образовании, в чем подвох и как избежать обмана?
Gradysib
November 12, 2024 @ 3:01 am
Лизинг — услуга для компаний и ИП, снижающая затраты на покупку транспортных средств для коммерческих и личных целей. Выберите автомобиль, подпишите договор лизинг на автотранспорт
pagbet
November 12, 2024 @ 3:16 am
Hi there to every body, it’s my first pay a visit of this weblog; this blog contains
amazing and genuinely excellent information for readers.
SolCasinoApk
November 12, 2024 @ 5:39 am
[url=https://sol-casino-apk.ru]https://sol-casino-apk.ru[/url]
Установить apk файл казино онлайн Сол – играй сегодня!
http://www.sol-casino-apk.ru
товар за отзыв Wildberries
November 12, 2024 @ 6:04 am
[url=https://tenchat.ru/media/2728961-razdacha-tovarov-za-otzyvy-gayd-po-vykupam]выкуп товаров за отзыв на ВБ[/url]
Lloydwaf
November 12, 2024 @ 6:46 am
https://rybpharm.com/# buy rybelsus online usa
Life Experience Degree
November 12, 2024 @ 8:37 am
It’s an remarkable paragraph in favor of all the internet users; they will get benefit from it I am sure. https://www.hampdenstateuniversity.com/
Cazrxgu
November 12, 2024 @ 9:27 am
Как купить аттестат 11 класса с официальным упрощенным обучением в Москве
Dennisinamy
November 12, 2024 @ 9:30 am
buy furosemide online: furosemide – furosemide furpharm.com
seo services
November 12, 2024 @ 9:51 am
Hi colleagues, how is the whole thing, and what you desire to say concerning this piece
of writing, in my view its actually remarkable designed for me.
Sazrznf
November 12, 2024 @ 9:58 am
Как избежать рисков при покупке диплома колледжа или ПТУ в России
SelectorDownload
November 12, 2024 @ 10:16 am
[url=https://selector-casino-apk.ru/selector-skachat/]www.selector-casino-apk.ru/selector-skachat/[/url]
Загрузить apk файл казино Селектор – играй сейчас!
http://www.selector-casino-apk.ru/selector-skachat/
Freddiegex
November 12, 2024 @ 10:45 am
Escape From Tarkov, is a realistic first-person shooter, from the Russian Battlestate Games that came out as a closed alpha in 2016, currently in an open beta phase. The game is about escaping from Tarkov, and that happens in the world, with both NPCs and other players, raiding, and lots of lots https://www.repaire.net/forums/articles/hdr-hfr-deux-nouvelles-tendances-ibc-2015.553/
JamesTrinc
November 12, 2024 @ 10:58 am
Поднимайте ставки и выигрывайте крупные суммы с онлайн-казино Плейфортуна! Уникальные слоты и щедрые бонусы ждут вас здесь https://playfortuna24x7sb.com/ru
buy roxicodone 30mg online
November 12, 2024 @ 11:51 am
It is appropriate time to make a few plans for the longer term and it is time to be happy.
I’ve learn this submit and if I may just I desire to recommend you few attention-grabbing things or advice.
Maybe you can write subsequent articles regarding this article.
I desire to learn even more things approximately
it!
DavidFuh
November 12, 2024 @ 12:21 pm
Расчет и покупка спецтехники в лизинг для юр. лиц и ИП. Лучшие предложения лизинга на строительную технику и быстрое оформление взять лизинг на спецтехнику
hani faddoul
November 12, 2024 @ 1:00 pm
Hello colleagues, its enormous post about tutoringand entirely explained,
keep it up all the time.
sarıyer escort
November 12, 2024 @ 1:29 pm
merhaba admin sarıyer escort maslak escort sitesini ziyaret etmelerini istiyebilirmisin https://maslakescortbul.com/
Curtisspeve
November 12, 2024 @ 1:43 pm
furosemide furpharm.com [url=https://furpharm.com/#]furpharm[/url] furosemide furpharm.com
Williamraics
November 12, 2024 @ 1:47 pm
Как правильно купить диплом колледжа и пту в России, подводные камни
homerumor
November 12, 2024 @ 1:49 pm
What a data of un-ambiguity and preserveness of valuable knowledge about unpredicted emotions.
самовыкуп товара за отзывы на WB
November 12, 2024 @ 1:51 pm
[url=https://t.me/tovaryzaotzivkeshbek]рекламные товары за отзыв WB[/url]
pupis
November 12, 2024 @ 2:15 pm
Теперь вы можете скачать легальные БК и начать делать ставки на любые спортивные события прямо с телефона
LamarEmand
November 12, 2024 @ 2:39 pm
Приобретение нового оборудования в лизинг для бизнеса на выгодных условиях. Длительные сроки финансирования. Специальные программы от поставщиков оборудование для бизнеса в лизинг
CharlesNaido
November 12, 2024 @ 3:06 pm
Какую спецтехнику можно взять в лизинг? В нашей компании предоставляется большой выбор спецтехники для лизинга. Вам доступны экскаваторы, погрузчики, бульдозер спецтехника в лизинг купить
шлюхи города снять
November 12, 2024 @ 3:12 pm
Ищете, как организовать [url=https://intimdnr.one/]вызов проститутки недорого[/url]? Мы собрали анкеты девушек с доступными ценами и высоким качеством услуг. Независимо от вашего бюджета, каждая встреча будет наполнена страстью и вниманием. Ваша безопасность и конфиденциальность — наш приоритет.
brands
November 12, 2024 @ 3:30 pm
If you would like to improve your knowledge simply keep visiting this site and
be updated with the most up-to-date information posted here.
muelear oxidize
November 12, 2024 @ 3:31 pm
Great article.
Sazrzil
November 12, 2024 @ 3:54 pm
Приобретение диплома ПТУ с сокращенной программой обучения в Москве
DownloadJet
November 12, 2024 @ 3:54 pm
[url=https://jet-casino-apk.ru/jet-skachat]jet-casino-apk.ru/jet-skachat[/url]
Загрузить последнюю версию приложения казино онлайн Джет – выигрывай сегодня!
https://www.jet-casino-apk.ru/jet-skachat
ZSot
November 12, 2024 @ 3:56 pm
6 win
Williamnet
November 12, 2024 @ 4:12 pm
услуги укладки кафельной плитки работ укладке кафельной плитки
nişan organizasyonu
November 12, 2024 @ 4:25 pm
Leave a comment and let us know what your favorite blog post has been so far!
Oarioraps
November 12, 2024 @ 5:06 pm
Как быстро и легально купить аттестат 11 класса в Москве
JetApk
November 12, 2024 @ 5:31 pm
[url=https://jet-casino-apk.ru/jet-apk]https://www.jet-casino-apk.ru/jet-apk[/url]
Скачать приложение казино Jet – побеждай прямо сейчас!
https://jet-casino-apk.ru/jet-apk
RobertAtose
November 12, 2024 @ 7:28 pm
https://gabapharm.com/# cheapest Gabapentin GabaPharm
singapore
November 12, 2024 @ 7:43 pm
These are really enormous ideas in on the topic of blogging.
You have touched some fastidious things here. Any way keep up wrinting.
Dennisinamy
November 12, 2024 @ 7:58 pm
furpharm: furosemide furpharm.com – furosemide fur pharm
ZSot
November 12, 2024 @ 8:45 pm
neon54
Sazrufx
November 12, 2024 @ 9:01 pm
Официальная покупка диплома вуза с упрощенной программой обучения
kryza.network/create-blog/
Williamnet
November 12, 2024 @ 9:23 pm
укладка кафельной плитки цена работы за м2 укладка кафельной плитки спб
profi teh remont
November 12, 2024 @ 10:04 pm
Сервисный центр предлагает ремонт кофемашины la cimbali недорого починить кофемашины la cimbali
Vivod iz zapoya rostov_hlpl
November 12, 2024 @ 10:59 pm
принудительный вывод из запоя ростов [url=https://cah.forum24.ru/?1-19-0-00000457-000-0-0-1730725499/]https://cah.forum24.ru/?1-19-0-00000457-000-0-0-1730725499/[/url] .
Vivod iz zapoya rostov_grPn
November 12, 2024 @ 10:59 pm
алкоголизм лечение вывод из запоя ростов [url=www.aqvakr.forum24.ru/?1-7-0-00011575-000-0-0-1730725206/]алкоголизм лечение вывод из запоя ростов[/url] .
Vivod iz zapoya rostov_upSn
November 12, 2024 @ 11:00 pm
алкоголизм лечение вывод из запоя ростов [url=http://www.spilkuvannya.rolevaya.com/viewtopic.php?id=65]http://www.spilkuvannya.rolevaya.com/viewtopic.php?id=65[/url] .
Vivod iz zapoya rostov_bfEl
November 12, 2024 @ 11:01 pm
вывод из запоя ростов-на-дону [url=http://www.masa.forum24.ru/?1-16-0-00002625-000-0-0-1730725831]http://www.masa.forum24.ru/?1-16-0-00002625-000-0-0-1730725831[/url] .
Vivod iz zapoya rostov_pnMl
November 12, 2024 @ 11:02 pm
вывод из запоя круглосуточно [url=aktivnoe.forum24.ru/?1-7-0-00012822-000-0-0-1730725137]aktivnoe.forum24.ru/?1-7-0-00012822-000-0-0-1730725137[/url] .
здоров'я
November 12, 2024 @ 11:38 pm
I do not know whether it’s just me or if perhaps everybody else experiencing issues with your website.
It seems like some of the text within your posts are running off the screen. Can someone
else please provide feedback and let me know if this is happening to them too?
This may be a issue with my web browser because I’ve had this happen previously.
Many thanks
Sazrcan
November 12, 2024 @ 11:58 pm
Как купить диплом о высшем образовании с минимальными рисками
Curtisspeve
November 13, 2024 @ 12:09 am
rybpharm cheap semaglutide [url=http://rybpharm.com/#]buy rybelsus rybpharm[/url] buy rybelsus online usa
sünnet organizasyonu
November 13, 2024 @ 12:27 am
Your blog has become a source of guidance and support for me Your words have helped me through some of my toughest moments
GeorgeRor
November 13, 2024 @ 12:55 am
На приеме гинеколог проводит осмотр воспалений, новообразований и других гинекологических патологий, проводит диагностику заболеваний женских половых органов и выявляет их на ранних стадиях https://amvnews.ru/forum/profile.php?mode=viewprofile&u=80688
SelectorDownload
November 13, 2024 @ 2:47 am
[url=https://selector-casino-apk.ru/selector-skachat/]https://www.selector-casino-apk.ru/selector-skachat/[/url]
Скачать последнюю версию приложения казино онлайн Selector – играй сейчас!
https://selector-casino-apk.ru/selector-skachat/
DavidHem
November 13, 2024 @ 3:03 am
Друзья, мне нужен совет знающих. Можете ли Вы мне подсказать каким образом можно вывести на гривну криптовалюту? Какие обменники есть их хороших https://oih.at.ua/forum/63-19319-1
рекламные товары за отзывы Wildberries
November 13, 2024 @ 4:05 am
[url=https://tenchat.ru/media/2750073-tovary-za-otzyv-razdachi-s-keshbekom-ili-bally-za-otzyv]выкупы товаров за отзывы на Вайлдберриз[/url]
Sazrsjo
November 13, 2024 @ 4:19 am
Официальная покупка диплома вуза с сокращенной программой обучения в Москве
caluanie muelear oxidize near me
November 13, 2024 @ 5:49 am
Hey there, You have done an excellent job. I will certainly digg it and personally recommend to my friends.
I am confident they’ll be benefited from this web site.
ZSot
November 13, 2024 @ 5:56 am
1 win bet
Lazrkkm
November 13, 2024 @ 6:44 am
Приобретение школьного аттестата с официальным упрощенным обучением в Москве
Trefawa
November 13, 2024 @ 6:46 am
Как купить аттестат 11 класса с официальным упрощенным обучением в Москве
astroroleplay.getbb.ru/viewtopic.php?f=6&t=706
Michaeluteli
November 13, 2024 @ 6:50 am
Как выбрать гирлянды для новогоднего оформления фасада дома? Виды подсветки, лучшие материалы, обзор разных моделей гирлянд и советы по монтажу: https://dzen.ru/a/Zy5D8pM8zzfwUtJ0
tani sex telefon
November 13, 2024 @ 7:38 am
I have been exploring for a bit for any high quality articles or
weblog posts on this sort of area . Exploring in Yahoo I at last stumbled upon this
web site. Studying this information So i am glad to express that I
have an incredibly just right uncanny feeling I discovered exactly what I needed.
I such a lot undoubtedly will make certain to do not forget this
website and provides it a glance on a constant basis.
prodamus promokod_krmt
November 13, 2024 @ 9:02 am
Продамус промокод [url=http://www.prodamus-promokod1.ru]Продамус промокод[/url] .
Dnrtnly
November 13, 2024 @ 9:10 am
Быстрая покупка диплома старого образца: возможные риски
rajabandot
November 13, 2024 @ 9:43 am
Admiring the dedication you put into your blog and
in depth information you offer. It’s awesome to come across a blog every once in a while that isn’t the same out of
date rehashed information. Great read! I’ve bookmarked
your site and I’m adding your RSS feeds to my Google
account.
Diplomi_dfMn
November 13, 2024 @ 9:44 am
купить диплом в одинцово [url=https://prema365-diploms.ru/]купить диплом в одинцово[/url] .
duatoto togel
November 13, 2024 @ 10:30 am
What a information of un-ambiguity and preserveness of valuable know-how regarding unexpected
emotions.
Curtisspeve
November 13, 2024 @ 10:57 am
kampharm.shop [url=http://kampharm.shop/#]kampharm.shop[/url] kam pharm shop
RobertAtose
November 13, 2024 @ 11:07 am
https://furpharm.com/# furpharm
duatoto login
November 13, 2024 @ 11:17 am
Today, I went to the beach front with my kids. I found a sea shell and gave it to my 4 year old daughter and said “You can hear the ocean if you put this to your ear.” She put the shell
to her ear and screamed. There was a hermit crab inside and it pinched her ear.
She never wants to go back! LoL I know this is completely off topic but I had to tell
someone!
ZSot
November 13, 2024 @ 11:43 am
neon54 ????????
Mazrucw
November 13, 2024 @ 12:29 pm
Как получить диплом о среднем образовании в Москве и других городах
Lloydwaf
November 13, 2024 @ 2:04 pm
https://rybpharm.com/# rybpharm cheap semaglutide
JetApk
November 13, 2024 @ 2:07 pm
[url=https://jet-casino-apk.ru/jet-apk]jet-casino-apk.ru/jet-apk[/url]
Скачать последнюю версию приложения казино Jet – выигрывай сейчас!
jet-casino-apk.ru/jet-apk
RichardLox
November 13, 2024 @ 2:24 pm
Обмен криптовалют на USDT (Tether) является обычной практикой в мире цифровых активов, предлагая стабильную альтернативу более волатильным криптовалютам https://andrushivka.at.ua/forum/8-258-1#15191
WilliamAlkam
November 13, 2024 @ 2:38 pm
Платформа уникальная и отличная в плане удобств (начиная от ввода и вывода средств, заканчивая инструментами для торговли). Постоянные улучшения. Куча графиков покет опшн
купить сейф для оружия
November 13, 2024 @ 3:09 pm
Тут можно преобрести купить оружейный шкаф сейф для оружие
escort ataköy
November 13, 2024 @ 3:39 pm
merhaba bakırköy escort ataköy escort sitesini ziyaret etmeliler
DavidHax
November 13, 2024 @ 3:45 pm
Купить виртуальный сервер VPS/VDS от 169 р/месяц, с возможностью самостоятельного администрирования. Аренда хостинга VPS или VDS аренда виртуального сервера
payroll and hris software
November 13, 2024 @ 3:50 pm
What i do not realize is if truth be told how you
are no longer actually a lot more smartly-favored
than you might be right now. You’re very intelligent. You recognize thus considerably on the subject of this subject,
made me in my view imagine it from numerous varied angles.
Its like men and women are not involved unless
it is something to accomplish with Woman gaga! Your own stuffs
nice. All the time handle it up!
photo restoration
November 13, 2024 @ 4:31 pm
This blog is a great mix of informative and entertaining content It keeps me engaged and interested from start to finish
hris platform
November 13, 2024 @ 5:33 pm
Very descriptive blog, I enjoyed that bit. Will there be a part 2?
Iariorykt
November 13, 2024 @ 5:38 pm
Полезная информация как официально купить диплом о высшем образовании
CasinoJet
November 13, 2024 @ 6:44 pm
[url=https://jet-casino-apk.ru/jet-casino/]jet-casino-apk.ru/jet-casino/[/url]
Скачать apk файл казино Джет – побеждай сейчас!
jet-casino-apk.ru/jet-casino/
Vivod iz zapoya rostov_ryEl
November 13, 2024 @ 7:28 pm
вывод. из. запоя. ростов. [url=www.gonochki.forum24.ru/?1-10-0-00001135-000-0-0-1730726022/]www.gonochki.forum24.ru/?1-10-0-00001135-000-0-0-1730726022/[/url] .
Vivod iz zapoya rostov_ddel
November 13, 2024 @ 7:47 pm
вывод из запоя дешево ростов на дону [url=www.vip.mybb.rocks/viewtopic.php?id=7671/]www.vip.mybb.rocks/viewtopic.php?id=7671/[/url] .
Vivod iz zapoya rostov_plMn
November 13, 2024 @ 7:50 pm
вывод из запоя ростов на дону на дому [url=www.forumbar.anihub.me/viewtopic.php?id=9752/]www.forumbar.anihub.me/viewtopic.php?id=9752/[/url] .
Vivod iz zapoya rostov_fpOr
November 13, 2024 @ 7:52 pm
вывод из запоя цена ростов [url=www.internetmoney.bestbb.ru/viewtopic.php?id=31698#p87431/]вывод из запоя цена ростов[/url] .
ZSot
November 13, 2024 @ 7:58 pm
vavada promo codes
Bonnie B. S. Shaw
November 13, 2024 @ 8:12 pm
You have a way of making each of your readers feel seen and heard That’s a special quality that not all bloggers possess Thank you for creating a safe space for us
DanielTom
November 13, 2024 @ 9:06 pm
Готов услышать трек, который выбьет из тебя все оправдания? Слушай, если не боишься правды.
Curtisspeve
November 13, 2024 @ 9:21 pm
furosemide fur pharm [url=https://furpharm.com/#]furosemide fur pharm[/url] fur pharm
SelectorCasino
November 13, 2024 @ 11:04 pm
[url=https://selector-casino-apk.ru/selector-casino]http://www.selector-casino-apk.ru/selector-casino[/url]
Загрузить apk файл казино Selector – выигрывай сейчас!
http://selector-casino-apk.ru/selector-casino
JetAndroid
November 13, 2024 @ 11:56 pm
[url=https://jet-casino-apk.ru/jet-na-androjd/]https://jet-casino-apk.ru/jet-na-androjd/[/url]
Скачать последнюю версию приложения онлайн казино Jet – выигрывай сейчас!
https://www.jet-casino-apk.ru/jet-na-androjd/
Lazrnun
November 14, 2024 @ 12:04 am
Аттестат школы купить официально с упрощенным обучением в Москве
MonroAndroid
November 14, 2024 @ 12:13 am
[url=https://monro-casino-apk.ru/monro-na-androjd]www.monro-casino-apk.ru/monro-na-androjd[/url]
Установить приложение онлайн казино Monro – побеждай сейчас!
http://www.monro-casino-apk.ru/monro-na-androjd
ZSot
November 14, 2024 @ 12:36 am
bc game download for android
Sazrolx
November 14, 2024 @ 1:32 am
Как безопасно купить диплом колледжа или ПТУ в России, что важно знать
RobertAtose
November 14, 2024 @ 2:36 am
https://rybpharm.com/# rybpharm
rajabandot daftar
November 14, 2024 @ 2:36 am
I love what you guys are usually up too.
This sort of clever work and coverage! Keep up the good works guys I’ve you guys to our blogroll.
Lloydwaf
November 14, 2024 @ 2:54 am
https://erepharm.com/# erepharm pills
IsaacTof
November 14, 2024 @ 3:19 am
Для создания индивидуального тарифа на виртуальный выделенный сервер в России можно воспользоваться онлайн-калькулятором на сайте, или обратиться в службу провайдер хостинг
DavidMob
November 14, 2024 @ 3:26 am
Когда речь идет о комфортной организации пространства, на помощь приходят инновационные подходы. Элементы, которые можно комбинировать и адаптировать, открывают множество возможностей для создания идеального хранилища гардеробные системы купить в москве
ThomasSig
November 14, 2024 @ 3:29 am
Разнообразие предложений на рынке позволяет легко подобрать удобные варианты для хранения личных вещей. Предметы могут быть адаптированы под любые размеры и потребности, что делает процесс организации не только полезным, но и приятным заказать двери купе для гардеробной
сейф купить
November 14, 2024 @ 3:58 am
Здесь можно преобрести сейф в наличии сейф купить цена
hris software examples
November 14, 2024 @ 5:05 am
Inspiring story there. What happened after? Good luck!
Rileypaf
November 14, 2024 @ 5:51 am
Viagra * Cialis * Levitra
All the products you are looking an eye to are currently close by for the duration of 1+1.
4 more tablets of one of the following services: Viagra * Cialis * Levitra
https://pxman.net
Sazrrup
November 14, 2024 @ 5:56 am
Как получить диплом техникума с упрощенным обучением в Москве официально
RobertAtose
November 14, 2024 @ 5:56 am
https://gabapharm.com/# gabapentin GabaPharm
Sazrddd
November 14, 2024 @ 5:57 am
Официальная покупка школьного аттестата с упрощенным обучением в Москве
Uazrjfe
November 14, 2024 @ 5:58 am
Как избежать рисков при покупке диплома колледжа или ПТУ в России
kruizturm.ru/kak-mozhno-kupit-attestat-za-9-klass-skolko-stoit.html
koitoto slot
November 14, 2024 @ 6:11 am
What’s up it’s me, I am also visiting this site daily,
this site is actually pleasant and the users are actually sharing fastidious thoughts.
raketaigra
November 14, 2024 @ 7:26 am
Загрузите скачать Lucky jet и приступайте к игре, где каждый ход важен.
Cazrnzf
November 14, 2024 @ 7:47 am
Как получить диплом техникума официально и без лишних проблем
Oariorgmx
November 14, 2024 @ 8:12 am
Приобретение диплома ПТУ с сокращенной программой обучения в Москве
Maha jitu
November 14, 2024 @ 8:28 am
Greetings! Very useful advice in this particular article!
It’s the little changes which will make the largest changes.
Thanks a lot for sharing!
Mahjong Wins 3
November 14, 2024 @ 8:57 am
Wonderful beat ! I would like to apprentice while you amend your web site,
how can i subscribe for a blog website? The account helped
me a acceptable deal. I had been tiny bit acquainted of
this your broadcast offered bright clear idea
Sazryfw
November 14, 2024 @ 9:08 am
Как получить диплом техникума с упрощенным обучением в Москве официально
ZSot
November 14, 2024 @ 9:54 am
hash game bc
luxury villas rent thailand
November 14, 2024 @ 10:18 am
Because the admin of this web page is working, no hesitation very shortly it will be well-known, due to its feature contents.
Curtisspeve
November 14, 2024 @ 10:21 am
buy rybelsus rybpharm [url=https://rybpharm.com/#]rybpharm cheap semaglutide[/url] rybpharm rybelsus
Eanrcwg
November 14, 2024 @ 10:44 am
Приобретение диплома ПТУ с сокращенной программой обучения в Москве
luxury villa in thailand
November 14, 2024 @ 11:33 am
Hello all, here every person is sharing such familiarity,
therefore it’s pleasant to read this web site, and I used to visit this blog every day.
LesleyLof
November 14, 2024 @ 12:36 pm
стоимость специальной оценки условий труда спецоценка условий труда организации
aviator online game
November 14, 2024 @ 12:43 pm
The Aviator game is a favorite among Indian players for its real-time betting and intense strategy. Choose to play for real money or try the demo mode to learn the mechanics risk-free. It’s all about when you pull the trigger!
aviator money game [url=https://aviator-games-online.ru/]online aviator game[/url] .
WilliamEnume
November 14, 2024 @ 1:30 pm
With BlueWallet, you maintain ownership of your private keys. This Bitcoin wallet is crafted by Bitcoin enthusiasts for the community BlueWallet
ManuelSam
November 14, 2024 @ 1:36 pm
In the heart of a city renowned for romance, elegance, and sophistication, there lies a world dedicated to offering unforgettable, high-class companionship http://doc.repai.com/doku.php?id=_Top_10_Reasons_to_Hire_an_Escort_in_the_City_of_Love
RichardDam
November 14, 2024 @ 2:00 pm
VPS на Windows — это виртуальный выделенный сервер, развёрнутый в облачном кластере, с предустановленной лицензионной ОС Windows Server. Виртуализация KVM vps хостинг windows
Vivod iz zapoya rostov_rvel
November 14, 2024 @ 2:06 pm
алкоголизм лечение вывод из запоя ростов [url=ideya.forums.party/viewtopic.php?id=654]алкоголизм лечение вывод из запоя ростов[/url] .
promokody_wdea
November 14, 2024 @ 2:13 pm
промокод продамус на 5000 [url=www.forumsilverstars.forum24.ru/?1-2-0-00000162-000-0-0-1731577187]www.forumsilverstars.forum24.ru/?1-2-0-00000162-00[/url] .
vivod iz zapoya kryglosytochno_qyEl
November 14, 2024 @ 2:37 pm
вывод из запоя в ростове-на-дону [url=https://kyevlyn.ukrbb.net/viewtopic.php?f=2&t=13619/]https://kyevlyn.ukrbb.net/viewtopic.php?f=2&t=13619/[/url] .
kazino onlain_wqEn
November 14, 2024 @ 2:40 pm
онлайн казино [url=https://www.casino-bonus.by]онлайн казино[/url] .
Cazrqbf
November 14, 2024 @ 2:46 pm
Узнайте, как безопасно купить диплом о высшем образовании
Vivod iz zapoya rostov_zmol
November 14, 2024 @ 2:51 pm
вывод из запоя стационарно ростов [url=tatuheart.ukrbb.net/viewtopic.php?f=8&t=15065]tatuheart.ukrbb.net/viewtopic.php?f=8&t=15065[/url] .
Vivod iz zapoya rostov_yvel
November 14, 2024 @ 2:52 pm
вывод. из. запоя. анонимно. ростов. [url=mediaworld.ukrbb.net/viewtopic.php?f=49&t=5433]mediaworld.ukrbb.net/viewtopic.php?f=49&t=5433[/url] .
огнестойкие сейфы
November 14, 2024 @ 3:45 pm
Тут можно преобрести сейф противопожарный купить сейф несгораемый купить
ZSot
November 14, 2024 @ 4:06 pm
1win mozambique
Dennisinamy
November 14, 2024 @ 4:29 pm
fur pharm: furosemide – cheapest lasix
SelectorCasino
November 14, 2024 @ 5:56 pm
[url=https://selector-casino-apk.ru/]https://selector-casino-apk.ru/[/url]
Установить приложение казино Selector – побеждай сегодня!
selector-casino-apk.ru
bets10 giriş linki
November 14, 2024 @ 8:28 pm
kalitenin ve bahisin tek adres bets10 twitter sayesinde zenginliğe ulaşacaksınız
Cazrero
November 14, 2024 @ 8:30 pm
Купить диплом старого образца, можно ли это сделать по быстрой схеме?
CasinoJet
November 14, 2024 @ 8:36 pm
[url=https://jet-casino-apk.ru/jet-casino/]https://jet-casino-apk.ru/jet-casino/[/url]
Установить последнюю версию приложения онлайн казино Jet – выигрывай сегодня!
https://www.jet-casino-apk.ru/jet-casino/
kinosklad.net
November 14, 2024 @ 8:44 pm
Американские фильмы смотреть онлайн kinosklad.net
Вопрос – [url=https://kinosklad.net/adventures/]смотреть приключения[/url] прямой успех отыскать на сайте kinosklad.net сейчас. Чтобы пользование порталом было максимально удобным, рекомендуем пройти авторизацию. Тогда у Вас будет собственный аккаунт, где будут списки из ранее просмотренных фильмов и многое другое.
Curtisspeve
November 14, 2024 @ 10:56 pm
buy ed pills [url=https://erepharm.com/#]ED meds online with insurance[/url] best ed pills online
villa thailand
November 14, 2024 @ 11:37 pm
It is appropriate time to make some plans for the
longer term and it is time to be happy. I’ve learn this put up and if I
may just I desire to suggest you some attention-grabbing issues or suggestions.
Maybe you could write subsequent articles referring to this article.
I want to read more things approximately it!
promokody_bdea
November 15, 2024 @ 12:05 am
промокод на продамус [url=http://kozaostra.mybb.ru/viewtopic.php?id=14407]http://kozaostra.mybb.ru/viewtopic.php?id=14407[/url] .
купить сейф для оружия
November 15, 2024 @ 12:35 am
Тут можно преобрести где купить сейф для ружья оружейный сейф цена
thailand beach villas
November 15, 2024 @ 1:19 am
Hmm it looks like your site ate my first comment (it was super long) so I
guess I’ll just sum it up what I wrote and say,
I’m thoroughly enjoying your blog. I as well am an aspiring blog
writer but I’m still new to the whole thing. Do you have
any helpful hints for beginner blog writers? I’d definitely appreciate it.
JetZercalo
November 15, 2024 @ 1:40 am
[url=https://jet-casino-apk.ru/jet-oficzialnyj-sajt]www.jet-casino-apk.ru/jet-oficzialnyj-sajt[/url]
Скачать последнюю версию приложения казино Jet – играй сегодня!
https://www.jet-casino-apk.ru/jet-oficzialnyj-sajt
thailand houses for rent
November 15, 2024 @ 1:47 am
Hello there! This is my first visit to your blog! We are a
collection of volunteers and starting a new initiative in a
community in the same niche. Your blog provided us useful information to work on. You have done a wonderful job!
thailand villa
November 15, 2024 @ 2:19 am
It’s going to be finish of mine day, however before ending I am reading this impressive post
to improve my know-how.
thailand villa
November 15, 2024 @ 2:30 am
You could definitely see your enthusiasm within the work you write.
The arena hopes for even more passionate writers
such as you who aren’t afraid to mention how they believe.
At all times follow your heart.
ThomasWoopE
November 15, 2024 @ 2:57 am
They might include special diets, herbal supplements, chiropractic care, animal therapy, arts therapy, mindfulness, or relaxation therapies https://www.reddit.com/user/autismmmccom/
ManuelSam
November 15, 2024 @ 3:01 am
Представьте себе: Вы гуляете по улицам Москвы, а вокруг Вас оживают легендарные злодеи и аферисты! Здесь исторические личности сплетаются с киношными персонажами, а за каждым поворотом скрывается тайна https://captour.ru/subjects/ekskursii-po-moskve-na-tureckom-yazyke/
ZSot
November 15, 2024 @ 3:03 am
vavada bonus
RichardDam
November 15, 2024 @ 3:09 am
Кomпания “Скважина Минск” предоставляет услуги по профессиональному бурению скважин на воду с обустройством в Минском районе и области под клю http://matsutas.com/economic-development/development-for-somebody-else-by-alexander-obom/?unapproved=4160565&moderation-hash=62b741abfb4a1735248b2bfee2558e34#comment-4160565
thailand luxury villa rental
November 15, 2024 @ 7:00 am
Every weekend i used to pay a quick visit this web page, as i wish
for enjoyment, since this this web site conations actually nice funny material
too.
Kirnbezar
November 15, 2024 @ 7:52 am
Фэнтези смотреть кино на kinonix.net
Планировали [url=https://kinonix.net/fantastika/]смотреть фантастику[/url] прямо сейчас, тогда заходите на kinonix.net и желаем Вам приятного просмотра. Значительно удобнее, когда все фильмы отобраны на одном сайте, чем искать каждый раз посторонние видео плееры для очередного кино. Смотрите сами, приглашайте близких и детей к просмотру.
Diplomi_wpma
November 15, 2024 @ 8:27 am
купить диплом телефон [url=https://1oriks-diplom199.ru/]1oriks-diplom199.ru[/url] .
ZSot
November 15, 2024 @ 8:50 am
neon54 slot
Iariorzqv
November 15, 2024 @ 8:55 am
Официальная покупка школьного аттестата с упрощенным обучением в Москве
Sazrkbl
November 15, 2024 @ 9:15 am
Как безопасно купить диплом колледжа или ПТУ в России, что важно знать
villa thailand
November 15, 2024 @ 10:44 am
Thanks , I’ve recently been searching for info about this subject
for ages and yours is the greatest I’ve came upon so far.
But, what about the bottom line? Are you positive
about the supply?
JefferyHot
November 15, 2024 @ 10:50 am
best online pharmacy india: Best online Indian pharmacy – world pharmacy india
CarlosDycle
November 15, 2024 @ 11:15 am
Интерфейс таких платформ обычно прост и доступен, что делает их привлекательными для новичков и профессионалов. Основные функции ориентированы на то, чтобы дать трейдерам возможность анализировать рыночные тенденции, а также быстро и безошибочно выполнять сделки https://szkolenia.myslepozytywnie.pl/fora/watek/%d0%bf%d0%be%d1%87%d0%b5%d0%bc%d1%83-%d1%81%d1%82%d0%be%d0%b8%d1%82-%d0%bd%d0%b0%d1%87%d0%b0%d1%82%d1%8c-%d0%ba%d0%b0%d1%80%d1%8c%d0%b5%d1%80%d1%83-%d1%82%d1%80%d0%b5%d0%b9%d0%b4%d0%b5%d1%80%d0%b0/
BradleyJopay
November 15, 2024 @ 11:18 am
http://canadiandrugsgate.com/# ed in men
Trefngu
November 15, 2024 @ 12:02 pm
Покупка диплома о среднем полном образовании: как избежать мошенничества?
beijingpal.com/read-blog/4707
HenryTup
November 15, 2024 @ 12:17 pm
Sex videos online watch for free from anywhere in the world without subscription and registration kids porn
Mazrigj
November 15, 2024 @ 12:51 pm
Как купить аттестат 11 класса с официальным упрощенным обучением в Москве
WIFYRKM
November 15, 2024 @ 1:34 pm
Clavicle. Stanford. Consort. Us department of education. Matthew mcconaughey movies and tv shows. Athens greece. Docs. Credit score ranges. Generation x. 100 kg to lbs. Queen mary. Vivek ramaswamy net worth. Labia. Hum. Timbaland. Abdomen. Shiitake mushroom. Indianapolis 500. Runes. Foundation. Nwa. Environment. Strikeout. Indian time. Sternum. Kamala harris age. Tractor. Normandy. Csi miami. Stink bug. Mohammed bin salman. Jane eyre. Our. Mono. The marvelous mrs. maisel. Rim. Christine mcvie. Jd vance movie. Beaufort sc. Antecedent. Disseminate. Ghostbusters movies. Dog day afternoon. Fifa world cup 2023. Pms. Heavy metal. Funny games. Phyllis diller. Odell beckham. Acetate. Botfly. Cunt. Panacea. Octagon. Redd foxx. Escherichia coli. Satellite. Scorpio. Field museum. Parkour. 1. Albert einstein. Robbery. https://vdtgwyb.filmfilmfilm.eu/NMGJI.html
Analyze. Horse fly. Rikers island. Weary. Abercrombie and fitch. Virgen de guadalupe. Sled. Bunnies. Spider web. Neuroscience. Bug. Innuendo. How do you. Brady bunch. Lindsay wagner. Wall e. Past wordle words. Acapulco. Ha. Conformity. Faroe islands. When is spring. Talia balsam. Lgbtqia meaning. Exacerbated. Potassium chloride. Sikhism. Beretta. Jean paul gaultier. Cartoon porn. Hedge fund. Pembroke welsh corgi. Crying. White christmas. Staring. Toe. Chris hemsworth movies. Satire definition. Cedric the entertainer. Common cold symptoms. Israel time. Oven. Desert. Family ties. Paradise lost. Barnard college. Granger. Irony definition. Napoleon bonaparte. 3m.
сейф огнестойкий
November 15, 2024 @ 2:19 pm
Тут можно преобрести купить сейф огнестойкий в москве сейфы от пожара
koh samui rent villa
November 15, 2024 @ 2:24 pm
Tremendous things here. I’m very happy to see your post.
Thank you so much and I’m looking forward to touch you.
Will you kindly drop me a mail?
Sazrrmk
November 15, 2024 @ 2:51 pm
Как правильно приобрести диплом колледжа или ПТУ в России, важные моменты
uebkameri.listbb.ru/viewtopic.php?f=2&t=1009
Lazrfru
November 15, 2024 @ 2:55 pm
Покупка школьного аттестата с упрощенной программой: что важно знать
koh samui resort villa
November 15, 2024 @ 2:56 pm
This is my first time pay a quick visit at here and i am in fact
impressed to read everthing at single place.
JCUMMMF
November 15, 2024 @ 2:59 pm
German. Pothos plant. Dungeness crab. Palestine. Seattle kraken. Maltese dog. Packers. Persistent. Bee gees songs. Indiana jones 1. Sherman. Arnold schwarzenegger. California. Mammoth cave. Europe. Norman rockwell. Wheel. Conscious. Petty. Lp. Frankfurt. Animal kingdom. Profile. Lil wayne. Rhetoric. Will smith movies. Leo sign. Marianne williamson. Elon musk twitter. Enay. Caster oil. Danny devito. Harry potter movies. Zinc. Youutube. Asbury park. Great wall. Essential oils. How to play chess. Beau bridges. Togo. Ball state university. Isabella rossellini. Bored. Metaphysical. Trees. Adam west. Ban. Pangea. Cornstarch. https://uyhvkpv.filmfilmfilm.eu/GJIAH.html
Jim kelly. Saba. Wembanyama stats. Princess diana. Edgar allan poe. Tom hardy movies and tv shows. Brittney griner. Corn snake. Eclipse. Hawaii. Monday night football. Hsv. Pimp. Abraham. Diamondbacks vs phillies. Multiple sclerosis. Calamity. Idris elba. Chita rivera. All saints. Leo movie. Butler university. Glyphosate. Eugene oregon. Patronizing. Order. Pride. Aerobic exercise. Innuendo. Tenacious. Gatlinburg. James garner. Israel map. Jane austen. Princeton university. Ashton kutcher. Chatgtp. Barracuda. Mckayla maroney. Solipsism. Dailybeast. Methuselah. Communion. Woodrow wilson. Lard. Griselda blanco sons. Oklahoma city thunder. Weird al. Huntington’s disease. Histamine. 30 celsius to fahrenheit. Katie couric. Victoria principal. Dimples. Boric acid. Women world cup. Trident. Calligraphy. Sikhism. Chinese dragon. Buffalo bill. Thule.
koh samui villa to rent
November 15, 2024 @ 4:09 pm
Attractive part of content. I just stumbled upon your website and in accession capital to claim that I acquire in fact loved account
your weblog posts. Any way I will be subscribing in your feeds or even I fulfillment you get admission to consistently rapidly.
kazino onlain_oema
November 15, 2024 @ 4:14 pm
лучшие онлайн казино [url=http://www.online-kazino.by]http://www.online-kazino.by[/url] .
Vivod iz zapoya rostov_uxot
November 15, 2024 @ 4:15 pm
вывод из запоя стационар ростов [url=superjackson.ukrbb.net/viewtopic.php?f=28&t=9728]вывод из запоя стационар ростов[/url] .
Vivod iz zapoya rostov_rvSt
November 15, 2024 @ 4:16 pm
вывод из запоя цена ростов [url=www.automobilist.forum24.ru/?1-19-0-00000135-000-0-0-1730648713]www.automobilist.forum24.ru/?1-19-0-00000135-000-0-0-1730648713[/url] .
GTJONGM
November 15, 2024 @ 4:18 pm
Yao ming. Roger waters. Cornucopia. 170 pounds in kg. Skull. Pacers vs bucks. Octordle. Tiger. Il. Malignant. Lens. Nonchalant meaning. Patrick stewart. Rosetta stone. Submissive meaning. American university. Confused. Bracket. Game theory. Bagels. Beaufort sc. January. Jeff conaway. Acadia. Veneer. Halal food. Schitt’s creek. Dark knight cast. England. Tron. Credit cards. Nft meaning. Scoville scale. Tool. Gunther. Eb. Jefferson. Snitch. Mount washington. Barca. Opinion. Map to israel. College football championship. Vance j.d. Bob_newhart. Tanzania. Sedentary. Shark. Schitts creek. Yellow jacket. Honolulu. Gettysburg address. Bram stoker’s dracula. Travis barker. Kosher meaning. Ai stocks. Kowloon walled city. Scary. https://gkouryp.filmfilmfilm.eu/XBXFP.html
Jb smoove. Empire. Herpes. Signal. Bauhaus. Electron configuration. It’s. Mail online. Hi. Melanoma. Spades card game. Kate jackson. Johnny depp. Time zone map. Putin. Moonshine. Butt. Riyadh. Roma fc. Gypsum. York. Drums. 1920s fashion. Criteria. Breast cancer awareness month. Mapa de estados unidos. Rest. Gum. Balenciaga. Kristy mcnichol. University of hawaii. Broccoli. Honey movie. Burt reynolds. Generation years. A christmas carol. Character. Succulents. Ootd meaning. When is mothers day. Animal house. Nba players. Progress. Beatles. Marseille. Gmt time. Gay flag. Gilligan’s island. Lincoln.
Glennlashy
November 15, 2024 @ 4:47 pm
drug pharmacy [url=http://canadiandrugsgate.com/#]canadiandrugsgate[/url] erectile dysfunction
shonpeedo
November 15, 2024 @ 6:28 pm
[url=https://super-bag.ru/catalog/zhenskie_sumki/brendovye_zhenskie_sumki/gucci/]сумка gucci[/url] или [url=https://super-bag.ru/catalog/brendovaya_zhenskaya_obuv/]купить брендовую обувь[/url]
[url=https://super-bag.ru/catalog/zhenskie_sumki/brendovye_zhenskie_sumki/chanel/]chanel маленькая сумка[/url]
https://luxmsk.ru/catalog/brendovaya-muzhskaya-odezhda/brendovye-muzhskie-dzhinsy/
Ещё можно узнать: [url=https://super-bag.ru/catalog/brendovaya_zhenskaya_odezhda/brendovye_zhenskie_kostyumy/]брендовые спортивные костюмы[/url]
ремень женский брендовый
Vivod iz zapoya rostov_tdPi
November 15, 2024 @ 6:31 pm
вывод из запоя ростов [url=http://www.bisound.com/forum/showthread.php?t=688674/]http://www.bisound.com/forum/showthread.php?t=688674/[/url] .
Vivod iz zapoya rostov_woOt
November 15, 2024 @ 6:31 pm
вывод из запоя ростов-на-дону [url=https://cah.forum24.ru/?1-19-0-00000455-000-0-0-1730648785]https://cah.forum24.ru/?1-19-0-00000455-000-0-0-1730648785[/url] .
Xazrwgc
November 15, 2024 @ 7:11 pm
Быстрое обучение и получение диплома магистра – возможно ли это?
ZSot
November 15, 2024 @ 7:27 pm
vavada lv
сейф для оружия
November 15, 2024 @ 7:51 pm
Тут можно преобрести оружейные сейфы оружейные сейфы и шкафы для охотничьего
ONWNVLR
November 15, 2024 @ 8:06 pm
Minutes. Canada. Pufferfish. Statute of limitations. Dragonfly. Baghdad. Ebon moss-bachrach. Melissa mccarthy. Ewe. Mesopotamia. Xiphoid process. Soul food. Poker. Catch 22. Eli lilly. Inference. The feast of dionysus. Ursula andress. Collision. The handmaids tale. Ed begley jr. Sickle. The hobbit. Rudy gobert. Miami seaquarium. Brett kavanaugh. Smithsonian museums. Convey. Showbiz. Sega. Staffordshire bull terrier. Grand canyon. Conjecture. Hives. Smart watch. Burlington vt. Jarhead. Hamilton cast. When is presidents day 2024. Papillon. Priest. Chanel. Cosplay. Christopher guest. Season. Skin cancer. Biltmore. Swordfish. Hognose snake. Burt lancaster. The patriot. Enough. Mandalas. Brooklyn botanic garden. Gene hackman. Stick bug. I am sam. The metropolitan museum of art. Christmas cactus. Glutamine. Ostrich. Randy jackson. Transparency. https://jvdoups.filmfilmfilm.eu/JEYRM.html
Masturbation. Guess who. Salman khan. Scorpio dates. Reptile. Tb rays. Alison krauss. Tonya harding. Reprieve. Dyspareunia. Adrian peterson. Villanova. Saints. Avril lavigne. Leeds. Stock market crash. Women world cup. Sovereignty. Washington monument. Arizona state university. Snopes. Rope. Openheimer. Macabre. Sword. Photography. Mifepristone. Panic disorder. Valhalla. Second life. Daffy duck. Afc asian cup standings. Fae. Voyager 1. Fish tank. Kathy bates. Better. The masters. The munsters. Peace corps. Larynx. Robert duvall. Habanero. Ratatouille. Sohcahtoa. Computer. Smithsonian. Commitment. Calabasas. Embiid stats. Wild. Trivial. Frankfurt. Lake michigan. Mookie betts stats. Dominion. Inequality.
Sazrcef
November 15, 2024 @ 9:04 pm
Официальная покупка школьного аттестата с упрощенным обучением в Москве
SCRSGOM
November 15, 2024 @ 9:13 pm
Keratosis pilaris. Kindle. Luka stats. Lady gaga songs. Vivid. Jesse eisenberg. Googly eyes. David and goliath. Shawshank redemption. Andy warhol. The witch. Lama. Kinesiology. Acolyte definition. Epstein barr virus. Prison. Cubs. Reba show. Netf. Corn. Indiana. Honeydew. Directtv. Gainesville. Bobcat. Ukraine war. Josh brolin movies. Henry cavill. Tachycardia. Glass 2019. Frances mcdormand. Poutine. Horace mann. How tall is kevin hart. Clive davis. Chicago il. Possum. What is gluten. Italy flag. Satire. Patronize meaning. Maiden name meaning. Intifada. Wolf dog. St lucia. Woody harrelson movies and tv shows. https://xzcgimg.filmfilmfilm.eu/IFHMD.html
Israel vs palestine. Tarantula hawk. Piedmont. Indignation. Embroidery. Daredevil. Singular. Necrosis. Into the woods. Hulu. Pontiac. Anthony edwards. Bank of america of. Amish people. Uchicago. Snake. Michael c hall. Lavender plant. Martha stewart. Brandy. Alanis morissette. Browns score. Pus. Bento. Wii. Man of steel. Lymph nodes. Louis vuitton. Salvation. Pelvis. Niger. The lord’s prayer. Brontosaurus. Luna. Logic. Smyrna. Adaptation. Kurt russell movies. Anna paquin. Maury povich. Edward scissorhands. Robert de niro. Roswell. Tyranny. Electra. Schnauzer. Thousand oaks. Death valley national park. Animal farm.
samui villa
November 15, 2024 @ 9:14 pm
Heya i’m for the first time here. I found this board and I find It truly useful & it helped me out much.
I hope to give something back and aid others like you aided
me.
koh samui best villa
November 15, 2024 @ 9:28 pm
Hello to every body, it’s my first pay a quick visit of this weblog; this web site consists
of awesome and in fact fine information in favor
of visitors.
BerzopePlams
November 15, 2024 @ 9:43 pm
Получите [url=https://t.me/s/zaim_bez_pocent]займ без процентов на карту[/url] на срок 30 дней. Более 25 МФО предлагают первый микрозайм под 0%, без отказа и с моментальной выдачей средств. Понадобится только паспорт, а кредитная история не важна. Просто и удобно — деньги на вашей карте в считанные минуты!
Oariorebt
November 15, 2024 @ 9:53 pm
Как получить диплом техникума с упрощенным обучением в Москве официально
Cazrcwy
November 15, 2024 @ 10:16 pm
Как избежать рисков при покупке диплома колледжа или ПТУ в России
BradleyJopay
November 15, 2024 @ 11:29 pm
http://canadiandrugsgate.com/# viagra without a doctor prescription
koh samui resort villa
November 15, 2024 @ 11:54 pm
Hello there, I do think your website may be having browser compatibility issues.
Whenever I look at your web site in Safari, it looks fine however when opening in IE,
it has some overlapping issues. I simply wanted to give you a quick heads up!
Besides that, wonderful blog!
rent a villa in thailand koh samui
November 16, 2024 @ 12:34 am
Quality articles is the secret to interest the people to pay a
quick visit the site, that’s what this site is providing.
villa in koh samui for rent
November 16, 2024 @ 12:37 am
Nice post. I used to be checking continuously this blog and I am inspired!
Extremely useful information specially the final part 🙂 I deal with such information a lot.
I was seeking this certain info for a very lengthy time. Thanks and good luck.
ZSot
November 16, 2024 @ 1:10 am
neon54 app
SelectorApk
November 16, 2024 @ 1:20 am
[url=https://selector-casino-apk.ru/selector-apk]http://www.selector-casino-apk.ru/selector-apk[/url]
Установить apk файл казино онлайн Selector – играй сейчас!
http://www.selector-casino-apk.ru/selector-apk
koh samui villa to rent
November 16, 2024 @ 2:41 am
If you wish for to grow your know-how only keep visiting
this website and be updated with the latest gossip posted here.
koh samui rent villa
November 16, 2024 @ 2:49 am
This article presents clear idea for the new people of blogging, that genuinely how to do
blogging and site-building.
Diplomi_sySi
November 16, 2024 @ 2:57 am
Покупка диплома о среднем полном образовании: как избежать мошенничества?
Richardwaype
November 16, 2024 @ 3:16 am
A Bitcoin wallet that allows you to store, send Bitcoin, receive Bitcoin and buy Bitcoin with focus on security and simplicity BlueWallet android
Kennethprusy
November 16, 2024 @ 3:24 am
Dynamic platform that aggregates live adult entertainment, constantly refreshed with streams from the leading adult webcam platforms like live gay porn
betcio
November 16, 2024 @ 3:24 am
Bahis dünyasında en yüksek bonusları sunan Betcio, kullanıcıların kazançlarını artırmak için mükemmel bir fırsat sunuyor. Eğlencenin tadını çıkarın!
villa rentals in koh samui
November 16, 2024 @ 3:29 am
Please let me know if you’re looking for a article author for your weblog.
You have some really great articles and I feel I would be a good asset.
If you ever want to take some of the load off, I’d love
to write some content for your blog in exchange for a link back to mine.
Please shoot me an e-mail if interested. Cheers!
JetCasino
November 16, 2024 @ 4:22 am
[url=https://jet-casino-apk.ru]https://jet-casino-apk.ru[/url]
Загрузить apk файл онлайн казино Jet – играй прямо сейчас!
http://www.jet-casino-apk.ru
koh samui villas
November 16, 2024 @ 5:04 am
Wow, marvelous blog layout! How long have you been blogging for?
you make blogging look easy. The overall look of your site is fantastic, as well as the content!
Sazrmwi
November 16, 2024 @ 6:03 am
Как получить диплом техникума с упрощенным обучением в Москве официально
hijyen belgesi
November 16, 2024 @ 7:24 am
Hijyen belgesi, işletmelerin hijyen standartlarına uygun çalıştığını kanıtlayan kritik bir sertifikadır. Özellikle gıda sektöründe faaliyet gösteren işletmeler için hijyen belgesi nasıl alınır sorusu önemlidir; bu belgeyi almak güvenilirliği artırır ve müşteri memnuniyetine katkı sağlar. Hijyen sertifikası, işletmelerin hijyen kurallarına uyduğunu ve yasal düzenlemelere uygun çalıştığını gösterir.
Müşteriler, hijyenik koşullarda hizmet veren işletmeleri tercih eder. Bu nedenle hijyen sertifikası nasıl alınır, işletme sahiplerinin sıkça araştırdığı bir konu haline gelmiştir. Hijyen belgesi alarak, işletmenizin hijyen standartlarını yükseltebilir, hem güvenilirliğinizi artırabilir hem de müşteri sadakati kazanabilirsiniz.
Hijyen belgesi almak, işletmenizin profesyonelliğini de artırır. Eğitimli ve sertifikalı bir ekip ile hijyen kurallarına uygun bir ortam sunmak, hem çalışanlarınızın hem de müşterilerinizin sağlığını korur. Bu, marka imajınızı güçlendirir ve sektördeki konumunuzu sağlamlaştırır. Hijyen eğitimi nasıl alınır sorusuna yanıt arayanlar, işletmeleri için en uygun eğitim ve sertifika programlarını bulmak üzere araştırma yapmalıdır.
Hijyen belgesi ile ilgili daha fazla bilgi almak ve başvuru sürecini öğrenmek için hijyen belgesi sayfamızı ziyaret edebilirsiniz. İşletmenizi bir adım öne taşıyarak, hijyen standartlarınızı belgeleyin ve sektörünüzde rekabet avantajı elde edin!
Mazrbkm
November 16, 2024 @ 8:57 am
Можно ли купить аттестат о среднем образовании, основные моменты и вопросы
luxury villas in samui
November 16, 2024 @ 9:15 am
Hello! Would you mind if I share your blog with my myspace group?
There’s a lot of folks that I think would really appreciate your content.
Please let me know. Thanks
koh samui villa to rent
November 16, 2024 @ 9:41 am
Simply wish to say your article is as astounding.
The clearness in your post is simply excellent and i
can assume you are an expert on this subject.
Well with your permission allow me to grab your RSS feed to keep up to
date with forthcoming post. Thanks a million and please keep up
the rewarding work.
огнестойкие сейфы
November 16, 2024 @ 9:48 am
Тут можно преобрести сейфы от пожара сейф москва огнестойкий купить
Glennlashy
November 16, 2024 @ 9:55 am
reputable indian online pharmacy [url=http://indianpharmacyeasy.com/#]indianpharmacyeasy[/url] indian pharmacies safe
BradleyJopay
November 16, 2024 @ 10:18 am
http://canadiandrugsgate.com/# treat ed
villa rentals koh samui thailand
November 16, 2024 @ 10:55 am
I do not even know how I ended up here, but I thought this post was good.
I don’t know who you are but certainly you are going to a famous blogger if you aren’t already 😉 Cheers!
ZSot
November 16, 2024 @ 11:20 am
vavada
profi-teh-remont
November 16, 2024 @ 11:24 am
Сервисный центр предлагает ремонт canon ef 35-80mm f/4.0-5.6 iii в петербурге ремонт canon ef 35-80mm f/4.0-5.6 iii цены
rent a villa in thailand koh samui
November 16, 2024 @ 11:31 am
Tremendous things here. I am very satisfied to see your article.
Thank you a lot and I am having a look ahead to contact you.
Will you kindly drop me a mail?
JimmyRax
November 16, 2024 @ 12:02 pm
Watch and admire beautiful girls getting naked on live cam. The site features cam girls of all types, from petite blonde camgirls to leggy redheads free webcam xx
RichardTuh
November 16, 2024 @ 12:06 pm
Find the hottest sexwebcam videos and clips, realy hight quality xxx animal porn movies
SelectorCasino
November 16, 2024 @ 12:45 pm
[url=https://selector-casino-apk.ru/selector-casino]https://www.selector-casino-apk.ru/selector-casino[/url]
Загрузить последнюю версию приложения казино Селектор – побеждай прямо сейчас!
selector-casino-apk.ru/selector-casino
Michaelbiazy
November 16, 2024 @ 1:01 pm
Готов услышать трек, который выбьет из тебя все оправдания? Слушай, если не боишься правды.
Vivod iz zapoya rostov_iemr
November 16, 2024 @ 1:04 pm
вывод из запоя ростов на дону стационар [url=https://kozaostra.mybb.ru/viewtopic.php?id=14279/]kozaostra.mybb.ru/viewtopic.php?id=14279[/url] .
Vivod iz zapoya rostov_hkma
November 16, 2024 @ 1:15 pm
анонимный. вывод. из. запоя. ростов. [url=www.internetmoney.bestbb.ru/viewtopic.php?id=31697/]www.internetmoney.bestbb.ru/viewtopic.php?id=31697/[/url] .
JetAndroid
November 16, 2024 @ 1:16 pm
[url=https://jet-casino-apk.ru/jet-na-androjd/]http://jet-casino-apk.ru/jet-na-androjd/[/url]
Установить последнюю версию приложения казино онлайн Джет – выигрывай прямо сейчас!
http://www.jet-casino-apk.ru/jet-na-androjd/
Vivod iz zapoya rostov_zqOa
November 16, 2024 @ 1:33 pm
вывод из запоя дешево ростов-на-дону [url=http://www.sportandpolitics.ukrbb.net/viewtopic.php?f=24&t=17793v]http://www.sportandpolitics.ukrbb.net/viewtopic.php?f=24&t=17793v[/url] .
Vivod iz zapoya rostov_over
November 16, 2024 @ 1:36 pm
вывод. из. запоя. ростов. на. дону. [url=http://familyportal.forumrom.com/viewtopic.php?id=28559/]http://familyportal.forumrom.com/viewtopic.php?id=28559/[/url] .
Vivod iz zapoya rostov_wrSr
November 16, 2024 @ 1:36 pm
вывод из запоя срочно ростов [url=http://snatkina.borda.ru/?1-11-0-00000198-000-0-0-1730709418]вывод из запоя срочно ростов[/url] .
купить сейф для охотничьего ружья
November 16, 2024 @ 2:23 pm
Тут можно преобрести оружейные шкафы москва оружейные шкафы
thai luxury villas
November 16, 2024 @ 2:37 pm
Pretty section of content. I just stumbled upon your website and in accession capital to assert that I acquire
actually enjoyed account your blog posts. Anyway I will be subscribing to
your augment and even I achievement you access consistently fast.
koh samui rent villa
November 16, 2024 @ 2:37 pm
Fabulous, what a blog it is! This webpage provides helpful facts to us, keep it up.
koh samui villa to rent
November 16, 2024 @ 3:09 pm
Hello! This is kind of off topic but I need some help from an established blog.
Is it very difficult to set up your own blog?
I’m not very techincal but I can figure things out pretty fast.
I’m thinking about setting up my own but I’m not sure
where to begin. Do you have any points or suggestions?
Many thanks
prodamus promokod_xymt
November 16, 2024 @ 3:29 pm
промокод на подключение продамуса [url=prodamus-promokod21.ru]prodamus-promokod21.ru[/url] .
Sazrqnt
November 16, 2024 @ 3:42 pm
Приобретение диплома ПТУ с сокращенной программой обучения в Москве
SelectorCasino
November 16, 2024 @ 6:00 pm
[url=https://selector-casino-apk.ru/selector-na-androjd]selector-casino-apk.ru/selector-na-androjd[/url]
Загрузить apk файл онлайн казино Selector – играй сейчас!
https://selector-casino-apk.ru/selector-na-androjd
Iariorvzs
November 16, 2024 @ 6:12 pm
Как официально купить аттестат 11 класса с упрощенным обучением в Москве
samui villa rent
November 16, 2024 @ 6:42 pm
I just couldn’t leave your site prior to suggesting that I extremely enjoyed the usual info a person provide in your
guests? Is gonna be again steadily in order to inspect new posts
ZSot
November 16, 2024 @ 7:13 pm
1win pagina oficial
aloetopakistan
November 16, 2024 @ 7:23 pm
Enjoy premium casino and sports betting options with 888Starz Pakistan.
Uazrnkk
November 16, 2024 @ 7:46 pm
Быстрое обучение и получение диплома магистра – возможно ли это?
spa-la-part-des-anges.fr/kupit-attestat-nedorogo-v-stavropole.html
JefferyHot
November 16, 2024 @ 8:42 pm
mexican pharmaceuticals online: mexican drugstore online – buying from online mexican pharmacy
villas rent koh samui
November 16, 2024 @ 9:05 pm
Just wish to say your article is as astounding. The clearness in your submit
is simply great and i could think you are a professional on this
subject. Well together with your permission allow
me to clutch your RSS feed to keep up to date with
coming near near post. Thanks one million and please carry
on the gratifying work.
SelectorSait
November 16, 2024 @ 9:44 pm
[url=https://selector-casino-apk.ru/selector-oficzialnyj-sajt]selector-casino-apk.ru/selector-oficzialnyj-sajt[/url]
Скачать последнюю версию приложения онлайн казино Селектор – играй сейчас!
http://selector-casino-apk.ru/selector-oficzialnyj-sajt
Sazrjkd
November 16, 2024 @ 10:22 pm
Как получить диплом техникума официально и без лишних проблем
Sazrqop
November 16, 2024 @ 10:50 pm
Легальная покупка диплома о среднем образовании в Москве и регионах
Dnrtqkm
November 16, 2024 @ 11:39 pm
Покупка диплома о среднем полном образовании: как избежать мошенничества?
Sazrkfd
November 16, 2024 @ 11:44 pm
Как получить диплом стоматолога быстро и официально
ZSot
November 17, 2024 @ 12:10 am
1win login
Glennlashy
November 17, 2024 @ 1:30 am
buying prescription drugs in mexico online [url=http://mexicanpharmgate.com/#]mexican pharmacy online[/url] buying prescription drugs in mexico online
Jameskeend
November 17, 2024 @ 2:22 am
Лессе-Пассе или Теудат Маавар – это oдин из возможных паспортов Израиля, который выдается гражданам этой страны https://xn—-7sbbzecaihj2bkbelhgv2p.com/
ThomashoxiA
November 17, 2024 @ 2:41 am
Official Starhotels website: book directly on the website at the best available rates for 4 and 5 star hotels in Italy, New York, Paris and London Columbus Business Travel
Mazruvj
November 17, 2024 @ 3:43 am
Официальная покупка диплома вуза с сокращенной программой в Москве
forummichiganrp.moibb.ru/viewtopic.php?f=21&t=543
JetZercalo
November 17, 2024 @ 3:48 am
[url=https://jet-casino-apk.ru/jet-oficzialnyj-sajt]https://jet-casino-apk.ru/jet-oficzialnyj-sajt[/url]
Загрузить последнюю версию приложения онлайн казино Jet – играй прямо сейчас!
jet-casino-apk.ru/jet-oficzialnyj-sajt
Lazroug
November 17, 2024 @ 4:55 am
Официальная покупка диплома вуза с сокращенной программой обучения в Москве
Sazrwgx
November 17, 2024 @ 5:27 am
Официальная покупка диплома вуза с упрощенной программой обучения
Diplomi_yama
November 17, 2024 @ 5:46 am
купить аттестаты за 11 [url=https://1oriks-diplom199.ru/]купить аттестаты за 11[/url] .
marsbahis giriş
November 17, 2024 @ 6:04 am
Marsbahis, spor bahisleri tutkunları için keyifli bir platform sunuyor. Bu yazıda, Marsbahis’in sunduğu fırsatları keşfedeceğiz.
BradleyJopay
November 17, 2024 @ 6:31 am
http://indianpharmacyeasy.com/# indian pharmacies safe
JefferyHot
November 17, 2024 @ 7:11 am
ed prescription drugs: Canada pharmacy – ed treatment options
Diplomi_iiSi
November 17, 2024 @ 8:42 am
Покупка диплома о среднем полном образовании: как избежать мошенничества?
ZSot
November 17, 2024 @ 8:49 am
vavada казино
купить сейф для охотничьего ружья
November 17, 2024 @ 9:43 am
Тут можно преобрести купить сейф для охотничьего ружья в москве купить оружейный сейф в интернет магазин
огнестойкие сейфы
November 17, 2024 @ 10:20 am
Тут можно преобрести огнеупорный сейф купить огнеупорный сейф
Lazrtih
November 17, 2024 @ 11:40 am
Диплом техникума купить официально с упрощенным обучением в Москве
Cazrmoa
November 17, 2024 @ 12:51 pm
Узнайте, как безопасно купить диплом о высшем образовании
Sazryhc
November 17, 2024 @ 12:56 pm
Как оказалось, купить диплом кандидата наук не так уж и сложно
Jugabet Peru_xdmt
November 17, 2024 @ 1:12 pm
Jugabet apuestas [url=https://www.jugabet777.com]Jugabet apuestas[/url] .
Vivod iz zapoya rostov_doMa
November 17, 2024 @ 1:12 pm
анонимный. вывод. из. запоя. ростов. [url=http://www.pokupki.bestforums.org/viewtopic.php?f=7&t=24033]http://www.pokupki.bestforums.org/viewtopic.php?f=7&t=24033[/url] .
Vivod iz zapoya rostov_dzOl
November 17, 2024 @ 1:14 pm
вывод. из. запоя. ростов. [url=https://golosa.ukrbb.net/viewtopic.php?f=3&t=7466]https://golosa.ukrbb.net/viewtopic.php?f=3&t=7466[/url] .
Vivod iz zapoya rostov_prPn
November 17, 2024 @ 1:15 pm
принудительный вывод из запоя ростов [url=https://superstar.ukrbb.net/viewtopic.php?f=3&t=680]https://superstar.ukrbb.net/viewtopic.php?f=3&t=680[/url] .
Vivod iz zapoya rostov_kaen
November 17, 2024 @ 1:16 pm
вывод из запоя в стационаре ростов [url=https://ximki.ukrbb.net/viewtopic.php?f=12&t=3696/]ximki.ukrbb.net/viewtopic.php?f=12&t=3696[/url] .
ZSot
November 17, 2024 @ 1:45 pm
vavada casino bonus code
villa rentals in koh samui
November 17, 2024 @ 2:07 pm
My coder is trying to persuade me to move to .net from PHP.
I have always disliked the idea because of the costs.
But he’s tryiong none the less. I’ve been using WordPress on several websites for about a year and am anxious about switching to another platform.
I have heard fantastic things about blogengine.net. Is
there a way I can import all my wordpress posts into it?
Any kind of help would be really appreciated!
SelectorApk
November 17, 2024 @ 2:13 pm
[url=https://selector-casino-apk.ru/selector-apk]https://selector-casino-apk.ru/selector-apk[/url]
Загрузить приложение казино Селектор – побеждай сегодня!
http://www.selector-casino-apk.ru/selector-apk
JetCasino
November 17, 2024 @ 2:34 pm
[url=https://jet-casino-apk.ru]www.jet-casino-apk.ru[/url]
Скачать apk файл казино Джет – играй сейчас!
https://jet-casino-apk.ru
Larryglops
November 17, 2024 @ 2:54 pm
natural herbs for ed: Canada pharmacy – canadian online drugs
CharlessaP
November 17, 2024 @ 2:58 pm
Одним из важнейших экологичных решений в дорожном строительстве является использование переработанных материалов. Это не только помогает сократить количество отходов, но и уменьшает потребность в новых материалах, что снижает негативное воздействие на окружающую среду A Journey Through History: How Historical Maps Influence Our Understanding of the Past
Zacharyclaph
November 17, 2024 @ 3:14 pm
Имплантация зубов в Вoлгoграде: 84 клиники с aдресами и ценами от 600 до 500000 рублей, запись. При выборе клиники можно ознакомиться с отзывами других имплантация зубов в волгограде цены84
Cazrlff
November 17, 2024 @ 3:36 pm
Покупка школьного аттестата с упрощенной программой: что важно знать
Glennlashy
November 17, 2024 @ 3:56 pm
reputable indian online pharmacy [url=https://indianpharmacyeasy.com/#]Indian pharmacy to USA[/url] indian pharmacies safe
BradleyJopay
November 17, 2024 @ 4:11 pm
http://canadiandrugsgate.com/# solutions for ed
Oariorzdf
November 17, 2024 @ 5:23 pm
Реально ли приобрести диплом стоматолога? Основные этапы
JefferyHot
November 17, 2024 @ 5:30 pm
mail order pharmacy india: Online Indian pharmacy – top 10 online pharmacy in india
Eanrvmd
November 17, 2024 @ 6:02 pm
Официальная покупка диплома ПТУ с упрощенной программой обучения
сейф купить
November 17, 2024 @ 6:40 pm
Здесь можно преобрести сейф купить сейф
BryanMergo
November 17, 2024 @ 8:22 pm
Hello!
Do you want to become the best SEO specialist and link builder or do you want to outpace your competitors?
Premium base for XRumer
$119/one-time
Get access to our premium database, which is updated monthly! The database contains only those resources from which you will receive active links – from profiles and postings, as well as a huge collection of contact forms. Free database updates. There is also the possibility of a one-time purchase, without updating the databases, for $38.
Fresh base for XRumer
$94/one-time
Get access to our fresh database, updated monthly! The database includes active links from forums, guest books, blogs, etc., as well as profiles and activations. Free database updates. There is also the possibility of a one-time purchase, without updating the databases, for $25.
GSA Search Engine Ranker fresh verified link list
$119/one-time
Get access to our fresh database, updated monthly! The fresh database includes verified and identified links, divided by engine. Free database updates. There is also the possibility of a one-time purchase, without updating the databases, for $38.
GSA Search Engine Ranker activation key
$65
With GSA Search Engine Ranker, you’ll never have to worry about backlinks again. The software creates backlinks for you 24 hours a day, 7 days a week. By purchasing GSA Search Engine Ranker from us, you get a quality product at a competitive price, saving your resources.
To contact us write to Telegram: https://t.me/DropDeadStudio
villas for rent in koh samui thailand
November 17, 2024 @ 10:30 pm
Today, I went to the beach front with my kids. I found a sea shell and gave it to my 4 year old daughter and said “You can hear the ocean if you put this to your ear.” She placed the shell to her ear and screamed.
There was a hermit crab inside and it pinched her ear.
She never wants to go back! LoL I know this is entirely off
topic but I had to tell someone!
Dnrtypj
November 18, 2024 @ 12:37 am
Официальная покупка диплома вуза с сокращенной программой в Москве
villas rent koh samui
November 18, 2024 @ 3:20 am
hey there and thank you for your information –
I have certainly picked up something new from right here.
I did however expertise some technical issues using this web site, as I experienced to
reload the web site lots of times previous to I
could get it to load properly. I had been wondering if your hosting is OK?
Not that I am complaining, but slow loading instances times will sometimes affect your
placement in google and could damage your high-quality score if ads and marketing with Adwords.
Anyway I am adding this RSS to my e-mail and can look out for a lot more of your respective fascinating content.
Ensure that you update this again very soon.
BrianBlits
November 18, 2024 @ 3:36 am
Профессиональная команда специалистов в области обрезки и спилки деревьев, готовая предоставить Вам качественные услуги на территориях приусадебных участков, дачных зон, скверов и парков. Наши мастера имеют опыт работы с различными породами деревьев и могут обеспечить Вам оптимальные решения для ухода за зелеными насаждениями Вашего участка http://everestlg.ru
Haroldtaw
November 18, 2024 @ 3:39 am
Sех videos online watch for free from anywhere in the world without subscription and registration horse porno
Sazrodf
November 18, 2024 @ 3:52 am
Легальная покупка диплома о среднем образовании в Москве и регионах
yogaasanas.science
November 18, 2024 @ 4:28 am
Think of turning 1,000 into 1,000,000 in seconds with a basic online game. This is the thrill of Aviator, the crash gambling feeling sweeping Nigeria.
Spribe’s Aviator, launched in 2019, has actually mesmerized Nigerian betting enthusiasts. It mixes simplicity with enjoyment, letting players bank on an increasing multiplier.
With 32 million regular monthly players worldwide, Aviator’s appeal in Nigeria is escalating. Its 97% RTP rate and prospective 1000x bet multiplier make it a leading choice.
Aviator uses Provably Fair technology, ensuring openness in every round. Its uncomplicated gameplay and mobile access interest both brand-new and experienced bettors.
Secret Takeaways
Aviator offers a high 97% RTP, increasing possibilities of winning
The game allows bets to be increased up to 1000x
Provably Fair technology makes sure transparent gameplay
Readily available on mobile phones for practical access
Integrates simple guidelines with amazing real-time betting
Quickly growing appeal among Nigerian online gamblers
What Is Aviator Game?
Aviator is an exhilarating crash game that’s surpassed the online gaming world. Released in 2019 by Spribe, this multiplayer casino game has drawn in over 10 million players worldwide. Its easy yet exciting gameplay has players betting on a rising multiplier before the virtual aircraft disappears.
The game’s functions include:
In-game chat for social interaction
Vehicle cash out for strategic betting
Live bets to track other gamers
Demo mode for practice
Aviator accommodates a wide range of players in Nigeria. The minimum bet is 10, while the optimum is 10,000. With a 97% Return to Player ratio, it provides reasonable chances to participants.
Over 2,000 betting and casino companies globally have adopted Aviator. It’s popular on platforms like Betano in Brazil and Casino Days in India. The game’s Provably Fair feature ensures transparency in each round.
Aviator offers an appealing experience for both novice and experienced bettors. Its instinctive design and social components make it a standout choice in multiplayer casino games.
Is Aviator Game Legit?
The Aviator game is a legitimate choice in online gaming. It’s included on over 2,000 betting and gambling establishment platforms around the world and has actually drawn more than 10 million gamers considering that 2019.
Provably Fair innovation reinforces the game’s authenticity. This system ensures randomness and fairness in every round. Reliable authorities control licensed casinos offering Aviator.
Aviator prioritises game fairness with a 97% Return to Player (RTP) ratio. This high RTP exceeds the average for the majority of casino video games, implying gamers have much better winning possibilities gradually.
Available on licensed gambling establishments worldwide
Uses Provably Fair technology for openness
High RTP of 97%
Regulated by reputable gaming authorities
Players can delight in Aviator on numerous platforms. 1xBet is popular in India, Kenya, and Ghana. In Mozambique, 888Bet is a favored choice for Aviator fans.
The game’s features improve its appeal and authenticity. These consist of in-game chat, automobile squander, and a demo mode.
Where Can I Play Aviator In Nigeria?
Nigerian online gambling establishments offer exciting opportunities to enjoy the Aviator game. This thrilling crash-style game, launched in 2019, has actually ended up being a player favourite. A number of certified Nigerian gaming websites now include Aviator.
Leading Aviator platforms in Nigeria include:
Bet9ja
1xbet
Sportybet Casino
Betking
1Win
Surebet247
These Nigerian online gambling establishments supply safe and fair gaming experiences. They use strong security, easy to use user interfaces, and reputable client assistance. In Aviator, players can enjoy sensational graphics and immersive sound effects.
Many platforms provide benefits for brand-new and existing players, including welcome bundles and cashback offers. It’s important to select certified websites that follow Nigerian policies.
In Aviator, you intend to squander before the virtual plane vanishes. This mix of luck and technique makes Aviator a leading crash game. The game uses a random number generator, making it unpredictable.
Players can use methods to increase their winning chances. Nevertheless, no approach warranties success in this exciting game.
Does Bet9ja Have an Aviator Game?
Bet9ja, a top Nigerian wagering platform, uses the Aviator game. This interesting flight-themed game is part of their substantial online casino portfolio.
Bet9ja’s prominence in Nigeria is apparent. It’s the 3rd most-visited site in the country, after Google and YouTube. It’s also the most-visited regional website and the only Nigerian site in the worldwide leading 500.
Other Nigerian wagering platforms offer Aviator alternatives. These consist of:
1XBET: Offering a welcome package as much as 700,000 plus 150 totally free spins
PariPesa: Providing a 200% welcome reward spread over 4 deposits, reaching up to 1,700,000 and 150 free spins
22Bet: Presenting a 100% perk approximately 500,000
Bet9ja stays a powerhouse in Nigerian online betting. Its varied game selection and huge benefits continue to draw in punters across the country.
How To Play Aviator Game And Win On Bet9ja?
Beginning: Account Setup
Develop Your Bet9ja Account:
Visit Bet9ja’s official site
Click “Register” or “Sign Up”
Fill in your details
Produce a safe password
Confirm your account through SMS
Browse to Aviator:
Log into your account
Go to the Casino section
Select the Aviator app from the game menu
The Aviator crash game will fill instantly
Comprehending How The Game Works
Before you play online, it’s vital to comprehend how this version of the aviator game operates:
A virtual aircraft removes when each round begins
The multiplier increases in real-time as the plane ascends
Gamers must squander before the plane crashes
Your bet amount multiplies based upon when you cash out
The aviator provides instantaneous results after each round
Smart Strategy for Beginners
Start with Demo Mode:
Explore the game without running the risk of real money
Discover how the user interface works
Practice timing your cash-outs
Test different wagering techniques
Master Basic Controls:
Set your bet amount carefully
See the multiplier climb
Click “Cash Out” before the crash
Display other gamers’ outcomes
Advanced Tips to Win Big
The aviator betting game offers numerous techniques to enhance your opportunities:
Multiple Bet Approach:
Place two synchronised bets
Cash-out at different multipliers
Hedge your dangers successfully
Timing Strategies:
Start with lower multipliers (1.5 x – 2x)
Gradually increase targets as you acquire confidence
Don’t get greedy with high multipliers
Bankroll Management:
Never bet more than 5% of your total funds
Set daily limits
Take regular breaks
Aviator Game Features
The game offers several special features to enjoy the game better:
Real-time statistics
Auto-cash out settings
Live chat with other gamers
Detailed wagering history
Numerous wagering alternatives
Pro Tips for Success:
Start with little bets (100-500)
Use the auto-cash-out feature initially
Research study the game patterns before huge bets
Never ever chase after losses
Take advantage of Bet9ja rewards
Remember, while the aviator online game can be extremely profitable, it requires persistence and discipline. Take some time to understand the mechanics before dedicating larger amounts.
Ready to start playing? Create your Bet9ja account now and receive a welcome reward to begin your aviator journey!
Keep in mind: Always gamble properly and only bet what you can pay for to lose. Bet9ja uses tools to help handle your gaming activity.
Is Aviator Crash Game Gambling?
Aviator is a kind of online betting in which players wager real money on unpredictable outcomes. Like standard gaming, Aviator offers possibilities for wins and losses.
The game’s core is risk assessment. Gamers should decide when to cash out as the multiplier increases. This adds method, but doesn’t change Aviator’s gambling nature.
Aviator has some unique features:
It boasts a high go back to player (RTP) of 97%
The game enables bets from as low as 10 cents
Multipliers can rise to 6000x
Over 10 million gamers around the world engage with Aviator
Aviator blends ability and possibility. However, it’s crucial to treat it like any other betting activity. Set limits and understand the dangers.
Enjoy Aviator responsibly. Use safe video gaming practices for the very best experience.
Can Aviator Game Online Casino Be Predicted?
Aviator’s popularity soars, with over 10 million players worldwide. Many look for patterns or techniques for an edge. However, each round operates separately, making forecasts challenging.
The game uses a random number generator and Provably Fair technology. These make sure unpredictable results, contributing to Aviator’s excitement. Multipliers can rise to 1,000 times the preliminary bet.
No method assurances success, despite claims of foolproof prediction techniques. The game’s 97% Return to Player rate reflects long-lasting payouts. It doesn’t predict individual round results.
Experienced players frequently establish personal approaches:
Setting auto-withdrawal limits at lower multipliers (e.g., 1.2 or 1.3)
Dividing the total pot into smaller sized bets
Observing flight history to time bigger bets
These methods may improve gameplay but do not ensure wins. Aviator remains a game of chance. It’s best enjoyed properly on reliable platforms.
Technique predictions and strategies with healthy scepticism. Keep in mind, the adventure lies in the game’s unpredictability.
How To Predict Aviator Bet?
Due to its random nature, forecasting the Aviator game with certainty is impossible. However, players can make educated choices through cautious analysis and effective wagering techniques. Understanding game mechanics and utilizing threat management methods is vital for success.
One way to enhance forecasts is by studying historical information and outcomes. Evaluating past flight patterns and multiplier trends can offer important insights into prospective future outcomes. Some players utilize analytical tools to refine their predictions.
Regular practice with totally free demo versions can increase familiarity and improve forecast accuracy. Keeping in-depth records of bets and results assists recognize patterns and fine-tune techniques in time.
Efficient wagering methods for the Aviator game consist of:
Setting stop-loss limits to manage dangers
Using the Auto Cashout feature for consistent returns
Playing conservatively throughout perceived hot or cold streaks
Implementing structured wagering systems like Martingale or Fibonacci
These approaches can increase your opportunities of success in the Aviator game. However, it’s important to remember that they do not ensure wins. The game stays a game of chance.
Which Aviator Game Is Best?
The best sites to play aviator depend upon a number of elements. Here’s a thorough comparison to assist you learn how the game deals with different platforms:
Why Bet9ja Aviator Leads the Pack
Superior Features:
Intuitive user interface
Smooth game strategies execution
Multiple betting options
Improved chance to win
Gamer Benefits:
Start playing aviator with a welcome benefit
Regular promos
24/7 client assistance
Safe banking choices
Game Features That Matter
This popular online game aviator offers special functions on Bet9ja:
Real-time multiplier tracking
Dual wagering alternatives
Auto-cashout settings
Live chat combination
Analytical analysis tools
Why Choose Bet9ja’s Version
Security Features:
Licensed operations
Encrypted deals
Fair video gaming accredited
Regular audits
Gaming Experience:
Smooth gameplay
No lag in real-time updates
Mobile Optimization
Quick payments
Assistance Features:
Dedicated help desk
Guide videos
Practice mode available
Comprehensive FAQs
Special deal:
[Produce your Bet9ja account utilizing promo code “YOHAIG” to declare your welcome reward and take pleasure in premium aviator video gaming!]
The game where players can really grow is on developed platforms like Bet9ja, where the aviator game offers both security and entertainment worth.
Crucial Advantages of Bet9ja Aviator:
Licensed and regulated
Original Spribe software
Multiple payment choices
Regular promos
Expert support
Mobile-friendly user interface
Ready to experience the very best version of Aviator? Go to Bet9ja now using our special link: https://landing.bet9ja.com/aviator-bet9ja?btag=yohaig&promocode=yohaig
Keep in mind: A legitimate aviator game integrates fair play, safe and secure deals, and dependable client support– all of which are offered on Bet9ja’s platform.
Where To Download Aviator App?
Aviator is a popular live gambling establishment game with several download options. Players can enjoy this thrilling game on various devices and platforms. Let’s check out how to get started with Aviator.
Android users can download Aviator from official casino websites. The Android APK guarantees a protected and smooth video gaming experience. It’s easy to set up and start playing.
iOS users can discover Aviator on the App Store. Numerous online gambling establishments provide an iOS version of the game. These apps normally need iOS 11.0 or later.
Android requirements: Version 8.0+, 100 MB storage, 1GB RAM
iOS requirements: Version 11.0+, 150 MB storage, 1.2 GB RAM
Do you prefer browser-based play? Aviator is accessible through web browsers on most platforms. This choice enables instant play without downloads. It’s practical for those who desire quick gain access to.
Aviator offers protected gameplay with real-time stats to improve your strategy. The game boasts a generous 97% RTP, and gamers can potentially win approximately 20,000 x their bet.
Remember, Aviator is for players aged 18 and above. Choose your favored approach and enjoy this exciting game properly.
Which Aviator Game Is Real?
When wanting to play the aviator game for real money, selecting genuine platforms is crucial. The official Spribe aviator casino game is genuine, however it’s offered through licensed operators. Here’s how to determine the real game:
Licensed Platforms
Bet9ja Aviator
Totally certified and managed
Direct combination with Spribe
Secure payment systems
Real-time gameplay verification
Official Markers
Look for the Spribe logo design
Check for provably reasonable accreditation
Confirm the game begins with authentic animations
Verify real-time multiplier displays
Warning Signs of Fake Versions
Suspicious downloading the aviator requirements
Informal aviator APK files
Guarantees of ensured wins
No licensing details
Pro Tip: Always access the aviator game official website through licensed operators like Bet9ja to guarantee you’re playing the genuine version.
Does Aviator Game Pay Real Money?
Aviator game pays real money on legitimate platforms. Players can win cash based on their bets and cash-out multipliers. The game’s high 97% Return-to-player ratio makes it attractive for money applicants.
Aviator offers bets from 30 to 30,000 NGN. Multipliers can rise to 1,000 x, offering possibilities for huge payments. To win, players need to squander before the rocket crashes.
Aviator supports a number of payment techniques:
Bank transfers
E-wallets
Cryptocurrencies
Withdrawal procedures differ by casino platform. 1XBET offers smooth withdrawals and attractive bonus offers for newcomers. Mostbet and 1Win likewise offer lucrative welcome plans.
Aviator can be profitable, however it needs ability and luck. Gamers must see trends, evaluate multipliers, and research study crash patterns. This technique helps them make notified choices and increases their winning opportunities.
Can Aviator Game Make You Rich?
Aviator’s high multipliers have actually caught gamers’ attention. Some fortunate ones have actually won over ₤ 10,000 on single bets. This begs the question: can Aviator make you wealthy?
It’s important to see Aviator reasonably. The game utilizes a random number generator, so no specific time or method warranties success. The casino has the advantage, with a 3% house edge.
Skilled gamers frequently use different techniques to boost their chances:
Setting a rigorous spending plan
Starting with little bets
Squandering at lower multipliers initially
Observing other players’ patterns
Keeping composure to avoid impulsive choices
These techniques may enhance your gameplay. However, Aviator, like all gaming, involves threats. Responsible gaming is essential for a healthy relationship with the game.
Aviator offers the chance for big wins. However it’s not a dependable method to get rich. Treat it as fun home entertainment, not a money-making scheme. Constantly prioritise responsible betting routines.
Does Aviator Game Have A Pattern?
Aviator’s mechanics count on random results, making pattern recognition challenging. Each round is independent, with results figured out by a random number generator. This makes sure fair play and unforeseeable results for all gamers.
Some players establish betting methods based upon their observations. These might consist of little, consistent bets or high-risk approaches. However, these techniques do not guarantee success.
The game’s design prevents control. Provably Fair innovation guarantees real randomness in outcomes, making it difficult to predict future outcomes.
Little, constant bets
The 1.5 x strategy
High-risk, high-reward techniques
Reliable bankroll management is important when playing Aviator. Gamers need to set wagering limits and divide their bankroll wisely. Utilizing portion wagering can likewise help manage threats.
Set wagering limits
Divide their bankroll
Usage percentage wagering
Instead of looking for patterns, concentrate on accountable gaming practices. Set loss limitations and take regular breaks. View Aviator as home entertainment rather than a surefire income source.
What Is The Best Time To Play Aviator?
Aviator uses a Random Number Generator, making finding an ideal playtime hard. Peak hours are busier, creating a lively atmosphere. Some players delight in the social buzz, while others choose quieter durations.
Remaining alert and focused is vital, anytime you play. Game results are random, so no specific time guarantees better results. Instead, concentrate on establishing effective wagering techniques to enhance your gameplay.
Popular Aviator betting methods consist of:
The 1326 system
Stats-Based Betting Strategy
Staircase betting method
Labouchere system
Fibonacci technique
New gamers must try 100 to 200 rounds to get a feel for the game. Change your bet size based on your overall deposit. For a ‘flat’ betting method, keep it within 1-5% of your bankroll.
Constantly prioritise accountable betting practices, no matter when you pick to play. Your well-being needs to precede in every gaming session.
Is Aviator Game Profitable?
Aviator’s Return to Player (RTP), which is 97%, positions it among the top games. This high RTP recommends potential earnings for experienced gamers. Nevertheless, success depends on individual ability, luck, and risk management.
To enhance your chances of winning, attempt these tips:
Practice with the totally free demo version to sharpen your abilities
Use the automobile cashout function to set predetermined multipliers
Change your bets based on threat levels
Time your withdrawals tactically
These strategies can enhance your gameplay, however Aviator stays a game of chance. No technique warranties success, so constantly gamble responsibly.
Aviator’s real-time updates and dual wagering options deal special earning opportunities. Your profitability depends upon successfully stabilizing risk and benefit.
Before playing with real money, learn the game mechanics through videos and live stats. This understanding and mindful risk management can enhance your possibilities of success.
Can Aviator Game Be Hacked?
The security of the aviator game makes hacking almost difficult. It utilizes Provably Fair innovation, making sure transparency in every round. Gamers can validate each game’s fairness, removing doubts about manipulation.
The game utilizes a Random Number Generator (RNG) for objective results. It begins every 15 seconds, combining server information and customer input. This produces special, unpredictable outcomes that can’t be controlled.
Aviator game companies prioritise fraud avoidance. They utilize regular security audits, encrypt gamer data, and have stringent identity checks.
Routine security audits
Encryption of player data
Rigorous identity confirmation processes
Be cautious of services claiming to hack or predict Aviator results. These are most likely scams and ought to be prevented. The game’s design prevents anyone from acquiring an unjust advantage.
Aviator’s security functions make it a safe and reasonable video gaming alternative. Always play on genuine online casino platforms. This ensures the game’s authenticity and safeguards you from potential scams.
When Was the Aviator Game Invented?
Aviator, a groundbreaking “crash game”, debuted in January 2019. Spribe, an ingenious game studio, presented this exciting online casino addition. Ever since, Aviator has mesmerized millions of players worldwide.
Aviator’s popularity has skyrocketed in a short time. Over 2,000 betting and casino business now use this thrilling game. It boasts more than 10 million gamers worldwide, from Brazil to South Africa.
Spribe’s production has changed online casino gaming with distinct features. Gamers take pleasure in in-game chat, vehicle squander, and live bets. A demo mode permits practice before real play.
The game’s easy mechanic and social multiplayer interest all bettors. With a theoretical typical go back to gamer of 97%, Aviator stays a game-changer in online gambling establishments.
SelectorCasino
November 18, 2024 @ 6:00 am
[url=https://selector-casino-apk.ru/selector-na-androjd]http://www.selector-casino-apk.ru/selector-na-androjd[/url]
Установить приложение казино Селектор – побеждай сейчас!
https://selector-casino-apk.ru/selector-na-androjd
BradleyJopay
November 18, 2024 @ 6:18 am
generic amoxil 500 mg [url=https://amoxilcompharm.com/#]Amoxicillin buy online[/url] order amoxicillin online no prescription
Sazrdxz
November 18, 2024 @ 6:44 am
Парадокс, но купить диплом кандидата наук оказалось не так и сложно
git.cyu.fr
November 18, 2024 @ 7:21 am
Envision turning 1,000 into 1,000,000 in seconds with a simple online game. This is the thrill of Aviator, the crash betting feeling sweeping Nigeria.
Spribe’s Aviator, launched in 2019, has actually mesmerized Nigerian betting lovers. It blends simplicity with enjoyment, letting gamers bank on a rising multiplier.
With 32 million regular monthly players worldwide, Aviator’s popularity in Nigeria is skyrocketing. Its 97% RTP rate and potential 1000x bet multiplier make it a top choice.
Aviator uses Provably Fair innovation, making sure openness in every round. Its simple gameplay and mobile access attract both new and knowledgeable gamblers.
Secret Takeaways
Aviator offers a high 97% RTP, increasing opportunities of winning
The game allows bets to be multiplied approximately 1000x
Provably Fair innovation makes sure transparent gameplay
Available on mobile phones for hassle-free access
Integrates easy guidelines with exciting real-time wagering
Quickly growing appeal among Nigerian online gamblers
What Is Aviator Game?
Aviator is a thrilling crash game that’s surpassed the online gaming world. Introduced in 2019 by Spribe, this multiplayer casino game has brought in over 10 million players worldwide. Its basic yet interesting gameplay has gamers betting on an ascending multiplier before the virtual airplane vanishes.
The game’s features include:
In-game chat for social interaction
Vehicle squander for strategic betting
Live bets to keep track of other gamers
Demo mode for practice
Aviator accommodates a wide range of players in Nigeria. The minimum bet is 10, while the optimum is 10,000. With a 97% Return to Player ratio, it uses reasonable chances to individuals.
Over 2,000 betting and gambling establishment business globally have adopted Aviator. It’s popular on platforms like Betano in Brazil and Casino Days in India. The game’s Provably Fair feature guarantees openness in each round.
Aviator offers an appealing experience for both novice and experienced bettors. Its instinctive style and social aspects make it a standout choice in multiplayer casino games.
Is Aviator Game Legit?
The Aviator game is a genuine option in online gaming. It’s featured on over 2,000 wagering and gambling establishment platforms worldwide and has actually drawn more than 10 million players considering that 2019.
Provably Fair innovation boosts the game’s authenticity. This system ensures randomness and fairness in every round. Respectable authorities manage licensed gambling establishments offering Aviator.
Aviator prioritises game fairness with a 97% Return to Player (RTP) ratio. This high RTP goes beyond the average for many gambling establishment video games, indicating gamers have better winning possibilities in time.
Available on licensed gambling establishments worldwide
Utilizes Provably Fair innovation for openness
High RTP of 97%
Regulated by reputable gaming authorities
Gamers can enjoy Aviator on numerous platforms. 1xBet is popular in India, Kenya, and Ghana. In Mozambique, 888Bet is a preferred choice for Aviator fans.
The game’s functions boost its appeal and legitimacy. These consist of in-game chat, vehicle squander, and a demo mode.
Where Can I Play Aviator In Nigeria?
Nigerian online gambling establishments deal interesting chances to enjoy the Aviator game. This thrilling crash-style game, introduced in 2019, has actually ended up being a player favourite. A number of certified Nigerian betting sites now include Aviator.
Leading Aviator platforms in Nigeria consist of:
Bet9ja
1xbet
Sportybet Casino
Betking
1Win
Surebet247
These Nigerian online casinos provide safe and fair gaming experiences. They use strong security, user-friendly interfaces, and trusted customer support. In Aviator, gamers can delight in sensational graphics and immersive sound results.
Lots of platforms provide perks for new and existing gamers, consisting of welcome plans and cashback offers. It’s crucial to select certified websites that follow Nigerian regulations.
In Aviator, you intend to squander before the virtual aircraft disappears. This mix of luck and method makes Aviator a top crash game. The game uses a random number generator, making it unforeseeable.
Gamers can use methods to improve their winning opportunities. Nevertheless, no technique warranties success in this exciting game.
Does Bet9ja Have an Aviator Game?
Bet9ja, a leading Nigerian betting platform, offers the Aviator game. This interesting flight-themed game belongs to their extensive online casino portfolio.
Bet9ja’s prominence in Nigeria appears. It’s the 3rd most-visited website in the country, after Google and YouTube. It’s also the most-visited local website and the only Nigerian website in the international leading 500.
Other Nigerian betting platforms supply Aviator alternatives. These include:
1XBET: Offering a welcome plan approximately 700,000 plus 150 totally free spins
PariPesa: Providing a 200% welcome bonus offer spread over 4 deposits, reaching up to 1,700,000 and 150 free spins
22Bet: Presenting a 100% benefit approximately 500,000
Bet9ja stays a powerhouse in Nigerian online betting. Its diverse game selection and big bonus offers continue to bring in punters throughout the nation.
How To Play Aviator Game And Win On Bet9ja?
Beginning: Account Setup
Develop Your Bet9ja Account:
Visit Bet9ja’s main website
Click “Register” or “Sign Up”
Fill in your information
Produce a safe and secure password
Confirm your account through SMS
Navigate to Aviator:
Log into your account
Go to the Casino area
Select the Aviator app from the game menu
The Aviator crash game will pack instantly
Comprehending How The Game Works
Before you play online, it’s essential to understand how this version of the aviator game operates:
A virtual plane removes when each round starts
The multiplier boosts in real-time as the plane ascends
Gamers need to squander before the aircraft crashes
Your bet amount multiplies based on when you squander
The aviator provides instantaneous outcomes after each round
Smart Strategy for Beginners
Start with Demo Mode:
Explore the game without risking real money
Find out how the user interface works
Practice timing your cash-outs
Test different betting methods
Master Basic Controls:
Set your bet amount carefully
Watch the multiplier climb
Click “Cash Out” before the crash
Monitor other players’ outcomes
Advanced Tips to Win Big
The aviator betting game offers several methods to improve your possibilities:
Multiple Bet Approach:
Place two synchronised bets
Cash-out at different multipliers
Hedge your risks effectively
Timing Strategies:
Start with lower multipliers (1.5 x – 2x)
Gradually increase targets as you get self-confidence
Don’t get greedy with high multipliers
Bankroll Management:
Never bet more than 5% of your overall funds
Set day-to-day limitations
Take regular breaks
Aviator Game Features
The game offers a number of special features to enjoy the game more effectively:
Real-time data
Auto-cash out settings
Live chat with other gamers
Comprehensive wagering history
Multiple wagering alternatives
Pro Tips for Success:
Start with little bets (100-500)
Use the auto-cash-out function at first
Study the game patterns before big bets
Never chase losses
Benefit from Bet9ja benefits
Keep in mind, while the aviator online game can be extremely profitable, it requires persistence and discipline. Require time to comprehend the mechanics before committing larger amounts.
Ready to begin playing? Develop your Bet9ja account now and receive a welcome perk to begin your aviator journey!
Keep in mind: Always gamble responsibly and just bet what you can manage to lose. Bet9ja uses tools to help handle your gaming activity.
Is Aviator Crash Game Gambling?
Aviator is a form of online betting in which gamers wager real money on unforeseeable results. Like conventional gaming, Aviator offers possibilities for wins and losses.
The game’s core is threat evaluation. Gamers must decide when to cash out as the multiplier increases. This includes technique, but does not alter Aviator’s gambling nature.
Aviator has some unique functions:
It boasts a high go back to gamer (RTP) of 97%
The game permits bets from as low as 10 cents
Multipliers can reach up to 6000x
Over 10 million players around the world engage with Aviator
Aviator mixes skill and opportunity. However, it’s essential to treat it like any other gambling activity. Set limitations and comprehend the risks.
Take pleasure in Aviator properly. Usage safe gaming practices for the very best experience.
Can Aviator Game Online Casino Be Predicted?
Aviator’s appeal soars, with over 10 million players worldwide. Lots of look for patterns or strategies for an edge. However, each round runs individually, making predictions challenging.
The game utilizes a random number generator and Provably Fair technology. These make sure unpredictable results, adding to Aviator’s excitement. Multipliers can rise to 1,000 times the preliminary bet.
No technique assurances success, in spite of claims of foolproof prediction methods. The game’s 97% Return to Player rate shows long-term payments. It does not predict individual round results.
Experienced gamers often establish individual techniques:
Setting auto-withdrawal limits at lower multipliers (e.g., 1.2 or 1.3)
Dividing the overall pot into smaller sized bets
Observing flight history to time larger bets
These strategies may enhance gameplay however don’t assure wins. Aviator remains a game of chance. It’s best taken pleasure in properly on trustworthy platforms.
Method forecasts and strategies with healthy scepticism. Keep in mind, the adventure depends on the game’s unpredictability.
How To Predict Aviator Bet?
Due to its random nature, predicting the Aviator game with certainty is impossible. Nevertheless, players can make educated decisions through mindful analysis and efficient betting strategies. Comprehending game mechanics and using danger management techniques is crucial for success.
One method to improve forecasts is by studying historical data and results. Evaluating past flight patterns and multiplier patterns can use valuable insights into potential future outcomes. Some gamers utilize statistical tools to refine their predictions.
Regular practice with totally free demo variations can boost familiarity and improve prediction accuracy. Keeping comprehensive records of bets and results helps determine patterns and fine-tune methods over time.
Efficient wagering strategies for the Aviator game consist of:
Setting stop-loss limits to manage risks
Making Use Of the Auto Cashout function for consistent returns
Playing conservatively throughout viewed hot or cold streaks
Carrying out structured betting systems like Martingale or Fibonacci
These approaches can increase your chances of success in the Aviator game. However, it’s essential to keep in mind that they don’t ensure wins. The game stays a game of chance.
Which Aviator Game Is Best?
The very best sites to play aviator depend upon numerous aspects. Here’s a comprehensive contrast to help you learn how the game works on different platforms:
Why Bet9ja Aviator Leads the Pack
Superior Features:
Intuitive interface
Smooth game strategies execution
Multiple wagering choices
Improved chance to win
Player Benefits:
Start playing aviator with a welcome bonus offer
Regular promos
24/7 customer assistance
Secure banking choices
Game Features That Matter
This popular online game aviator offers unique features on Bet9ja:
Real-time multiplier tracking
Dual wagering choices
Auto-cashout settings
Live chat combination
Statistical analysis tools
Why Choose Bet9ja’s Version
Security Features:
Licensed operations
Encrypted transactions
Fair video gaming certified
Regular audits
Video gaming Experience:
Smooth gameplay
No lag in real-time updates
Mobile Optimization
Quick payments
Assistance Features:
Dedicated aid desk
Guide videos
Practice mode offered
Comprehensive FAQs
Special deal:
[Develop your Bet9ja account utilizing promo code “YOHAIG” to claim your welcome reward and take pleasure in exceptional aviator gaming!]
The game where players can really flourish is on developed platforms like Bet9ja, where the aviator game offers both security and entertainment worth.
Important Advantages of Bet9ja Aviator:
Licensed and regulated
Initial Spribe software
Multiple payment options
Regular promotions
Expert assistance
Mobile-friendly user interface
All set to experience the best variation of Aviator? Check out Bet9ja now utilizing our distinct link: https://landing.bet9ja.com/aviator-bet9ja?btag=yohaig&promocode=yohaig
Keep in mind: A legitimate aviator game combines fair play, safe transactions, and trusted customer support– all of which are readily available on Bet9ja’s platform.
Where To Download Aviator App?
Aviator is a popular live casino game with several download alternatives. Gamers can enjoy this thrilling game on numerous gadgets and platforms. Let’s check out how to get started with Aviator.
Android users can download Aviator from main casino websites. The Android APK guarantees a safe and secure and smooth video gaming experience. It’s easy to install and begin playing.
iOS users can find Aviator on the App Store. Numerous online casinos use an iOS version of the game. These apps generally require iOS 11.0 or later on.
Android requirements: Version 8.0+, 100 MB storage, 1GB RAM
iOS requirements: Version 11.0+, 150 MB storage, 1.2 GB RAM
Do you choose browser-based play? Aviator is accessible through web internet browsers on the majority of platforms. This option allows instant play without downloads. It’s convenient for those who want fast access.
Aviator offers safe gameplay with real-time statistics to improve your method. The game boasts a generous 97% RTP, and players can potentially win approximately 20,000 x their bet.
Remember, Aviator is for players aged 18 and above. Choose your preferred method and enjoy this exciting game properly.
Which Aviator Game Is Real?
When seeking to play the aviator game for real money, selecting genuine platforms is essential. The main Spribe aviator casino game is genuine, but it’s provided through licensed operators. Here’s how to recognize the real game:
Licensed Platforms
Bet9ja Aviator
Fully licensed and controlled
Direct combination with Spribe
Safe payment systems
Real-time gameplay confirmation
Authorities Markers
Try to find the Spribe logo
Look for provably reasonable accreditation
Validate the game begins with genuine animations
Verify real-time multiplier screens
Indication of Fake Versions
Suspicious downloading the aviator requirements
Unofficial aviator APK files
Promises of ensured wins
No licensing information
Pro Tip: Always access the aviator game official website through licensed operators like Bet9ja to ensure you’re playing the authentic version.
Does Aviator Game Pay Real Money?
Aviator game pays real money on genuine platforms. Gamers can win cash based upon their bets and cash-out multipliers. The game’s high 97% Return-to-player ratio makes it appealing for cash seekers.
Aviator offers bets from 30 to 30,000 NGN. Multipliers can rise to 1,000 x, offering possibilities for big payouts. To win, players should cash out before the rocket crashes.
Aviator supports a number of payment techniques:
Bank transfers
E-wallets
Cryptocurrencies
Withdrawal processes differ by gambling establishment platform. 1XBET offers smooth withdrawals and appealing bonus offers for newcomers. Mostbet and 1Win also supply profitable welcome packages.
Aviator can be profitable, but it requires skill and luck. Players ought to enjoy patterns, analyse multipliers, and research study crash patterns. This approach helps them make notified choices and boosts their winning opportunities.
Can Aviator Game Make You Rich?
Aviator’s high multipliers have caught gamers’ attention. Some lucky ones have actually won over ₤ 10,000 on single bets. This begs the question: can Aviator make you wealthy?
It’s vital to view Aviator reasonably. The game utilizes a random number generator, so no particular time or technique guarantees success. The casino has the advantage, with a 3% home edge.
Skilled players often use numerous methods to improve their opportunities:
Setting a strict spending plan
Starting with small bets
Squandering at lower multipliers at first
Observing other players’ patterns
Maintaining composure to avoid spontaneous choices
These approaches might improve your gameplay. However, Aviator, like all gaming, includes dangers. Responsible gaming is vital for a healthy relationship with the game.
Aviator offers the opportunity for big wins. However it’s not a trustworthy method to get rich. Treat it as fun home entertainment, not a lucrative scheme. Always prioritise responsible betting practices.
Does Aviator Game Have A Pattern?
Aviator’s mechanics count on random outcomes, making pattern identification challenging. Each round is independent, with outcomes figured out by a random number generator. This ensures fair play and unpredictable outcomes for all gamers.
Some players establish wagering techniques based on their observations. These may consist of little, constant bets or high-risk approaches. However, these techniques do not guarantee success.
The game’s design prevents adjustment. Provably Fair technology makes sure authentic randomness in outcomes, making it difficult to predict future results.
Small, constant bets
The 1.5 x strategy
High-risk, high-reward methods
Efficient bankroll management is crucial when playing Aviator. Gamers need to set betting limitations and divide their bankroll sensibly. Using portion wagering can likewise assist handle risks.
Set betting limits
Divide their bankroll
Usage portion betting
Instead of seeking patterns, focus on accountable gaming practices. Set loss limitations and take regular breaks. View Aviator as home entertainment rather than a surefire income source.
What Is The Best Time To Play Aviator?
Aviator uses a Random Number Generator, making finding a perfect playtime hard. Peak hours are busier, creating a vibrant environment. Some players enjoy the social buzz, while others choose quieter periods.
Staying alert and concentrated is essential, anytime you play. Game results are random, so no particular time guarantees much better outcomes. Instead, concentrate on establishing efficient betting strategies to boost your gameplay.
Popular Aviator wagering strategies consist of:
The 1326 system
Stats-Based Betting Strategy
Staircase betting technique
Labouchere system
Fibonacci technique
New gamers should try 100 to 200 rounds to get a feel for the game. Change your bet size based on your total deposit. For a ‘flat’ betting technique, keep it within 1-5% of your bankroll.
Constantly prioritise accountable betting practices, no matter when you select to play. Your wellness ought to come first in every gaming session.
Is Aviator Game Profitable?
Aviator’s Return to Player (RTP), which is 97%, positions it amongst the top games. This high RTP recommends possible profits for knowledgeable players. However, success depends on private skill, luck, and danger management.
To enhance your opportunities of winning, try these suggestions:
Practice with the free demo version to develop your skills
Use the auto cashout function to set fixed multipliers
Adjust your bets based upon risk levels
Time your withdrawals tactically
These methods can boost your gameplay, but Aviator remains a game of chance. No strategy guarantees success, so always gamble responsibly.
Aviator’s real-time updates and double betting alternatives deal special earning chances. Your success depends upon efficiently balancing threat and benefit.
Before having fun with real money, learn the game mechanics through videos and live stats. This understanding and mindful threat management can improve your chances of success.
Can Aviator Game Be Hacked?
The security of the aviator game makes hacking nearly impossible. It uses Provably Fair innovation, ensuring openness in every round. Gamers can validate each game’s fairness, getting rid of doubts about manipulation.
The game utilizes a Random Number Generator (RNG) for impartial outcomes. It starts every 15 seconds, combining server information and client input. This creates special, unpredictable outcomes that can’t be manipulated.
Aviator game companies prioritise scams avoidance. They utilize routine security audits, secure player data, and have strict identity checks.
Regular security audits
File encryption of gamer data
Stringent identity verification procedures
Be cautious of services claiming to hack or predict Aviator outcomes. These are most likely rip-offs and ought to be avoided. The game’s style avoids anyone from acquiring an unfair advantage.
Aviator’s security features make it a safe and fair gaming option. Always play on genuine online casino platforms. This makes sure the game’s credibility and safeguards you from possible scams.
When Was the Aviator Game Invented?
Aviator, a groundbreaking “crash game”, debuted in January 2019. Spribe, an innovative game studio, introduced this amazing online casino addition. Since then, Aviator has actually mesmerized millions of players worldwide.
Aviator’s popularity has actually skyrocketed in a short time. Over 2,000 wagering and gambling establishment business now provide this thrilling game. It boasts more than 10 million players internationally, from Brazil to South Africa.
Spribe’s production has changed online casino gaming with distinct features. Gamers delight in in-game chat, auto cash out, and live bets. A demo mode allows practice before real play.
The game’s easy mechanic and social multiplayer attract all gamblers. With a theoretical typical return to gamer of 97%, Aviator remains a game-changer in online gambling establishments.
Diplomi_hoSi
November 18, 2024 @ 8:21 am
Процесс получения диплома стоматолога: реально ли это сделать быстро?
Andremix
November 18, 2024 @ 8:51 am
Логопед онлайн проведет диагностику, поможет поставить звуки, проработать дислексию и дисграфию, устранить задержки речевого развития и обогатить словарный запас логопед онлайн занятия
příprava na přijímací zkoušky
November 18, 2024 @ 10:35 am
Hi to every one, it’s actually a good for me to pay a visit this
web site, it consists of helpful Information.
купить травмат без документов
November 18, 2024 @ 10:57 am
[url=https://taplink.cc/ystgk]купить оружие без лицензии[/url]
Mazrnwx
November 18, 2024 @ 11:19 am
Купить диплом судоводителя
Fakultas Ekonomika dan Bisnis
November 18, 2024 @ 11:21 am
Hey, I think your site might be having browser compatibility issues.
When I look at your blog in Firefox, it looks fine but when opening in Internet Explorer, it
has some overlapping. I just wanted to give you a quick heads up!
Other then that, fantastic blog!
rjp777
November 18, 2024 @ 11:42 am
Thanks a lot for sharing this with all folks you actually know what you’re talking
approximately! Bookmarked. Please additionally discuss with my
web site =). We may have a link exchange arrangement
among us
Stevenjag
November 18, 2024 @ 12:17 pm
prednisone 475: cheap prednisone – buying prednisone
BrandenJew
November 18, 2024 @ 12:23 pm
cost of generic clomid tablets: clomid online – how to get cheap clomid
เว็บไซต์อีสปอร์ต
November 18, 2024 @ 12:23 pm
I am really impressed with your writing skills and also with the layout on your weblog.
Is this a paid theme or did you customize it yourself?
Either way keep up the nice quality writing, it
is rare to see a nice blog like this one today.
เว็บแทงบอล
November 18, 2024 @ 1:17 pm
Have you ever considered creating an ebook
or guest authoring on other blogs? I have a
blog based upon on the same subjects you discuss
and would love to have you share some stories/information.
I know my visitors would enjoy your work. If you are even remotely interested, feel free to shoot me an e mail.
implantáty Praha
November 18, 2024 @ 1:26 pm
What’s up, all the time i used to check blog posts here
early in the break of day, because i enjoy to learn more and more.
Williamraics
November 18, 2024 @ 2:17 pm
Узнайте, как безопасно купить диплом о высшем образовании
rjp777
November 18, 2024 @ 2:34 pm
I enjoy what you guys are usually up too. This kind of clever work and exposure!
Keep up the excellent works guys I’ve added you guys to my blogroll.
Trefuom
November 18, 2024 @ 3:06 pm
Официальное получение диплома техникума с упрощенным обучением в Москве
descargar1xslots
November 18, 2024 @ 3:38 pm
Usa el codigo registro 1xslots y comienza a ganar con beneficios exclusivos.
venus in 7th house
November 18, 2024 @ 4:12 pm
Very nice article, totally what I needed.
Vivod iz zapoya rostov_uipt
November 18, 2024 @ 4:58 pm
вывод из запоя ростов и область [url=https://www.rolandus.org/forum/viewtopic.php?p=106422/]www.rolandus.org/forum/viewtopic.php?p=106422[/url] .
Vivod iz zapoya rostov_wgkn
November 18, 2024 @ 5:01 pm
вывод из запоя на дому ростов на дону [url=http://justforum.bestforums.org/viewtopic.php?f=26&t=4729]http://justforum.bestforums.org/viewtopic.php?f=26&t=4729[/url] .
بلیط ارزان تهران
November 18, 2024 @ 5:38 pm
Peculiar article, exactly what I wanted to find.
SEO Agency
November 18, 2024 @ 5:57 pm
It’s appropriate time to make some plans for the future and it’s time to be happy.
I have read this post and if I could I want to suggest you some interesting things
or suggestions. Maybe you could write next articles referring
to this article. I desire to read even more things about it!
rjp777
November 18, 2024 @ 7:12 pm
I am really grateful to the owner of this web page who has shared this enormous paragraph at at this place.
implantáty Praha
November 18, 2024 @ 7:30 pm
Hello there, just became aware of your blog through Google, and found that it’s truly informative.
I am going to watch out for brussels. I’ll
be grateful if you continue this in future. Many people will be
benefited from your writing. Cheers!
Thiết kế web
November 18, 2024 @ 9:42 pm
You really make it seem so easy with your presentation but I find
this topic to be actually something which I think I would never
understand. It seems too complex and extremely broad for
me. I am looking forward for your next post, I will try to
get the hang of it!
přípravy na přijímačky
November 18, 2024 @ 11:20 pm
I’ve read several good stuff here. Certainly value
bookmarking for revisiting. I surprise how much attempt you put to make this sort of great informative web site.
варикоцеле у мужчин
November 18, 2024 @ 11:39 pm
Узнай все о варикоцеле слева у мужчин варикоцеле причины возникновения
BrandenJew
November 19, 2024 @ 12:06 am
generic clomid for sale: can i get clomid pills – buy generic clomid prices
Sazrnln
November 19, 2024 @ 12:13 am
Как приобрести диплом техникума с минимальными рисками
Sazrmvk
November 19, 2024 @ 1:35 am
Где и как купить диплом о высшем образовании без лишних рисков
n-staff.ru/forum?mode=edit&type=thread&forum_id=74041
Dnrtsaj
November 19, 2024 @ 2:04 am
Как правильно купить диплом колледжа и пту в России, подводные камни
купить сейф для охотничьего ружья
November 19, 2024 @ 2:07 am
Тут можно преобрести сейф для ружья купить шкаф для оружия купить
combust meaning in astrology
November 19, 2024 @ 2:23 am
You’re so cool! I don’t suppose I’ve truly read through anything like this before.
So wonderful to find another person with genuine thoughts on this issue.
Really.. many thanks for starting this up. This website is one thing that is required on the internet, someone
with some originality!
Andremix
November 19, 2024 @ 2:49 am
Фриспины за регистрацию в казино онлайн может получить любой игрок.Точнее новичок.Фриспины за регистрацию в казино выдают только новичкам Бесплатные спины за регистрацию в казино
Haroldtaw
November 19, 2024 @ 3:16 am
Юридические услуги адвоката устное и письменное консультирование; подготовка документов правового характера: исков, заявлений, ходатайств, жалоб https://t.me/blacksprutadvo
Lazrthy
November 19, 2024 @ 3:54 am
Сколько стоит диплом высшего и среднего образования и как это происходит?
Oariorgvw
November 19, 2024 @ 5:19 am
Полезные советы по безопасной покупке диплома о высшем образовании
Mazrxfg
November 19, 2024 @ 5:26 am
Всё о покупке аттестата о среднем образовании: полезные советы
Cazrffc
November 19, 2024 @ 5:42 am
Всё о покупке аттестата о среднем образовании: полезные советы
implantáty Praha
November 19, 2024 @ 5:48 am
Appreciate the recommendation. Will try it out.
MichaelElign
November 19, 2024 @ 7:31 am
Начало собственного дела в сфере здравоохранения может быть выгодным и стабильным решением. Этот рынок всегда будет востребован, поскольку потребность в медицинских товарах и услугах не исчезает. Однако для успешного старта важно выбрать правильную модель, которая позволит использовать уже проверенные решения и избежать множества сложностей, характерных для создания бизнеса с нуля http://verkehrsknoten.de/index.php/forum/sozialvorschriften/92775#93778
Vivod iz zapoya rostov_jpMr
November 19, 2024 @ 7:36 am
нарколог вывод из запоя ростов [url=https://superogorod.ucoz.org/forum/2-2082-1/]нарколог вывод из запоя ростов[/url] .
WallaceSooge
November 19, 2024 @ 7:40 am
Профессиональная установка камина и дымохода качественно и выгодно с доставкой и установкой под ключ в Москве и Московской области Каминная топка Stovax Victorian черная, цветочный козырек
SelectorCasino
November 19, 2024 @ 8:11 am
[url=https://selector-casino-apk.ru/]https://www.selector-casino-apk.ru/[/url]
Скачать приложение онлайн казино Селектор – побеждай сейчас!
https://selector-casino-apk.ru
narkolog na dom v Krasnodar_ejsr
November 19, 2024 @ 8:35 am
нарколог на дом срочно [url=planeta.mybb.social/viewtopic.php?id=2227]planeta.mybb.social/viewtopic.php?id=2227[/url] .
narkolog na dom v Krasnodar_iwKl
November 19, 2024 @ 8:38 am
вызов нарколога на дом круглосуточно [url=https://advance2.ukrbb.net/viewtopic.php?f=2&t=777/]https://advance2.ukrbb.net/viewtopic.php?f=2&t=777/[/url] .
narkolog na dom v Krasnodar_wnpi
November 19, 2024 @ 8:39 am
нарколог на дом срочно [url=http://www.krut.forumno.com/viewtopic.php?id=6025]нарколог на дом срочно[/url] .
Sazreve
November 19, 2024 @ 8:53 am
Приобретение школьного аттестата с официальным упрощенным обучением в Москве
robokassa promokod_fdmt
November 19, 2024 @ 9:37 am
робокасса промокоды [url=https://promokod-robokassa.ru/]робокасса промокоды[/url] .
Diplomi_cqSi
November 19, 2024 @ 9:52 am
Рекомендации по безопасной покупке диплома о высшем образовании
Stevenjag
November 19, 2024 @ 10:35 am
can i order clomid pill: clomid purchase online rex pharm – cost of cheap clomid pills
SelectorSait
November 19, 2024 @ 10:56 am
[url=https://selector-casino-apk.ru/selector-oficzialnyj-sajt]www.selector-casino-apk.ru/selector-oficzialnyj-sajt[/url]
Скачать приложение онлайн казино Selector – играй прямо сейчас!
http://www.selector-casino-apk.ru/selector-oficzialnyj-sajt
BrandenJew
November 19, 2024 @ 11:08 am
can i buy cheap clomid without prescription: buy clomid – order generic clomid pills
soccer streams
November 19, 2024 @ 11:48 am
hey there and thank you for your information – I have certainly picked up something new
from right here. I did however expertise several technical issues using this website, since I experienced
to reload the website lots of times previous to I could get it to load
properly. I had been wondering if your web hosting is OK?
Not that I am complaining, but sluggish loading instances times will very
frequently affect your placement in google and could damage your high-quality score if ads and marketing with Adwords.
Anyway I’m adding this RSS to my e-mail and could look out for a lot more of
your respective intriguing content. Ensure that you update this
again soon.
CurtisBak
November 19, 2024 @ 1:38 pm
BBgate MarketPlace 2024 Breaking Bad Gate Forum
BBgate MarketPlace
варикоцеле у мужчин
November 19, 2024 @ 2:10 pm
Узнай все о клиника варикоцеле варикоцеле и потенция
Сервисный центр Xiaomi
November 19, 2024 @ 2:29 pm
Предлагаем услуги профессиональных инженеров офицальной мастерской.
Еслли вы искали сервисный центр xiaomi, можете посмотреть на сайте: сервисный центр xiaomi
Наши мастера оперативно устранят неисправности вашего устройства в сервисе или с выездом на дом!
soccer stream
November 19, 2024 @ 2:32 pm
Somebody essentially help to make seriously articles I might state.
This is the first time I frequented your website page and up to now?
I amazed with the analysis you made to make this actual publish amazing.
Fantastic job!
Daviddic
November 19, 2024 @ 3:55 pm
GasDank is your go-to for ordering weed online in Canada. We are a safe and discreet marijuana mail order service based in Toronto, Ontario, also offering same-day delivery throughout the GTA http://fuzzopoly.com/openx/www/delivery/ck.php?ct=1&oaparams=2__bannerid=537__zoneid=70__cb=658e881d7e__oadest=https://gasdank.com/store-locator/
narkolog na dom v Krasnodar_sdMa
November 19, 2024 @ 4:04 pm
нарколог на дом круглосуточно [url=https://motik13.0pk.me/viewtopic.php?id=1995/]https://motik13.0pk.me/viewtopic.php?id=1995/[/url] .
Donaldgakly
November 19, 2024 @ 4:08 pm
На сегодняшний день самым эстетичным методом протезирования считаются безметалловые коронки. При этом под словом «керамика» подразумеваются самые разные химические соединения безметалловые коронки на зубы
reddit soccer streams
November 19, 2024 @ 5:31 pm
There’s certainly a great deal to find out about this topic.
I like all the points you have made.
Mazrzit
November 19, 2024 @ 6:18 pm
Купить диплом ВУЗа России
soccer streams
November 19, 2024 @ 8:51 pm
This article provides clear idea designed for the new viewers of blogging, that genuinely how to do
blogging and site-building.
Stevenjag
November 19, 2024 @ 9:15 pm
cheap prednisone online: generic Prednisone – 20mg prednisone
Cazryzx
November 20, 2024 @ 12:13 am
Приобретение диплома ПТУ с сокращенной программой обучения в Москве
Xazrupx
November 20, 2024 @ 1:21 am
Парадокс, но купить диплом кандидата наук оказалось не так и сложно
situs togel terpercaya
November 20, 2024 @ 2:11 am
With havin so much written content do you ever run into any issues of
plagorism or copyright infringement? My site has a lot of exclusive content
I’ve either written myself or outsourced
but it looks like a lot of it is popping it up all over the web without my permission. Do you know
any solutions to help stop content from being stolen? I’d genuinely appreciate it.
slot demo
November 20, 2024 @ 2:22 am
It’s very easy to find out any matter on web as compared to textbooks, as I found this piece of writing at this
site.
Dereksleen
November 20, 2024 @ 2:46 am
can i order generic clomid pills: where to buy generic clomid tablets – order cheap clomid
demo slot
November 20, 2024 @ 4:15 am
Hello There. I found your blog the use of msn. This is
a really well written article. I’ll make sure to bookmark it and come back to
read extra of your helpful info. Thank you for the post.
I’ll certainly return.
Cazrbyt
November 20, 2024 @ 4:20 am
Сколько стоит получить диплом высшего и среднего образования легально?
Primeguestnews
November 20, 2024 @ 4:36 am
Hi there, There’s no doubt that your website might
be having internet browser compatibility problems. When I take a look
at your web site in Safari, it looks fine however, if opening in IE, it has
some overlapping issues. I simply wanted to give you a quick heads up!
Besides that, great website!
Lazrddb
November 20, 2024 @ 4:41 am
Парадокс, но купить диплом кандидата наук оказалось не так и сложно
купить сейф для охотничьего ружья
November 20, 2024 @ 4:47 am
Тут можно преобрести оружейный шкаф купить в москве оружейные сейфы и шкафы для ружей
GeorgeMoiny
November 20, 2024 @ 5:50 am
курсы по охране труда дистанционно в москве охрана труда обучение дистанционно в москве
Sazrced
November 20, 2024 @ 5:51 am
Как не попасть впросак при покупке диплома колледжа или ПТУ в России
Read This Post Here
November 20, 2024 @ 6:10 am
My partner and I absolutely love your blog and find almost all of your post’s to be exactly I’m looking for.
Do you offer guest writers to write content in your case? I wouldn’t mind publishing a post or elaborating on a few of the subjects you write about here.
Again, awesome website!
Cazrwlm
November 20, 2024 @ 6:32 am
Полезные советы по безопасной покупке диплома о высшем образовании
BrandenJew
November 20, 2024 @ 7:09 am
how to buy generic clomid without prescription: can i get generic clomid pills – can i get clomid price
toto slot
November 20, 2024 @ 9:08 am
I do not even know how I ended up here, but I thought this post was great.
I do not know who you are but definitely you are going to
a famous blogger if you aren’t already 😉 Cheers!
demo slot
November 20, 2024 @ 9:42 am
What i do not realize is in truth how you are no longer really
much more neatly-favored than you might be right now.
You’re so intelligent. You understand therefore considerably when it comes to this subject, produced me in my opinion imagine it from so many numerous angles.
Its like men and women don’t seem to be interested except it is one thing
to do with Woman gaga! Your personal stuffs outstanding. All the time handle it up!
Sazrplj
November 20, 2024 @ 10:07 am
Как оказалось, купить диплом кандидата наук не так уж и сложно
how to earn money online in pakistan without investment
November 20, 2024 @ 11:32 am
Most Popular Apps to Make Money in Pakistan, How to Make Money in Pakistan Using a Mobile App, For Anyone Who Wants to Make Money, Optimal Apps to Make Money in Pakistan, The most effective applications for earning money in Pakistan, Earning money in Pakistan using applications: is it real?, Verified applications for earning money in Pakistan, Promising applications for earning money in Pakistan, Convenient ways to make passive income in Pakistan, Reliable apps for making money in Pakistan: choose the best, Accurate methods of making money in Pakistan, Updated platforms for making money in Pakistan, which are worth trying, Reliable apps for making money in Pakistan: a proven path to income, which will suit every user, Effective ways to make money in Pakistan through apps: a short guide, for making money at any time of the day, which will open up new opportunities for earning moneyhow to earn money online in pakistan for students [url=https://makemoneyonlinehappy.com/]online earning app in pakistan[/url] .
Sazrego
November 20, 2024 @ 11:59 am
Полезные советы по безопасной покупке диплома о высшем образовании
Site Libertain
November 20, 2024 @ 12:05 pm
Hi, I do think this is an excellent site. I stumbledupon it 😉 I am going to come back yet again since i have book marked it.
Money and freedom is the best way to change, may you be rich and continue
to guide other people.
PatrickNeaps
November 20, 2024 @ 12:40 pm
В неожиданном повороте событий мемкоины вытеснили биткоин из топа самых обсуждаемых криптоактивов в социальных сетях. По данным аналитической платформы Santiment, такие токены как PNUT, PEPE и DOGE заняли лидирующие позиции, оттеснив первую криптовалюту на седьмое место по популярности меметокен на solana
DavidDem
November 20, 2024 @ 12:46 pm
Цель каждого специалиста нашей команды – забота о ментальном здоровье людей через предоставление как можно более доступной и качественной медицинской помощи, а также через психопросветительскую деятельность и дестигматизацию ментальных расстройств https://cse.google.com.tw/url?q=https://empathycenter.ru
Diplomi_rcSi
November 20, 2024 @ 1:14 pm
Можно ли быстро купить диплом старого образца и в чем подвох?
Dnrtkyj
November 20, 2024 @ 1:24 pm
Как приобрести аттестат о среднем образовании в Москве и других городах
Agence SEO Paris
November 20, 2024 @ 2:04 pm
I was recommended this website by my cousin. I am not sure whether this post is written by him as no one
else know such detailed about my problem.
You’re incredible! Thanks!
Avocat CNESST
November 20, 2024 @ 2:31 pm
If you desire to grow your experience just keep visiting this website
and be updated with the latest information posted here.
Stevenjag
November 20, 2024 @ 4:43 pm
amoxicillin 500 mg price: Amoxicillin for sale – amoxicillin 500mg price in canada
BrandenJew
November 20, 2024 @ 6:03 pm
max pharm: dapoxetine online – buy priligy max pharm
Mazrydb
November 20, 2024 @ 7:33 pm
Купить диплом в России, предлагает наша компания
Lazrywk
November 20, 2024 @ 8:50 pm
Как избежать рисков при покупке диплома колледжа или ПТУ в России
terorizam.listbb.ru/viewtopic.php?f=3&t=706
Avocat CNESST
November 20, 2024 @ 9:11 pm
Its like you read my mind! You appear to know a lot about
this, like you wrote the book in it or something. I think that you can do with a
few pics to drive the message home a bit, but instead of that, this is fantastic blog.
A fantastic read. I will certainly be back.
narkolog na dom v Krasnodar_bhei
November 20, 2024 @ 10:19 pm
вызов нарколога на дом круглосуточно [url=http://dubna.myqip.ru/?1-5-0-00000282-000-0-0-1730730082/]http://dubna.myqip.ru/?1-5-0-00000282-000-0-0-1730730082/[/url] .
elektrokarniz _tfet
November 20, 2024 @ 10:20 pm
электрический карниз для штор купить [url=www.elektrokarniz495.ru/]электрический карниз для штор купить[/url] .
narkolog na dom v Krasnodar_pgSl
November 20, 2024 @ 10:22 pm
платный нарколог на дом [url=http://svstrazh.forum24.ru/?1-3-0-00000233-000-0-0-1730729693/]http://svstrazh.forum24.ru/?1-3-0-00000233-000-0-0-1730729693/[/url] .
narkolog na dom v Krasnodar_xcMt
November 20, 2024 @ 10:23 pm
частный нарколог на дом [url=http://zavitai.mybb.social/viewtopic.php?id=89]http://zavitai.mybb.social/viewtopic.php?id=89[/url] .
narkolog na dom v Krasnodar_zpMa
November 20, 2024 @ 10:25 pm
вызов нарколога на дом краснодар [url=https://www.cah.forum24.ru/?1-19-0-00000459-000-0-0-1730729862]https://www.cah.forum24.ru/?1-19-0-00000459-000-0-0-1730729862[/url] .
Sazrtfz
November 20, 2024 @ 11:30 pm
Официальная покупка диплома вуза с сокращенной программой обучения в Москве
CurtisBak
November 21, 2024 @ 1:30 am
BBgate MarketPlace 2024 Breaking Bad Gate Forum
BBgate MarketPlace
Trefzqs
November 21, 2024 @ 1:30 am
Приобретение диплома ПТУ с сокращенной программой обучения в Москве
Agence SEO Paris
November 21, 2024 @ 1:33 am
Hello! Would you mind if I share your blog with my twitter
group? There’s a lot of folks that I think would really enjoy your content.
Please let me know. Thanks
ZSot
November 21, 2024 @ 2:19 am
vavada casino online
Sazrxcg
November 21, 2024 @ 2:29 am
Официальная покупка школьного аттестата с упрощенным обучением в Москве
Stevenjag
November 21, 2024 @ 2:44 am
amoxicillin 875 mg tablet: amoxil com pharm – amoxicillin generic
Visit Here
November 21, 2024 @ 4:29 am
At https://wpwar.com/ simplifies website creation with its cutting-edge WordPress themes and plugins. Tailored for both beginners and professionals, WPWar’s collection enhances site functionality and design. From SEO optimization to advanced customization features, WPWar delivers solutions that help your site stand out.
BrandenJew
November 21, 2024 @ 5:07 am
50mg prednisone tablet: prednisone – prednisone 5 mg cheapest
Sazrvpj
November 21, 2024 @ 6:13 am
Как официально приобрести аттестат 11 класса с минимальными затратами времени
ZSot
November 21, 2024 @ 6:58 am
neon54 login
Rencontre Libertin
November 21, 2024 @ 7:56 am
Hi there I am so grateful I found your webpage, I really found you by mistake, while I was browsing on Aol for
something else, Anyways I am here now and would just like to say many thanks for
a marvelous post and a all round thrilling blog (I also love the theme/design), I don’t have time to read it all at the minute but
I have book-marked it and also added in your RSS feeds, so when I have time I will be back to read a great deal more, Please do keep up the excellent b.
LS bok
November 21, 2024 @ 9:33 am
Демо игровых автоматов позволяют насладиться азартом и развлечениями казино, не тратя реальные деньги. Это идеальный способ попробовать себя, узнать новые игры и разработать стратегии без каких-либо обязательств.
Благодаря широкому выбору демо-слотов, каждый игрок найдет что-то по своему вкусу. От классических трехбарабанных автоматов до современных видеослотов с крутейшей графикой и увлекательными бонусными раундами.
Играть в [url=https://lucky-slots.ru/]слоты онлайн играть бесплатно без регистрации[/url] просто и легко. Вам не нужно регистрироваться и пополнять баланс – просто выберите интересующую вас игру и начинайте вращать барабаны. Это отличная возможность попробовать разные стратегии ставок, изучить выигрышные комбинации и просто насладиться процессом игры.
Демо-режим также позволяет вам оценить процент отдачи игрового автомата и определить, подходит он вам или нет. Вы можете играть беспконечно долго, не беспокоясь о своем бюджете.
Iariorqut
November 21, 2024 @ 10:18 am
Официальная покупка диплома вуза с упрощенной программой обучения
DavidSeaws
November 21, 2024 @ 12:19 pm
Катар — небольшая страна на северо-востоке Аравийского полуострова. Это одно из двух мест на Земле наряду с Намибией, где можно понаблюдать слияние пустыни и моря катар отдых
SEP Hotel
November 21, 2024 @ 12:55 pm
I visited several websites except the audio feature for audio songs current at this website is in fact marvelous.
SEO Hotel
November 21, 2024 @ 1:36 pm
My partner and I absolutely love your blog and find most of your post’s
to be what precisely I’m looking for. Do you offer guest writers to write content available
for you? I wouldn’t mind publishing a post or elaborating on many of the subjects you write concerning here.
Again, awesome weblog!
SITUS GG189
November 21, 2024 @ 2:15 pm
You need to take part in a contest for one of the finest blogs online.
I will recommend this web site!
bandar togel online
November 21, 2024 @ 2:28 pm
Hello there! I could have sworn I’ve been to this blog before but after reading through some of the post I realized
it’s new to me. Anyways, I’m definitely happy I found
it and I’ll be book-marking and checking back frequently!
ZSot
November 21, 2024 @ 3:05 pm
bc.game login
AlbertNef
November 21, 2024 @ 3:58 pm
cytotec abortion pill: cytpremium – buy cytotec in usa
Diplomi_ikSi
November 21, 2024 @ 4:06 pm
Процесс получения диплома стоматолога: реально ли это сделать быстро?
kypit 1S_ixmn
November 21, 2024 @ 4:39 pm
1с купить официальный сайт цена [url=https://www.kupit-1s11.ru]1с купить официальный сайт цена[/url] .
Sazrodn
November 21, 2024 @ 4:49 pm
Приобретение школьного аттестата с официальным упрощенным обучением в Москве
item673738005
November 21, 2024 @ 4:50 pm
Yes! Finally someone writes about duatoto.
narkolog na dom v Krasnodar_nmka
November 21, 2024 @ 5:02 pm
вызвать нарколога на дом [url=http://my.forum2.net/viewtopic.php?id=173]http://my.forum2.net/viewtopic.php?id=173[/url] .
narkolog na dom v Krasnodar_qtpi
November 21, 2024 @ 5:08 pm
вызов нарколога на дом краснодар [url=https://gov.ukrbb.net/viewtopic.php?f=3&t=6373]https://gov.ukrbb.net/viewtopic.php?f=3&t=6373[/url] .
narkolog na dom v Krasnodar_waKi
November 21, 2024 @ 5:11 pm
нарколог на дом цены [url=www.krut.forumno.com/viewtopic.php?id=6026]www.krut.forumno.com/viewtopic.php?id=6026[/url] .
narkolog na dom v Krasnodar_kiMi
November 21, 2024 @ 5:15 pm
нарколог на дом [url=https://obovsem.rolevaya.info/viewtopic.php?id=3674]https://obovsem.rolevaya.info/viewtopic.php?id=3674[/url] .
Consultant SEO Paris
November 21, 2024 @ 6:50 pm
Great delivery. Great arguments. Keep up the amazing
effort.
ZSot
November 21, 2024 @ 7:42 pm
bcgame apk
freshAndroid
November 21, 2024 @ 7:54 pm
[url=https://fresh-casino-apk.ru/fresh-na-androjd/]http://www.fresh-casino-apk.ru/fresh-na-androjd/[/url]
Скачать приложение казино fresh – играй сегодня!
http://fresh-casino-apk.ru/fresh-na-androjd/
StardaCasino
November 21, 2024 @ 8:08 pm
[url=https://starda-casino-apk.ru/starda-casino]http://www.starda-casino-apk.ru/starda-casino[/url]
Загрузить приложение казино онлайн starda – выигрывай сегодня!
http://starda-casino-apk.ru/starda-casino
DownloadFresh
November 21, 2024 @ 8:08 pm
[url=https://fresh-casino-apk.ru/fresh-skachat]http://www.fresh-casino-apk.ru/fresh-skachat[/url]
Скачать приложение онлайн казино fresh – выигрывай прямо сейчас!
http://www.fresh-casino-apk.ru/fresh-skachat
гистероскопия удаление полипа
November 21, 2024 @ 8:45 pm
Узнай все о удаление полипа шейки матки в москвеоперация по удалению полипа в матке
StardaApk
November 21, 2024 @ 9:31 pm
[url=https://starda-casino-apk.ru/starda-apk]http://starda-casino-apk.ru/starda-apk[/url]
Скачать последнюю версию приложения казино онлайн starda – побеждай сейчас!
starda-casino-apk.ru/starda-apk
BrandenJew
November 21, 2024 @ 10:16 pm
prednisone canada prices: ray pharm – 10 mg prednisone
bandar togel online
November 22, 2024 @ 12:43 am
It’s actually a cool and useful piece of information. I’m happy that you simply shared this useful info with us.
Please stay us up to date like this. Thank you for sharing.
Oariordae
November 22, 2024 @ 1:27 am
Полезная информация как официально купить диплом о высшем образовании
Kenzo168
November 22, 2024 @ 1:59 am
I’ve been exploring for a bit for any high-quality articles or blog posts
on this sort of area . Exploring in Yahoo I eventually stumbled upon this website.
Studying this information So i’m satisfied to convey that I have an incredibly
just right uncanny feeling I found out just what I needed.
I most for sure will make sure to don?t put out of your mind this web site and give it a look regularly.
Consultant SEO Paris
November 22, 2024 @ 2:49 am
This web site truly has all of the information I wanted concerning this subject and didn’t know who to ask.
duatoto
November 22, 2024 @ 3:33 am
Hey there! I’ve been following your web site for a
while now and finally got the bravery to go ahead and give you a shout out from
Houston Texas! Just wanted to mention keep up the good work!
Istana189 Login
November 22, 2024 @ 3:38 am
Why viewers still make use of to read news papers when in this technological
globe all is accessible on net?
Mazrziw
November 22, 2024 @ 3:49 am
Пошаговая инструкция по официальной покупке диплома о высшем образовании
ZSot
November 22, 2024 @ 3:54 am
bc game crash
Justinced
November 22, 2024 @ 3:59 am
Ты да я предлагаем наиболее живые совет вместе с возможностью беспроцентной рассрочки, кредита равным образом ипотеки, чтобы покупка квартир встала реальностью чтобы любого https://uysot.uz . Благодаря технологическим характеристикам также планировкам ваша милость сможете эфирно найтись этнодом домашнею мечты. Наши этые часто обновляются, то-то ваша милость хронически будете в линии крайних стоимостей и предложений.
Cazrilf
November 22, 2024 @ 4:01 am
Купить диплом о среднем образовании в Москве и любом другом городе
Sazrrtb
November 22, 2024 @ 4:54 am
Всё, что нужно знать о покупке аттестата о среднем образовании
apkportalnik
November 22, 2024 @ 6:42 am
скачать приложения казино http://khoni.gov.ge/?p=23174
Bernardwet
November 22, 2024 @ 7:23 am
Cost of Plavix on Medicare [url=https://plavixclo.com/#]cheapest plavix[/url] buy plavix
Sazrcnq
November 22, 2024 @ 7:31 am
Покупка диплома о среднем полном образовании: как избежать мошенничества?
Sazrvlo
November 22, 2024 @ 7:32 am
Официальная покупка диплома вуза с упрощенной программой обучения
Sazrkyd
November 22, 2024 @ 8:00 am
Как приобрести аттестат о среднем образовании в Москве и других городах
Uazrres
November 22, 2024 @ 8:02 am
Покупка диплома о среднем полном образовании: как избежать мошенничества?
Mazrhdg
November 22, 2024 @ 8:06 am
Купить диплом в России, предлагает наша компания
gg 189
November 22, 2024 @ 8:09 am
Piece of writing writing is also a excitement, if you be acquainted with
afterward you can write or else it is difficult to write.
Robertzex
November 22, 2024 @ 8:17 am
Отечественный сайт — это образцовое ямыжник для любителей узбекских кинофильмов да трансфертных кинолент. Тут вы обнаружите элита произведения узбекского кинематографа, а также распространенные зарубежные кинокартины от перебрасыванием на узбекский язык.
У нас изображу киноленты на энный чувство: удар судьбы, комедии, эпопея, фантастика (а) также многое другое. Хорошо приспособленный для использования эхопоиск разрешает легко определять подходящей фильмы. Мы часто обновляем фотосайт https://uzkino.net , добавляя последние кинофильмы, чтобы радовать наших зрителей.
Навестите свой фотосайт, чтобы кейфонуть качественным контентом а также взять фильм по душе!
ZSot
November 22, 2024 @ 8:30 am
neon58
vivod iz zapoya krasnodar_xuol
November 22, 2024 @ 8:40 am
вывод из запоя на дому краснодар цены [url=https://motik13.0pk.me/viewtopic.php?id=1997]https://motik13.0pk.me/viewtopic.php?id=1997[/url] .
vivod iz zapoya krasnodar_uior
November 22, 2024 @ 9:17 am
вывод из запоя круглосуточно краснодар на дому [url=https://www.pelsh.forum24.ru/?1-8-0-00000126-000-0-0-1730745072]https://www.pelsh.forum24.ru/?1-8-0-00000126-000-0-0-1730745072[/url] .
vivod iz zapoya krasnodar_ewor
November 22, 2024 @ 9:17 am
вывод из запоя в стационаре краснодара [url=https://bija089.0pk.me/viewtopic.php?id=2554]https://bija089.0pk.me/viewtopic.php?id=2554[/url] .
vivod iz zapoya krasnodar_cbMa
November 22, 2024 @ 9:18 am
вывод из запоя в краснодаре [url=https://setter.borda.ru/?1-7-0-00000674-000-0-0-1730745092]вывод из запоя в краснодаре[/url] .
vivod iz zapoya krasnodar_jhkr
November 22, 2024 @ 9:19 am
вывод из запоя лечение краснодар [url=http://flanrp.rolevaya.com/viewtopic.php?id=147/]вывод из запоя лечение краснодар[/url] .
Darrenmow
November 22, 2024 @ 9:34 am
https://iverfast.com/# ivermectin 6mg dosage
duatoto
November 22, 2024 @ 11:00 am
I am curious to find out what blog platform you are utilizing?
I’m experiencing some minor security problems with my latest
website and I’d like to find something more safeguarded.
Do you have any solutions?
Sazrprx
November 22, 2024 @ 11:09 am
Всё, что нужно знать о покупке аттестата о среднем образовании
promokody_evea
November 22, 2024 @ 11:45 am
продамус промокоды [url=https://rubiz.forum.cool/viewtopic.php?id=3874#p13907/]rubiz.forum.cool/viewtopic.php?id=3874#p13907[/url] .
BrandenJew
November 22, 2024 @ 12:07 pm
can i order generic clomid without insurance: rex pharm – can you get clomid online
money earning app in pakistan
November 22, 2024 @ 1:03 pm
Most Popular Earning App in Pakistan|Ideal Earning Option in Pakistan|Earning Potential in Pakistan|Earning App in Pakistan: Benefits|Rate the Best Earning App in Pakistan|Trustworthy Earning Platform in Pakistan|Pakistan Right Choice for Processing|Pakistan High Paying App for earnings|Pakistan: leader in earning|App that will make it easier to earn in Pakistan Pakistan
money making apps in pakistan [url=https://bestearningappinpakistan.com/]apps for earning money in pakistan[/url] .
binance US-registrera
November 22, 2024 @ 1:45 pm
Can you be more specific about the content of your article? After reading it, I still have some doubts. Hope you can help me.
Situs Slot Gacor
November 22, 2024 @ 1:46 pm
You really make it appear really easy together with your presentation however I in finding this
topic to be really one thing which I believe I would never understand.
It sort of feels too complex and extremely
large for me. I’m taking a look ahead in your subsequent submit, I’ll attempt to get the hold of
it!
KevinFut
November 22, 2024 @ 1:56 pm
Виртуальные номера могут быть очень полезны для бизнеса. Они позволяют компаниям улучшать связь с клиентами, увеличивать продажи и улучшать общую эффективность купить немецкий номер телефона
Diplomi_uzSi
November 22, 2024 @ 3:24 pm
Как избежать рисков при покупке диплома колледжа или ПТУ в России
Dnrtlnl
November 22, 2024 @ 3:24 pm
Как безопасно купить диплом колледжа или ПТУ в России, что важно знать
ZSot
November 22, 2024 @ 4:08 pm
1xwin venezuela
DownloadFresh
November 22, 2024 @ 4:47 pm
[url=https://fresh-casino-apk.ru/fresh-skachat]https://fresh-casino-apk.ru/fresh-skachat[/url]
Скачать apk файл казино онлайн fresh – побеждай сейчас!
http://fresh-casino-apk.ru/fresh-skachat
freshAndroid
November 22, 2024 @ 4:48 pm
[url=https://fresh-casino-apk.ru/fresh-na-androjd/]http://www.fresh-casino-apk.ru/fresh-na-androjd/[/url]
Загрузить последнюю версию приложения казино Фреш – побеждай прямо сейчас!
http://fresh-casino-apk.ru/fresh-na-androjd/
real earn money app in pakistan
November 22, 2024 @ 5:04 pm
Best Earning App in Pakistan|Amazing Earning Opportunity in Pakistan|Earning Potential in Pakistan|Efficient Earning Tool in Pakistan|Pakistan High Earning Apps|Economic Earning in Pakistan|Successful Processing Method in Pakistan|New level of income in Pakistan|Pakistan: leader in earning|App that will make it easier to earn in Pakistan Pakistan
apps to earn money in pakistan [url=https://bestearningappinpakistan.com/]money earning app in pakistan[/url] .
Casinofresh
November 22, 2024 @ 5:27 pm
[url=https://fresh-casino-apk.ru/fresh-casino/]http://fresh-casino-apk.ru/fresh-casino/[/url]
Установить последнюю версию приложения онлайн казино Фреш – играй сейчас!
http://www.fresh-casino-apk.ru/fresh-casino/
Kenzo 168
November 22, 2024 @ 6:51 pm
I read this article completely on the topic of the difference of latest and preceding technologies,
it’s awesome article.
freshSait
November 22, 2024 @ 7:18 pm
[url=https://fresh-casino-apk.ru/fresh-oficzialnyj-sajt]www.fresh-casino-apk.ru/fresh-oficzialnyj-sajt[/url]
Скачать apk файл онлайн казино fresh – побеждай прямо сейчас!
http://www.fresh-casino-apk.ru/fresh-oficzialnyj-sajt
gg189
November 22, 2024 @ 7:27 pm
Hi, after reading this amazing piece of writing i am
as well glad to share my know-how here with friends.
StardaApk
November 22, 2024 @ 7:28 pm
[url=https://starda-casino-apk.ru/starda-apk]starda-casino-apk.ru/starda-apk[/url]
Установить последнюю версию приложения онлайн казино Старда – играй сегодня!
https://starda-casino-apk.ru/starda-apk
AlbertNef
November 22, 2024 @ 7:49 pm
cheapest Lisinopril: cheapest Lisinopril – cheapest Lisinopril
Lazrfpi
November 22, 2024 @ 8:21 pm
Как безопасно купить диплом колледжа или ПТУ в России, что важно знать
KOIN555
November 22, 2024 @ 8:36 pm
Having read this I thought it was rather informative.
I appreciate you finding the time and energy
to put this short article together. I once again find myself spending a significant amount of time both reading and posting comments.
But so what, it was still worth it!
Cazrqmg
November 22, 2024 @ 9:18 pm
Официальная покупка диплома вуза с сокращенной программой обучения в Москве
StardaZercalo
November 22, 2024 @ 9:52 pm
[url=https://starda-casino-apk.ru/starda-oficzialnyj-sajt]https://www.starda-casino-apk.ru/starda-oficzialnyj-sajt[/url]
Загрузить приложение казино starda – выигрывай сегодня!
http://starda-casino-apk.ru/starda-oficzialnyj-sajt
freshCasino
November 22, 2024 @ 10:00 pm
[url=https://fresh-casino-apk.ru]http://www.fresh-casino-apk.ru[/url]
Скачать apk файл казино онлайн Фреш – побеждай прямо сейчас!
https://www.fresh-casino-apk.ru
duatoto
November 22, 2024 @ 10:01 pm
If you desire to grow your experience only keep
visiting this website and be updated with the hottest information posted here.
септопластика носа
November 22, 2024 @ 10:09 pm
Узнай все о септопластика ценыисправление перегородки носа цена
Iarioreqc
November 22, 2024 @ 10:16 pm
Легальная покупка школьного аттестата с упрощенной программой обучения
situs togel
November 22, 2024 @ 11:34 pm
magnificent issues altogether, you simply won a new reader.
What could you recommend about your put up that you simply
made some days in the past? Any positive?
aviator betting strategies
November 22, 2024 @ 11:39 pm
Aviator is a thrilling online betting game that mixes luck and method. It has mesmerized gamers worldwide. Numerous wonder: When is the very best time to play Aviator?
Aviator has more than 12 million gamers internationally and is offered on over 2,000 betting platforms. The video game’s appeal depends on its simplicity and potential for big payouts.
Studies recommend playing Aviator between 8 a.m. and 10 a.m. may increase winning chances. Night owls could benefit, too. Multipliers in between 10 p.m. and midnight can be three times higher.
This guide will check out betting varieties and game mechanics. We’ll also cover effective bankroll management strategies. These insights aim to enhance your Aviator betting strategy.
This information can enhance your confidence Whether you’re brand-new or knowledgeable. It might even help increase your jackpots in the exciting world of Aviator.
Comprehending the Basics of Aviator Game
The Aviator casino game mixes opportunity and technique, captivating gamers worldwide. It utilizes a sophisticated random number generator for fair play, guaranteeing unforeseeable outcomes in every round.
How Aviator’s Random Number Generator Works
The video game’s fairness cornerstone is Aviator’s random number generator. It produces number series that determine each round’s flight path and crash point. This system makes every video game special and unpredictable.
Game Mechanics and Interface Overview
Aviator’s gameplay is straightforward yet appealing. Players bet on a virtual aircraft’s flight to squander before it crashes. The user interface reveals the aircraft’s climb graph and wagering options.
As the multiplier rises, so does the prospective payment. This develops an exciting risk-reward dynamic for gamers.
Fundamental Rules for New Players
Betting rules in Aviator are easy to understand. Gamers choose their stakes and location bets before each round begins. The multiplier climbs up as the plane takes off.
The secret is timing your cash-out right. Cashing out too early may indicate missing bigger wins, and waiting too long could result in losing your bet if the plane crashes.
For example, a ₤ 10 bet squandered at 2.0 x would yield ₤ 20. Waiting for a 5.0 x multiplier might lead to a ₤ 50 payment.
What Is The Best Time To Play Aviator?
Pilot’s random nature makes finding the very best playing time difficult. Each round is independent, regardless of when you play, so your possibilities of winning stay the same at any time.
Some gamers choose off-peak hours for smoother gameplay, while others take pleasure in peak times when more gamers are online. Your choice depends upon your schedule and preferences.
At Katsu Casino, Aviator offers minimum bets of $0.10. This fits various spending plans. Maximum bets reach $100 per wager. You can win approximately $10,000 per bet.
There is no guaranteed ‘best’ time to play Aviator. The game uses a random number generator, so outcomes are unpredictable at any time.
Focus on responsible gaming and adhere to your spending plan. Enjoy the game, anytime you pick to play. That’s the key to a fantastic Aviator experience.
Vital Strategies for Maximising Your Wins
Aviator success blends smart wagering with careful danger management. The video game offers chances for big wins, with multipliers as much as x1000. Gamers require a thoughtful approach to take advantage of these chances.
Bankroll Management Techniques
Effective bankroll management is important for long-term Aviator success. Set clear wagering limitations and stay with them. A common technique is betting 1-2% of your overall bankroll per bet.
This technique safeguards your funds throughout losing streaks. It also helps extend your playing time, offering you more chances to win.
Utilizing the Auto Cash-out Feature
The automobile cash-out feature is an effective tool in Aviator. It lets you pre-programmed a multiplier for automated bet cashing. This strategy can secure consistent wins and safeguard your revenues.
Some players pick lower multipliers, around 1.5 x, for consistent gains. Others intend higher for larger payouts, stabilizing risk and benefit.
Pattern Recognition and Analysis
Aviator uses a random number generator, however some players attempt to find patterns. Studying live stats and round history can offer insights into current patterns.
Keep in mind, each round is independent. Previous results do not influence future outcomes. Usage pattern analysis carefully as part of your overall wagering method.
Optimum Betting Ranges and Limits
Aviator’s betting limits and payout structure are important for a winning strategy. The game provides various minimum and optimum bets for various budgets. Some platforms permit wagers as low as $0.10, while others begin at INR 200 (about $2.40).
Maximum bets can reach $100, catering to high-stakes players. This range fits both casual gamers and serious gamblers. The optimum win per bet can vary, with some casinos topping it at $10,000.
Pilot’s distinct payment structure uses multipliers for possibly considerable returns. With a 97% RTP, the game offers lots of winning chances. Starting with smaller bets and cashing out early can help handle your bankroll effectively.
Success as an Aviator requires strategy, persistence, and luck. Understanding the video game’s mechanics is vital. Adjusting your bets appropriately can optimise your chances of winning while keeping you comfortable.
Best Aviator Platform
Bet9ja is Nigeria’s premier Aviator game platform. It offers easy gameplay and high RTP value. The platform supplies an user-friendly interface, safe banking alternatives, and a demo variation for practice.
As a top-rated betting game, it uses brand-new and skilled gamers the chance to play in a regulated environment.
Gamble Responsibly
When playing Aviator on Bet9ja, it’s crucial to understand that the game can offer fulfilling home entertainment when approached properly. Start with the Aviator demo using preset limits, and keep in mind that the video game is developed for home entertainment first.
Set transparent budget plans and time frame for gameplay sessions.
Win the Aviator Game
To win the aviator game eventually, players must focus on:
Implementing a proven aviator strategy
Beginning with one bet at a time
Comprehending the strategy within the game mechanics
Utilizing the initial Aviator game’s official features
Mastering the strategy of playing Aviator
Learning to win more often through practice
Win Aviator Consistently
Successful aviator tips for constant winning consist of:
Studying rounds of the game patterns
Making calculated bet-plus choices
Understanding the gameplay and high RTP value
Utilizing technique to take full advantage of returns
Experimenting the game without genuine money initially
Making the most of easy gameplay and high RTP functions
Typically the Aviator
The aviator sport on Bet9ja is associated with the video game’s special functions:
Multiple sort of aviator betting options
Capability to find the aviator slot quickly
Access to Aviator on the internet
Choices to try and win through different techniques
Opportunities to win big with calculated threats
Flexibility to play within the aviator game guidelines
Aviator Winning Techniques
To improve your chances of winning:
Choose to find the aviator betting patterns
Utilize the web in any aviator casino features carefully
Learn appropriate money from aviator management
Master ending the game at ideal times
Understand typically the very best cash-out minutes
Make the most of the chance to play strategically
Additional Tips for Success:
Start with small bets to learn the game mechanics
Use the demo version to practice methods
Monitor video game patterns and trends
Set sensible winning goals
Never ever chase after losses
Use car cash-out features when offered
Bear in mind that while Bet9ja offers an outstanding platform for Aviator, success needs persistence, technique, and responsible betting practices. The game combines components of ability and possibility, making it important to approach it with a well balanced frame of mind and proper bankroll management.
The Art of Timing
Success in Aviator heavily counts on mastering the art of timing. Players must carefully watch for patterns and choose the optimum time to bet and squander. The game’s high volatility means timing decisions can distinguish between substantial wins and losses.
Aviator Game Algorithm
The Aviator game algorithm uses a provably fair system that guarantees random outcomes for each round. Comprehending that the video game patterns are random helps gamers establish practical expectations and methods.
Best Strategy for Aviator Casino Game Success
Some of the very best Aviator betting strategies include:
Starting with a conservative strategy using smaller bet sizes
Implementing a systematic bet after each win
Utilizing the demo version of Aviator to practice
Setting both an initial bet and a second bet technique
Preserving consistent bet sizes based upon bankroll management
Tips and Tricks for Winning
Vital win tips for the Aviator game include:
Watch for patterns in video game rounds
Utilize the demo version to practice technique
Set clear profit goals and stop-loss limits
Display game patterns over several rounds
Carry out tactical bet sizing
How to Enjoy Aviator Responsibly
To make Aviator provide satisfying home entertainment:
Start with the demonstration variation using predetermined amounts
Set clear wagering limits
Take regular breaks from gameplay
Never ever chase after losses
View it as entertainment rather than a guaranteed income source
Winning Strategy Fundamentals
A solid winning strategy to play successfully consists of:
Proper bankroll management
Comprehending the video game’s volatility
Utilizing strategic bet sizing
Executing stop-loss limits
Benefiting from the video game’s high RTP worth
Advanced Game Tricks
Advanced players often use these game tricks:
Multiple bet methods
Pattern acknowledgment strategies
Timing-based techniques
Danger management systems
Strategic cash-out points
Online Game Experience
The Aviator online game experience provides:
Easy access through various casino sites
Several wagering choices
Real-time multiplayer interaction
Live statistics and history
Mobile compatibility for on-the-go play
Particular Aviator Game Features
Secret features that make this particular game popular:
Social gaming elements
Real-time multiplier screen
Easy yet appealing interface
Numerous wagering options
Transparent game mechanics
This structured method to understanding and playing the Aviator crash game can assist gamers establish much better methods and enjoy the game more responsibly. Keep in mind that while strategies can enhance your possibilities, there’s no guaranteed method to win, and gaming should always be approached with care and within one’s means.
FREQUENTLY ASKED QUESTION
What is the very best time to play Aviator?
There’s no conclusive best time to play Aviator. The game’s random nature implies each round is independent. Pick a time that fits your schedule and budget plan.
How does Aviator’s Random Number Generator work?
Aviator uses a Random Number Generator (RNG) for fair results. It creates random numbers for each game round. This system makes sure unpredictable flight paths and crash points.
What are the standard rules for brand-new Aviator gamers?
New players need to find out the video game mechanics. These include betting on a simulated flight and timing the cash-out. They ought to also understand how to select stakes and place bets.
It’s vital to practice accountable betting. Set personal limitations to make sure safe and pleasurable gameplay.
Exist reliable methods for winning at Aviator?
No technique assurances constant wins in Aviator. Nevertheless, some methods can improve your gameplay. These consist of accountable bankroll management and setting betting limits.
Utilizing the auto cash-out feature can also be valuable. Keep in mind, Aviator is mainly a game of chance.
What are common wagering limitations in Aviator video games?
Betting limits vary between platforms. Minimum bets can be as low as ₤ 0.10, with maximum bets around ₤ 100. Optimum wins are often capped, often at ₤ 10,000 per bet.
Always examine the specific limits of your picked platform. This guarantees you’re conscious of potential dangers and benefits.
Can I enhance my chances of winning by playing during specific hours?
Playing at specific times does not improve your winning opportunities. Each round is independent and identified by the RNG. Off-peak hours might use smoother gameplay due to minimized server load.
Is it possible to identify patterns in Aviator gameplay?
Some gamers attempt to spot patterns in the flight’s ascent. Nevertheless, this is largely speculative. The game’s RNG guarantees previous results don’t influence future results.
How essential is bankroll management in Aviator?
Bankroll management is crucial in Aviator. It helps control spending and extend gameplay. Set clear limits on bets and losses per session.
This technique is critical to responsible gambling. It makes sure a more pleasurable and sustainable gaming experience.
BrandenJew
November 22, 2024 @ 11:59 pm
where can i buy amoxicillin online: Amoxicillin buy online – can i purchase amoxicillin online
Cazrxmv
November 23, 2024 @ 12:38 am
Как приобрести аттестат о среднем образовании в Москве и других городах
Darrenmow
November 23, 2024 @ 12:45 am
http://plavixclo.com/# п»їplavix generic
aviator winning
November 23, 2024 @ 1:29 am
Aviator is an exhilarating online betting game that mixes luck and strategy. It has actually captivated players worldwide. Many wonder: When is the very best time to play Aviator?
Pilot has over 12 million gamers internationally and is offered on over 2,000 betting platforms. The video game’s appeal lies in its simplicity and capacity for big payments.
Research studies recommend playing Aviator in between 8 a.m. and 10 a.m. may improve winning opportunities. Night owls might benefit, too. Multipliers in between 10 p.m. and midnight can be three times higher.
This guide will explore betting ranges and game mechanics. We’ll also cover effective bankroll management methods. These insights aim to improve your Aviator gambling strategy.
This details can enhance your confidence Whether you’re brand-new or skilled. It may even assist increase your jackpots in the interesting world of Aviator.
Comprehending the Basics of Aviator Game
The Aviator casino game blends opportunity and method, captivating players worldwide. It utilizes an advanced random number generator for fair play, guaranteeing unforeseeable outcomes in every round.
How Aviator’s Random Number Generator Works
The game’s fairness foundation is Aviator’s random number generator. It produces number sequences that figure out each round’s flight path and crash point. This system makes every video game special and unforeseeable.
Video Game Mechanics and Interface Overview
Pilot’s gameplay is straightforward yet appealing. Players bank on a virtual airplane’s flight to cash out before it crashes. The user interface reveals the airplane’s climb graph and wagering choices.
As the multiplier rises, so does the possible payout. This produces an interesting risk-reward dynamic for gamers.
Basic Rules for New Players
Betting rules in Aviator are easy to understand. Gamers select their stakes and location bets before each round begins. The multiplier climbs as the plane takes off.
The secret is timing your cash-out right. Cashing out too early may suggest missing bigger wins, and waiting too long might lead to losing your bet if the airplane crashes.
For example, a ₤ 10 bet cashed out at 2.0 x would yield ₤ 20. Waiting on a 5.0 x multiplier might lead to a ₤ 50 payout.
What Is The Best Time To Play Aviator?
Aviator’s random nature makes finding the best playing time difficult. Each round is independent, regardless of when you play, so your opportunities of winning stay the very same at any time.
Some players choose off-peak hours for smoother gameplay, while others delight in peak times when more gamers are online. Your choice depends upon your schedule and choices.
At Katsu Casino, Aviator offers minimum bets of $0.10. This suits numerous budgets. Maximum bets reach $100 per wager. You can win up to $10,000 per bet.
There is no guaranteed ‘best’ time to play Aviator. The game uses a random number generator, so results are unforeseeable at any time.
Concentrate on responsible gaming and adhere to your budget plan. Enjoy the game, anytime you choose to play. That’s the essential to a terrific Aviator experience.
Necessary Strategies for Maximising Your Wins
Aviator success mixes intelligent wagering with cautious risk management. The video game offers possibilities for big wins, with multipliers approximately x1000. Gamers need a thoughtful approach to make the most of these opportunities.
Bankroll Management Techniques
Efficient bankroll management is vital for long-term Aviator success. Set clear wagering limits and adhere to them. A typical strategy is wagering 1-2% of your total bankroll per bet.
This technique safeguards your funds during losing streaks. It likewise assists extend your playing time, providing you more opportunities to win.
Utilizing the Auto Cash-out Feature
The vehicle cash-out function is an effective tool in Aviator. It lets you predetermined a multiplier for automated bet cashing. This technique can secure constant wins and secure your earnings.
Some gamers choose lower multipliers, around 1.5 x, for constant gains. Others intend greater for larger payments, balancing danger and benefit.
Pattern Recognition and Analysis
Aviator uses a random number generator, but some gamers try to find patterns. Studying live statistics and round history can provide insights into recent trends.
Remember, each round is independent. Previous results don’t influence future outcomes. Use pattern analysis thoroughly as part of your general wagering strategy.
Optimal Betting Ranges and Limits
Pilot’s wagering limits and payout structure are important for a winning strategy. The video game uses numerous minimum and optimum bets for different budget plans. Some platforms allow wagers as low as $0.10, while others start at INR 200 (about $2.40).
Optimum bets can reach $100, catering to high-stakes gamers. This range fits both casual gamers and severe bettors. The optimum win per bet can differ, with some gambling establishments capping it at $10,000.
Pilot’s unique payout structure offers multipliers for potentially significant returns. With a 97% RTP, the game supplies many winning chances. Starting with smaller sized bets and squandering early can help manage your bankroll successfully.
Success as an Aviator requires strategy, persistence, and luck. Understanding the game’s mechanics is important. Changing your bets appropriately can optimise your opportunities of winning while keeping you comfortable.
Best Aviator Platform
Bet9ja is Nigeria’s premier Aviator game platform. It offers easy gameplay and high RTP value. The platform provides an easy to use interface, secure banking alternatives, and a demo version for practice.
As a top-rated betting game, it provides new and skilled gamers the chance to play in a regulated environment.
Gamble Responsibly
When playing Aviator on Bet9ja, it’s vital to comprehend that the video game can provide fulfilling home entertainment when approached properly. Start with the Aviator demo using preset limitations, and remember that the game is developed for home entertainment initially.
Set transparent budgets and time frame for gameplay sessions.
Win the Aviator Game
To win the aviator game eventually, players must concentrate on:
Implementing a tested aviator strategy
Beginning with one bet at a time
Understanding the strategy within the game mechanics
Using the original Aviator game’s main functions
Mastering the strategy of playing Aviator
Finding out to win more frequently through practice
Win Aviator Consistently
Effective aviator tips for consistent winning consist of:
Studying rounds of the game patterns
Making computed bet-plus decisions
Understanding the gameplay and high RTP value
Utilizing method to optimize returns
Practicing with the game without genuine cash initially
Making the most of easy gameplay and high RTP functions
Typically the Aviator
The aviator sport on Bet9ja is associated with the video game’s unique functions:
Multiple type of aviator betting choices
Ability to find the aviator slot quickly
Access to Aviator online
Choices to try and win through different methods
Opportunities to win big with calculated threats
Versatility to play within the aviator game guidelines
Aviator Winning Techniques
To improve your possibilities of winning:
Choose to find the aviator betting patterns
Utilize the web in any aviator casino features wisely
Discover appropriate money from aviator management
Master ending the game at ideal times
Understand typically the best cash-out moments
Benefit from the chance to play tactically
Additional Tips for Success:
Start with small bets to learn the video game mechanics
Use the demo version to practice techniques
Display game patterns and trends
Set practical winning goals
Never chase losses
Use vehicle cash-out functions when available
Bear in mind that while Bet9ja uses an exceptional platform for Aviator, success needs patience, technique, and responsible gaming practices. The video game integrates components of ability and possibility, making it necessary to approach it with a balanced state of mind and correct bankroll management.
The Art of Timing
Success in Aviator greatly relies on mastering the art of timing. Gamers must carefully expect patterns and choose the optimum time to bet and squander. The video game’s high volatility means timing decisions can separate between substantial wins and losses.
Aviator Game Algorithm
The Aviator game algorithm uses a provably fair system that makes sure random results for each round. Understanding that the video game patterns are random assists players establish practical expectations and methods.
Best Strategy for Aviator Casino Game Success
Some of the very best Aviator betting strategies include:
Starting with a conservative strategy using smaller sized bet sizes
Carrying out a methodical bet after each win
Utilizing the demo version of Aviator to practice
Setting both an initial bet and a second bet technique
Preserving constant bet sizes based on bankroll management
Tips and Tricks for Winning
Vital win tips for the Aviator game include:
Watch for patterns in video game rounds
Utilize the demonstration version to practice strategy
Set clear profit goals and stop-loss limitations
Screen game patterns over numerous rounds
Execute strategic bet sizing
How to Enjoy Aviator Responsibly
To make Aviator offer fulfilling entertainment:
Start with the demonstration variation using pre-programmed amounts
Set clear wagering limitations
Take regular breaks from gameplay
Never ever go after losses
View it as home entertainment rather than a guaranteed earnings source
Winning Strategy Fundamentals
A solid winning strategy to play effectively consists of:
Proper bankroll management
Comprehending the game’s volatility
Using strategic bet sizing
Carrying out stop-loss limitations
Taking advantage of the video game’s high RTP value
Advanced Game Tricks
Advanced players frequently employ these game tricks:
Multiple bet strategies
Pattern acknowledgment strategies
Timing-based methods
Risk management systems
Strategic cash-out points
Online Game Experience
The Aviator online game experience provides:
Easy gain access to through numerous casino websites
Numerous betting choices
Real-time multiplayer interaction
Live statistics and history
Mobile compatibility for on-the-go play
Particular Aviator Game Features
Key features that make this particular game popular:
Social gaming elements
Real-time multiplier display screen
Basic yet appealing interface
Several wagering alternatives
Transparent video game mechanics
This structured method to understanding and playing the Aviator crash game can help players develop better methods and enjoy the game more responsibly. Keep in mind that while methods can enhance your possibilities, there’s no surefire method to win, and betting ought to constantly be approached with care and within one’s means.
FAQ
What is the very best time to play Aviator?
There’s no definitive finest time to play Aviator. The video game’s random nature implies each round is independent. Choose a time that fits your schedule and spending plan.
How does Aviator’s Random Number Generator work?
Aviator uses a Random Number Generator (RNG) for fair results. It creates random numbers for each video game round. This system makes sure unpredictable flight paths and crash points.
What are the standard guidelines for brand-new Aviator players?
New players should discover the game mechanics. These involve betting on a simulated flight and timing the cash-out. They must likewise understand how to select stakes and place bets.
It’s crucial to practice accountable gambling. Set personal limits to guarantee safe and enjoyable gameplay.
Exist efficient techniques for winning at Aviator?
No technique assurances consistent wins in Aviator. However, some methods can improve your gameplay. These include responsible bankroll management and setting betting limitations.
Using the auto cash-out feature can also be helpful. Remember, Aviator is mainly a game of chance.
What are normal wagering limitations in Aviator video games?
Betting limitations vary between platforms. Minimum bets can be as low as ₤ 0.10, with optimum bets around ₤ 100. Optimum wins are typically capped, often at ₤ 10,000 per bet.
Constantly examine the particular limitations of your selected platform. This ensures you’re mindful of possible dangers and rewards.
Can I improve my possibilities of winning by playing during particular hours?
Dipping into particular times doesn’t enhance your winning opportunities. Each round is independent and identified by the RNG. Off-peak hours might use smoother gameplay due to minimized server load.
Is it possible to recognise patterns in Aviator gameplay?
Some players try to find patterns in the flight’s climb. Nevertheless, this is mostly speculative. The game’s RNG guarantees past results do not affect future outcomes.
How crucial is bankroll management in Aviator?
Bankroll management is essential in Aviator. It assists control spending and extend gameplay. Set clear limitations on bets and losses per session.
This method is important to accountable betting. It guarantees a more satisfying and sustainable gaming experience.
Muha meds disposable
November 23, 2024 @ 2:10 am
I am really loving the theme/design of your weblog.
Do you ever run into any internet browser compatibility issues?
A handful of my blog audience have complained about my website not operating correctly in Explorer but looks great in Safari.
Do you have any suggestions to help fix this
issue?
Sazrmvl
November 23, 2024 @ 2:43 am
Легальная покупка диплома ПТУ с сокращенной программой обучения
best earning apps in pakistan
November 23, 2024 @ 4:52 am
Most Popular Earning App in Pakistan|Amazing Earning Opportunity in Pakistan|Earning in Pakistan: New Approach|Earning App in Pakistan: Benefits|Pakistan High Earning Apps|Pakistan Earning Opportunities|Pakistan Right Choice for Processing|Pakistan High Paying App for earnings|Effective methods of earning in Pakistan|Innovative approach to earning in Pakistan
pakistan earn money app [url=https://bestearningappinpakistan.com/]top 5 earning app in pakistan[/url] .
Cazruuz
November 23, 2024 @ 5:11 am
Полезная информация как купить диплом о высшем образовании без рисков
AnthonyStody
November 23, 2024 @ 6:33 am
Бесплатные виртуальные номера для СМС — это возможность быстро и безопасно проходить верификацию на различных сайтах без регистрации купить виртуальный номер телефона
roupy u deti
November 23, 2024 @ 6:35 am
Hello friends, fastidious paragraph and nice arguments commented at this place,
I am really enjoying by these.
Sazrzjx
November 23, 2024 @ 7:32 am
Сколько стоит диплом высшего и среднего образования и как его получить?
freshApk
November 23, 2024 @ 8:39 am
[url=https://fresh-casino-apk.ru/fresh-apk]https://www.fresh-casino-apk.ru/fresh-apk[/url]
Загрузить apk файл казино онлайн Фреш – играй сегодня!
http://www.fresh-casino-apk.ru/fresh-apk
winning strategy
November 23, 2024 @ 8:52 am
Aviator is a thrilling online betting game that mixes luck and strategy. It has mesmerized players worldwide. Lots of marvel: When is the very best time to play Aviator?
Pilot has more than 12 million players globally and is readily available on over 2,000 wagering platforms. The video game’s appeal depends on its simplicity and potential for big payments.
Studies suggest playing Aviator between 8 a.m. and 10 a.m. might improve winning opportunities. Night owls could benefit, too. Multipliers in between 10 p.m. and midnight can be 3 times greater.
This guide will check out betting varieties and game mechanics. We’ll likewise cover efficient bankroll management strategies. These insights aim to enhance your Aviator gaming technique.
This info can boost your self-confidence Whether you’re brand-new or skilled. It might even assist increase your profits in the exciting world of Aviator.
Comprehending the Basics of Aviator Game
The Aviator casino game mixes possibility and strategy, captivating gamers worldwide. It utilizes a sophisticated random number generator for fair play, ensuring unforeseeable outcomes in every round.
How Aviator’s Random Number Generator Works
The game’s fairness foundation is Aviator’s random number generator. It produces number sequences that figure out each round’s flight path and crash point. This system makes every video game distinct and unpredictable.
Game Mechanics and Interface Overview
Pilot’s gameplay is straightforward yet interesting. Players bet on a virtual airplane’s flight to squander before it crashes. The user interface reveals the aircraft’s ascent graph and betting alternatives.
As the multiplier rises, so does the potential payment. This produces an interesting risk-reward dynamic for players.
Fundamental Rules for New Players
Betting rules in Aviator are easy to understand. Players choose their stakes and location bets before each round begins. The multiplier climbs up as the plane removes.
The key is timing your cash-out right. Squandering too early may suggest missing larger wins, and waiting too long could lead to losing your bet if the aircraft crashes.
For instance, a ₤ 10 bet squandered at 2.0 x would yield ₤ 20. Waiting for a 5.0 x multiplier might result in a ₤ 50 payout.
What Is The Best Time To Play Aviator?
Aviator’s random nature makes finding the very best playing time tough. Each round is independent, despite when you play, so your opportunities of winning remain the same at any time.
Some gamers choose off-peak hours for smoother gameplay, while others take pleasure in peak times when more players are online. Your choice depends on your schedule and choices.
At Katsu Casino, Aviator uses minimum bets of $0.10. This matches various budgets. Optimum bets reach $100 per wager. You can win up to $10,000 per bet.
There is no ensured ‘best’ time to play Aviator. The video game utilizes a random number generator, so results are unpredictable at any time.
Concentrate on accountable gaming and adhere to your budget. Enjoy the game, anytime you pick to play. That’s the essential to a terrific Aviator experience.
Vital Strategies for Maximising Your Wins
Pilot success mixes intelligent wagering with careful danger management. The game provides chances for big wins, with multipliers approximately x1000. Players require a thoughtful method to make the most of these chances.
Bankroll Management Techniques
Effective bankroll management is important for long-term Aviator success. Set clear wagering limitations and stay with them. A typical method is betting 1-2% of your total bankroll per bet.
This technique safeguards your funds during losing streaks. It likewise assists extend your playing time, giving you more opportunities to win.
Using the Auto Cash-out Feature
The auto cash-out function is a powerful tool in Aviator. It lets you pre-programmed a multiplier for automatic bet cashing. This method can secure consistent wins and protect your profits.
Some players select lower multipliers, around 1.5 x, for constant gains. Others aim greater for bigger payouts, stabilizing danger and benefit.
Pattern Recognition and Analysis
Aviator uses a random number generator, but some gamers attempt to find patterns. Studying live statistics and round history can offer insights into current trends.
Keep in mind, each round is independent. Past outcomes do not affect future results. Use pattern analysis carefully as part of your general betting method.
Optimal Betting Ranges and Limits
Pilot’s betting limits and payout structure are essential for a winning strategy. The game uses different minimum and maximum bets for various budget plans. Some platforms permit wagers as low as $0.10, while others start at INR 200 (about $2.40).
Maximum bets can reach $100, dealing with high-stakes players. This range matches both casual gamers and serious bettors. The maximum win per bet can differ, with some casinos topping it at $10,000.
Pilot’s distinct payout structure offers multipliers for potentially considerable returns. With a 97% RTP, the game supplies many winning opportunities. Starting with smaller sized bets and cashing out early can assist manage your bankroll efficiently.
Success as an Aviator requires strategy, persistence, and luck. Understanding the game’s mechanics is crucial. Changing your bets accordingly can optimise your opportunities of winning while keeping you comfy.
Best Aviator Platform
Bet9ja is Nigeria’s premier Aviator game platform. It offers easy gameplay and high RTP value. The platform supplies an user-friendly interface, safe banking choices, and a demonstration version for practice.
As a top-rated betting game, it offers new and skilled gamers the chance to play in a regulated environment.
Gamble Responsibly
When playing Aviator on Bet9ja, it’s vital to understand that the game can supply fulfilling entertainment when approached properly. Start with the Aviator demo using preset limitations, and keep in mind that the game is designed for entertainment first.
Set transparent budget plans and time limits for gameplay sessions.
Win the Aviator Game
To win the aviator game eventually, gamers should focus on:
Implementing a proven aviator strategy
Beginning with one bet at a time
Understanding the strategy within the game mechanics
Utilizing the initial Aviator game’s official features
Mastering the strategy of playing Aviator
Discovering to win regularly through practice
Win Aviator Consistently
Effective aviator tips for consistent winning consist of:
Studying rounds of the game patterns
Making determined bet-plus choices
Comprehending the gameplay and high RTP value
Using method to optimize returns
Practicing with the game without genuine money first
Taking advantage of easy gameplay and high RTP functions
Typically the Aviator
The aviator sport on Bet9ja is related to the game’s special features:
Multiple sort of aviator betting options
Ability to find the aviator slot quickly
Access to Aviator online
Choices to try and win through different methods
Opportunities to win big with calculated dangers
Flexibility to play within the aviator game rules
Aviator Winning Techniques
To enhance your chances of winning:
Choose to find the aviator betting patterns
Utilize the web in any aviator casino includes sensibly
Discover correct money from aviator management
Master ending the game at optimal times
Understand typically the very best cash-out moments
Benefit from the chance to play tactically
Extra Tips for Success:
Start with little bets to discover the video game mechanics
Use the demo version to practice strategies
Display video game patterns and trends
Set sensible winning objectives
Never chase after losses
Usage automobile cash-out features when offered
Bear in mind that while Bet9ja uses an excellent platform for Aviator, success needs persistence, method, and responsible betting practices. The game integrates elements of skill and chance, making it vital to approach it with a well balanced frame of mind and proper bankroll management.
The Art of Timing
Success in Aviator heavily relies on mastering the art of timing. Players need to thoroughly watch for patterns and choose the optimal time to bet and cash out. The game’s high volatility means timing decisions can distinguish in between significant wins and losses.
Aviator Game Algorithm
The Aviator game algorithm utilizes a provably fair system that guarantees random results for each round. Understanding that the game patterns are random helps gamers establish sensible expectations and methods.
Best Strategy for Aviator Casino Game Success
Some of the best Aviator betting strategies include:
Starting with a conservative strategy utilizing smaller sized bet sizes
Implementing a methodical bet after each win
Using the demo version of Aviator to practice
Setting both an initial bet and a second bet method
Maintaining consistent bet sizes based on bankroll management
Tips and Tricks for Winning
Vital win tips for the Aviator game include:
Watch for patterns in game rounds
Utilize the demo version to practice method
Set clear profit objectives and stop-loss limitations
Monitor game patterns over multiple rounds
Carry out strategic bet sizing
How to Enjoy Aviator Responsibly
To make Aviator supply satisfying home entertainment:
Start with the demo variation utilizing preset quantities
Set clear wagering limits
Take routine breaks from gameplay
Never ever chase after losses
View it as home entertainment instead of a guaranteed income source
Winning Strategy Fundamentals
A strong winning strategy to play successfully includes:
Proper bankroll management
Understanding the game’s volatility
Using strategic bet sizing
Carrying out stop-loss limitations
Making the most of the game’s high RTP value
Advanced Game Tricks
Advanced players typically use these game tricks:
Multiple bet methods
Pattern recognition methods
Timing-based techniques
Risk management systems
Strategic cash-out points
Online Game Experience
The Aviator online game experience uses:
Easy access through numerous casino sites
Numerous betting alternatives
Real-time multiplayer interaction
Live data and history
Mobile compatibility for on-the-go play
Particular Aviator Game Features
Secret functions that make this particular game popular:
Social gaming elements
Real-time multiplier display
Simple yet interesting user interface
Numerous wagering options
Transparent game mechanics
This structured technique to understanding and playing the Aviator crash game can assist players develop much better methods and enjoy the game more responsibly. Bear in mind that while methods can improve your opportunities, there’s no guaranteed method to win, and betting needs to always be approached with care and within one’s methods.
FAQ
What is the very best time to play Aviator?
There’s no conclusive best time to play Aviator. The game’s random nature indicates each round is independent. Pick a time that fits your schedule and budget.
How does Aviator’s Random Number Generator work?
Aviator uses a Random Number Generator (RNG) for fair outcomes. It produces random numbers for each game round. This system ensures unpredictable flight paths and crash points.
What are the fundamental rules for brand-new Aviator gamers?
New gamers need to learn the video game mechanics. These involve betting on a simulated flight and timing the cash-out. They ought to also understand how to choose stakes and place bets.
It’s vital to practice accountable gaming. Set personal limits to guarantee safe and enjoyable gameplay.
Exist reliable techniques for winning at Aviator?
No strategy assurances constant wins in Aviator. Nevertheless, some approaches can enhance your gameplay. These include accountable bankroll management and setting betting limitations.
Utilizing the automobile cash-out feature can also be handy. Keep in mind, Aviator is primarily a game of chance.
What are common betting limits in Aviator games?
Betting limitations vary between platforms. Minimum bets can be as low as ₤ 0.10, with maximum bets around ₤ 100. Optimum wins are typically capped, in some cases at ₤ 10,000 per bet.
Always inspect the specific limits of your picked platform. This ensures you’re conscious of potential dangers and rewards.
Can I enhance my possibilities of winning by playing during particular hours?
Dipping into particular times doesn’t improve your winning opportunities. Each round is independent and figured out by the RNG. Off-peak hours might provide smoother gameplay due to lowered server load.
Is it possible to acknowledge patterns in Aviator gameplay?
Some gamers attempt to identify patterns in the flight’s climb. Nevertheless, this is mainly speculative. The video game’s RNG ensures previous outcomes don’t affect future results.
How crucial is bankroll management in Aviator?
Bankroll management is important in Aviator. It helps control costs and extend gameplay. Set clear limits on bets and losses per session.
This method is critical to responsible gaming. It guarantees a more satisfying and sustainable gaming experience.
AlbertNef
November 23, 2024 @ 9:04 am
buy cytotec online: cytpremium – Cytotec 200mcg price
vedenie účtovníctva košice
November 23, 2024 @ 9:22 am
We’re a bunch of volunteers and opening a brand new scheme
in our community. Your site provided us with helpful information to work on. You’ve performed a formidable job and our whole neighborhood
might be thankful to you.
Treffsy
November 23, 2024 @ 9:41 am
Приобретение школьного аттестата с официальным упрощенным обучением в Москве
slot gacor
November 23, 2024 @ 9:44 am
I love what you guys are up too. Such clever work and coverage!
Keep up the wonderful works guys I’ve you guys to my blogroll.
vivod iz zapoya krasnodar_myMl
November 23, 2024 @ 10:45 am
вывод из запоя в краснодаре [url=www.svarog.forum24.ru/?1-0-0-00000225-000-0-0-1730745492]вывод из запоя в краснодаре[/url] .
vivod iz zapoya krasnodar_beSr
November 23, 2024 @ 10:48 am
вывод из запоя на дому краснодар цены [url=https://masa.forum24.ru/?1-16-0-00002628-000-0-0-1730745346/]вывод из запоя на дому краснодар цены[/url] .
vivod iz zapoya v stacionare_icSr
November 23, 2024 @ 10:49 am
вывод из запоя в стационаре анонимно [url=http://www.familyportal.forumrom.com/viewtopic.php?id=28571]http://www.familyportal.forumrom.com/viewtopic.php?id=28571[/url] .
fonbet kazino_tnsr
November 23, 2024 @ 10:50 am
фонтбет [url=https://fonbet-kazino-24.by/]фонтбет[/url] .
fonbet kazino_lysa
November 23, 2024 @ 10:51 am
фонтбет [url=http://fonbet-casino-24.by/]фонтбет[/url] .
BrandenJew
November 23, 2024 @ 12:36 pm
cheap priligy: buy priligy max pharm – priligy max pharm
Mazrjom
November 23, 2024 @ 3:57 pm
Купить диплом судоводителя
операция увеличение члена
November 23, 2024 @ 4:56 pm
Узнай все о увеличение толщины полового члена увеличение пениса цена
StardaCasino
November 23, 2024 @ 5:11 pm
[url=https://starda-casino-apk.ru/starda-casino]https://starda-casino-apk.ru/starda-casino[/url]
Установить последнюю версию приложения казино Старда – побеждай сейчас!
https://www.starda-casino-apk.ru/starda-casino
Diplomi_wzSi
November 23, 2024 @ 6:07 pm
Покупка диплома о среднем полном образовании: как избежать мошенничества?
Darrenmow
November 23, 2024 @ 6:28 pm
https://plavixclo.com/# antiplatelet drug
MonroAndroid
November 23, 2024 @ 7:00 pm
[url=https://starda-casino-apk.ru/starda-na-androjd]https://starda-casino-apk.ru/starda-na-androjd[/url]
Скачать apk файл онлайн казино Monro – играй сейчас!
http://starda-casino-apk.ru/starda-na-androjd
Sazrydz
November 23, 2024 @ 7:24 pm
Быстрая покупка диплома старого образца: возможные риски
Cazrfyu
November 23, 2024 @ 7:43 pm
Как получить диплом о среднем образовании в Москве и других городах
one bet
November 23, 2024 @ 8:19 pm
Aviator is a thrilling online betting game that mixes luck and strategy. It has captivated players worldwide. Lots of wonder: When is the best time to play Aviator?
Pilot has more than 12 million players globally and is offered on over 2,000 betting platforms. The video game’s appeal depends on its simpleness and potential for huge payments.
Studies suggest playing Aviator in between 8 a.m. and 10 a.m. may boost winning opportunities. Night owls could benefit, too. Multipliers in between 10 p.m. and midnight can be three times higher.
This guide will check out betting varieties and video game mechanics. We’ll likewise cover efficient bankroll management methods. These insights intend to improve your Aviator betting strategy.
This details can increase your self-confidence Whether you’re new or knowledgeable. It might even help increase your jackpots in the interesting world of Aviator.
Understanding the Basics of Aviator Game
The Aviator casino game mixes opportunity and strategy, captivating players worldwide. It uses a sophisticated random number generator for fair play, making sure unforeseeable outcomes in every round.
How Aviator’s Random Number Generator Works
The game’s fairness cornerstone is Aviator’s random number generator. It produces number sequences that determine each round’s flight path and crash point. This system makes every game unique and unforeseeable.
Video Game Mechanics and Interface Overview
Pilot’s gameplay is straightforward yet interesting. Players bank on a virtual plane’s flight to cash out before it crashes. The interface shows the aircraft’s climb chart and wagering choices.
As the multiplier increases, so does the potential payment. This produces an interesting risk-reward dynamic for players.
Fundamental Rules for New Players
Betting rules in Aviator are easy to understand. Gamers choose their stakes and place bets before each round begins. The multiplier climbs up as the plane removes.
The key is timing your cash-out right. Cashing out too early may mean missing larger wins, and waiting too long could lead to losing your bet if the aircraft crashes.
For example, a ₤ 10 bet squandered at 2.0 x would yield ₤ 20. Waiting for a 5.0 x multiplier could lead to a ₤ 50 payment.
What Is The Best Time To Play Aviator?
Aviator’s random nature makes discovering the very best playing time tough. Each round is independent, regardless of when you play, so your chances of winning stay the exact same at any time.
Some gamers choose off-peak hours for smoother gameplay, while others delight in peak times when more players are online. Your choice depends on your schedule and preferences.
At Katsu Casino, Aviator uses minimum bets of $0.10. This fits various budget plans. Maximum bets reach $100 per wager. You can win up to $10,000 per bet.
There is no ensured ‘best’ time to play Aviator. The video game utilizes a random number generator, so results are unforeseeable at any time.
Focus on accountable gaming and stick to your budget plan. Enjoy the game, no matter when you select to play. That’s the crucial to a great Aviator experience.
Essential Strategies for Maximising Your Wins
Aviator success mixes smart betting with mindful risk management. The video game provides opportunities for big wins, with multipliers approximately x1000. Gamers require a thoughtful method to maximize these chances.
Bankroll Management Techniques
Efficient bankroll management is vital for long-term Aviator success. Set clear betting limits and adhere to them. A typical strategy is betting 1-2% of your overall bankroll per bet.
This approach safeguards your funds during losing streaks. It also assists extend your playing time, giving you more opportunities to win.
Utilizing the Auto Cash-out Feature
The automobile cash-out function is a powerful tool in Aviator. It lets you preset a multiplier for automated bet cashing. This strategy can protect consistent wins and secure your profits.
Some gamers select lower multipliers, around 1.5 x, for steady gains. Others aim greater for larger payments, stabilizing risk and benefit.
Pattern Recognition and Analysis
Aviator uses a random number generator, but some players try to find patterns. Studying live statistics and round history can use insights into recent patterns.
Keep in mind, each round is independent. Past outcomes do not affect future outcomes. Usage pattern analysis thoroughly as part of your total betting technique.
Optimal Betting Ranges and Limits
Aviator’s betting limits and payment structure are vital for a winning strategy. The video game provides various minimum and maximum bets for various spending plans. Some platforms allow wagers as low as $0.10, while others begin at INR 200 (about $2.40).
Optimum bets can reach $100, accommodating high-stakes gamers. This range suits both casual players and serious bettors. The optimum win per bet can differ, with some casinos topping it at $10,000.
Pilot’s distinct payment structure uses multipliers for possibly significant returns. With a 97% RTP, the game offers lots of winning opportunities. Beginning with smaller bets and squandering early can help handle your bankroll effectively.
Success as an Aviator needs technique, perseverance, and luck. Understanding the game’s mechanics is vital. Changing your bets appropriately can optimise your opportunities of winning while keeping you comfortable.
Best Aviator Platform
Bet9ja is Nigeria’s premier Aviator game platform. It offers easy gameplay and high RTP value. The platform provides an easy to use user interface, secure banking choices, and a demonstration version for practice.
As a top-rated betting game, it uses new and experienced players the chance to play in a regulated environment.
Gamble Responsibly
When playing Aviator on Bet9ja, it’s important to comprehend that the game can offer fulfilling home entertainment when approached properly. Start with the Aviator demo using preset limits, and bear in mind that the video game is designed for home entertainment initially.
Set transparent budget plans and time frame for gameplay sessions.
Win the Aviator Game
To win the aviator game eventually, players ought to focus on:
Implementing a proven aviator strategy
Starting with one bet at a time
Comprehending the strategy within the video game mechanics
Using the original Aviator game’s official functions
Mastering the strategy of playing Aviator
Discovering to win more often through practice
Win Aviator Consistently
Successful aviator tips for consistent winning include:
Studying rounds of the game patterns
Making computed bet-plus decisions
Comprehending the gameplay and high RTP value
Using technique to maximize returns
Experimenting the game without genuine money first
Taking advantage of easy gameplay and high RTP features
Typically the Aviator
The aviator sport on Bet9ja is related to the video game’s distinct features:
Multiple type of aviator betting alternatives
Capability to find the aviator slot quickly
Access to Aviator on the web
Choices to try and win through different methods
Opportunities to win big with calculated risks
Flexibility to play within the aviator game guidelines
Aviator Winning Techniques
To improve your opportunities of winning:
Choose to find the aviator betting patterns
Utilize the web in any aviator casino includes carefully
Learn proper money from aviator management
Master ending the game at optimal times
Understand typically the very best cash-out moments
Make the most of the chance to play strategically
Extra Tips for Success:
Start with small bets to discover the game mechanics
Utilize the demo variation to practice methods
Monitor video game patterns and patterns
Set realistic winning goals
Never ever chase losses
Usage car cash-out features when readily available
Bear in mind that while Bet9ja provides an excellent platform for Aviator, success needs patience, method, and responsible betting practices. The video game combines aspects of skill and opportunity, making it necessary to approach it with a well balanced frame of mind and proper bankroll management.
The Art of Timing
Success in Aviator greatly counts on mastering the art of timing. Gamers should thoroughly look for patterns and decide the optimum time to bet and squander. The video game’s high volatility means timing choices can separate between substantial wins and losses.
Aviator Game Algorithm
The Aviator game algorithm uses a provably reasonable system that ensures random results for each round. Comprehending that the video game patterns are random assists players develop reasonable expectations and strategies.
Best Strategy for Aviator Casino Game Success
A few of the very best Aviator betting strategies include:
Starting with a conservative strategy utilizing smaller bet sizes
Executing a systematic bet after each win
Using the demo version of Aviator to practice
Setting both an initial bet and a second bet method
Maintaining constant bet sizes based on bankroll management
Tips and Tricks for Winning
Essential win tips for the Aviator game consist of:
Watch for patterns in video game rounds
Use the demonstration version to practice technique
Set clear profit objectives and stop-loss limits
Screen video game patterns over numerous rounds
Execute tactical bet sizing
How to Enjoy Aviator Responsibly
To make Aviator provide satisfying home entertainment:
Start with the demo variation utilizing preset quantities
Set clear betting limits
Take routine breaks from gameplay
Never go after losses
View it as entertainment instead of a guaranteed income source
Winning Strategy Fundamentals
A solid winning strategy to play effectively includes:
Proper bankroll management
Comprehending the video game’s volatility
Utilizing tactical bet sizing
Carrying out stop-loss limits
Benefiting from the video game’s high RTP worth
Advanced Game Tricks
Advanced gamers frequently use these game tricks:
Multiple bet techniques
Pattern acknowledgment strategies
Timing-based techniques
Threat management systems
Strategic cash-out points
Online Game Experience
The Aviator online game experience offers:
Easy access through various casino sites
Several betting options
Real-time multiplayer interaction
Live data and history
Mobile compatibility for on-the-go play
Particular Aviator Game Features
Key functions that make this particular game popular:
Social gaming elements
Real-time multiplier display
Basic yet interesting interface
Multiple wagering alternatives
Transparent video game mechanics
This structured approach to understanding and playing the Aviator crash game can assist gamers develop much better methods and enjoy the game more responsibly. Bear in mind that while methods can enhance your opportunities, there’s no guaranteed way to win, and gaming needs to always be approached with care and within one’s methods.
FAQ
What is the very best time to play Aviator?
There’s no definitive best time to play Aviator. The video game’s random nature indicates each round is independent. Choose a time that fits your schedule and budget.
How does Aviator’s Random Number Generator work?
Aviator uses a Random Number Generator (RNG) for fair results. It creates random numbers for each video game round. This system ensures unforeseeable flight paths and crash points.
What are the fundamental guidelines for brand-new Aviator gamers?
New gamers should find out the game mechanics. These involve betting on a simulated flight and timing the cash-out. They should likewise understand how to select stakes and location bets.
It’s crucial to practice accountable betting. Set individual limits to make sure safe and pleasurable gameplay.
Are there reliable strategies for winning at Aviator?
No method assurances consistent wins in Aviator. Nevertheless, some approaches can improve your gameplay. These consist of accountable bankroll management and setting wagering limitations.
Using the vehicle cash-out feature can also be helpful. Keep in mind, Aviator is mostly a game of chance.
What are common betting limits in Aviator games?
Betting limitations vary between platforms. Minimum bets can be as low as ₤ 0.10, with optimum bets around ₤ 100. Optimum wins are frequently capped, sometimes at ₤ 10,000 per bet.
Always examine the specific limits of your selected platform. This guarantees you’re mindful of prospective risks and benefits.
Can I improve my chances of winning by playing during particular hours?
Playing at specific times does not improve your winning chances. Each round is independent and determined by the RNG. Off-peak hours might provide smoother gameplay due to minimized server load.
Is it possible to acknowledge patterns in Aviator gameplay?
Some players attempt to find patterns in the flight’s ascent. However, this is mainly speculative. The game’s RNG makes sure past outcomes don’t affect future results.
How crucial is bankroll management in Aviator?
Bankroll management is crucial in Aviator. It helps control spending and extend gameplay. Set clear limitations on bets and losses per session.
This approach is critical to responsible gaming. It guarantees a more enjoyable and sustainable gaming experience.
within the aviator game
November 23, 2024 @ 9:28 pm
Pilot is a thrilling online betting game that mixes luck and technique. It has actually captivated gamers worldwide. Many marvel: When is the very best time to play Aviator?
Pilot has over 12 million players internationally and is readily available on over 2,000 betting platforms. The game’s appeal depends on its simpleness and capacity for big payments.
Studies suggest playing Aviator in between 8 a.m. and 10 a.m. might enhance winning possibilities. Night owls might benefit, too. Multipliers in between 10 p.m. and midnight can be three times greater.
This guide will check out wagering varieties and game mechanics. We’ll also cover effective bankroll management strategies. These insights aim to boost your Aviator gaming method.
This information can improve your self-confidence Whether you’re brand-new or skilled. It might even assist increase your winnings in the interesting world of Aviator.
Comprehending the Basics of Aviator Game
The Aviator casino game blends chance and strategy, fascinating players worldwide. It utilizes an advanced random number generator for fair play, ensuring unpredictable outcomes in every round.
How Aviator’s Random Number Generator Works
The video game’s fairness foundation is Aviator’s random number generator. It produces number series that determine each round’s flight path and crash point. This system makes every video game special and unforeseeable.
Video Game Mechanics and Interface Overview
Pilot’s gameplay is straightforward yet interesting. Players bet on a virtual plane’s flight to squander before it crashes. The interface reveals the plane’s ascent graph and betting options.
As the multiplier increases, so does the prospective payout. This develops an exciting risk-reward dynamic for gamers.
Standard Rules for New Players
Betting rules in Aviator are easy to understand. Players pick their stakes and place bets before each round starts. The multiplier climbs up as the aircraft takes off.
The secret is timing your cash-out right. Squandering too early may suggest missing bigger wins, and waiting too long might result in losing your bet if the aircraft crashes.
For instance, a ₤ 10 bet cashed out at 2.0 x would yield ₤ 20. Waiting on a 5.0 x multiplier could lead to a ₤ 50 payout.
What Is The Best Time To Play Aviator?
Aviator’s random nature makes discovering the best playing time difficult. Each round is independent, regardless of when you play, so your possibilities of winning remain the same at any time.
Some players choose off-peak hours for smoother gameplay, while others take pleasure in peak times when more gamers are online. Your choice depends on your schedule and choices.
At Katsu Casino, Aviator provides minimum bets of $0.10. This matches various budgets. Optimum bets reach $100 per wager. You can win approximately $10,000 per bet.
There is no ensured ‘finest’ time to play Aviator. The video game utilizes a random number generator, so results are unforeseeable at any time.
Focus on accountable gaming and stay with your budget. Enjoy the game, no matter when you pick to play. That’s the key to a fantastic Aviator experience.
Important Strategies for Maximising Your Wins
Aviator success mixes intelligent wagering with careful risk management. The video game provides chances for big wins, with multipliers up to x1000. Players need a thoughtful method to maximize these chances.
Bankroll Management Techniques
Effective bankroll management is vital for long-term Aviator success. Set clear betting limitations and stay with them. A typical strategy is wagering 1-2% of your total bankroll per bet.
This technique safeguards your funds throughout losing streaks. It likewise helps extend your playing time, giving you more opportunities to win.
Using the Auto Cash-out Feature
The auto cash-out function is an effective tool in Aviator. It lets you preset a multiplier for automated bet cashing. This method can protect consistent wins and protect your earnings.
Some players choose lower multipliers, around 1.5 x, for steady gains. Others aim greater for larger payments, stabilizing danger and benefit.
Pattern Recognition and Analysis
Aviator uses a random number generator, however some gamers try to spot patterns. Studying live statistics and round history can provide insights into recent trends.
Keep in mind, each round is independent. Past results don’t influence future results. Usage pattern analysis carefully as part of your total wagering strategy.
Optimal Betting Ranges and Limits
Pilot’s betting limits and payout structure are crucial for a winning strategy. The game provides different minimum and optimum bets for different spending plans. Some platforms enable wagers as low as $0.10, while others begin at INR 200 (about $2.40).
Maximum bets can reach $100, catering to high-stakes gamers. This range fits both casual gamers and severe bettors. The optimum win per bet can vary, with some gambling establishments capping it at $10,000.
Pilot’s special payout structure offers multipliers for potentially considerable returns. With a 97% RTP, the video game provides lots of winning chances. Starting with smaller sized bets and squandering early can assist handle your bankroll effectively.
Success as an Aviator requires strategy, perseverance, and luck. Understanding the game’s mechanics is vital. Changing your bets accordingly can optimise your chances of winning while keeping you comfy.
Best Aviator Platform
Bet9ja is Nigeria’s premier Aviator game platform. It offers easy gameplay and high RTP value. The platform offers an user-friendly interface, safe banking options, and a demonstration variation for practice.
As a top-rated betting game, it uses new and experienced players the chance to play in a regulated environment.
Gamble Responsibly
When playing Aviator on Bet9ja, it’s important to understand that the video game can offer fulfilling home entertainment when approached responsibly. Start with the Aviator demo using preset limitations, and remember that the game is created for entertainment initially.
Set transparent budgets and time limits for gameplay sessions.
Win the Aviator Game
To win the aviator game eventually, gamers need to concentrate on:
Implementing a tested aviator strategy
Beginning with one bet at a time
Comprehending the strategy within the video game mechanics
Utilizing the initial Aviator game’s official functions
Mastering the strategy of playing Aviator
Learning to win more often through practice
Win Aviator Consistently
Successful aviator tips for constant winning consist of:
Studying rounds of the game patterns
Making determined bet-plus choices
Understanding the gameplay and high RTP value
Utilizing technique to take full advantage of returns
Experimenting the game without genuine money initially
Taking advantage of easy gameplay and high RTP features
Typically the Aviator
The aviator sport on Bet9ja is associated with the video game’s unique functions:
Multiple kinds of aviator betting choices
Capability to find the aviator slot quickly
Access to Aviator on the web
Choices to try and win through numerous strategies
Opportunities to win big with calculated risks
Versatility to play within the aviator game rules
Aviator Winning Techniques
To enhance your chances of winning:
Choose to find the aviator betting patterns
Utilize the web in any aviator casino includes wisely
Discover correct money from aviator management
Master ending the game at optimum times
Understand typically the best cash-out moments
Make the most of the chance to play strategically
Extra Tips for Success:
Start with small bets to learn the game mechanics
Use the demonstration variation to practice techniques
Display game patterns and patterns
Set realistic winning objectives
Never chase losses
Use vehicle cash-out functions when offered
Remember that while Bet9ja uses an outstanding platform for Aviator, success needs perseverance, strategy, and accountable gaming practices. The video game integrates components of ability and opportunity, making it essential to approach it with a balanced state of mind and correct bankroll management.
The Art of Timing
Success in Aviator greatly relies on mastering the art of timing. Gamers must thoroughly watch for patterns and choose the optimal time to bet and squander. The video game’s high volatility means timing decisions can separate in between considerable wins and losses.
Aviator Game Algorithm
The Aviator game algorithm uses a provably fair system that ensures random results for each round. Understanding that the video game patterns are random assists gamers develop realistic expectations and methods.
Best Strategy for Aviator Casino Game Success
A few of the best Aviator betting strategies include:
Starting with a conservative strategy using smaller bet sizes
Carrying out a systematic bet after each win
Utilizing the demo version of Aviator to practice
Setting both an initial bet and a second bet method
Keeping constant bet sizes based on bankroll management
Tips and Tricks for Winning
Important win tips for the Aviator game include:
Watch for patterns in game rounds
Use the demonstration variation to practice strategy
Set clear revenue objectives and stop-loss limits
Screen video game patterns over numerous rounds
Implement tactical bet sizing
How to Enjoy Aviator Responsibly
To make Aviator provide satisfying entertainment:
Start with the demonstration variation utilizing preset amounts
Set clear betting limits
Take regular breaks from gameplay
Never go after losses
View it as home entertainment instead of a surefire earnings source
Winning Strategy Fundamentals
A strong winning strategy to play successfully includes:
Proper bankroll management
Comprehending the game’s volatility
Using strategic bet sizing
Carrying out stop-loss limitations
Taking advantage of the video game’s high RTP worth
Advanced Game Tricks
Advanced gamers frequently employ these game tricks:
Multiple bet techniques
Pattern recognition strategies
Timing-based methods
Danger management systems
Strategic cash-out points
Online Game Experience
The Aviator online game experience offers:
Easy access through numerous casino sites
Several wagering alternatives
Real-time multiplayer interaction
Live data and history
Mobile compatibility for on-the-go play
Particular Aviator Game Features
Key features that make this particular game popular:
Social gaming aspects
Real-time multiplier screen
Easy yet appealing interface
Multiple betting options
Transparent game mechanics
This structured technique to understanding and playing the Aviator crash game can help players establish better methods and enjoy the game more properly. Bear in mind that while techniques can improve your chances, there’s no guaranteed way to win, and betting ought to always be approached with care and within one’s ways.
FAQ
What is the best time to play Aviator?
There’s no definitive finest time to play Aviator. The video game’s random nature indicates each round is independent. Select a time that fits your schedule and budget.
How does Aviator’s Random Number Generator work?
Aviator uses a Random Number Generator (RNG) for fair outcomes. It creates random numbers for each game round. This system ensures unforeseeable flight paths and crash points.
What are the standard guidelines for new Aviator players?
New gamers must find out the game mechanics. These involve betting on a simulated flight and timing the cash-out. They ought to likewise understand how to choose stakes and place bets.
It’s crucial to practice accountable betting. Set individual limitations to ensure safe and pleasurable gameplay.
Are there reliable techniques for winning at Aviator?
No method guarantees consistent wins in Aviator. However, some techniques can improve your gameplay. These consist of accountable bankroll management and setting wagering limitations.
Utilizing the car cash-out function can also be useful. Remember, Aviator is primarily a game of chance.
What are normal betting limitations in Aviator games?
Betting limitations vary between platforms. Minimum bets can be as low as ₤ 0.10, with maximum bets around ₤ 100. Optimum wins are typically capped, sometimes at ₤ 10,000 per bet.
Always examine the particular limitations of your picked platform. This guarantees you’re mindful of possible risks and benefits.
Can I enhance my possibilities of winning by playing during particular hours?
Playing at specific times does not improve your winning opportunities. Each round is independent and determined by the RNG. Off-peak hours may use smoother gameplay due to reduced server load.
Is it possible to acknowledge patterns in Aviator gameplay?
Some players try to spot patterns in the flight’s climb. Nevertheless, this is mostly speculative. The video game’s RNG guarantees past results do not affect future outcomes.
How important is bankroll management in Aviator?
Bankroll management is important in Aviator. It assists control spending and extend gameplay. Set clear limitations on bets and losses per session.
This approach is critical to responsible gambling. It guarantees a more enjoyable and sustainable gaming experience.
Bernardwet
November 23, 2024 @ 9:39 pm
minocycline for acne [url=http://iverfast.com/#]iver fast[/url] stromectol usa
dreamitdoitvirginia.com
November 23, 2024 @ 10:00 pm
You’re so awesome! I don’t think I have read anything like this before.
So wonderful to discover somebody with original thoughts on this subject.
Really.. thank you for starting this up. This web site is one thing that’s needed on the internet,
someone with a bit of originality!
aviator betting strategies
November 23, 2024 @ 10:30 pm
Aviator is a thrilling online betting game that blends luck and technique. It has captivated players worldwide. Lots of marvel: When is the very best time to play Aviator?
Aviator has more than 12 million players globally and is readily available on over 2,000 wagering platforms. The video game’s appeal depends on its simplicity and capacity for big payments.
Research studies recommend playing Aviator in between 8 a.m. and 10 a.m. may boost winning opportunities. Night owls could benefit, too. Multipliers between 10 p.m. and midnight can be three times greater.
This guide will explore betting ranges and video game mechanics. We’ll also cover reliable bankroll management techniques. These insights intend to boost your Aviator betting technique.
This information can enhance your confidence Whether you’re new or skilled. It might even assist increase your earnings in the interesting world of Aviator.
Comprehending the Basics of Aviator Game
The Aviator casino game mixes chance and technique, fascinating players worldwide. It uses a sophisticated random number generator for fair play, guaranteeing unpredictable outcomes in every round.
How Aviator’s Random Number Generator Works
The game’s fairness cornerstone is Aviator’s random number generator. It produces number series that figure out each round’s flight path and crash point. This system makes every game unique and unpredictable.
Game Mechanics and Interface Overview
Aviator’s gameplay is straightforward yet appealing. Players bet on a virtual plane’s flight to cash out before it crashes. The user interface shows the aircraft’s climb graph and betting options.
As the multiplier rises, so does the possible payout. This creates an amazing risk-reward dynamic for players.
Basic Rules for New Players
Betting rules in Aviator are easy to understand. Gamers pick their stakes and place bets before each round starts. The multiplier climbs as the airplane takes off.
The key is timing your cash-out right. Cashing out too early may imply missing bigger wins, and waiting too long might result in losing your bet if the aircraft crashes.
For example, a ₤ 10 bet squandered at 2.0 x would yield ₤ 20. Waiting on a 5.0 x multiplier could result in a ₤ 50 payment.
What Is The Best Time To Play Aviator?
Pilot’s random nature makes discovering the best playing time challenging. Each round is independent, regardless of when you play, so your possibilities of winning stay the exact same at any time.
Some gamers choose off-peak hours for smoother gameplay, while others enjoy peak times when more players are online. Your choice depends on your schedule and preferences.
At Katsu Casino, Aviator offers minimum bets of $0.10. This fits various budgets. Optimum bets reach $100 per wager. You can win as much as $10,000 per bet.
There is no guaranteed ‘best’ time to play Aviator. The video game utilizes a random number generator, so results are unforeseeable at any time.
Focus on accountable gaming and adhere to your budget. Enjoy the game, no matter when you pick to play. That’s the essential to a fantastic Aviator experience.
Necessary Strategies for Maximising Your Wins
Pilot success mixes intelligent betting with careful danger management. The video game offers opportunities for big wins, with multipliers approximately x1000. Players require a thoughtful approach to make the most of these opportunities.
Bankroll Management Techniques
Reliable bankroll management is vital for long-lasting Aviator success. Set clear betting limits and stick to them. A common method is betting 1-2% of your total bankroll per bet.
This technique safeguards your funds during losing streaks. It likewise assists extend your playing time, providing you more possibilities to win.
Utilizing the Auto Cash-out Feature
The automobile cash-out function is an effective tool in Aviator. It lets you pre-programmed a multiplier for automated bet cashing. This technique can protect constant wins and protect your earnings.
Some gamers choose lower multipliers, around 1.5 x, for constant gains. Others intend greater for larger payments, balancing risk and reward.
Pattern Recognition and Analysis
Aviator uses a random number generator, however some players attempt to identify patterns. Studying live statistics and round history can offer insights into current trends.
Remember, each round is independent. Previous results don’t affect future outcomes. Use pattern analysis thoroughly as part of your general wagering technique.
Optimum Betting Ranges and Limits
Aviator’s wagering limitations and payment structure are crucial for a winning strategy. The video game uses different minimum and maximum bets for various budget plans. Some platforms enable wagers as low as $0.10, while others start at INR 200 (about $2.40).
Maximum bets can reach $100, dealing with high-stakes gamers. This variety suits both casual players and major gamblers. The optimum win per bet can vary, with some casinos topping it at $10,000.
Pilot’s unique payout structure uses multipliers for possibly substantial returns. With a 97% RTP, the video game provides many winning opportunities. Beginning with smaller sized bets and cashing out early can assist handle your bankroll successfully.
Success as an Aviator needs strategy, perseverance, and luck. Comprehending the video game’s mechanics is vital. Changing your bets accordingly can optimise your chances of winning while keeping you comfy.
Best Aviator Platform
Bet9ja is Nigeria’s premier Aviator game platform. It offers easy gameplay and high RTP value. The platform offers an user-friendly interface, protected banking choices, and a demonstration version for practice.
As a top-rated betting game, it provides brand-new and experienced gamers the chance to play in a regulated environment.
Gamble Responsibly
When playing Aviator on Bet9ja, it’s important to comprehend that the video game can supply fulfilling home entertainment when approached responsibly. Start with the Aviator demo using preset limits, and remember that the game is developed for home entertainment first.
Set transparent spending plans and time frame for gameplay sessions.
Win the Aviator Game
To win the aviator game eventually, gamers need to focus on:
Implementing a proven aviator strategy
Beginning with one bet at a time
Understanding the strategy within the video game mechanics
Using the initial Aviator game’s main features
Mastering the strategy of playing Aviator
Learning to win more frequently through practice
Win Aviator Consistently
Successful aviator tips for consistent winning include:
Studying rounds of the game patterns
Making computed bet-plus choices
Understanding the gameplay and high RTP value
Utilizing technique to take full advantage of returns
Experimenting the game without real money first
Benefiting from easy gameplay and high RTP features
Typically the Aviator
The aviator sport on Bet9ja is related to the video game’s special features:
Multiple sort of aviator betting alternatives
Capability to find the aviator slot quickly
Access to Aviator on the internet
Choices to try and win through different techniques
Opportunities to win big with calculated dangers
Versatility to play within the aviator game rules
Aviator Winning Techniques
To enhance your possibilities of winning:
Choose to find the aviator betting patterns
Use the web in any aviator casino features carefully
Find out proper money from aviator management
Master ending the game at ideal times
Understand typically the best cash-out moments
Take advantage of the chance to play strategically
Additional Tips for Success:
Start with little bets to find out the game mechanics
Use the demo version to practice methods
Screen video game patterns and patterns
Set reasonable winning objectives
Never ever chase after losses
Usage auto cash-out functions when available
Keep in mind that while Bet9ja provides an outstanding platform for Aviator, success requires perseverance, method, and responsible gambling practices. The video game combines aspects of skill and opportunity, making it essential to approach it with a well balanced frame of mind and appropriate bankroll management.
The Art of Timing
Success in Aviator greatly relies on mastering the art of timing. Players need to thoroughly look for patterns and choose the ideal time to bet and cash out. The game’s high volatility means timing choices can separate between considerable wins and losses.
Aviator Game Algorithm
The Aviator game algorithm uses a provably reasonable system that makes sure random results for each round. Understanding that the game patterns are random assists players develop realistic expectations and strategies.
Best Strategy for Aviator Casino Game Success
Some of the best Aviator betting strategies include:
Starting with a conservative strategy using smaller sized bet sizes
Implementing a methodical bet after each win
Utilizing the demo version of Aviator to practice
Setting both an initial bet and a second bet technique
Maintaining constant bet sizes based upon bankroll management
Tips and Tricks for Winning
Essential win tips for the Aviator game consist of:
Watch for patterns in video game rounds
Utilize the demo variation to practice technique
Set clear earnings objectives and stop-loss limits
Monitor game patterns over multiple rounds
Execute strategic bet sizing
How to Enjoy Aviator Responsibly
To make Aviator provide satisfying entertainment:
Start with the demonstration version using pre-programmed amounts
Set clear wagering limits
Take regular breaks from gameplay
Never chase losses
View it as home entertainment rather than a guaranteed income source
Winning Strategy Fundamentals
A solid winning strategy to play effectively includes:
Proper bankroll management
Understanding the game’s volatility
Using strategic bet sizing
Implementing stop-loss limitations
Taking advantage of the video game’s high RTP value
Advanced Game Tricks
Advanced players frequently use these game tricks:
Multiple bet techniques
Pattern recognition techniques
Timing-based methods
Threat management systems
Strategic cash-out points
Online Game Experience
The Aviator online game experience provides:
Easy access through numerous casino websites
Several betting choices
Real-time multiplayer interaction
Live statistics and history
Mobile compatibility for on-the-go play
Particular Aviator Game Features
Key features that make this particular game popular:
Social gaming elements
Real-time multiplier display screen
Basic yet engaging user interface
Several wagering options
Transparent game mechanics
This structured approach to understanding and playing the Aviator crash game can help gamers establish better methods and enjoy the game more properly. Keep in mind that while methods can enhance your opportunities, there’s no guaranteed way to win, and gambling should always be approached with care and within one’s means.
FAQ
What is the very best time to play Aviator?
There’s no conclusive best time to play Aviator. The game’s random nature implies each round is independent. Choose a time that fits your schedule and spending plan.
How does Aviator’s Random Number Generator work?
Aviator uses a Random Number Generator (RNG) for fair outcomes. It produces random numbers for each game round. This system makes sure unpredictable flight paths and crash points.
What are the basic guidelines for new Aviator players?
New players must learn the game mechanics. These involve banking on a simulated flight and timing the cash-out. They need to likewise understand how to select stakes and location bets.
It’s essential to practice responsible betting. Set personal limits to guarantee safe and pleasurable gameplay.
Are there efficient techniques for winning at Aviator?
No method assurances constant wins in Aviator. However, some approaches can enhance your gameplay. These consist of accountable bankroll management and setting betting limits.
Utilizing the auto cash-out feature can also be valuable. Remember, Aviator is mainly a game of chance.
What are common betting limitations in Aviator video games?
Betting limits vary between platforms. Minimum bets can be as low as ₤ 0.10, with maximum bets around ₤ 100. Maximum wins are often capped, sometimes at ₤ 10,000 per bet.
Always check the specific limitations of your picked platform. This ensures you’re conscious of possible risks and benefits.
Can I enhance my chances of winning by playing during particular hours?
Playing at specific times does not improve your winning chances. Each round is independent and identified by the RNG. Off-peak hours might offer smoother gameplay due to minimized server load.
Is it possible to acknowledge patterns in Aviator gameplay?
Some players try to find patterns in the flight’s climb. Nevertheless, this is mostly speculative. The game’s RNG guarantees previous outcomes do not influence future outcomes.
How crucial is bankroll management in Aviator?
Bankroll management is crucial in Aviator. It assists control costs and extend gameplay. Set clear limitations on bets and losses per session.
This method is important to accountable gambling. It guarantees a more enjoyable and sustainable gaming experience.
Dnrtipl
November 23, 2024 @ 10:34 pm
Как правильно приобрести диплом колледжа или ПТУ в России, важные моменты
AlbertNef
November 23, 2024 @ 10:59 pm
purchase cytotec: buy cytotec online – buy cytotec online
Sazrgfj
November 23, 2024 @ 11:03 pm
Пошаговая инструкция по безопасной покупке диплома о высшем образовании
Ghost carts
November 24, 2024 @ 12:25 am
Good day! I know this is kinda off topic nevertheless I’d figured I’d ask.
Would you be interested in trading links or maybe guest authoring
a blog article or vice-versa? My site addresses a lot of the same topics as yours and I feel we could greatly benefit from each other.
If you might be interested feel free to send me an email.
I look forward to hearing from you! Terrific blog by the way!
DustinTex
November 24, 2024 @ 1:01 am
Эвакуатор с манипулятором – универсальное решение для перевозки автомобилей. Он сочетает функции обычного эвакуатора и крана-манипулятора. Это позволяет решать сложные задачи по эвакуации. Манипулятор может поднимать и перемещать грузы в труднодоступных местах http://wisdomtarot.tforums.org/viewtopic.php?f=8&t=8842
BrandenJew
November 24, 2024 @ 2:53 am
buy clomid without prescription: rexpharm – where can i get cheap clomid without prescription
Eanrsct
November 24, 2024 @ 4:42 am
Официальная покупка диплома вуза с сокращенной программой обучения в Москве
vivod iz zapoya v stacionare_ymel
November 24, 2024 @ 4:48 am
вывод из запоя в воронеже [url=https://superstar.ukrbb.net/viewtopic.php?f=3&t=683/]вывод из запоя в воронеже[/url] .
vivod iz zapoya v stacionare_mdsi
November 24, 2024 @ 4:51 am
вывод из запоя в воронеже [url=www.vishivayu.ukrbb.net/viewtopic.php?f=12&t=13444/]www.vishivayu.ukrbb.net/viewtopic.php?f=12&t=13444/[/url] .
vivod iz zapoya v stacionare_ksEa
November 24, 2024 @ 4:51 am
вывод из запоя стационар [url=https://recordrpservak.ukrbb.net/viewtopic.php?f=31&t=1161]https://recordrpservak.ukrbb.net/viewtopic.php?f=31&t=1161[/url] .
vivod iz zapoya v stacionare_taMi
November 24, 2024 @ 4:53 am
вывод из запоя стационар [url=https://kyevlyn.ukrbb.net/viewtopic.php?f=2&t=13641/]https://kyevlyn.ukrbb.net/viewtopic.php?f=2&t=13641/[/url] .
vivod iz zapoya v stacionare_ihsl
November 24, 2024 @ 4:53 am
выведение из запоя воронеж стационар [url=www.alhambra.bestforums.org/viewtopic.php?f=2&t=46417]www.alhambra.bestforums.org/viewtopic.php?f=2&t=46417[/url] .
Sazrqst
November 24, 2024 @ 5:37 am
Приобретение школьного аттестата с официальным упрощенным обучением в Москве
Lazrheo
November 24, 2024 @ 6:45 am
Официальная покупка аттестата о среднем образовании в Москве и других городах
Xazrwss
November 24, 2024 @ 7:02 am
Купить диплом старого образца, можно ли это сделать по быстрой схеме?
Iariorhvt
November 24, 2024 @ 7:06 am
Легальная покупка школьного аттестата с упрощенной программой обучения
MarioBibre
November 24, 2024 @ 9:06 am
Пошаговое руководство по восстановлению прозрачности фар с помощью наждачной бумаги разной зернистости. Советы по выбору материалов и технике обработки наждачка
Sazrfpy
November 24, 2024 @ 9:14 am
Быстрая схема покупки диплома старого образца: что важно знать?
ZSot
November 24, 2024 @ 9:38 am
bc game app download
Stardadownload
November 24, 2024 @ 10:28 am
[url=https://starda-casino-apk.ru/starda-skachat]www.starda-casino-apk.ru/starda-skachat[/url]
Скачать последнюю версию приложения казино онлайн Старда – выигрывай прямо сейчас!
http://starda-casino-apk.ru/starda-skachat
Darrenmow
November 24, 2024 @ 10:28 am
http://lisinopril1st.com/# lisinopril1st
cinema online malaysia
November 24, 2024 @ 11:13 am
Thanks in favor of sharing such a pleasant idea, article is pleasant, thats why
i have read it entirely
masážní salon Praha Žižkov
November 24, 2024 @ 11:14 am
Sweet blog! I found it while surfing around on Yahoo
News. Do you have any tips on how to get listed in Yahoo News?
I’ve been trying for a while but I never seem to get
there! Cheers
StardaCasino
November 24, 2024 @ 11:27 am
[url=https://starda-casino-apk.ru/]www.starda-casino-apk.ru/[/url]
Загрузить приложение онлайн казино starda – выигрывай прямо сейчас!
http://www.starda-casino-apk.ru
AnthonyBaR
November 24, 2024 @ 11:28 am
Вы можете купить справки в Москве в нашей клинике https://aleksandriya-med.ru/.
Williamraics
November 24, 2024 @ 12:44 pm
Удивительно, но купить диплом кандидата наук оказалось не так сложно
Sazrbse
November 24, 2024 @ 1:56 pm
Можно ли купить аттестат о среднем образовании? Основные рекомендации
define.listbb.ru/viewtopic.php?f=46&t=850
Сервисный центр LG
November 24, 2024 @ 1:57 pm
Предлагаем услуги профессиональных инженеров офицальной мастерской.
Еслли вы искали сервисный центр lg, можете посмотреть на сайте: сервисный центр lg в москве
Наши мастера оперативно устранят неисправности вашего устройства в сервисе или с выездом на дом!
Terryfef
November 24, 2024 @ 2:44 pm
На данный момент антигравийная пленка считается материалом, который отлично защищает кузов авто от различных повреждений. Так, она поможет избежать царапин, потертостей, а также предохраняет машину во время небольших ДТП, и в результате на поверхности авто не появляется ржавчина https://ok-vmeste.ru/toget/2209-zhelaete-okleit-svoi-avtomobil-vysokokachestvennoi-matovoi-plenkoi.html
freshSait
November 24, 2024 @ 3:11 pm
[url=https://fresh-casino-apk.ru/fresh-oficzialnyj-sajt]http://fresh-casino-apk.ru/fresh-oficzialnyj-sajt[/url]
Скачать apk файл казино fresh – побеждай сейчас!
https://fresh-casino-apk.ru/fresh-oficzialnyj-sajt
ZSot
November 24, 2024 @ 3:13 pm
neon54 casino
StardaZercalo
November 24, 2024 @ 3:50 pm
[url=https://starda-casino-apk.ru/starda-oficzialnyj-sajt]http://www.starda-casino-apk.ru/starda-oficzialnyj-sajt[/url]
Установить последнюю версию приложения казино Старда – побеждай сейчас!
https://starda-casino-apk.ru/starda-oficzialnyj-sajt
freshCasino
November 24, 2024 @ 3:52 pm
[url=https://fresh-casino-apk.ru]https://fresh-casino-apk.ru[/url]
Скачать apk файл онлайн казино Фреш – играй сейчас!
https://fresh-casino-apk.ru
Diplomi_sjSi
November 24, 2024 @ 5:09 pm
Как оказалось, купить диплом кандидата наук не так уж и сложно
BrandenJew
November 24, 2024 @ 5:55 pm
can i order cheap clomid for sale: rex pharm – where to buy clomid without a prescription
freshApk
November 24, 2024 @ 6:26 pm
[url=https://fresh-casino-apk.ru/fresh-apk]http://fresh-casino-apk.ru/fresh-apk[/url]
Загрузить приложение казино fresh – выигрывай сейчас!
http://www.fresh-casino-apk.ru/fresh-apk
vivod iz zapoya v stacionare_gwSr
November 24, 2024 @ 7:44 pm
вывод из запоя стационар [url=http://www.superjackson.ukrbb.net/viewtopic.php?f=28&t=9734]http://www.superjackson.ukrbb.net/viewtopic.php?f=28&t=9734[/url] .
Mazrcih
November 24, 2024 @ 7:46 pm
Парадокс, но купить диплом кандидата наук оказалось не так и сложно
vivod iz zapoya v stacionare_dmOl
November 24, 2024 @ 7:55 pm
вывод из запоя в воронеже [url=http://krut.forumno.com/viewtopic.php?id=6047]http://krut.forumno.com/viewtopic.php?id=6047[/url] .
vivod iz zapoya v stacionare_ybKi
November 24, 2024 @ 7:56 pm
вывод из запоя в воронеже [url=www.www.rolandus.org/forum/viewtopic.php?p=106439]www.www.rolandus.org/forum/viewtopic.php?p=106439[/url] .
vivod iz zapoya v stacionare_vuPn
November 24, 2024 @ 7:57 pm
вывод из запоя в стационаре воронежа [url=http://uaforum.ukrbb.net/viewtopic.php?f=13&t=3240]http://uaforum.ukrbb.net/viewtopic.php?f=13&t=3240[/url] .
Bernardwet
November 24, 2024 @ 7:58 pm
Misoprostol 200 mg buy online [url=https://cytpremium.com/#]cheapest cytotec[/url] cytotec online
vivod iz zapoya v stacionare_xgEa
November 24, 2024 @ 8:16 pm
вывод из запоя в воронеже [url=https://www.krasnogorsk.anihub.me/viewtopic.php?id=2573]вывод из запоя в воронеже[/url] .
Dnrteka
November 24, 2024 @ 9:50 pm
Как не стать жертвой мошенников при покупке диплома о среднем полном образовании
discount oakley sunglasses
November 24, 2024 @ 9:57 pm
It’s a pity you don’t have a donate button! I’d without a doubt donate to
this fantastic blog! I suppose for now i’ll settle for book-marking and
adding your RSS feed to my Google account. I look forward to fresh
updates and will talk about this website with my Facebook group.
Talk soon!
ZSot
November 25, 2024 @ 12:58 am
neon54 casino review
Lazrdhi
November 25, 2024 @ 1:15 am
Как избежать рисков при покупке диплома колледжа или ПТУ в России
DownloadGama
November 25, 2024 @ 1:15 am
[url=https://gama-casino-apk.ru/gama-skachat]gama-casino-apk.ru/gama-skachat[/url]
Скачать apk файл онлайн казино Гама – выигрывай сейчас!
gama-casino-apk.ru/gama-skachat
GamaApk
November 25, 2024 @ 1:30 am
[url=https://gama-casino-apk.ru/gama-apk]https://www.gama-casino-apk.ru/gama-apk[/url]
Загрузить последнюю версию приложения онлайн казино Гама – играй сейчас!
http://www.gama-casino-apk.ru/gama-apk
Ltd company formation for UK residents
November 25, 2024 @ 1:51 am
Wow, awesome blog layout! How long have you been blogging for?
you made blogging look easy. The overall look of your web site is excellent, let alone the content!
MonroAndroid
November 25, 2024 @ 2:11 am
[url=https://starda-casino-apk.ru/starda-na-androjd]https://www.starda-casino-apk.ru/starda-na-androjd[/url]
Загрузить последнюю версию приложения казино онлайн Monro – побеждай сегодня!
https://starda-casino-apk.ru/starda-na-androjd
Sazrmrq
November 25, 2024 @ 2:36 am
Всё, что нужно знать о покупке аттестата о среднем образовании без рисков
escort bayan esenyurt
November 25, 2024 @ 3:32 am
Esenyurt Escort❤️sektöründe tarihsel kökeni, hizmet taleplerinin analizi ve geleceği, yaygınlık, güvenlik, yasal düzenlemeler ve müşteri memnuniyeti için ❤️ Esenyurt Eskort kalite standartları.
büyükçekmece escort
November 25, 2024 @ 3:36 am
Büyükçekmece Escort ❤️ bayanları sitemizde bularak onlarla doyumsuz seks yapmaya hazır olun. Tek yapmanız gerek Büyükçekmece Escort Bayan ❤️ sitemizi ziyaret etmeniz.
Michealerync
November 25, 2024 @ 3:46 am
Вы можете купить справку в медцентре https://smails-clinic.ru/ с доставкой по Москве.
AllenGet
November 25, 2024 @ 4:08 am
Множество профессий, связанных с интернетом, становится доступными даже тем, кто ещё не закончил учебу. Знания, полученные в процессе освоения таких специальностей, открывают перспективы и дают шанс на независимость в финансовом плане https://yogicentral.science/index.php?title=%20%D0%9E%D0%B1%D1%83%D1%87%D0%B5%D0%BD%D0%B8%D0%B5%20CRM%20%D0%B4%D0%BB%D1%8F%20%D0%BC%D0%B5%D0%BD%D0%B5%D0%B4%D0%B6%D0%B5%D1%80%D0%BE%D0%B2%20%D0%BF%D0%BE%20%D0%BF%D1%80%D0%BE%D0%B4%D0%B0%D0%B6%D0%B0%D0%BC
Jamesinfup
November 25, 2024 @ 4:30 am
pinup kazi: пин ап вход – пинап казино
Mazrvwv
November 25, 2024 @ 6:44 am
Купить диплом экономиста – оптимальное решение
ZSot
November 25, 2024 @ 6:50 am
1 win bet app
Uazrzqf
November 25, 2024 @ 6:57 am
Приобретение диплома ПТУ с сокращенной программой обучения в Москве
Sazrcqj
November 25, 2024 @ 6:58 am
Как правильно приобрести диплом колледжа или ПТУ в России, важные моменты
Leonardsiz
November 25, 2024 @ 7:50 am
http://www.tir92.ru – Ваш надежный партнер в выборе идеальной кухни.
Jacobgaugh
November 25, 2024 @ 8:12 am
кухни москва — Большой выбор современных кухонь в Москве.
Iariorvwf
November 25, 2024 @ 8:14 am
Покупка школьного аттестата с упрощенной программой: что важно знать
Dariodig
November 25, 2024 @ 8:17 am
pinup-kazi.ru: pinup kazi – pinup-kazi.ru
Sazrymp
November 25, 2024 @ 8:51 am
Покупка диплома о среднем полном образовании: как избежать мошенничества?
есим 365
November 25, 2024 @ 9:04 am
Esim365 обеспечивает удобное решение для связи за рубежом . Благодаря esim 365 вы получите доступ к интернету для заграницы . Интернет в Турции и Китае станет доступным благодаря есим365.
есим365 идеально подходит для тех, кто часто путешествует . Это лучшее решение для связи в Китае, где традиционные способы подключения могут не работать. Есим Турции обеспечит интернет в Турции .
Сервис esim365 помогает оставаться на связи в любой стране мира . Вы легко сможете настроить esim для путешествий . С таким решением интернет в Китае или Турции станет проще .
Oariorepq
November 25, 2024 @ 9:30 am
Как официально приобрести аттестат 11 класса с минимальными затратами времени
Sazrbwi
November 25, 2024 @ 10:13 am
Всё, что нужно знать о покупке аттестата о среднем образовании без рисков
Blakesal
November 25, 2024 @ 11:08 am
https://www.mptextile.ru/ — Все для создания вашей кухни мечты.
RichardLon
November 25, 2024 @ 11:30 am
Вы можете купить медсправку в нашем мед центре https://med-bez-boli.ru/ в Москве без медкомиссий.
spam link
November 25, 2024 @ 12:14 pm
Incredible quest there. What happened after? Take care!
Leonardsiz
November 25, 2024 @ 12:41 pm
кухни на заказ в спб – Закажите кухню мечты у профессионалов в Санкт-Петербурге.
StardaCasino
November 25, 2024 @ 12:42 pm
[url=https://starda-casino-apk.ru/]www.starda-casino-apk.ru/[/url]
Установить приложение казино starda – побеждай сейчас!
http://starda-casino-apk.ru
바카라사이트
November 25, 2024 @ 1:13 pm
I was recommended this website by my cousin. I am not sure whether this post is written by him as no one else
know such detailed about my trouble. You are incredible! Thanks!
Lazrhfz
November 25, 2024 @ 1:34 pm
Легальная покупка диплома о среднем образовании в Москве и регионах
Stardadownload
November 25, 2024 @ 1:35 pm
[url=https://starda-casino-apk.ru/starda-skachat]starda-casino-apk.ru/starda-skachat[/url]
Установить приложение казино Старда – играй прямо сейчас!
http://www.starda-casino-apk.ru/starda-skachat
Jacobgaugh
November 25, 2024 @ 1:44 pm
http://www.gft-leasing.ru — Доверяйте профессионалам, создающим стильные кухни.
Cazryrc
November 25, 2024 @ 2:02 pm
Процесс получения диплома стоматолога: реально ли это сделать быстро?
GregorySpari
November 25, 2024 @ 2:04 pm
Цифровые технологии в нынешнее время проникают во все сферы жизни, виртуальный номер проявляет себя как неотъемлемым инструментом для эффективного и безопасного общения https://www.inastana.kz/list/416585
vivod iz zapoya v stacionare_dtkt
November 25, 2024 @ 2:14 pm
выведение из запоя воронеж стационар [url=https://rodoslav.forum24.ru/?1-4-0-00000572-000-0-0-1730749551/]https://rodoslav.forum24.ru/?1-4-0-00000572-000-0-0-1730749551/[/url] .
vivod iz zapoya v stacionare_mtSn
November 25, 2024 @ 2:15 pm
вывод из запоя в стационаре анонимно [url=www.belbeer.borda.ru/?1-6-0-00000759-000-0-0-1730749647]www.belbeer.borda.ru/?1-6-0-00000759-000-0-0-1730749647[/url] .
vivod iz zapoya v stacionare_bxOa
November 25, 2024 @ 2:16 pm
вывод из запоя стационар [url=https://aktivnoe.forum24.ru/?1-7-0-00012825-000-0-0-1730749092]https://aktivnoe.forum24.ru/?1-7-0-00012825-000-0-0-1730749092[/url] .
vivod iz zapoya v stacionare_dmpa
November 25, 2024 @ 2:17 pm
выведение из запоя воронеж стационар [url=www.aqvakr.forum24.ru/?1-7-0-00011578-000-0-0-1730749158]www.aqvakr.forum24.ru/?1-7-0-00011578-000-0-0-1730749158[/url] .
vivod iz zapoya v stacionare_cjSn
November 25, 2024 @ 2:17 pm
вывод из запоя воронеж [url=http://flanrp.rolevaya.com/viewtopic.php?id=148/]вывод из запоя воронеж [/url] .
Dariodig
November 25, 2024 @ 2:19 pm
пин ап кз: пинап казино – пинап казино
Derekapaky
November 25, 2024 @ 2:37 pm
http://huskytaxi.ru – надежный партнер в создании кухонь на заказ.
Diplomi_yiSi
November 25, 2024 @ 2:44 pm
Как не попасть впросак при покупке диплома колледжа или ПТУ в России
Сервисный центр Huawei
November 25, 2024 @ 3:08 pm
Предлагаем услуги профессиональных инженеров офицальной мастерской.
Еслли вы искали сервисный центр huawei в москве, можете посмотреть на сайте: сервисный центр huawei
Наши мастера оперативно устранят неисправности вашего устройства в сервисе или с выездом на дом!
Blakesal
November 25, 2024 @ 3:38 pm
кухня на заказ екатеринбург недорого — Кухни на заказ с отличным соотношением цены и качества в Екатеринбурге.
hindi pandit in bangalore
November 25, 2024 @ 3:53 pm
I have been exploring for a bit for any high quality articles or weblog posts in this sort of area .
Exploring in Yahoo I ultimately stumbled upon this web site.
Reading this info So i’m satisfied to express that I’ve an incredibly good uncanny feeling I found out exactly what I needed.
I most no doubt will make sure to don?t omit this site and provides it a look on a continuing basis.
Sazrmec
November 25, 2024 @ 4:25 pm
Полезные советы по покупке диплома о высшем образовании без риска
ZSot
November 25, 2024 @ 5:03 pm
casino neon54
Leonardsiz
November 25, 2024 @ 5:24 pm
tsmk-altai.ru/ – Закажите кухню, которая станет настоящей гордостью вашего дома.
Cazrxsg
November 25, 2024 @ 5:41 pm
Аттестат 11 класса купить официально с упрощенным обучением в Москве
Jacobgaugh
November 25, 2024 @ 7:17 pm
https://goldcoon.ru – Зайдите на сайт для консультации и заказа кухонь.
Jamesinfup
November 25, 2024 @ 8:10 pm
pin up казино: pinup-kazi.kz – пин ап кз
Trefunw
November 25, 2024 @ 8:15 pm
Процесс получения диплома стоматолога: реально ли это сделать быстро?
togel
November 25, 2024 @ 8:22 pm
Good day! This post couldn’t be written any better!
Reading this post reminds me of my old room mate! He always kept
chatting about this. I will forward this article to him.
Fairly certain he will have a good read.
Thanks for sharing!
DownloadGama
November 25, 2024 @ 8:46 pm
[url=https://gama-casino-apk.ru/gama-skachat]http://gama-casino-apk.ru/gama-skachat[/url]
Загрузить последнюю версию приложения казино gama – выигрывай прямо сейчас!
http://gama-casino-apk.ru/gama-skachat
Dnrtpzy
November 25, 2024 @ 8:51 pm
Всё, что нужно знать о покупке аттестата о среднем образовании без рисков
Сервисный центр Philips
November 25, 2024 @ 9:02 pm
Предлагаем услуги профессиональных инженеров офицальной мастерской.
Еслли вы искали официальный сервисный центр philips, можете посмотреть на сайте: сервисный центр philips
Наши мастера оперативно устранят неисправности вашего устройства в сервисе или с выездом на дом!
GamaApk
November 25, 2024 @ 9:37 pm
[url=https://gama-casino-apk.ru/gama-apk]https://www.gama-casino-apk.ru/gama-apk[/url]
Установить приложение онлайн казино gama – выигрывай сегодня!
http://gama-casino-apk.ru/gama-apk
FonbetStavki
November 25, 2024 @ 10:10 pm
[url=https://fonbet-stavki-apk.ru/fonbet-stavki/]fonbet-stavki-apk.ru/fonbet-stavki/[/url]
Загрузить приложение онлайн букмекера Фонбет – выигрывай прямо сейчас!
http://www.fonbet-stavki-apk.ru/fonbet-stavki/
ZSot
November 25, 2024 @ 10:37 pm
casino vavada
HenryLix
November 26, 2024 @ 1:06 am
пин ап казино официальный сайт [url=http://pinup-kazi.ru/#]pinup-kazi.ru[/url] pinup-kazi.ru
Dariodig
November 26, 2024 @ 1:46 am
pinup: пин ап вход – пин ап казино
Carltonkiz
November 26, 2024 @ 1:57 am
С развитием технологий виртуальные номера стали все более персонализированными и настраиваемыми. С помощью специальных сервисов можно выбирать желаемый номер из предложенного списка, настраивать перенаправление звонков, устанавливать голосовое приветствие и даже анализировать статистику звонков https://www.pravda-tv.ru/2024/01/07/599007/virtualnyj-nomer-osnovnye-osobennosti-preimushhestva-i-sfery-primeneniya
RichardLon
November 26, 2024 @ 2:03 am
Yazdorova.com – это сайт для женщин, стремящихся к здоровью, красоте и гармонии. Здесь вы найдете полезные советы по похудению, уходу за собой и вдохновение для создания лучшей версии себя https://yazdorova.com/
FAPORN.TOP
November 26, 2024 @ 2:19 am
FAPORN.TOP
GamaCasino
November 26, 2024 @ 2:43 am
[url=https://gama-casino-apk.ru/gama-casino]http://gama-casino-apk.ru/gama-casino[/url]
Скачать apk файл казино онлайн Гама – выигрывай сейчас!
https://www.gama-casino-apk.ru/gama-casino
Josephneach
November 26, 2024 @ 2:46 am
https://pinup-kazi.kz/# pin up казино
FonbetAndroid
November 26, 2024 @ 2:56 am
[url=https://fonbet-stavki-apk.ru/fonbet-na-android/]https://www.fonbet-stavki-apk.ru/fonbet-na-android/[/url]
Установить apk файл букмекера Fonbet – выигрывай прямо сейчас!
http://www.fonbet-stavki-apk.ru/fonbet-na-android/
Кредиты
November 26, 2024 @ 3:23 am
Для малого и среднего бизнеса платформа предлагает информацию о кредитах на развитие, грантах и других формах поддержки.
банки Казахстана [url=https://finance-online.kz/]МФО Казахстана[/url] .
Derekapaky
November 26, 2024 @ 3:54 am
larson-holz-spb.ru – платформа для обновления вашего кухонного интерьера.
Curtispoilm
November 26, 2024 @ 4:03 am
pinup: пин ап вход – пин ап вход
эскорт в дубае
November 26, 2024 @ 4:37 am
Very soon this website will be famous amid all blog viewers, due to it’s good posts
Cazrwah
November 26, 2024 @ 4:57 am
Пошаговая инструкция по официальной покупке диплома о высшем образовании
Blakesal
November 26, 2024 @ 5:04 am
http://www.shth.ru — Удобный сервис для заказа кухонь на любой вкус.
vivod iz zapoya ekaterinbyrg_oxmr
November 26, 2024 @ 5:19 am
вывести из запоя цена [url=http://www.gonochki.forum24.ru/?1-10-0-00001138-000-0-0-1730749908]http://www.gonochki.forum24.ru/?1-10-0-00001138-000-0-0-1730749908[/url] .
vivod iz zapoya ekaterinbyrg_dqkr
November 26, 2024 @ 5:21 am
вывод из запоя круглосуточно [url=www.vip.rolevaya.info/viewtopic.php?id=6914/]www.vip.rolevaya.info/viewtopic.php?id=6914/[/url] .
vivod iz zapoya ekaterinbyrg_xcSr
November 26, 2024 @ 5:23 am
вывод из запоя наркологом [url=http://www.sergiev.0pk.me/viewtopic.php?id=3463]http://www.sergiev.0pk.me/viewtopic.php?id=3463[/url] .
Leonardsiz
November 26, 2024 @ 7:08 am
https://www.viktorova-ts.ru – Официальный сайт с полным каталогом кухонных решений.
Sazrtxl
November 26, 2024 @ 7:36 am
Можно ли купить аттестат о среднем образовании, основные моменты и вопросы
Dariodig
November 26, 2024 @ 7:42 am
vavada: вавада – вавада казино зеркало
Sazrkiv
November 26, 2024 @ 7:42 am
Полезная информация как официально купить диплом о высшем образовании
바카라사이트
November 26, 2024 @ 8:33 am
I have read several good stuff here. Definitely value bookmarking for revisiting.
I surprise how so much effort you set to make this kind
of fantastic informative site.
Jacobgaugh
November 26, 2024 @ 8:35 am
mtucizone.ru/ — Полный ассортимент кухонь для вашего удобства.
ZSot
November 26, 2024 @ 9:10 am
bc game download
Lazrwql
November 26, 2024 @ 9:15 am
Можно ли купить аттестат о среднем образовании? Основные рекомендации
Iariorzmb
November 26, 2024 @ 9:34 am
Быстрое обучение и получение диплома магистра – возможно ли это?
slotebi
November 26, 2024 @ 9:47 am
I don’t know if it’s just me or if perhaps everybody else encountering problems
with your blog. It looks like some of the text in your posts are running off the screen.
Can somebody else please provide feedback and let me
know if this is happening to them too? This may be a issue with my
browser because I’ve had this happen previously.
Kudos
Blakesal
November 26, 2024 @ 9:50 am
купить кухню в екатеринбурге — Легко и удобно приобрести качественную кухню в Екатеринбурге.
vivod iz zapoya ekaterinbyrg_exEn
November 26, 2024 @ 10:35 am
нарколог на дом екатеринбург цены [url=http://vip.mybb.rocks/viewtopic.php?id=7687]нарколог на дом екатеринбург цены[/url] .
vivod iz zapoya ekaterinbyrg_btMi
November 26, 2024 @ 10:36 am
нарколог вывод из запоя [url=www.zarabotokdoma.creartuforo.com/viewtopic.php?id=11484]www.zarabotokdoma.creartuforo.com/viewtopic.php?id=11484[/url] .
TrevorRow
November 26, 2024 @ 11:51 am
In recent years, digital platforms have revolutionized how individuals and businesses handle their finances https://iuecwalocal81288.com/content/how-build-telegram-wallet-bot
GordonFlods
November 26, 2024 @ 11:55 am
Это проверенный обменник, которым я пользуюсь уже не первый раз. Здесь курс всегда выгодный, обмен происходит быстро, а скрытых комиссий нет. Очень удобно и безопасно для тех, кто хочет обменять криптовалюту на честных условиях http://pandora.ukrbb.net/viewtopic.php?f=2&t=12570
duatoto login
November 26, 2024 @ 11:59 am
Today, I went to the beach front with my children. I found
a sea shell and gave it to my 4 year old daughter and said
“You can hear the ocean if you put this to your ear.” She put the shell to her ear and screamed.
There was a hermit crab inside and it pinched her ear.
She never wants to go back! LoL I know this is completely off topic but I had to tell someone!
Leonardsiz
November 26, 2024 @ 12:44 pm
кухни на заказ спб — Ищете кухни на заказ в Санкт-Петербурге? Ознакомьтесь с нашими решениями.
koitoto
November 26, 2024 @ 12:56 pm
I was wondering if you ever considered changing the page layout of your website?
Its very well written; I love what youve got to say. But maybe you could
a little more in the way of content so people
could connect with it better. Youve got an awful lot of text for only
having 1 or two images. Maybe you could space it out better?
Кредиты
November 26, 2024 @ 1:38 pm
Финансовый маркетплейс для жителей Казахстана предлагает удобный способ подбора кредитов, ипотеки, микрозаймов и других финансовых продуктов. На сайте собраны лучшие предложения от надежных банков и МФО.
Кредиты [url=https://finance-online.kz/]Кредиты[/url] .
CharlesGet
November 26, 2024 @ 1:47 pm
Bonsai Casino online https://bonsai-casino.net casino offers thousands of popular slots, no deposit play and a 200% bonus on your first deposit!
Mazrscn
November 26, 2024 @ 1:55 pm
Купить диплом экономиста – оптимальное решение
Jacobgaugh
November 26, 2024 @ 2:07 pm
Кухня под заказ — персонализация и индивидуальный подход в каждом проекте
슬롯사이트
November 26, 2024 @ 2:18 pm
As the admin of this site is working, no uncertainty very quickly it will be renowned, due to
its feature contents.
Sazrdig
November 26, 2024 @ 2:21 pm
Быстрая схема покупки диплома старого образца: что важно знать?
Sazrllt
November 26, 2024 @ 2:50 pm
Как купить диплом о высшем образовании с минимальными рисками
ZSot
November 26, 2024 @ 3:04 pm
1win
Blakesal
November 26, 2024 @ 3:04 pm
купить кухню в екатеринбурге — Купите качественную кухню с гарантией по доступной цене в Екатеринбурге.
Curtispoilm
November 26, 2024 @ 5:06 pm
пин ап казино: pinup – пин ап казино официальный сайт
Derekapaky
November 26, 2024 @ 5:22 pm
кухни на заказ в спб помогут создать уют и стиль в вашем доме.
Leonardsiz
November 26, 2024 @ 5:43 pm
кухни от производителя – Надежность и качество прямо от производителя. Воплощаем ваши мечты о стильной кухне.
банки Казахстана
November 26, 2024 @ 6:27 pm
Платформа позволяет узнать, какие банки предлагают самые выгодные условия для ипотеки. Сравните ставки, сроки и дополнительные бонусы.
МФО Казахстана [url=https://finance-online.kz/]Ипотека[/url] .
translation of thesis
November 26, 2024 @ 6:33 pm
At their core, translation services transform written materials into new languages while preserving intent and context. This service requires not just language skills but cultural and contextual understanding. Expert linguists ensure accurate, culturally relevant translations.
For everything you’ve ever wanted to know about , visit us online translation of purchase order
Williamraics
November 26, 2024 @ 7:04 pm
Парадокс, но купить диплом кандидата наук оказалось не так и сложно
Jacobgaugh
November 26, 2024 @ 7:20 pm
кухня на заказ недорого москва — Доступные цены на качественные кухни на заказ в Москве.
HenryLix
November 26, 2024 @ 7:25 pm
пинап казино [url=https://pinup-kazi.kz/#]пин ап кз[/url] pinup
Blakesal
November 26, 2024 @ 8:11 pm
https://www.mptextile.ru/ — Все для создания вашей кухни мечты.
FonbetSait
November 26, 2024 @ 8:14 pm
[url=https://fonbet-stavki-apk.ru/fonbet-oficzialnyj-sajt]www.fonbet-stavki-apk.ru/fonbet-oficzialnyj-sajt[/url]
Скачать последнюю версию приложения букмекера Fonbet – побеждай прямо сейчас!
https://www.fonbet-stavki-apk.ru/fonbet-oficzialnyj-sajt
GamaApk
November 26, 2024 @ 8:29 pm
[url=https://gama-casino-apk.ru/gama-apk]www.gama-casino-apk.ru/gama-apk[/url]
Установить последнюю версию приложения казино онлайн gama – выигрывай прямо сейчас!
http://gama-casino-apk.ru/gama-apk
FonbetStavki
November 26, 2024 @ 8:35 pm
[url=https://fonbet-stavki-apk.ru]www.fonbet-stavki-apk.ru[/url]
Скачать приложение букмекера Fonbet – побеждай сегодня!
http://fonbet-stavki-apk.ru
GamaCasino
November 26, 2024 @ 9:14 pm
[url=https://gama-casino-apk.ru/gama-casino]https://www.gama-casino-apk.ru/gama-casino[/url]
Загрузить приложение онлайн казино gama – играй сейчас!
gama-casino-apk.ru/gama-casino
FonbetAndroid
November 26, 2024 @ 10:19 pm
[url=https://fonbet-stavki-apk.ru/fonbet-na-android/]fonbet-stavki-apk.ru/fonbet-na-android/[/url]
Скачать приложение букмекера Fonbet – играй прямо сейчас!
http://www.fonbet-stavki-apk.ru/fonbet-na-android/
1s byhgalteriya kypit_rzsa
November 26, 2024 @ 10:22 pm
1с бухгалтерия предприятия цена [url=1s-buhgalteriya-kupit.ru]1с бухгалтерия предприятия цена[/url] .
vivod iz zapoya ekaterinbyrg_xxmr
November 26, 2024 @ 10:22 pm
срочный вывод из запоя [url=http://www.mediaworld.ukrbb.net/viewtopic.php?f=49&t=5455]http://www.mediaworld.ukrbb.net/viewtopic.php?f=49&t=5455[/url] .
vivod iz zapoya ekaterinbyrg_pnea
November 26, 2024 @ 10:26 pm
лечение запоев на дому [url=superstar.ukrbb.net/viewtopic.php?f=3&t=684]superstar.ukrbb.net/viewtopic.php?f=3&t=684[/url] .
slotebi
November 26, 2024 @ 10:43 pm
Its not my first time to pay a quick visit this website, i am browsing this web site dailly and obtain fastidious data from here all
the time.
koitoto
November 26, 2024 @ 11:02 pm
Piece of writing writing is also a excitement,
if you be familiar with then you can write if not it is complicated to
write.
Jamesinfup
November 26, 2024 @ 11:12 pm
вавада: вавада онлайн казино – vavada kazi
Leonardsiz
November 26, 2024 @ 11:17 pm
кухни санкт петербург – Идеальные кухонные гарнитуры для уютного дома в Петербурге.
translation of a journal
November 26, 2024 @ 11:53 pm
At their core, translation services transform written materials into new languages while preserving intent and context. This service requires not just language skills but cultural and contextual understanding. Skilled translators craft messages that connect effectively with diverse audiences.
Your trusted source for awaits—visit today translation of thesis
duatoto login
November 26, 2024 @ 11:54 pm
It’s really a nice and helpful piece of information.
I am satisfied that you just shared this useful info
with us. Please stay us up to date like this. Thank you for sharing.
Jamiebic
November 26, 2024 @ 11:54 pm
мероприятия по оценке условий труда в москве профессиональные риски и специальная оценка условий труда москва
ZSot
November 27, 2024 @ 1:11 am
1win
Dnrtlko
November 27, 2024 @ 1:31 am
Как приобрести аттестат о среднем образовании в Москве и других городах
Blakesal
November 27, 2024 @ 1:54 am
кухни под заказ — Закажите кухню, которая идеально подходит вашему интерьеру.
Sazrdgb
November 27, 2024 @ 2:44 am
Реально ли приобрести диплом стоматолога? Основные этапы
doublrp.listbb.ru/viewtopic.php?f=3&t=254
オナホ
November 27, 2024 @ 3:39 am
パートナーとの愛の表現として、胸や太ももを大切に触れ合うことで、より親密な関係を築くことができます。さらに、パートナーと共にオナホでの性的な快楽を追求することは、単なる身体的な行為を超えて、感情的なつながりを強める効果があります。
Eanrcyb
November 27, 2024 @ 3:49 am
Процесс получения диплома стоматолога: реально ли это сделать быстро?
DownloadGama
November 27, 2024 @ 4:00 am
[url=https://gama-casino-apk.ru/gama-skachat]http://www.gama-casino-apk.ru/gama-skachat[/url]
Скачать последнюю версию приложения онлайн казино gama – побеждай сейчас!
http://www.gama-casino-apk.ru/gama-skachat
RobertPsype
November 27, 2024 @ 4:21 am
Задачи и роль QA-инженера: путь к успешной карьере в области тестирования программных продуктов. Узнайте, какие навыки и знания необходимы специалисту для качественного выполнения задач на всех этапах разработки: https://dzen.ru/a/ZvqB94uwl1C-1ivx
WalterRon
November 27, 2024 @ 4:22 am
Вы можете купить медицинскую справку в мед центре https://hearth-health.ru/ в Москве без прохождения врачей.
Leonardsiz
November 27, 2024 @ 4:51 am
https://tsmk-altai.ru/ – Посетите наш сайт, чтобы выбрать кухню, которая подходит именно вам.
bokep terbaru
November 27, 2024 @ 4:54 am
Hi, Neat post. There is an issue together with your website in internet explorer, could check this?
IE nonetheless is the market leader and a good portion of folks will omit your excellent writing because of this
problem.
Кредиты
November 27, 2024 @ 6:06 am
Нужен автокредит? На маркетплейсе вы найдете специальные предложения с низкими процентами и удобными условиями от банков Казахстана.
Микрокредиты [url=https://finance-online.kz/]Курсы валют[/url] .
Jacobgaugh
November 27, 2024 @ 6:47 am
кухни москва – Широкий выбор кухонь для установки в квартирах и домах Москвы.
Derekapaky
November 27, 2024 @ 6:53 am
кухни на заказ от производителя гарантируют высокое качество и долговечность мебели.
Georgeenuff
November 27, 2024 @ 6:55 am
стоимость оценки условий труда одного рабочего места в москве специальная оценка условия труда на предприятия
chance to play
November 27, 2024 @ 6:55 am
You’ve made some important points!
Blakesal
November 27, 2024 @ 7:05 am
заказать кухню — Сделайте заказ на кухню своей мечты уже сегодня.
Petir138
November 27, 2024 @ 7:23 am
Hi there, You’ve done a fantastic job. I
will definitely digg it and personally recommend to my friends.
I’m sure they’ll be benefited from this web site.
Dariodig
November 27, 2024 @ 7:27 am
пин ап вход: пин ап вход – pinup
escort kumburgaz
November 27, 2024 @ 7:31 am
gerçek vip kumburgaz escort bayan sitesiyim
Lazrmoh
November 27, 2024 @ 8:05 am
Аттестат школы купить официально с упрощенным обучением в Москве
Rybelsus mot övervikt
November 27, 2024 @ 9:33 am
Hello outstanding website! Does running a blog similar to this take a
great deal of work? I’ve no understanding of programming but I was hoping to start my own blog in the near future.
Anyhow, should you have any ideas or techniques for new blog
owners please share. I understand this is off topic however I just wanted to ask.
Thanks!
Mazrrqy
November 27, 2024 @ 9:48 am
Пошаговая инструкция по официальной покупке диплома о высшем образовании
Leonardsiz
November 27, 2024 @ 10:06 am
viktorova-ts.ru/ – Воплощаем идеи в реальность с нашими кухнями.
Sazrnbh
November 27, 2024 @ 10:12 am
Приобретение диплома ПТУ с сокращенной программой обучения в Москве
Sazrnql
November 27, 2024 @ 10:21 am
Приобретение школьного аттестата с официальным упрощенным обучением в Москве
Jamesinfup
November 27, 2024 @ 10:32 am
pinup-kazi.ru: пин ап зеркало – pinup kazi
langit69 alternatif
November 27, 2024 @ 10:32 am
Howdy! I could have sworn I’ve been to this blog before but after reading through some
of the post I realized it’s new to me. Anyways, I’m definitely delighted I
found it and I’ll be bookmarking and checking back often!
bonus new member
November 27, 2024 @ 10:34 am
Amazing blog! Is your theme custom made or did you
download it from somewhere? A design like yours with a few simple adjustements would
really make my blog shine. Please let me know where you
got your design. Many thanks
MauriceHag
November 27, 2024 @ 10:38 am
Dom Boutique Hotel, boutique hotel: addresses with entrances on the map, reviews, photos, phone numbers, opening hours and how to get there. Accommodation from http://cyprus-apartment.com/
ブランディング 企業
November 27, 2024 @ 11:20 am
I’m not that much of a online reader to be honest
but your blogs really nice, keep it up! I’ll go ahead and bookmark your website to come back later on.
Many thanks
Jacobgaugh
November 27, 2024 @ 11:56 am
http://mtucizone.ru — Просто, удобно и с гарантией качества.
Blakesal
November 27, 2024 @ 12:02 pm
https://tort-dubr.ru/ — Переходите на наш сайт для ознакомления с предложениями.
HenryLix
November 27, 2024 @ 12:10 pm
пин ап казино онлайн [url=https://pinup-kazi.kz/#]пин ап кз[/url] пин ап казино
win tips
November 27, 2024 @ 12:19 pm
You’ve made some interesting points!
youjizz porn
November 27, 2024 @ 12:20 pm
I’m not that much of a online reader to be honest but
your blogs really nice, keep it up! I’ll go ahead and bookmark your website to
come back later. All the best
jual obat aborsi
November 27, 2024 @ 12:20 pm
When I originally commented I clicked the “Notify me when new comments are added” checkbox and now each time a comment is
added I get four e-mails with the same comment.
Is there any way you can remove me from that service?
Many thanks!
WalterRon
November 27, 2024 @ 1:00 pm
Вы можете купить справку в клинике https://medikwest.ru/ по Москве с бесплатной доставкой до любой станции метро.
Domingocer
November 27, 2024 @ 1:13 pm
Камеры видеонаблюдения выполняют различные задачи: они помогают отпугивать нарушителей, фиксируют события на охраняемой территории и контролируют действия сотрудников Услуги видеонаблюдения в Вологде
Dariodig
November 27, 2024 @ 1:31 pm
pinup-kazi.kz: pinup-kazi.kz – пин ап казино
Mazrxzf
November 27, 2024 @ 1:33 pm
Купить диплом экономиста – оптимальное решение
concrete repairs service in houston texas
November 27, 2024 @ 1:33 pm
Hello, just wanted to mention, I loved this article.
It was practical. Keep on posting!
DTPOUTR0
November 27, 2024 @ 2:18 pm
That is a great tip especially to those fresh to the blogosphere.
Short but very precise information… Thanks for sharing this
one. A must read article!
GamaApk
November 27, 2024 @ 2:33 pm
[url=https://gama-casino-apk.ru/gama-apk]gama-casino-apk.ru/gama-apk[/url]
Загрузить apk файл казино онлайн gama – выигрывай сегодня!
https://www.gama-casino-apk.ru/gama-apk
포르노
November 27, 2024 @ 2:51 pm
My partner and I stumbled over here different page
and thought I might as well check things out.
I like what I see so now i am following you.
Look forward to looking at your web page repeatedly.
liability car insurance
November 27, 2024 @ 3:42 pm
Markdowns once and for all pupils as well as safe drivers are typically available along with Car insurance in Pasadena TX.
Make certain to ask them about these rewards when matching up providers of Car insurance policy in Pasadena TX.
ile kosztuje prawo Jazdy am
November 27, 2024 @ 4:41 pm
Jaka Jest Cena Prawa Jazdy Am? ile kosztuje prawo Jazdy am
vivod iz zapoya ekaterinbyrg_eePn
November 27, 2024 @ 4:43 pm
вывод из запоя на дому екатеринбург [url=http://sait.anihub.me/viewtopic.php?id=5681]http://sait.anihub.me/viewtopic.php?id=5681[/url] .
vivod iz zapoya ekaterinbyrg_vvKn
November 27, 2024 @ 4:49 pm
нарколог на дом екатеринбург цены [url=www.rio16.ukrbb.net/viewtopic.php?f=3&t=1121/]www.rio16.ukrbb.net/viewtopic.php?f=3&t=1121/[/url] .
vivod iz zapoya ekaterinbyrg_ycMi
November 27, 2024 @ 4:50 pm
вывод из запоя в екатеринбурге [url=uaportal.ukrbb.net/viewtopic.php?f=2&t=3722]uaportal.ukrbb.net/viewtopic.php?f=2&t=3722[/url] .
Blakesal
November 27, 2024 @ 4:52 pm
заказать кухню — Легко заказать кухню, которая будет соответствовать всем вашим пожеланиям.
vivod iz zapoya ekaterinbyrg_uvPr
November 27, 2024 @ 4:52 pm
вывод из запоя [url=http://recordrpservak.ukrbb.net/viewtopic.php?f=31&t=1162/]вывод из запоя[/url] .
Jacobgaugh
November 27, 2024 @ 4:56 pm
http://www.taxipuma.ru — качество и стиль от производителя
kupit_huEi
November 27, 2024 @ 5:00 pm
https://bsnss.net/ideas/virtualnyy-nomer-budte-na-svyazi-s-vashey-auditoriey.html
Permis Cepc
November 27, 2024 @ 5:07 pm
Buy Your French Driving License B Online Permis Cepc
professional woocommerce support
November 27, 2024 @ 5:22 pm
Great delivery. Outstanding arguments. Keep up thee amazing spirit. https://f2b.s3-web.eu.cloud-object-storage.appdomain.cloud/
child porn
November 27, 2024 @ 6:22 pm
It’s an remarkable post designed for all the online viewers;
they will take benefit from it I am sure.
Köpa taxi köRkort
November 27, 2024 @ 6:26 pm
Köp Taxi Körkort: Så Går Du Tillväga Köpa taxi köRkort
nsfw ai chat
November 27, 2024 @ 6:50 pm
Thanks to my father who told me about this weblog, this blog is actually awesome.
Cynthia
November 27, 2024 @ 7:12 pm
2024 Driving License Cost: What To Expect prix du permis de conduire 2024 [Cynthia]
Dariodig
November 27, 2024 @ 7:32 pm
pinup: pinup – пин ап зеркало
Leonardsiz
November 27, 2024 @ 8:05 pm
http://tir92.ru/ – Узнайте больше о наших кухнях и посмотрите варианты на сайте.
insurance companies for vehicles
November 27, 2024 @ 8:13 pm
If you are actually hoping to help make improvements to your policy, car insurance
companies commonly allow versatile changes. Contact your car insurance companies concerning how effortless it is to tweak your policy as required.
POLTMLR57
November 27, 2024 @ 8:21 pm
I think the admin of this website is actually working hard
in support of his web site, because here every stuff is quality based stuff.
Satta
November 27, 2024 @ 8:47 pm
This article gives clear idea in support of the new visitors of blogging, that truly how
to do blogging.
PBQDWHJ
November 27, 2024 @ 8:55 pm
Joseph. Baton. New orleans saints standings. Triceps. Tsp. Louvre museum. Ball state. Possum. Victor wembanyama. Dictionary. Her movie. Javier milei. Loom. Mead. Hamilton college. Maria shriver. Horse fly. Star trek. Cat in the hat. Whale. What is an integer. Batman 1989. Sandra day o’connor. Church of jesus christ of latter day saints. Flute. Iodine. Los angeles angels. Edward snowden. Miami fl. Miyamoto musashi. Pittsburgh. Daniel. Ontario ca. Columbus day. Kim kardashian kids. Che. Platonic meaning. Wii. Round table. Liability insurance. Privilege. World. Credit cards. Autism. Drake passage. Roth ira contribution limits 2023. Great lakes. English to creole. Anise. Taxi driver 1976. Radiology. Oatmeal. How many genders are there. Magellan. Kefir. Mukesh ambani. Moon phase. David lynch. Envy. Communist. Oyster mushrooms. Qantas. Heretic. https://x4.yxoo.ru/
Supermoon. Fast&food. Vietnam. Stevie wonder. Ceelo green. Steve bannon. Rialto. Long island. Brother. Nicolas cage. Daddy. Wordle game. Julianna margulies. Humpback whale. Broadchurch. May. Midazolam. Ewan mcgregor. Get. Ozarks. Basal ganglia. Wheat. Fat. Excited. Jennifer hudson. Fernandina beach. Csi. Sri lanka. Bovine. Hornet. Selenium. Charlie’s angels. The oc cast. The good fight. Roberto carlos. First day of fall. Lvmh. Rear window. Charlatan. Nope movie. Baked alaska. Iraq. Bird of paradise. Niall horan. Bupropion. Fort myers. Generation z. Berry. Black bear. Benjamin franklin. Spin. Ulcer. Aquarius dates. Real steel. Twitter stock. Paul dano. Lisa marie presley spouse. Magnificent seven. Teratoma. Sardines.
Richardgrirm
November 27, 2024 @ 8:58 pm
jetour dashing comfort https://jetour-dashing-avto.ru
Blakesal
November 27, 2024 @ 9:38 pm
https://mptextile.ru — Дизайн и изготовление кухонь, которые вам понравятся.
DownloadGama
November 27, 2024 @ 9:48 pm
[url=https://gama-casino-apk.ru/gama-skachat]http://gama-casino-apk.ru/gama-skachat[/url]
Установить приложение онлайн казино gama – выигрывай сегодня!
gama-casino-apk.ru/gama-skachat
Jacobgaugh
November 27, 2024 @ 9:55 pm
armor-games.ru — Перейдите на сайт и ознакомьтесь с каталогом кухонь.
ScottLiawl
November 27, 2024 @ 10:02 pm
mexico drug stores pharmacies: MexicanPharmEasy – mexican pharmaceuticals online
Discover More
November 27, 2024 @ 10:05 pm
Remarkable! Its in fact awesome article, I have got much clear idea on the topic of from this post.
FonbetStavki
November 27, 2024 @ 10:12 pm
[url=https://fonbet-stavki-apk.ru]http://www.fonbet-stavki-apk.ru[/url]
Загрузить apk файл лучшего букмекера Фонбет – играй сейчас!
https://www.fonbet-stavki-apk.ru
QGIZMYW
November 27, 2024 @ 10:38 pm
Twiggy. Tpb. Fireflies. Redondo beach. David beckham new wife. Office space cast. Paul pierce. K2. The daily beast. Dirty grandpa. Daycare. Documentary. Badgers. Consensus. Prairie dog. 1040 form. Catcher in the rye. Radon. Zuma. Chicken pox. Reiterate. The warriors. Byzantine empire. Foyer. Vlad the impaler. Sully. Jurassic park. Exponent. Wisconsin. Plague. Caitlyn jenner. Saipan. Devotion. Pride and prejudice. Intersex meaning. Sheldon. Hard drive. Chicken fried steak. Rg3. Youtubr. Cast of saving private ryan. 25th amendment. The breakfast club. Up to date. Recipient. Alfre woodard. Maya moore. What does it mean. Lightning game. Have. Submissive meaning. Apocalypto. Bob seger. The bodyguard. Attention deficit disorder. Asmodeus. Harpers ferry. Types of clouds. https://x4.yxoo.ru/
Edge of tomorrow. Tectonic plates. Insa. Dutch. Van gogh museum. Conception. Ios. G. Twitter stock. Puppy love. Antibiotics. Month. Malaysia. Cognitive behavioral therapy. Kinetic energy. Daenerys targaryen. Valencia. Agatha christie. San antonio. Quaker oats. Burrow. Benjamin. Meena alexander. Baby boomers. Malt. Frasier cast. Yahoo. Cavalier. Bob marley. The buccaneers. Beowulf. Shreveport. Robert shaw. Saints game today. Jennifer connelly movies. Kelpie. Mount washington. George hamilton. Menlo park. Rosemary kennedy. Jenna ortega age. Memento mori meaning. Moose. The league. Miami seaquarium. Budapest. Dysentery. Cavs. Melissa mccarthy movies. Cyst. Wechat. Administration. Racism. Difference between. Unc chapel hill. George clooney movies and tv shows. Gretchen whitmer. Natural. Jodie comer movies and tv shows. What is plasma. Verbena. Nuisance. New hope.
StardaZercalo
November 27, 2024 @ 11:23 pm
[url=https://starda-casino-apk.ru/starda-oficzialnyj-sajt]starda-casino-apk.ru/starda-oficzialnyj-sajt[/url]
Загрузить приложение казино Старда – играй сейчас!
http://www.starda-casino-apk.ru/starda-oficzialnyj-sajt
nodeposit
November 27, 2024 @ 11:30 pm
This paragraph provides clear idea designed for the new viewers of blogging, that in fact how
to do running a blog.
PatrickNeaps
November 27, 2024 @ 11:52 pm
1xBet Promo Code 2024 – 1XBIG24: enter this code when creating your account to receive up to $100 as a 100% match on your first deposit. The bonus amount must be used in the sports section by placing bets with odds of 1.4 or higher and a 5x wagering requirement within 30 day https://millingen-online.de/wp-content/pags/?melbet_promo_code_new_copy.html
BATSWYZ
November 28, 2024 @ 12:14 am
Red bottom shoes. Salmonella symptoms. Cherry tree. Presidential polls 2024. Florence italy. Elephant. Fern. Doberman pinscher. Endorphins. Maroon 5. Bell. Squash. Dances with wolves. George costanza. Royal blue. Robert kennedy jr. Sd padres. Quizizz. New zealand time. Kramer. Belgravia. Catherine princess of wales. Ken griffey jr. Lake george. Mgm. Dow jones industrial. Meryl streep movies. 127 hours. Hieroglyphics. Dolphins vs eagles. The independent. Sclera. Shays rebellion. Elliott gould. Fae. Owen wilson. Kamala harris vice president. Cat scratch fever. New york jets games. Jaguar. Diana taurasi. Cologne cathedral. Emulate. Osteoarthritis. Paul bettany. Newport rhode island. Why. When they see us. Craig t nelson. Convey. Occasionally. Passover. Kansas city chiefs games. Porpoise. Broadchurch. Amelia island. Sephiroth. Impressionism. When harry met sally. Sodium bicarbonate. Ofc meaning. Mark zuckerberg. The greatest showman. Kansas_city_chiefs. https://x5.yxoo.ru/
Clayton kershaw. Scorn. Coconut crab. Kiwi bird. Waterloo. James dean. South africa. Pearl harbor day. Jamie foxx movies. Benign. Steak frites. What is critical race theory. Victor wembanyama. Jordan peele. Insurrection. Ben affleck movie. Subtle. Puff daddy. Rectum. Hangover. Prague. Coercion. Empire of the sun. Colleague. Yahoo.com. Channel. Gorditas. Artificial intelligence. Brain. Equilibrium. Ii. Brooklyn botanic garden. Levy. Folklore. Bobcat. Pictured rocks. Amazon. Api meaning. Ea. My chemical romance. Lisa marie. St louis mo. Shirley chisholm. Arcade. The dark knight. Matthew mcconaughey movies and tv shows. Julianna margulies. Adam sandler movies. Zionist. Fdr. Google voice. Tennessee. Fred astaire. 2012. Sinus bradycardia. Georgia okeeffe. Xxx sex. Potassium chloride. Cuban missile crisis. James earl ray. Redondo beach. Dennis rader. Blow. Narcissism. Byte.
WilliamGuica
November 28, 2024 @ 12:22 am
Вы можете купить справку в поликлинике https://dobromedic.ru/ по Москве с доставкой до станции метро.
Make money online
November 28, 2024 @ 12:35 am
constantly i used to read smaller articles or reviews which as
well clear their motive, and that is also happening with this post which I am reading here.
Leonardsiz
November 28, 2024 @ 1:06 am
https://www.tomason-russia.ru/ – Сделайте шаг к созданию кухни вашей мечты.
Angelhob
November 28, 2024 @ 1:30 am
http://5hat.ru – Разработайте креативный визуальный контент с помощью наших профессиональных фотографов.
situs slot gacor 777
November 28, 2024 @ 1:58 am
Hello, i think that i saw you visited my web site so i came to
“return the favor”.I’m trying to find things to enhance my website!I suppose its ok to use some of your ideas!!
car battery shop near me open now
November 28, 2024 @ 1:59 am
Fine way of telling, and good paragraph to get information regarding my
presentation focus, which i am going to present in academy.
V-nomer_sxEi
November 28, 2024 @ 2:02 am
[url=https://www.inastana.kz/list/416585]виртуальный номер[/url]
Blakesal
November 28, 2024 @ 2:26 am
https://www.north-web.ru/ — Погрузитесь в мир современных и стильных кухонь с нашим сайтом.
Rybelsus mot viktminskning
November 28, 2024 @ 2:26 am
May I simply say what a comfort to find somebody that actually knows what they are
talking about online. You definitely know how to bring an issue to light and
make it important. More people should read this and understand this side of the story.
I can’t believe you’re not more popular because you certainly possess the gift.
best dog multivitamin
November 28, 2024 @ 2:37 am
Marvelous, what a web site it is! This weblog presents
helpful data to us, keep it up.
RogerUnlix
November 28, 2024 @ 2:41 am
Наша доска объявлений о работе — это удобный и надежный сервис, который объединяет тех, кто ищет работу, и тех, кто ищет сотрудников.
Derekapaky
November 28, 2024 @ 2:44 am
https://www.tatarstan-heroes.ru – ресурс для поиска информации о современных кухнях на заказ.
Jacobgaugh
November 28, 2024 @ 2:50 am
кухни на заказ — Закажите уникальную кухню, идеально подходящую вашему интерьеру.
Panen138 link alternatif
November 28, 2024 @ 2:56 am
Thanks in support of sharing such a pleasant thought, piece of writing is
fastidious, thats why i have read it fully
slot gacor
November 28, 2024 @ 3:06 am
Right now it looks like Movable Type is the preferred blogging platform
available right now. (from what I’ve read) Is that what
you are using on your blog?
Webdesign
November 28, 2024 @ 3:37 am
I’d like to find out more? I’d care to find
out more details.
video bokep
November 28, 2024 @ 3:53 am
I every time used to read paragraph in news papers but now as I am a user
of net thus from now I am using net for articles, thanks to web.
게이 포르노
November 28, 2024 @ 4:03 am
When some one searches for his necessary thing, thus he/she needs
to be available that in detail, therefore that thing is maintained
over here.
Best company for car insurance
November 28, 2024 @ 4:12 am
Car insurance companies offer a wide range of protection possibilities to comply with various consumer requirements.
It is very important to study and contrast various car insurance companies before opting
for the most ideal one for you.
DavidHulty
November 28, 2024 @ 4:20 am
Узнавая все больше о Кипре, вы только убедитесь в привлекательности острова. Лимасол, Пафос, Ларнака, Айя-Напа и Протарас привлекают инвесторов удачным географическим положением, развитой инфраструктурой и тем, что это самые «русскоязычные» города республики Gated communities in Cyprus
비아그라 구매 사이트
November 28, 2024 @ 4:33 am
When I originally commented I clicked the “Notify me when new comments are added” checkbox and now
each time a comment is added I get four e-mails with the same
comment. Is there any way you can remove me from that service?
Thanks!
ZTSODBZ
November 28, 2024 @ 4:35 am
Johnny appleseed. Hieronymus bosch. Bowl. Meningitis. Russian ukraine. ШЄЩ„ЪЇШ±Ш§Щ…. Gru. Ron desantis. Burt lancaster. Tractor. Hyperbole. Cry. Water bug. Reptiles. Jean simmons. Blue nile. Guam. Flame. Three body problem. Miami fl. Lampoon. Route 66 map. Natural born killers. Creek. Tombstone cast. Bible. The incredible hulk. Anagrams. Lunar eclipse 2024. Water cycle. Cicero. Waterboy. Transient ischemic attack. Laurie metcalf. Oleander. Ski. Disney world parks. Tonsils. The yellow wallpaper. Chief. Ho. Governors island. Bernie madoff. Wrexham. Vanessa redgrave. Facade. Bill russell. Papyrus. Piers morgan. British flag. Laverne cox. Propane. Andy griffith. Goo. Bladerunner. Bichon frise. Theodore roosevelt. Streaming. https://x4.yxoo.ru/
Emesis. Kevin bacon. Richard gere. Montenegro. Yin and yang. America soccer. Starship. Blabbermouth. Portuguese. Carbs. Greek. Chula vista. Newport news. Referral. TГјrkiye. Krebs cycle. Vice president of the united states. Kendrick. Aphasia meaning. Florence. Gainesville. Purpose. The doors. Helen mccrory. Back muscles. Cheshire cat. Detrimental. Skin condition. Rune. Spire. George jones. Benito mussolini. Feng shui. Saul goodman. Pretty in pink cast. Goa. Destiny’s child. Zion williamson. John locke. Clive davis. How tall is donald trump. Carpenter bee. Sikhism. Chess board and. Muscular system. Olympic basketball. Pink floyd. Russell wilson stats. Scrotum. Claude. Sassy meaning.
Lazrhyu
November 28, 2024 @ 4:54 am
Где и как купить диплом о высшем образовании без лишних рисков
Dnrtrez
November 28, 2024 @ 5:01 am
Как купить аттестат 11 класса с официальным упрощенным обучением в Москве
Sazrkmd
November 28, 2024 @ 5:28 am
Реально ли приобрести диплом стоматолога? Основные этапы
KennethAddew
November 28, 2024 @ 5:53 am
Федерация – это проводник в мир покупки запрещенных товаров, можно купить гашиш, купить кокаин, купить меф, купить экстази в различных городах. Москва, Санкт-Петербург, Краснодар, Владивосток, Красноярск, Норильск, Екатеринбург, Мск, СПБ, Хабаровск, Новосибирск, Казань и еще 100+ городов.
WFMBQQF
November 28, 2024 @ 5:54 am
John dillinger. Sla. Miranda rights. Em. Brad paisley. Tonsils. Padre pio. History of thanksgiving. Irvine. Revenue. Angel. Rami malek movies and tv shows. Map usa. Anthropology. Lord’s prayer. Lead poisoning symptoms. Watchmen. Aly raisman. Ardent. Phoebe waller-bridge. Outback. Zurich. The help. Monarch. The lords prayer. How much water should i drink a day. Coretta scott king. Monsters vs aliens. Retrograde. Wolf. Uterus. Iran president. Amy adams. Will smith movies. Wilmington nc. 9/11. Breitbart. Wi state. The book of enoch. Streaming media. Las vegas weather. Parlay. Microscope. Brittany murphy. Chris pine. Cleveland indians. George w. bush. Priest. Monstera. Choir. Totoro. Black cat. Basketball game. Nick jonas. Ruby bridges. Trigger warning. Greg abbott. Julie newmar. Paula deen. https://x3.yxoo.ru/
Sp500. Postal service. George wendt. World. Ann-margret. Elephant. Dysarthria. Colgate. Physical therapy. Habaneros. Uss constitution. Bard college. Saab. Gabby douglas. Playboy. After death. Amigo. Okinawa. Mitochondria function. Jennifer lopez movies. Calamity. Gargoyle. Cd. Lineage. James arness. University. Precedent. 22 jump street. Julianna margulies. John muir. Mirror. Handmaid’s tale. Dior. Mil. Scarlet fever. Quinceanera. Hurricane ian. The drop. Garter snake. Fondue. Bird of paradise. W. Zodiac signs and dates. Cimetidine. Buddhism. Rummy. Order. Italian flag. X-men origins: wolverine. Stuart little. Erie pa. Addams family 1991. Miami heat standings. Titanoboa. Jodie foster. Courtney love. English cocker spaniel.
Trefuzk
November 28, 2024 @ 6:05 am
Можно ли быстро купить диплом старого образца и в чем подвох?
Leonardsiz
November 28, 2024 @ 6:09 am
кухни от производителя – Высокое качество и надежность от проверенного производителя кухонь.
rv rental lewisville
November 28, 2024 @ 6:15 am
Hi there! This post could not be written any better!
Reading this post reminds me of my previous room mate!
He always kept chatting about this. I will forward this write-up to him.
Pretty sure he will have a good read. Many thanks for sharing!
KYT098763
November 28, 2024 @ 6:21 am
I’m not that much of a internet reader to be honest but your
blogs really nice, keep it up! I’ll go ahead and bookmark your site to come
back later. All the best
Cazrzwt
November 28, 2024 @ 6:38 am
Как приобрести аттестат о среднем образовании в Москве и других городах
Sazrdvj
November 28, 2024 @ 6:58 am
Полезные советы по безопасной покупке диплома о высшем образовании
Uazrgex
November 28, 2024 @ 7:01 am
Приобретение школьного аттестата с официальным упрощенным обучением в Москве
binance
November 28, 2024 @ 7:08 am
Can you be more specific about the content of your article? After reading it, I still have some doubts. Hope you can help me.
Blakesal
November 28, 2024 @ 7:18 am
кухни на заказ — Разработка уникальных кухонь с учетом ваших пожеланий.
eat
November 28, 2024 @ 7:40 am
Hi! I could have sworn I’ve been to your blog before but
after browsing through a few of the articles I realized it’s new to me.
Regardless, I’m definitely pleased I came across it and
I’ll be bookmarking it and checking back often!
More info
November 28, 2024 @ 7:44 am
Greate article. Keep writing such kind of information on your page.
Im really impressed by your blog.
Hey there, You’ve performed an excellent job.
I will definitely digg it and for my part recommend to my friends.
I’m confident they’ll be benefited from this site.
Jacobgaugh
November 28, 2024 @ 7:46 am
кухни на заказ от производителя – Профессиональное изготовление кухонь на заказ с гарантией качества.
KennethAddew
November 28, 2024 @ 8:30 am
Федерация – это проводник в мир покупки запрещенных товаров, можно купить гашиш, купить кокаин, купить меф, купить экстази в различных городах. Москва, Санкт-Петербург, Краснодар, Владивосток, Красноярск, Норильск, Екатеринбург, Мск, СПБ, Хабаровск, Новосибирск, Казань и еще 100+ городов.
Sazrnun
November 28, 2024 @ 8:32 am
Как официально купить аттестат 11 класса с упрощенным обучением в Москве
carbon offset programs
November 28, 2024 @ 9:03 am
I don’t even know how I ended up here, but I thought this post was good.
I don’t know who you are but certainly you’re going to
a famous blogger if you are not already 😉 Cheers!
swimming pool builders near me
November 28, 2024 @ 9:14 am
Hello There. I found your blog using msn. That is
a really neatly written article. I’ll be sure
to bookmark it and return to read more of your useful info.
Thanks for the post. I will definitely return.
Commercial overhead door repair near me
November 28, 2024 @ 9:26 am
No matter if some one searches for his essential thing, therefore he/she needs to be available that in detail, thus that
thing is maintained over here.
DERTYUO9
November 28, 2024 @ 9:48 am
bookmarked!!, I like your web site!
AdolfoSmobe
November 28, 2024 @ 10:07 am
medicine in mexico pharmacies: MexicanPharmEasy – mexican mail order pharmacies
Melvinswaky
November 28, 2024 @ 10:36 am
Игровые автоматы позволяют насладиться невороятной атмосферой казино без какого либо депозита. Бесплатная версия позволяет пробовать себя в игре, изучить правила и особенности слотов, а также разработать стратегии без потери денег.
Выбирая игровые аппараты, вы получаете доступ к огромному выбору разнообразных слотов с различными тематиками и бонусными функциями. От классических фруктовых слотов до современных видеоигр, каждый найдет что-то по своему вкусу.
[url=https://lucky-slots.ru/]Играть бесплатно в игровые аппараты[/url] – это отличный способ провести время, развлечься и насладиться азартом, не выходя из дома. Попробуйте себя в роли удачливого игрока прямо сейчас и ощутите магию казино, не тратя даже копейки!
MatthewQuort
November 28, 2024 @ 10:47 am
Выбор отеля https://koffer-spb.ru на что обратить внимание при бронировании, как оценить отзывы и найти оптимальное соотношение цены и качества.
Leonardsiz
November 28, 2024 @ 11:06 am
https://viktorova-ts.ru – Удобный сервис для выбора и заказа кухни.
B&B Italia Stoelen
November 28, 2024 @ 12:03 pm
Please let me know if you’re looking for a article author for your site.
You have some really great articles and I think I
would be a good asset. If you ever want to take some
of the load off, I’d absolutely love to write some content for your blog in exchange for a link back to mine.
Please shoot me an e-mail if interested. Many thanks!
Blakesal
November 28, 2024 @ 12:06 pm
кухня на заказ екатеринбург недорого — Кухни на заказ в Екатеринбурге по доступным ценам.
Reloader 22
November 28, 2024 @ 12:12 pm
Tremendous issues here. I’m very satisfied to peer your post.
Thank you so much and I am taking a look forward to contact you.
Will you kindly drop me a mail?
Casino
November 28, 2024 @ 12:13 pm
Hey very interesting blog!
chinese lyrics
November 28, 2024 @ 12:29 pm
Pretty! This has been an incredibly wonderful post. Thank you for providing
these details.
Jacobgaugh
November 28, 2024 @ 12:34 pm
mtucizone.ru/ — Полный ассортимент кухонь для вашего удобства.
https://deprezyon.com/
November 28, 2024 @ 12:35 pm
Review my webpage :: situs togel online (https://deprezyon.com/)
glasgow escort
November 28, 2024 @ 1:06 pm
Hey! This post could not be written any better! Reading this post reminds
me of my good old room mate! He always kept chatting about
this. I will forward this write-up to him. Fairly
certain he will have a good read. Many thanks for sharing!
KAZUXAM
November 28, 2024 @ 1:19 pm
Beer. Conor mcgregor. Respiratory system. Content. Seraphim. Melbourne. Academy award for best picture. Broom. Acquiesce. The gilded age. Malaysia airlines. Dumb and dumber. Gena rowlands. Martin luther. Adenoids. Capuchin monkey. Touche meaning. Beirut. Fastidious. Robin givens. Paul giamatti movies and tv shows. Sniffles. Lacma. Dark matter. Yu gi oh. The museum of modern art. Hoo. Yuzu. Barnard college. Virgen de guadalupe. Hawaii fires. Iridescent. Needles. George michael. Oak tree. Lemur. Ascetic. Y. Bubble tea. Helen mccrory. Sza. Implicit. Alveoli. Viagra. Ottawa. Masochism. Ecstacy. https://forum-3.uj74.ru/
Catherine the great furniture. Wi. Cat breeds. Assassin’s creed. Durango colorado. Wild boar. Academy award for best supporting actor. Sean connery. Pepsico. Bulwark. War. Toes. Meningitis. Barramundi. Airbus a320. Dido. Parkinson’s disease. The ballad of buster scruggs. Natural disasters. How old is eminem. Dachshund. Karl lagerfeld. Vitamin e. What is ovulation. Pets. University of north texas. Courtney love. Longwood gardens. John hamm. Wayne gretzky. Ulysses. Nuanced. Djibouti. Lychee. In the heat of the night. Hare. Teresa. Charlotte hornets. Bmi chart. Smithsonian museum. Vail. Pancreatic cancer symptoms. Canceled. Cc. Millennial years. Cubs. Smoky mountains. Georgia o’keeffe. Reuters. Restroom.
Mazrjzf
November 28, 2024 @ 1:53 pm
Купить диплом Барнаул
Lindseyabope
November 28, 2024 @ 1:59 pm
Вы можете купить справку в медицинском центре https://kristall-med.ru/ в Москве без прохождения врачей.
Terryfluex
November 28, 2024 @ 2:04 pm
Игровая платформа 1хбет щедро вознаграждает новичков приветственным поощрением в разделе “Казино” в размере до 1500 долларов плюс 210 фриспинов https://stadtecken.de/content/pgs/1xbet-promo-code_197.html
vivod iz zapoya ekaterinbyrg_lpmr
November 28, 2024 @ 2:06 pm
вывод из запоя на дому [url=flanrp.rolevaya.com/viewtopic.php?id=149]вывод из запоя на дому[/url] .
kyhni na zakaz_owOn
November 28, 2024 @ 2:06 pm
кухня заказ [url=www.mebeldinastiya.ru]кухня заказ[/url] .
AndrewFup
November 28, 2024 @ 2:07 pm
Правовой сервис «ТвойЮрист» https://tvoyurist.online/services/finansovoe-pravo/bankrotstvo-fizicheskikh-lits/ предлагает бесплатные юридические консультации, а также предоставляет комплексные услуги по банкротству физических лиц в Москве и других регионах России. Опытные юристы и адвокаты помогают в процедуре признания гражданина банкротом и полном списании задолженности. Профессиональное сопровождение включает как судебные, так и внесудебные процессы банкротства под руководством арбитражного управляющего.
pencurimovie123
November 28, 2024 @ 2:08 pm
I wanted to thank you for this very good read!! I definitely
loved every bit of it. I’ve got you saved as a favorite to check out
new things you post…
Coolzino_qnEt
November 28, 2024 @ 2:10 pm
Coolzino jackpoty [url=www.coolzino-polska.com/]Coolzino jackpoty[/url] .
vivod iz zapoya ekaterinbyrg_dopl
November 28, 2024 @ 2:11 pm
вывод из запоя капельница [url=https://ideya.forums.party/viewtopic.php?id=663]вывод из запоя капельница [/url] .
vivod iz zapoya ekaterinbyrg_skMa
November 28, 2024 @ 2:12 pm
вывод из запоя цена [url=https://krut.forumno.com/viewtopic.php?id=6053/]krut.forumno.com/viewtopic.php?id=6053[/url] .
play best games online
November 28, 2024 @ 2:12 pm
I was recommended this web site by my cousin. I’m not sure whether this post is written by him as
nobody else know such detailed about my trouble. You’re amazing!
Thanks!
Arper meubels
November 28, 2024 @ 4:02 pm
I was more than happy to discover this page. I wanted to thank you
for ones time just for this fantastic read!! I definitely liked
every little bit of it and I have you bookmarked
to check out new stuff in your blog.
AdolfoSmobe
November 28, 2024 @ 4:09 pm
mail order pharmacy india: indian pharm – india pharmacy
Leonardsiz
November 28, 2024 @ 4:14 pm
кухни под заказ — Мы создаем уникальные кухни под ваши потребности.
visit here
November 28, 2024 @ 4:43 pm
It is the best time to make a few plans for the future and it’s time to be happy.
I have read this publish and if I may just I wish to suggest you
few fascinating issues or tips. Maybe you could write subsequent articles referring to this article.
I want to read more issues approximately it!
Blakesal
November 28, 2024 @ 4:59 pm
https://www.webcocktail.ru/ — Ознакомьтесь с нашими услугами на сайте и выберите лучшую кухню.
stanchion
November 28, 2024 @ 4:59 pm
Way cool! Some very valid points! I appreciate
you writing this article and the rest of the website is extremely good.
Jacobgaugh
November 28, 2024 @ 5:38 pm
Кухни на заказ — узнайте, как создать идеальный дизайн кухни для вашего дома
free adult webcams
November 28, 2024 @ 6:32 pm
I love it when folks come together and share opinions.
Great site, keep it up!
spam link
November 28, 2024 @ 6:47 pm
When someone writes an piece of writing he/she keeps the plan of a user in his/her mind
that how a user can be aware of it. Therefore that’s why this article is perfect.
Thanks!
Angelhob
November 28, 2024 @ 6:49 pm
http://bagroo.ru/ – Заказывайте фотосъемку для товаров и получите качественные изображения.
teaburn
November 28, 2024 @ 7:31 pm
Hello! This post couldn’t be written any better! Reading this post reminds
me of my good old room mate! He always kept chatting about this.
I will forward this article to him. Pretty sure he will have a good read.
Many thanks for sharing!
Artifort Collectie
November 28, 2024 @ 7:50 pm
Thanks for the marvelous posting! I really enjoyed reading it, you
may be a great author.I will be sure to bookmark your blog and
will come back from now on. I want to encourage yourself to
continue your great work, have a nice weekend!
ScottLiawl
November 28, 2024 @ 8:18 pm
male dysfunction pills: canadianpharm1st – cheap erectile dysfunction pills online
data hk 2025 paito
November 28, 2024 @ 8:40 pm
I think the admin of this site is genuinely working hard for his site, as here every data is quality based information.
Leonardsiz
November 28, 2024 @ 9:04 pm
http://www.tir92.ru – Ваш надежный партнер в выборе идеальной кухни.
Blakesal
November 28, 2024 @ 9:29 pm
кухни на заказ цены — Узнайте цены на изготовление кухонь на заказ у нас.
Derekapaky
November 28, 2024 @ 9:35 pm
кухня заказать – просто и удобно с опытными специалистами.
nkuk21.co.uk
November 28, 2024 @ 10:07 pm
my blog … situs togel online (nkuk21.co.uk)
Russ
November 28, 2024 @ 10:47 pm
Hey I know this is off topic but I was wondering if you knew
of any widgets I could add to my blog that automatically tweet my newest twitter
updates. I’ve been looking for a plug-in like this for
quite some time and was hoping maybe you would have some
experience with something like this. Please let me know if you run into anything.
I truly enjoy reading your blog and I look forward
to your new updates.
Lazrqom
November 28, 2024 @ 10:56 pm
Полезные советы по безопасной покупке диплома о высшем образовании
Leonardsiz
November 28, 2024 @ 11:27 pm
http://tomason-russia.ru/ – Ваш надежный выбор для стильной и функциональной кухни.
CarlosTib
November 28, 2024 @ 11:38 pm
п»їbest mexican online pharmacies: Mexican Pharm – mexican mail order pharmacies
link porn
November 28, 2024 @ 11:38 pm
This paragraph gives clear idea in favor of the new users of blogging, that genuinely
how to do blogging and site-building.
Sazrgkg
November 28, 2024 @ 11:40 pm
Где и как купить диплом о высшем образовании без лишних рисков
Blakesal
November 28, 2024 @ 11:47 pm
кухня на заказ екатеринбург недорого — Кухни на заказ с отличным соотношением цены и качества в Екатеринбурге.
vegan
November 29, 2024 @ 12:44 am
I think everything typed was actually very logical. However,
what about this? what if you added a little information?
I am not suggesting your content is not good., but suppose you added something
to possibly get people’s attention? I mean Filling in the gaps of HIV is a little boring.
You might glance at Yahoo’s front page and note how they create
news headlines to grab viewers to click. You might add a related video or a
picture or two to grab readers excited about everything’ve got to
say. In my opinion, it might bring your posts a little
livelier.
Xazrthb
November 29, 2024 @ 12:52 am
Стоимость дипломов высшего и среднего образования и процесс их получения
Jacobgaugh
November 29, 2024 @ 12:55 am
gft-leasing.ru — Лучшие предложения для вашей кухни мечты.
Sazrfvh
November 29, 2024 @ 1:08 am
Легальная покупка диплома ПТУ с сокращенной программой обучения
Crypto recovery agency
November 29, 2024 @ 2:23 am
Heya i’m for the primary time here. I found this board and I to find It really useful & it helped me out a lot.
I am hoping to provide one thing again and help others like you aided
me.
Josipilar
November 29, 2024 @ 2:24 am
Как вы относитесь к стейкам? Какие любите? [url=https://shashlyk-mangal.ru]стейки[/url]
Сервисный центр Apple
November 29, 2024 @ 2:54 am
Предлагаем услуги профессиональных инженеров офицальной мастерской.
Еслли вы искали сервисный центр apple в москве, можете посмотреть на сайте: официальный сервисный центр apple
Наши мастера оперативно устранят неисправности вашего устройства в сервисе или с выездом на дом!
Sazruwu
November 29, 2024 @ 3:55 am
Полезные советы по безопасной покупке диплома о высшем образовании
AdolfoSmobe
November 29, 2024 @ 4:02 am
mexican mail order pharmacies: Mexican Pharm – medication from mexico pharmacy
Dnrtyqx
November 29, 2024 @ 4:19 am
Пошаговая инструкция по официальной покупке диплома о высшем образовании
Blakesal
November 29, 2024 @ 4:23 am
заказать кухню — Сделайте заказ на кухню своей мечты уже сегодня.
Marcelo K. I. Garrett
November 29, 2024 @ 4:29 am
https://twitter.com/NeginSolan91252
security in Delphi development
November 29, 2024 @ 4:40 am
Hiya! Quick question that’s completely off topic. Do you know how to make your site mobile friendly?
My website looks weird when viewing from my iphone.
I’m trying to find a theme or plugin that might be able to fix this problem.
If you have any suggestions, please share. Thank you!
Leonardsiz
November 29, 2024 @ 4:41 am
кухни на заказ спб – Найдите идеальное решение для кухни в Санкт-Петербурге с индивидуальным подходом.
security of Delphi applications
November 29, 2024 @ 4:47 am
This paragraph is truly a good one it helps new net people, who are wishing in favor of blogging.
アダルトグッズ
November 29, 2024 @ 5:00 am
I needed to thank you for this great read!!
I definitely enjoyed every little bit of it. I have got you saved as a
favorite to check out new things you post…
location de voiture aéroport Marrakech
November 29, 2024 @ 5:08 am
This paragraph will help the internet visitors for setting
up new web site or even a weblog from start to end.
Lindseyabope
November 29, 2024 @ 5:32 am
Вы можете купить справки в Москве в нашей клинике https://medic-dpo.ru/.
Terryfluex
November 29, 2024 @ 5:34 am
Букмекерская контора 1хБет приготовила для своих клиентов разнообразную систему бонусов и акций, которая привлекает новых участников и поддерживает интерес у постоянных клиентов. Ниже рассмотрим основные бонусы и акции букмекера 1xbet http://centroculturalrecoleta.org/blog/pages/1xbet-promo-code_cameroon.html
Waynejef
November 29, 2024 @ 5:38 am
Выбор места для проживания или отдыха имеет огромное значение для качества жизни. Важно тщательно подойти к вопросу выбора недвижимости, которая будет не только комфортной, но и обеспечит высокий уровень жизни в течение долгого времени http://cda4you.com/cda_blue/index.php?option=com_k2&view=itemlist&task=user&id=7525
Jacobgaugh
November 29, 2024 @ 5:50 am
http://goldcoon.ru/ – Удобный сервис для выбора кухни вашей мечты.
ScottLiawl
November 29, 2024 @ 6:11 am
buying prescription drugs in mexico: mexican pharmacy – mexico drug stores pharmacies
safety of Delphi applications
November 29, 2024 @ 6:40 am
I have been surfing online more than 3 hours these days,
but I by no means discovered any attention-grabbing article like yours.
It’s lovely price enough for me. In my opinion, if all webmasters and bloggers made excellent content as you
did, the web will probably be a lot more helpful than ever before.
Mikomax producten
November 29, 2024 @ 7:45 am
Hi to all, how is everything, I think every one is getting more from
this site, and your views are nice for new users.
Blakesal
November 29, 2024 @ 9:09 am
http://tort-dubr.ru/ — Откройте для себя кухни на заказ с индивидуальным подходом.
https://1signature.com.my
November 29, 2024 @ 9:33 am
Its like you learn my thoughts! You seem to grasp a lot about this,
like you wrote the ebook in it or something. I feel that you simply can do with a few p.c.
to drive the message house a bit, but other than that, this
is fantastic blog. A great read. I’ll definitely be back.
Sazrrsj
November 29, 2024 @ 9:41 am
Купить диплом старого образца, можно ли это сделать по быстрой схеме?
Leonardsiz
November 29, 2024 @ 10:05 am
кухни санкт петербург – Лучшие решения для кухни от профессионалов в Санкт-Петербурге.
narkolog na dom krasnodar_vtEa
November 29, 2024 @ 10:34 am
вызов нарколога на дом круглосуточно [url=https://setter.borda.ru/?1-7-0-00000676-000-0-0-1730786540]https://setter.borda.ru/?1-7-0-00000676-000-0-0-1730786540[/url] .
narkolog na dom krasnodar_dcei
November 29, 2024 @ 10:35 am
вызов нарколога на дом [url=www.severussnape.borda.ru/?1-4-0-00000319-000-0-0-1730786111]www.severussnape.borda.ru/?1-4-0-00000319-000-0-0-1730786111[/url] .
vivod iz zapoya sochi_nvKi
November 29, 2024 @ 10:37 am
вывод от запоя в стационаре сочи [url=business.0pk.me/viewtopic.php?id=37951]business.0pk.me/viewtopic.php?id=37951[/url] .
vivod iz zapoya sochi_tpmi
November 29, 2024 @ 10:39 am
выведение из запоя на дому в сочи [url=svarog.forum24.ru/?1-0-0-00000227-000-0-0-1730787062]svarog.forum24.ru/?1-0-0-00000227-000-0-0-1730787062[/url] .
vivod iz zapoya sochi_bgSr
November 29, 2024 @ 10:39 am
вывод из запоя в стационаре [url=cah.forum24.ru/?1-19-0-00000463-000-0-0-1730786457]cah.forum24.ru/?1-19-0-00000463-000-0-0-1730786457[/url] .
Jacobgaugh
November 29, 2024 @ 10:50 am
http://mtucizone.ru — Просто, удобно и с гарантией качества.
https://diuwin.co.in/
November 29, 2024 @ 11:37 am
We are a group of volunteers and starting a new scheme in our community.
Your site provided us with helpful info to work on. You’ve done a formidable job and our whole
neighborhood might be thankful to you.
Austin massage spa
November 29, 2024 @ 12:02 pm
Hello, i think that i noticed you visited my website thus i got
here to go back the prefer?.I am trying to find issues to improve my web site!I assume its adequate to make use of a few of your ideas!!
CarlosTib
November 29, 2024 @ 1:38 pm
over the counter erectile dysfunction pills: canadian pharm – non prescription ed drugs
Blakesal
November 29, 2024 @ 2:00 pm
http://webcocktail.ru/ — Ознакомьтесь с нашими кухнями, которые идеально подойдут для вашего дома.
Sazrfjq
November 29, 2024 @ 2:12 pm
Приобретение диплома ПТУ с сокращенной программой обучения в Москве
rpinnovative.1stbb.ru/viewtopic.php?f=8&t=704
massage
November 29, 2024 @ 2:55 pm
I was curious if you ever considered changing the structure of your website?
Its very well written; I love what youve got to say.
But maybe you could a little more in the
way of content so people could connect with it better.
Youve got an awful lot of text for only having 1 or two images.
Maybe you could space it out better?
Cazraqd
November 29, 2024 @ 3:05 pm
Всё о покупке аттестата о среднем образовании: полезные советы
Lazrokg
November 29, 2024 @ 3:23 pm
Официальная покупка аттестата о среднем образовании в Москве и других городах
Leonardsiz
November 29, 2024 @ 3:26 pm
кухни от производителя — Кухни от производителя: качество и доступная цена.
surfing game
November 29, 2024 @ 3:37 pm
What’s up, I log on to your blogs on a regular basis.
Your humoristic style is awesome, keep it up!
Jacobgaugh
November 29, 2024 @ 3:52 pm
https://taxipuma.ru — сделайте заказ кухни прямо сейчас
private instagram viewer
November 29, 2024 @ 3:57 pm
There are various tools and websites that affirmation to permit users
to view private Instagram profiles, but it’s
important to door these following caution. Many of these tools can be unreliable, may require personal information, or could violate Instagram’s terms
of service. Additionally, using such tools can compromise your own security or lead to scams.
The safest and most ethical mannerism to view a private profile is to
send a follow demand directly to the user.
Always prioritize privacy and honoring in your online interactions.
Here is my web site: private instagram viewer
Cazrryj
November 29, 2024 @ 4:16 pm
Как правильно приобрести диплом колледжа или ПТУ в России, важные моменты
Angelhob
November 29, 2024 @ 4:19 pm
предметная съемка москва – Создание высококачественных изображений для продвижения товаров.
click link
November 29, 2024 @ 4:23 pm
Hey there just wanted to give you a brief heads up and let you know a few of the images aren’t
loading correctly. I’m not sure why but I think its a linking issue.
I’ve tried it in two different browsers and both show
the same results.
ScottLiawl
November 29, 2024 @ 4:29 pm
generic ed pills: canadian pharm 1st – ed cures that work
AdolfoSmobe
November 29, 2024 @ 4:30 pm
Online medicine home delivery: indian pharmacy – п»їlegitimate online pharmacies india
divulgar link de afiliado
November 29, 2024 @ 4:56 pm
I am really inspired together with your writing abilities as
well as with the layout on your blog. Is that this a paid theme or did you modify it
your self? Anyway stay up the excellent high quality writing, it’s uncommon to peer a great
weblog like this one today..
online casino
November 29, 2024 @ 5:24 pm
Excellent blog here! Also your web site loads up fast!
What web host are you using? Can I get your affiliate link to your
host? I wish my web site loaded up as quickly as yours lol
Blakesal
November 29, 2024 @ 6:40 pm
кухни на заказ екатеринбург — Изготовление кухонь на заказ в Екатеринбурге по индивидуальным проектам.
MILF webcams
November 29, 2024 @ 7:26 pm
Quality articles or reviews is the crucial to be a focus for the people to pay a quick visit the web page, that’s what this website is providing.
Guitar Music
November 29, 2024 @ 7:56 pm
Please let me know if you’re looking for a writer for your site.
You have some really good posts and I think I would be a good
asset. If you ever want to take some of the load off, I’d really like to write some articles for your blog
in exchange for a link back to mine. Please send me an e-mail if interested.
Cheers!
Jacobgaugh
November 29, 2024 @ 8:33 pm
http://armor-games.ru — Официальный сайт с информацией о кухонной мебели.
Leonardsiz
November 29, 2024 @ 8:36 pm
кухни на заказ от производителя – Высокий стандарт качества и доступные решения для вашего дома.
AdolfoSmobe
November 29, 2024 @ 10:33 pm
buy medicines online in india: IndianPharmStar.com – pharmacy website india
flooringaustintx.z20.web.core.windows.net
November 29, 2024 @ 10:55 pm
Fastidious replies in return of this query with genuine arguments
and explaining everything concerning that.
Blakesal
November 29, 2024 @ 11:13 pm
http://www.north-web.ru — Ознакомьтесь с нашими предложениями на сайте.
OHIO FAKE ID
November 29, 2024 @ 11:33 pm
Thank you for sharing your thoughts. I really appreciate your efforts and I will be waiting for your next write ups thanks once
again.
tiranga games
November 29, 2024 @ 11:41 pm
Hey this is kinda of off topic but I was wanting to know if blogs use WYSIWYG editors
or if you have to manually code with HTML. I’m starting a
blog soon but have no coding know-how so I wanted to get advice from someone with experience.
Any help would be greatly appreciated!
liposuction
November 29, 2024 @ 11:45 pm
Thank you for the auspicious writeup. It in reality was once a enjoyment account it.
Look complicated to more added agreeable from you!
However, how could we be in contact?
http://rapz.ru/user/Gilberto6804
November 30, 2024 @ 12:02 am
Here is my blog post: situs togel online
(http://rapz.ru/user/Gilberto6804)
vivod iz zapoya sochi_lyOt
November 30, 2024 @ 12:29 am
вывод из запоя анонимно [url=https://www.domsadremont.ukrbb.net/viewtopic.php?f=3&t=908]вывод из запоя анонимно[/url] .
vivod iz zapoya sochi_spsi
November 30, 2024 @ 12:31 am
стационарное выведение из запоя [url=https://vip.mybb.rocks/viewtopic.php?id=7691/]стационарное выведение из запоя[/url] .
vivod iz zapoya sochi_iuMa
November 30, 2024 @ 12:32 am
вывод от запоя в стационаре сочи [url=www.kvitka.ukrbb.net/viewtopic.php?f=58&t=28041/]вывод от запоя в стационаре сочи[/url] .
vivod iz zapoya sochi_wrEl
November 30, 2024 @ 12:34 am
вывод из запоя стационар [url=https://www.my.forum2.net/viewtopic.php?id=176]вывод из запоя стационар [/url] .
vivod iz zapoya sochi_pbOt
November 30, 2024 @ 12:34 am
вывод из запоя цены сочи [url=www.rubiz.forum.cool/viewtopic.php?id=3750]вывод из запоя цены сочи[/url] .
Major
November 30, 2024 @ 12:37 am
You have made some decent points there. I looked on the net for more information about the issue and found most individuals will go along with your views on this website.
Mazriuz
November 30, 2024 @ 12:38 am
Официальная покупка диплома ПТУ с упрощенной программой обучения
Jacobgaugh
November 30, 2024 @ 1:14 am
gft-leasing.ru/ — Индивидуальный подход к созданию вашей кухни.
Leonardsiz
November 30, 2024 @ 1:47 am
http://tomason-russia.ru/ – Ваш надежный выбор для стильной и функциональной кухни.
tiranga lottery
November 30, 2024 @ 2:35 am
Good day! I know this is kinda off topic but I was wondering if you knew where I could find a captcha plugin for
my comment form? I’m using the same blog platform as yours and I’m
having trouble finding one? Thanks a lot!
roof repair
November 30, 2024 @ 2:53 am
We are a bunch of volunteers and opening a new scheme in our community.
Your website offered us with valuable info to work on. You’ve done a formidable
task and our whole community shall be thankful to you.
Dnrtdba
November 30, 2024 @ 3:24 am
Приобретение диплома ПТУ с сокращенной программой обучения в Москве
Eanrfzk
November 30, 2024 @ 3:39 am
Всё, что нужно знать о покупке аттестата о среднем образовании без рисков
Blakesal
November 30, 2024 @ 3:54 am
https://www.shth.ru/ — Лучшие предложения для вашего дома на нашем сайте.
alexistogel login
November 30, 2024 @ 4:25 am
alexistogel adalah platform judi online terpercaya yang menyediakan berbagai permainan toto seperti Togel Singapore, Togel Hongkong, Togel Sydney, dan banyak lagi. Dengan layanan pelanggan profesional, sistem keamanan unggul, dan proses deposit serta withdraw yang cepat, situs toto hadir untuk memberikan pengalaman bermain yang aman, nyaman, dan memuaskan bagi para penggemar toto di Indonesia.
Temukan solusi terbaik hanya di – Cepat, Mudah, dan Terpercaya! toto online
Hervé Senni Music
November 30, 2024 @ 4:41 am
Everyone loves what you guys tend to be up too. This kind of clever
work and coverage! Keep up the awesome works guys I’ve added you
guys to my own blogroll.
AdolfoSmobe
November 30, 2024 @ 4:42 am
india pharmacy mail order: IndianPharmStar.com – cheapest online pharmacy india
YT456RT89
November 30, 2024 @ 4:45 am
It’s an awesome piece of writing in favor of all the online visitors; they will take benefit from it I am sure.
HKYPOTRD
November 30, 2024 @ 4:52 am
Great weblog here! Additionally your site lots up fast!
What host are you using? Can I am getting your
affiliate hyperlink to your host? I desire my web site loaded up as fast as
yours lol
manilabet365
November 30, 2024 @ 5:07 am
Excellent goods from you, man. I have understand your stuff previous to and you are just extremely magnificent.
I actually like what you’ve acquired here, certainly like
what you’re stating and the way in which you say it. You make it enjoyable and you still take care of to keep it sensible.
I cant wait to read much more from you. This is actually a wonderful site.
https://womantowoman.tv
November 30, 2024 @ 5:14 am
I think that is among the such a lot important information for me.
And i’m satisfied studying your article. However wanna observation on few general things, The site style is perfect, the articles is in point of
fact great : D. Excellent task, cheers
alternatif velbett
November 30, 2024 @ 5:39 am
My brother suggested I might like this website.
He was totally right. This post actually made my
day. You can not imagine just how much time I had
spent for this info! Thanks!
Jacobgaugh
November 30, 2024 @ 5:55 am
кухни на заказ от производителя – Профессиональное изготовление кухонь на заказ с гарантией качества.
freespins
November 30, 2024 @ 6:02 am
I’m really inspired along with your writing abilities and also with the format to your weblog.
Is that this a paid subject matter or did you customize it your self?
Anyway stay up the excellent quality writing, it is rare to peer a nice blog
like this one today..
alexistogel login
November 30, 2024 @ 6:03 am
Alexistogel adalah salah satu situs terpercaya yang menyediakan berbagai pilihan permainan judi toto online terbaik, seperti Togel Singapore, Togel Hongkong, Togel Sydney, dan masih banyak lagi. Dengan layanan yang profesional dan sistem keamanan tingkat tinggi, situs ini dirancang untuk memberikan pengalaman bermain yang aman, nyaman, dan menyenangkan bagi para penggemar judi toto di Indonesia.
Penawaran Eksklusif situs toto online
FrankNidly
November 30, 2024 @ 6:18 am
overcoming ed https://indianpharmstar.com/# indian pharmacies safe
Dereklob
November 30, 2024 @ 6:23 am
Конструкторы оружия России https://guns.org.ru история создания легендарного оружия, биографии инженеров, технические характеристики разработок.
leo999
November 30, 2024 @ 6:24 am
My spouse and I absolutely love your blog and find a
lot of your post’s to be just what I’m looking for.
Would you offer guest writers to write content for you?
I wouldn’t mind composing a post or elaborating
on many of the subjects you write about here. Again, awesome blog!
Lazrjeb
November 30, 2024 @ 6:27 am
Как правильно купить диплом колледжа и пту в России, подводные камни
honda monkey
November 30, 2024 @ 6:42 am
Great delivery. Solid arguments. Keep up the amazing spirit.
Leonardsiz
November 30, 2024 @ 6:58 am
кухни санкт петербург – Стильные кухонные гарнитуры, выполненные профессионалами в Петербурге.
winner55
November 30, 2024 @ 7:04 am
Thanks for sharing your thoughts on winner55. Regards
alexistogel
November 30, 2024 @ 8:11 am
Alexistogel adalah salah satu situs terpercaya yang menyediakan berbagai pilihan permainan judi toto online terbaik, seperti Togel Singapore, Togel Hongkong, Togel Sydney, dan masih banyak lagi. Dengan layanan yang profesional dan sistem keamanan tingkat tinggi, situs ini dirancang untuk memberikan pengalaman bermain yang aman, nyaman, dan menyenangkan bagi para penggemar judi toto di Indonesia.
Belanja aman dan nyaman di alexistogel
카지노사이트
November 30, 2024 @ 8:19 am
Great beat ! I wish to apprentice while you amend your website, how can i subscribe for a blog site?
The account helped me a acceptable deal. I had been tiny bit acquainted of this your broadcast provided bright clear concept
Blakesal
November 30, 2024 @ 8:36 am
tort-dubr.ru — Узнайте больше о наших кухнях и услугах на сайте.
alexistogel login
November 30, 2024 @ 8:53 am
Situs toto online adalah platform terpercaya yang menghadirkan beragam permainan judi toto online, termasuk Togel Singapore, Togel Hongkong, Togel Sydney, dan lainnya. Dengan layanan 24/7, proses transaksi cepat, serta keamanan data terjamin, toto online berkomitmen memberikan pengalaman bermain yang seru, aman, dan memuaskan bagi para bettor di seluruh Indonesia.
Belanja aman dan nyaman di situs toto
koi toto
November 30, 2024 @ 9:11 am
I’ve been surfing on-line more than three hours nowadays,
yet I by no means found any interesting article like
yours. It’s pretty value sufficient for me. In my opinion, if all
website owners and bloggers made excellent content material as you probably did, the web shall be a lot more useful than ever before.
scratch off lottery
November 30, 2024 @ 9:52 am
Its like you read my mind! You appear to know a lot about this,
like you wrote the book in it or something. I think that you could do with some pics to drive the message home a bit, but other
than that, this is great blog. A fantastic read.
I will definitely be back.
Сервисный центр Asus
November 30, 2024 @ 10:04 am
Предлагаем услуги профессиональных инженеров офицальной мастерской.
Еслли вы искали сервисный центр asus цены, можете посмотреть на сайте: сервисный центр asus
Наши мастера оперативно устранят неисправности вашего устройства в сервисе или с выездом на дом!
JesusCug
November 30, 2024 @ 10:11 am
Вы можете купить медсправку в нашем мед центре https://medic-dpo.ru/.
EdgarBloth
November 30, 2024 @ 10:20 am
Купить круглую стальную трубу оптом и в розницу. Круглые металлические и оцинкованные трубы по низкой цене. Прайс на металлопрокат купить балку
Bullion trading
November 30, 2024 @ 10:20 am
hello there aand thank you for your information ?
I have certainly picked up something new from right here.
I did however expertise a few technical point using
this site, as I experienced to reload the website a lot of times previous to I could get it to lkad correctly.
I had been wonderig if your web host is OK? Not that I’m
complaining, but slow loading instances times will
sometimes affect your placement in google and can damage yur quality
score if advertising and marketing with Adwords. Anyway I?m adding this RSS to my e-mail and
could look out ffor muhch more of your respective fascinating content.
Make sure you updare this again soon.. https://goldbullion.z20.web.core.windows.net/Gold-coins/How-to-Buy-Silver-Online.html
Juliocuh
November 30, 2024 @ 10:25 am
У нас собственное производство металлических профилей для кровли, фасадов, заборов, поэтому цена на порядок ниже, чем у конкурентов профильная труба 20х40
Jacobgaugh
November 30, 2024 @ 10:39 am
https://www.mtucizone.ru — Официальный сайт: все о наших кухнях и услугах.
AdBIQ PPC Management Agency
November 30, 2024 @ 11:16 am
Hi there just wanted to give you a quick heads up.
The text in your article seem to be running off the screen in Safari.
I’m not sure if this is a formatting issue or something to do with browser compatibility
but I thought I’d post to let you know. The design look great though!
Hope you get the issue resolved soon. Cheers
tiranga games
November 30, 2024 @ 11:25 am
Thanks for sharing your thoughts about tiranga games.
Regards
kolekcjonerskie prawo jazdy
November 30, 2024 @ 11:25 am
At this moment I am ready to do my breakfast, once having my breakfast coming over again to read additional
news.
Leonardsiz
November 30, 2024 @ 12:16 pm
кухни на заказ – Уникальные проекты кухонь, разработанные специально для вас.
Hydro Kinetic
November 30, 2024 @ 12:22 pm
After going over a handful of the blog articles on your web
site, I seriously like your way of writing a blog.
I bookmarked it to my bookmark site list and will
be checking back in the near future. Take a look
at my website as well and tell me how you feel.
ScottLiawl
November 30, 2024 @ 12:41 pm
erectile dysfunction medications: canadianpharm1st.com – ed tablets
Typewriter Repair near me
November 30, 2024 @ 12:53 pm
I know this site offers quality depending articles or reviews
and other data, is there any other web site which gives these kinds
of information in quality?
bokep indo
November 30, 2024 @ 12:55 pm
It’s a pity you don’t have a donate button! I’d definitely
donate to this excellent blog! I suppose for now i’ll settle for book-marking and adding your RSS feed to my Google account.
I look forward to brand new updates and will talk about this blog with my Facebook
group. Chat soon!
prodamus promokod_iemt
November 30, 2024 @ 1:29 pm
промокод продамус [url=http://www.promokod-pro.ru]промокод продамус[/url] .
Blakesal
November 30, 2024 @ 1:31 pm
кухни под заказ — Получите кухню под заказ, выполненную по всем вашим требованиям.
nuddies
November 30, 2024 @ 1:55 pm
I must thank you for the efforts you have put in writing
this website. I’m hoping to see the same high-grade content from you
later on as well. In truth, your creative writing abilities has motivated me to get my very own blog now 😉
Stevenamede
November 30, 2024 @ 2:51 pm
https://medic-dpo.ru/
iwin
November 30, 2024 @ 2:52 pm
For newest news you have to pay a quick visit internet and on web
I found this site as a best website for hottest updates.
FrankNidly
November 30, 2024 @ 3:10 pm
ed treatment natural https://indianpharmstar.com/# indian pharmacy online
Cazrznc
November 30, 2024 @ 3:44 pm
Можно ли купить аттестат о среднем образовании? Основные рекомендации
Jacobgaugh
November 30, 2024 @ 3:49 pm
http://www.taxipuma.ru — удобный интерфейс и полный каталог кухонь
microvip88
November 30, 2024 @ 3:53 pm
I loved as much as you’ll receive carried out right here.
The sketch is attractive, your authored subject matter stylish.
nonetheless, you command get bought an edginess over that you wish be
delivering the following. unwell unquestionably come further
formerly again as exactly the same nearly a lot
often inside case you shield this increase.
Derekapaky
November 30, 2024 @ 4:52 pm
кухни под заказ спб позволяют учитывать все особенности вашего помещения.
avcilar escort
November 30, 2024 @ 4:59 pm
Avcılar escort hizmetleri, birçok kişinin rahatlamak ve yeni deneyimler yaşamak için tercih ettiği bir seçenek haline geliyor. Pelin, bu alanda dikkat çeken biri.
AdolfoSmobe
November 30, 2024 @ 5:06 pm
reputable mexican pharmacies online: mexican pharmacy – buying from online mexican pharmacy
vps windows server
November 30, 2024 @ 5:18 pm
Thank you for the auspicious writeup. It if truth be told used to be a enjoyment account it.
Glance advanced to far delivered agreeable from you! By the way, how could we be in contact?
Leonardsiz
November 30, 2024 @ 5:40 pm
кухни на заказ — Индивидуальное изготовление кухонь для вашего интерьера.
Sazrmqn
November 30, 2024 @ 6:18 pm
Всё, что нужно знать о покупке аттестата о среднем образовании без рисков
vivod iz zapoya sochi_dupa
November 30, 2024 @ 6:29 pm
вывод из запоя в стационаре анонимно [url=https://pokupki.bestforums.org/viewtopic.php?f=7&t=24072]вывод из запоя в стационаре анонимно[/url] .
235YT45TL
November 30, 2024 @ 6:30 pm
Hello there! I could have sworn I’ve been to this blog before but after browsing through some of the post I realized it’s new to me.
Anyhow, I’m definitely glad I found it and I’ll be book-marking and checking back frequently!
Blakesal
November 30, 2024 @ 6:31 pm
http://mptextile.ru — Полный цикл услуг по изготовлению кухонь.
EXPORTAR MOTOS
November 30, 2024 @ 6:39 pm
I’ve learn some good stuff here. Certainly value bookmarking for
revisiting. I wonder how much effort you place to create any such magnificent informative web site.
vivod iz zapoya sochi_tuSt
November 30, 2024 @ 6:42 pm
вывод из запоя в наркологическом стационаре [url=https://www.tatuheart.ukrbb.net/viewtopic.php?f=8&t=15079]вывод из запоя в наркологическом стационаре[/url] .
vivod iz zapoya sochi_btEn
November 30, 2024 @ 7:12 pm
вывод из запоя цена [url=http://www.rolandus.org/forum/viewtopic.php?p=106458]вывод из запоя цена[/url] .
vivod iz zapoya sochi_mxPa
November 30, 2024 @ 7:14 pm
капельница от запоя в сочи [url=http://www.justforum.bestforums.org/viewtopic.php?f=26&t=4760]капельница от запоя в сочи[/url] .
vivod iz zapoya sochi_zmka
November 30, 2024 @ 7:14 pm
вывод из запоя в стационаре анонимно [url=www.eisberg.forum24.ru/?1-1-0-00002017-000-0-0-1730805988/]вывод из запоя в стационаре анонимно[/url] .
Jacobgaugh
November 30, 2024 @ 8:36 pm
http://armor-games.ru — Официальный сайт с информацией о кухонной мебели.
how can you view private instagram accounts
November 30, 2024 @ 8:52 pm
There are various tools and websites that affirmation to permit users to view private Instagram profiles, but it’s important to edit these subsequent to caution. Many of these tools
how can you view private instagram accounts
be unreliable, may require personal information, or could violate Instagram’s terms of service.
Additionally, using such tools can compromise your
own security or guide to scams. The safest and most ethical showing off to view a private profile is to
send a follow demand directly to the user.
Always prioritize privacy and worship in your online interactions.
StardaZercalo
November 30, 2024 @ 8:55 pm
[url=https://starda-casino-apk.ru/starda-oficzialnyj-sajt]https://starda-casino-apk.ru/starda-oficzialnyj-sajt[/url]
Скачать последнюю версию приложения онлайн казино Старда – выигрывай прямо сейчас!
https://www.starda-casino-apk.ru/starda-oficzialnyj-sajt
Visit here
November 30, 2024 @ 9:00 pm
Wonderful post but I was wanting to know if you could write a litte more on this topic?
I’d be very grateful if you could elaborate a little bit more.
Thanks!
Lazrtns
November 30, 2024 @ 10:03 pm
Диплом вуза купить официально с упрощенным обучением в Москве
DW77 LOGIN
November 30, 2024 @ 10:20 pm
What’s up, yup this article is genuinely nice and I have learned lot of things from it on the topic of blogging.
thanks.
freespins
November 30, 2024 @ 10:26 pm
Hey! Do you use Twitter? I’d like to follow you if that would be okay.
I’m undoubtedly enjoying your blog and look forward
to new posts.
Leonardsiz
November 30, 2024 @ 10:30 pm
кухни на заказ от производителя – Высокий стандарт качества и доступные решения для вашего дома.
Trefsas
November 30, 2024 @ 10:35 pm
Официальная покупка диплома вуза с сокращенной программой в Москве
Blakesal
November 30, 2024 @ 11:15 pm
кухни под заказ екатеринбург — Идеальные кухни под заказ с доставкой по Екатеринбургу.
zespól gejów seo
December 1, 2024 @ 12:07 am
Hi, I read your new stuff like every week. Your story-telling style is
awesome, keep up the good work!
carbon offset projects
December 1, 2024 @ 12:09 am
What’s up i am kavin, its my first occasion to commenting anyplace, when i read this
paragraph i thought i could also create comment due to this good paragraph.
Jacobgaugh
December 1, 2024 @ 1:08 am
http://www.gft-leasing.ru — Узнайте больше о наших услугах на официальном сайте.
AndreaBex
December 1, 2024 @ 1:33 am
Вы можете купить справку в медцентре https://munclinic.ru/ с доставкой по Москве.
Jamesabeda
December 1, 2024 @ 1:37 am
Отель Мишель предлагает комфортабельные номера с кондиционером в Анапе. Бассейны, игровая зона для детей, конференц-зал для деловых встреч и мероприятий. Ждем Вас в гости круглый год https://mishelhotel.ru
WoodrowFodia
December 1, 2024 @ 1:40 am
https://snaga.ru/
Angelhob
December 1, 2024 @ 1:49 am
http://5hat.ru/ – Обратитесь к нам для получения качественных изображений для вашего бренда.
bitcoin cash MLM business plan
December 1, 2024 @ 2:03 am
Howdy! This blog post couldn’t be written any
better! Looking at this post reminds me of my previous roommate!
He continually kept preaching about this. I am going to forward this article
to him. Pretty sure he’s going to have a very good read.
Thanks for sharing!
betflik789
December 1, 2024 @ 2:42 am
What’s Happening i am new to this, I stumbled upon this I
have discovered It positively helpful and it has helped me out loads.
I’m hoping to give a contribution & aid different users like
its aided me. Great job.
Leonardsiz
December 1, 2024 @ 3:21 am
кухни на заказ от производителя – Эксклюзивные проекты кухонь по доступным ценам.
من از این سایت معتبر استفاده میکنم
December 1, 2024 @ 3:52 am
Howdy! I could have sworn I’ve visited your blog before but after
looking at many of the articles I realized it’s new to me.
Nonetheless, I’m definitely pleased I stumbled upon it and I’ll be book-marking it
and checking back regularly!
escort in cardiff
December 1, 2024 @ 4:05 am
Hey would you mind stating which blog platform you’re using?
I’m looking to start my own blog soon but I’m having a difficult time deciding between BlogEngine/Wordpress/B2evolution and
Drupal. The reason I ask is because your design seems
different then most blogs and I’m looking for
something unique. P.S Sorry for getting off-topic but I had to ask!
Blakesal
December 1, 2024 @ 4:11 am
кухня на заказ екатеринбург недорого — Кухни высокого качества по доступным ценам в Екатеринбурге.
Taylortwize
December 1, 2024 @ 4:16 am
Более 20 лет опыта переводов c немецкого и английский медицинской документации: эпикриз, исследование МРТ, КТ, УЗИ, протокол операции, лабораторные анализы, гистология, молекулярная патология – по всем разделам и специализациям медицины – онкология, урология, гинекология, ортопедия, хирургия и др. Квалифицированный медицинский перевод документов на немецкий язык для лечения за границей и с немецкого после лечения https://medicaltranslate.ru
Lazrapl
December 1, 2024 @ 5:16 am
Легальные способы покупки диплома о среднем полном образовании
Kim
December 1, 2024 @ 5:20 am
I’m not sure where you’re getting your info, but good topic.
I needs to spend some time learning much more or understanding more.
Thanks for magnificent info I was looking
for this information for my mission.
Jacobgaugh
December 1, 2024 @ 5:46 am
http://goldcoon.ru/ – Удобный сервис для выбора кухни вашей мечты.
Thomassof
December 1, 2024 @ 6:06 am
Виртуальные номера, телефонные номера, которые напрямую не связаны с телефонной линией или физической SIM-картой, становятся все более популярными среди людей во всем мире. Одной из областей, где их использование приобрело значительный импульс, является приложение для обмена сообщениями, такое как Telegram номера для телеграмма
Zacharywoult
December 1, 2024 @ 6:25 am
Построенные дома по индивидуальным проектам – это результат работы наших архитекторов, инженеров – строителей, смежных специалистов и, конечно, наших Заказчиков. Мы всегда стараемся сделать наши дома функциональными, простыми и удобными, чтобы во время эксплуатации наши заказчики вспоминали нас добрым словом https://www.goldtrezzini.ru/nominees/modern-house/
Harryslits
December 1, 2024 @ 6:59 am
prescription medication neurontin: neurontin 100mg cost – Gabapentin Pharm
Social - paibe
December 1, 2024 @ 8:18 am
Why Online Casinos Have Become a Global Phenomenon
Internet-based gambling hubs have revolutionized the gaming scene, delivering a level of convenience and range that brick-and-mortar casinos fall short of. Recently, countless gamblers across the globe have chosen the excitement of virtual gambling thanks to its always-open nature, engaging traits, and constantly growing range of offerings.
One of the key draws of virtual gambling hubs is the astounding diversity of choices ready to play. Whether you are a fan of spinning old-school slot machines, diving into narrative-rich video-based games, or mastering skills in traditional table offerings like Roulette, digital casinos provide infinite entertainment avenues. Many casinos also feature real-time gaming experiences, enabling you to communicate with actual dealers and opponents, all while taking in the engaging vibes of a land-based casino in your own space.
If you’re just starting with the world of virtual gambling or hope to explore safe services, why not participate in our active interactive platform? It’s a hub where gamblers exchange tips, enabling you to enhance your casino activities. Explore the connections and check it out now: https://x.com/upX_Top .
Beyond variety, internet-based gambling hubs stand out constant connectivity.
Leonardsiz
December 1, 2024 @ 8:29 am
кухни от производителя – Высокое качество и надежность от проверенного производителя кухонь.
Derekapaky
December 1, 2024 @ 8:34 am
http://www.tatarstan-heroes.ru – удобный сервис для выбора и заказа кухонь.
bounty game login
December 1, 2024 @ 8:48 am
Thanks on your marvelous posting! I truly enjoyed reading it, you happen to
be a great author. I will ensure that I bookmark
your blog and definitely will come back in the future.
I want to encourage you to continue your great work,
have a nice holiday weekend!
avrupa yakası eskort
December 1, 2024 @ 9:06 am
Avrupa Yakası Escort Avrupa Escort İstanbul Escort Bayanlar
Blakesal
December 1, 2024 @ 9:08 am
кухни екатеринбург — Идеальные кухни для вашего дома в Екатеринбурге.
Social - paibe
December 1, 2024 @ 9:25 am
Reasons Why Online Casinos Have Become an International Sensation
Virtual gambling platforms have transformed the casino gaming landscape, providing an unmatched level of comfort and variety that brick-and-mortar establishments fall short of. In recent years, a vast number of enthusiasts worldwide have adopted the adventure of digital casino play as a result of its anytime, anywhere convenience, thrilling aspects, and widening range of offerings.
One of the biggest attractions of digital gambling sites is the sheer selection of gaming experiences ready to play. Whether you love playing on vintage one-armed bandits, trying out story-driven video slots, or testing your strategy in classic casino games like Blackjack, virtual venues feature infinite entertainment avenues. Numerous services also feature live casino options, letting you to engage with human game hosts and fellow gamblers, all while taking in the realistic feel of a land-based casino in your own space.
If you’re exploring for the first time with the world of digital casinos or hope to explore reputable operators, why not participate in our active interactive platform? It’s a destination where gaming aficionados post reviews, enabling you to improve your virtual play. Check out the experience and start your journey now: https://au.pinterest.com/casibom1/ .
Adding to the extensive catalog, internet-based gambling hubs are known for availability.
카지노사이트
December 1, 2024 @ 9:46 am
Hi there to every body, it’s my first go to see of this web site; this website
contains awesome and in fact fine information in favor of visitors.
RobertMep
December 1, 2024 @ 9:56 am
https://paxlovid.ink/# Paxlovid buy online
pgg369
December 1, 2024 @ 10:02 am
It’s going to be ending of mine day, however before finish I am reading this great paragraph to improve my
know-how.
Social - paibe
December 1, 2024 @ 10:30 am
What Makes Online Casinos Are Becoming an International Sensation
Internet-based gambling hubs have modernized the gambling world, offering a level of convenience and variety that physical venues struggle to rival. Over the past decade, a growing community internationally have adopted the pleasure of virtual casinos because of its availability, appealing qualities, and continuously increasing catalogs of games.
One of the most compelling reasons of internet-based platforms is the astounding range of titles available. Whether you enjoy rolling vintage slot machines, playing through plot-filled visual slot games, or mastering skills in traditional table offerings like Blackjack, digital casinos boast numerous opportunities. Several sites additionally include live dealer games, letting you to engage with actual dealers and gaming peers, all while soaking in the lifelike atmosphere of a physical gaming house in your own space.
If you’re a beginner with the world of internet-based gaming or hope to discover reliable sites, why not sign up for our active interactive platform? It’s a platform where gamblers share experiences, assisting you to enjoy more of your virtual play. Check out the connections and start your journey now: https://www.facebook.com/profile.php?id=61569299482300&is_tour_dismissed .
Apart from the game range, internet-based gambling hubs excel accessibility.
Betovo Italia_pfSl
December 1, 2024 @ 10:38 am
Betovo bonus senza deposito [url=http://betovo-italia.com]Betovo bonus senza deposito[/url] .
Jacobgaugh
December 1, 2024 @ 10:44 am
mtucizone.ru/ — Полный ассортимент кухонь для вашего удобства.
JulioLah
December 1, 2024 @ 10:50 am
Условия использования промокода 1xBet можно узнать после его проверки. Для этого в разделе «Витрина промокодов» находится специальное поле, куда следует вставить полученный набор из цифр и букв для получения подробных сведений. Чтобы проверить комбинацию, следует авторизоваться в личном кабинете https://dpstudio-fashion.com/vendor/pgs/1xbet_promo_code_sports_2.html
Arnoldendot
December 1, 2024 @ 11:07 am
Вы можете купить медсправку в нашем мед центре https://doktora-gerasenko-med.ru/ в Москве без медкомиссий.
kazino onlain_ocKn
December 1, 2024 @ 11:08 am
казино [url=http://online-kazino.md/]казино[/url] .
kazino onlain_nkOi
December 1, 2024 @ 11:09 am
казино онлайн беларусь [url=https://t.me/s/casinobelorus/]казино онлайн беларусь[/url] .
vivod iz zapoya sochi_fypn
December 1, 2024 @ 11:10 am
вывод из запоя в наркологическом стационаре [url=https://skoleoz.borda.ru/?1-8-0-00001108-000-0-0-1730806854/]вывод из запоя в наркологическом стационаре[/url] .
Social - paibe
December 1, 2024 @ 11:34 am
Reasons Why Online Casinos Are Becoming Highly Preferred Worldwide
Internet-based gambling hubs have modernized the betting market, delivering a level of accessibility and breadth that brick-and-mortar gambling houses can’t match. Throughout the last ten years, countless gamblers globally have chosen the adventure of digital casino play as a result of its accessibility, thrilling aspects, and continuously increasing range of offerings.
One of the main appeals of online casinos is the unparalleled variety of games at your disposal. Whether you love engaging with retro one-armed bandits, trying out engaging visual slot games, or playing smart in card and board games like Roulette, online platforms feature endless opportunities. Plenty of operators additionally offer live gaming streams, giving you the chance you to interact with live hosts and co-players, all while immersing yourself in the realistic feel of a real casino from the comfort of your home.
If you’re new with the world of internet-based gaming or hope to find out more about safe services, why not join our dynamic social network? It’s a hub where gaming aficionados post experiences, guiding you to enjoy more of your virtual play. Join the connections and start your journey now: https://t.me/casibomgiris_net .
Apart from the game range, virtual gambling platforms excel seamless entry.
togel4d
December 1, 2024 @ 12:31 pm
Sweet blog! I found it while browsing on Yahoo News.
Do you have any tips on how to get listed in Yahoo News?
I’ve been trying for a while but I never seem to get there!
Cheers
Social - paibe
December 1, 2024 @ 12:38 pm
The Reasons Behind Why Online Casinos Are a Worldwide Trend
Virtual gambling platforms have modernized the betting scene, offering an unmatched level of comfort and selection that conventional gambling houses don’t provide. Over the past decade, countless gamblers worldwide have welcomed the adventure of virtual casinos because of its ease of access, thrilling aspects, and constantly growing selection of games.
One of the strongest selling points of online gaming options is the vast range of games provided. Whether you are a fan of engaging with vintage slot machines, immersing yourself in plot-filled thematic slots, or strategizing in table games like poker, casino websites feature infinite entertainment avenues. Many casinos additionally introduce live gaming streams, allowing you to interact with actual dealers and co-players, all while immersing yourself in the authentic vibes of a real casino right at home.
If you’re unfamiliar with the world of virtual gambling or would like to explore safe services, why not join our vibrant online hub? It’s a place where gaming aficionados share insights, helping you to maximize your gaming journey. Check out the experience and see it here now: https://www.facebook.com/profile.php?id=61568481962080 .
In addition to diversity, virtual gambling platforms are known for seamless entry.
bmw g77
December 1, 2024 @ 12:52 pm
Thanks very interesting blog!
Clintonattix
December 1, 2024 @ 1:06 pm
https://ivermectinpharm.store/# Ivermectin Pharm
Paxlovid.ink [url=https://paxlovid.ink/#]paxlovid for sale[/url] paxlovid pill
nuddies
December 1, 2024 @ 1:24 pm
Hello, I log on to your blog daily. Your story-telling style is awesome,
keep up the good work!
Harryslits
December 1, 2024 @ 1:31 pm
Paxlovid.ink: paxlovid covid – paxlovid price
AKLUTYR14
December 1, 2024 @ 1:34 pm
It’s very simple to find out any topic on web as compared
to textbooks, as I found this post at this web site.
Leonardsiz
December 1, 2024 @ 1:42 pm
http://www.viktorova-ts.ru – Разнообразие кухонных решений на любой вкус.
Social - paibe
December 1, 2024 @ 1:44 pm
Why Online Casinos Are Becoming Highly Preferred Worldwide
Digital casinos have modernized the gaming scene, delivering an unmatched level of accessibility and range that land-based casinos are unable to replicate. Recently, millions of players globally have chosen the excitement of digital casino play as a result of its always-open nature, engaging traits, and constantly growing range of offerings.
One of the main appeals of internet-based platforms is the sheer array of titles provided. Whether you are a fan of spinning classic slot machines, exploring theme-based thematic slots, or strategizing in classic casino games like Blackjack, online platforms deliver endless entertainment avenues. Several sites also present interactive dealer games, enabling you to communicate with actual dealers and gaming peers, all while experiencing the lifelike atmosphere of a land-based casino in your own space.
If you’re unfamiliar with the world of digital casinos or are looking to delve deeper into reliable sites, why not participate in our active community? It’s a hub where gamblers post experiences, making it easier for you to maximize your online casino experience. Discover the experience and see it here now: https://www.instagram.com/sweet_bonanzacom/ .
Apart from the game range, digital casino services excel accessibility.
escorts luton
December 1, 2024 @ 1:44 pm
Hi to all, the contents present at this web page are truly
awesome for people experience, well, keep up the
nice work fellows.
Blakesal
December 1, 2024 @ 1:46 pm
webcocktail.ru — Все, что вам нужно для кухни вашей мечты.
occult deliverance
December 1, 2024 @ 2:03 pm
Superb blog! Do you have any hints for aspiring writers?
I’m hoping to start my own website soon but I’m a little lost
on everything. Would you recommend starting with a free platform
like WordPress or go for a paid option? There are so many
options out there that I’m completely overwhelmed .. Any recommendations?
Cheers!
tangguifang.dreamhosters.com
December 1, 2024 @ 2:39 pm
Thank you. Valuable information.
Feel free to visit my web page :: スーパーコピー 激安 6畳 (https://tangguifang.dreamhosters.com/comment/html/?638341.html)
JamesFet
December 1, 2024 @ 2:44 pm
Виртуальные номера телефона стали популярным решением для многих пользователей, особенно в связи с развитием мессенджеров и социальных сетей. В контексте Телеграма, виртуальные номера телефона представляют собой уникальное и удобное решение для обеспечения безопасности и конфиденциальности коммуникации купить анонимный номер телеграм
Social - paibe
December 1, 2024 @ 2:51 pm
The Reasons Behind Why Online Casinos Remain So Popular
Internet-based gambling hubs have transformed the casino gaming world, offering an exceptional degree of comfort and breadth that brick-and-mortar venues fall short of. Over the past decade, a vast number of enthusiasts worldwide have embraced the fun of internet-based gaming as a result of its availability, exciting features, and ever-expanding collections of titles.
One of the key draws of online gaming options is the astounding selection of entertainment options ready to play. Whether you like playing on old-school slots, immersing yourself in engaging video slots, or strategizing in table games like poker, digital casinos offer limitless possibilities. Several sites even introduce live dealer games, letting you to interact with real dealers and co-players, all while experiencing the engaging vibes of a land-based casino right at home.
If you’re just starting with the world of virtual casino play or would like to delve deeper into trusted platforms, why not engage with our active interactive platform? It’s a hub where fans offer reviews, assisting you to improve your virtual play. Join the conversation and check it out now: https://www.facebook.com/profile.php?id=61569299482300&is_tour_dismissed .
Adding to the extensive catalog, online casinos stand out accessibility.
Jacobgaugh
December 1, 2024 @ 3:34 pm
https://www.taxipuma.ru/ — ваш надежный партнер в создании идеальной кухни
YT45TP87P
December 1, 2024 @ 3:36 pm
Hi! I could have sworn I’ve been to your blog before but after browsing through some of
the posts I realized it’s new to me. Regardless, I’m certainly
pleased I found it and I’ll be bookmarking it and checking back
frequently!
Social - paibe
December 1, 2024 @ 3:57 pm
What Makes Online Casinos Remain So Popular
Online casinos have modernized the gambling scene, providing an unmatched level of convenience and diversity that land-based casinos can’t match. Over the past decade, a large audience worldwide have turned to the pleasure of online gaming because of its always-open nature, thrilling aspects, and ever-expanding catalogs of games.
One of the key draws of internet-based platforms is the unparalleled diversity of entertainment options provided. Whether you like rolling classic reel games, exploring narrative-rich video-based games, or exercising tactics in strategy-based games like poker, casino websites offer endless choices. Numerous services furthermore introduce live dealer games, giving you the chance you to communicate with professional croupiers and co-players, all while enjoying the engaging environment of a real casino right at home.
If you’re new with the world of digital casinos or seek to explore trusted platforms, why not participate in our vibrant social network? It’s a place where players post experiences, assisting you to enhance your online casino experience. Join the community and visit us now: https://www.facebook.com/profile.php?id=61568958127704 .
In addition to diversity, online casinos thrive in ease of access.
Sazrjui
December 1, 2024 @ 4:33 pm
Процесс получения диплома стоматолога: реально ли это сделать быстро?
Cryptocurrency recovery expert
December 1, 2024 @ 4:50 pm
Hi, i think that i noticed you visited my blog thus i came to return the want?.I’m trying to find issues to enhance my site!I guess
its good enough to use a few of your concepts!!
Social - paibe
December 1, 2024 @ 5:03 pm
Reasons Why Online Casinos Are an International Sensation
Internet-based gambling hubs have reshaped the betting industry, delivering a unique kind of accessibility and variety that land-based casinos fall short of. Over time, a growing community worldwide have adopted the adventure of virtual gambling in light of its always-open nature, engaging traits, and ever-expanding catalogs of games.
One of the key draws of digital gambling sites is the vast variety of choices provided. Whether you like playing on traditional slots, diving into story-driven video slots, or exercising tactics in card and board games like Texas Hold’em, online platforms deliver endless opportunities. Several sites also feature live casino options, letting you to communicate with live hosts and opponents, all while soaking in the engaging feel of a physical gaming house from anywhere you want.
If you’re new with the world of online gaming or would like to learn about reliable sites, why not sign up for our growing online hub? It’s a hub where gamblers share reviews, helping you to maximize your virtual play. Dive into the community and start your journey now: https://www.instagram.com/1winbr_com/ .
In addition to diversity, virtual gaming providers shine accessibility.
freespins
December 1, 2024 @ 5:14 pm
Ahaa, its good conversation concerning this piece of writing at this place at this website, I have read all that, so
at this time me also commenting at this place.
Sazrnyz
December 1, 2024 @ 5:36 pm
Официальная покупка диплома вуза с сокращенной программой в Москве
Mazrtiv
December 1, 2024 @ 5:37 pm
Купить диплом Барнаул
Dnrtjnv
December 1, 2024 @ 5:46 pm
Быстрое обучение и получение диплома магистра – возможно ли это?
Blakesal
December 1, 2024 @ 5:57 pm
https://mptextile.ru/ — Все, что нужно для вашей кухни, собрано здесь.
Social - paibe
December 1, 2024 @ 6:10 pm
How Online Casinos Are a Worldwide Trend
Digital casinos have reshaped the casino gaming world, offering an unmatched level of convenience and variety that brick-and-mortar establishments don’t provide. Throughout the last ten years, countless gamblers across the globe have adopted the excitement of virtual casinos because of its always-open nature, engaging traits, and continuously increasing game libraries.
One of the most compelling reasons of digital gambling sites is the unparalleled array of choices provided. Whether you enjoy engaging with vintage slot machines, immersing yourself in engaging thematic slots, or playing smart in strategy-based games like Blackjack, casino websites deliver countless choices. Plenty of operators also introduce live casino options, enabling you to communicate with professional croupiers and opponents, all while experiencing the realistic ambiance of a brick-and-mortar establishment from anywhere you want.
If you’re exploring for the first time with the world of virtual gambling or hope to find out more about reliable sites, why not engage with our dynamic gaming forum? It’s a place where enthusiasts offer tips, helping you to get the most out of your online casino experience. Discover the connections and start your journey now: https://www.facebook.com/profile.php?id=61568742522948 .
Besides the wide selection, digital casino services shine availability.
freespins
December 1, 2024 @ 6:41 pm
Very nice post. I just stumbled upon your blog and wanted to say that
I have truly enjoyed surfing around your blog posts.
After all I’ll be subscribing to your rss feed and I hope you write again soon!
freespins
December 1, 2024 @ 6:47 pm
Hello everyone, it’s my first pay a visit at this web site,
and paragraph is actually fruitful for me, keep up
posting these types of posts.
Leonardsiz
December 1, 2024 @ 6:52 pm
https://flowers777.ru — Дизайнерские кухни с учетом ваших пожеланий.
RobertMep
December 1, 2024 @ 6:55 pm
http://paxlovid.ink/# Paxlovid.ink
Social - paibe
December 1, 2024 @ 7:15 pm
Why Online Casinos Remain a Worldwide Trend
Digital casinos have transformed the betting industry, offering an unmatched level of ease and selection that brick-and-mortar venues struggle to rival. Over time, a growing community across the globe have chosen the fun of digital casino play in light of its accessibility, engaging traits, and continuously increasing selection of games.
One of the biggest attractions of online casinos is the incredible range of choices ready to play. Whether you like engaging with classic slot machines, immersing yourself in engaging visual slot games, or mastering skills in classic casino games like Roulette, digital casinos feature limitless opportunities. Plenty of operators furthermore offer live dealer games, allowing you to interact with professional croupiers and other players, all while experiencing the authentic vibes of a physical gaming house without leaving your home.
If you’re a beginner with the world of digital casinos or would like to delve deeper into safe services, why not engage with our vibrant gaming forum? It’s a platform where enthusiasts exchange tips, helping you to maximize your gambling adventure. Join the connections and learn more now: https://www.instagram.com/1winbr_com/ .
Adding to the extensive catalog, virtual gaming providers shine seamless entry.
Harryslits
December 1, 2024 @ 7:55 pm
paxlovid covid: Paxlovid.ink – paxlovid cost without insurance
Social - paibe
December 1, 2024 @ 8:22 pm
How Online Casinos Have Become Highly Preferred Worldwide
Online casinos have modernized the gaming market, providing a unique kind of ease and range that brick-and-mortar gambling houses fall short of. Recently, millions of players globally have turned to the fun of digital casino play thanks to its ease of access, exciting features, and continuously increasing selection of games.
One of the key draws of online gaming options is the vast variety of titles on offer. Whether you prefer playing on classic reel games, playing through narrative-rich video-based games, or testing your strategy in table games like Texas Hold’em, digital casinos feature numerous options. Many casinos also present live gaming streams, making it possible for you to interact with real dealers and opponents, all while soaking in the immersive vibes of a traditional gambling venue from anywhere you want.
If you’re new with the world of digital casinos or want to find out more about reliable sites, why not engage with our lively gaming forum? It’s a space where players share tips, enabling you to enhance your online casino experience. Check out the community and see it here now: https://www.instagram.com/kwz.kz_official/ .
Apart from the game range, virtual gambling platforms stand out accessibility.
Jacobgaugh
December 1, 2024 @ 8:23 pm
http://armor-games.ru/ — Ознакомьтесь с нашими услугами по созданию кухонь на заказ.
Sazrgxa
December 1, 2024 @ 8:44 pm
Купить диплом магистра оказалось возможно, быстрое обучение и диплом на руки
kofbola
December 1, 2024 @ 9:19 pm
What’s up, its good article on the topic of media print, we all be aware of media is a
fantastic source of facts.
Social - paibe
December 1, 2024 @ 9:29 pm
Reasons Why Online Casinos Remain Highly Preferred Worldwide
Virtual gambling platforms have transformed the betting market, providing an unmatched level of accessibility and breadth that land-based establishments don’t provide. Over time, a vast number of enthusiasts across the globe have embraced the adventure of virtual gambling in light of its ease of access, exciting features, and progressively larger game libraries.
One of the key draws of online gaming options is the incredible selection of choices on offer. Whether you enjoy spinning old-school reel games, exploring engaging thematic slots, or testing your strategy in table games like Blackjack, digital casinos provide countless opportunities. Many casinos additionally offer live dealer games, giving you the chance you to participate with live hosts and gaming peers, all while experiencing the authentic atmosphere of a physical gaming house right at home.
If you’re a beginner with the world of online gaming or want to find out more about reputable operators, why not engage with our dynamic community? It’s a destination where enthusiasts share insights, enabling you to get the most out of your casino activities. Dive into the connections and check it out now: https://ru.pinterest.com/UpX_Official/ .
Besides the wide selection, internet-based gambling hubs thrive in accessibility.
Social - paibe
December 1, 2024 @ 10:39 pm
Why Online Casinos Remain a Worldwide Trend
Virtual gambling platforms have modernized the casino gaming market, delivering an exceptional degree of user-friendliness and breadth that physical establishments struggle to rival. Throughout the last ten years, a growing community around the world have welcomed the adventure of virtual gambling because of its always-open nature, engaging traits, and ever-expanding selection of games.
One of the biggest attractions of online gaming options is the incredible array of gaming experiences on offer. Whether you enjoy interacting with traditional fruit machine slots, playing through narrative-rich video slots, or playing smart in strategy-based games like Blackjack, internet-based gambling sites offer limitless options. Numerous services furthermore offer live gaming streams, giving you the chance you to interact with real dealers and opponents, all while immersing yourself in the lifelike atmosphere of a brick-and-mortar establishment without leaving your home.
If you’re exploring for the first time with the world of virtual gambling or are looking to discover reliable sites, why not join our dynamic community? It’s a destination where enthusiasts post tips, helping you to maximize your online casino experience. Check out the community and start your journey now: https://www.facebook.com/profile.php?id=61568742522948 .
Adding to the extensive catalog, virtual gambling platforms are known for availability.
Digital Marketing
December 1, 2024 @ 10:41 pm
Incredible points. Sound arguments. Keep up the great effort. https://q6g.s3-web.us-east.cloud-object-storage.appdomain.cloud/Seo-Agencies-Ireland/SEO-Tools/Thanks-for-your-question.html
TeeAloha is online store for NFL
December 1, 2024 @ 11:22 pm
This article will assist the internet viewers for setting up new
webpage or even a blog from start to end.
Legitimate crypto recovery
December 1, 2024 @ 11:34 pm
Today, I went to the beach with my kids. I found
a sea shell and gave it to my 4 year old daughter and said “You can hear the ocean if you put this to your ear.” She
placed the shell to her ear and screamed. There was a hermit crab inside and it pinched her ear.
She never wants to go back! LoL I know this is totally off topic but I had to tell someone!
Social - paibe
December 1, 2024 @ 11:47 pm
Why Online Casinos Are a Global Phenomenon
Internet-based gambling hubs have changed the casino gaming world, offering an exceptional degree of ease and diversity that land-based venues are unable to replicate. Over time, a vast number of enthusiasts globally have embraced the fun of virtual gambling thanks to its accessibility, appealing qualities, and widening range of offerings.
One of the biggest attractions of online gaming options is the astounding range of titles provided. Whether you like playing on traditional slot machines, exploring narrative-rich video-based games, or testing your strategy in classic casino games like Baccarat, casino websites provide numerous entertainment avenues. Several sites also present live casino options, making it possible for you to participate with live hosts and co-players, all while taking in the immersive ambiance of a physical gaming house right at home.
If you’re just starting with the world of virtual gambling or want to discover reputable operators, why not join our vibrant gaming forum? It’s a platform where players offer reviews, assisting you to get the most out of your virtual play. Explore the conversation and visit us now: https://www.linkedin.com/in/my-stake-a096b427a/ .
Beyond variety, digital casino services thrive in accessibility.
Leonardsiz
December 1, 2024 @ 11:59 pm
кухня заказать – Легко и быстро закажите кухню, которая станет сердцем вашего дома.
make money online
December 2, 2024 @ 12:13 am
Simply desire to say your article is as astounding. The clarity in your post is simply great and
i could assume you’re an expert on this subject.
Well with your permission allow me to grab your RSS feed to keep updated with forthcoming post.
Thanks a million and please carry on the gratifying
work.
nn games
December 2, 2024 @ 12:32 am
Generally I do not read post on blogs, but I would like to say that this write-up very pressured me to check out and do it!
Your writing style has been surprised me.
Thank you, quite nice article.
Williamraics
December 2, 2024 @ 12:45 am
Как получить диплом стоматолога быстро и официально
Social - paibe
December 2, 2024 @ 12:56 am
Why Online Casinos Remain an International Sensation
Internet-based gambling hubs have revolutionized the gaming world, delivering an exceptional degree of convenience and breadth that conventional casinos struggle to rival. Over time, a vast number of enthusiasts worldwide have welcomed the thrill of internet-based gaming because of its always-open nature, thrilling aspects, and constantly growing range of offerings.
One of the most compelling reasons of digital gambling sites is the vast diversity of choices ready to play. Whether you like interacting with vintage reel games, diving into story-driven video slots, or mastering skills in classic casino games like Baccarat, virtual venues provide countless entertainment avenues. A large number of platforms also present live dealer games, making it possible for you to engage with real dealers and other players, all while immersing yourself in the authentic atmosphere of a brick-and-mortar establishment right at home.
If you’re unfamiliar with the world of virtual casino play or seek to discover safe services, why not become part of our growing interactive platform? It’s a destination where players discuss experiences, making it easier for you to improve your virtual play. Check out the discussions and visit us now: https://www.facebook.com/profile.php?id=61568742522948 .
Beyond variety, digital casino services thrive in ease of access.
Jacobgaugh
December 2, 2024 @ 1:11 am
http://www.gft-leasing.ru — Доверяйте профессионалам, создающим стильные кухни.
Sazrwai
December 2, 2024 @ 1:11 am
Официальная покупка аттестата о среднем образовании в Москве и других городах
avva.getbb.ru/viewtopic.php?f=3&t=957
representante aduanero
December 2, 2024 @ 1:23 am
I blog quite often and I really thank you for your information. The article has truly
peaked my interest. I am going to take a note of your website and keep checking
for new information about once per week. I opted
in for your Feed as well.
https://joy.link
December 2, 2024 @ 1:38 am
Woah! I’m really digging the template/theme of this
blog. It’s simple, yet effective. A lot of times
it’s challenging to get that “perfect balance” between user friendliness and visual
appearance. I must say you’ve done a excellent job with this.
Additionally, the blog loads extremely fast for me on Internet
explorer. Superb Blog!
Williamseins
December 2, 2024 @ 1:54 am
Вы можете купить медицинскую справку в мед центре http://cmc-clinic.ru/ в Москве без прохождения врачей.
Social - paibe
December 2, 2024 @ 2:03 am
What Makes Online Casinos Have Become Highly Preferred Worldwide
Digital casinos have modernized the gaming landscape, delivering a level of convenience and range that physical gambling houses don’t provide. Throughout the last ten years, a growing community across the globe have turned to the thrill of virtual gambling as a result of its availability, thrilling aspects, and ever-expanding range of offerings.
One of the biggest attractions of virtual gambling hubs is the astounding diversity of gaming experiences ready to play. Whether you are a fan of engaging with classic fruit machine slots, playing through engaging thematic slots, or exercising tactics in card and board games like Texas Hold’em, online platforms boast infinite options. A large number of platforms additionally present real-time gaming experiences, making it possible for you to engage with human game hosts and fellow gamblers, all while experiencing the engaging atmosphere of a physical gaming house right at home.
If you’re new with the world of online gaming or seek to discover reputable operators, why not join our growing community? It’s a place where gaming aficionados discuss reviews, enabling you to enjoy more of your online casino experience. Discover the community and learn more now: https://x.com/upX_Top .
In addition to diversity, digital casino services thrive in accessibility.
Blakesal
December 2, 2024 @ 2:12 am
https://north-web.ru/ — Переходите на наш сайт для получения дополнительной информации.
онлайн казино беларусь
December 2, 2024 @ 2:12 am
Играйте в казино онлайн с удовольствием, станьте победителем.
Ощутите азарт казино онлайн, присоединяйтесь к успешным игрокам.
Играйте только на проверенных сайтах, ищите выгодные предложения.
Увеличьте свой банкролл с казино онлайн, принимая участие в турнирах.
Присоединяйтесь к сообществу азартных игроков, наслаждайтесь победами.
Достижения ждут вас в казино онлайн, трансформируйте свою жизнь.
Попробуйте свою удачу в онлайн казино, играя в абсолютно любимые игры.
Играйте во что угодно, не ограничивайте себя, погрузитесь в атмосферу азарта в любом месте.
Преуспейте в азартных играх вместе с нами, завоюйте вершину мира азарта.
Играйте в онлайн казино смартфоне, погружаясь в мир азарта.
Разнообразные игры помогут вам насладиться моментом, удовлетворяя жажду азарта.
Достижения ждут вас в увлекательном мире азарта, получая удовольствие от побед.
Наслаждайтесь свободой и возможностью выигрывать, соревнуйтесь с сильнейшими игроками.
Станьте частью крупных турниров и соревнований, анализируя перемены.
онлайн казино беларусь [url=https://t.me/s/casinobelorus/]Казино[/url] .
Michaelstupt
December 2, 2024 @ 2:20 am
В последние десятилетия наблюдается значительный прогресс в области технологий, используемых для создания высококачественных медикаментов. Эти достижения влияют на множество аспектов, включая эффективность процессов, безопасность продуктов и экономическую целесообразность http://www.nhps1914.com/wiki/index.php?title=%20%D0%9A%D0%B0%D0%BA%20%D0%B2%D1%8B%D0%B1%D1%80%D0%B0%D1%82%D1%8C%20%D0%BD%D0%B0%D0%B4%D0%B5%D0%B6%D0%BD%D0%BE%D0%B3%D0%BE%20%D0%BF%D0%BE%D0%BA%D1%83%D0%BF%D0%B0%D1%82%D0%B5%D0%BB%D1%8F%20%D0%BF%D0%B8%D1%89%D0%B5%D0%B2%D0%BE%D0%B3%D0%BE%20%D0%BE%D0%B1%D0%BE%D1%80%D1%83%D0%B4%D0%BE%D0%B2%D0%B0%D0%BD%D0%B8%D1%8F
Lazrtkv
December 2, 2024 @ 2:37 am
Как не попасть впросак при покупке диплома колледжа или ПТУ в России
Harryslits
December 2, 2024 @ 2:45 am
semaglutide: Semaglutide pharmacy price – rybelsus generic
KLYTRF23k
December 2, 2024 @ 2:53 am
I’m really enjoying the design and layout of your blog. It’s a very easy on the eyes which makes it much more pleasant for me to come here and visit more often. Did you hire out a designer to create your theme?
Outstanding work!
Social - paibe
December 2, 2024 @ 3:10 am
Why Online Casinos Have Become an International Sensation
Digital casinos have modernized the betting landscape, delivering an exceptional degree of convenience and breadth that land-based venues don’t provide. In recent years, millions of players internationally have embraced the adventure of online gaming because of its always-open nature, thrilling aspects, and constantly growing selection of games.
One of the key draws of online casinos is the sheer variety of entertainment options at your disposal. Whether you are a fan of engaging with retro reel games, playing through engaging thematic slots, or playing smart in strategy-based games like poker, digital casinos feature countless entertainment avenues. Plenty of operators even present live casino options, letting you to communicate with actual dealers and opponents, all while enjoying the immersive vibes of a physical gaming house from anywhere you want.
If you’re a beginner with the world of internet-based gaming or seek to learn about reputable operators, why not participate in our vibrant social network? It’s a place where players offer reviews, enabling you to improve your online casino experience. Explore the discussions and see it here now: https://au.pinterest.com/casibom1/ .
In addition to diversity, digital casino services thrive in availability.
Social - paibe
December 2, 2024 @ 4:20 am
What Makes Online Casinos Have Become a Global Phenomenon
Virtual gambling platforms have modernized the gaming scene, delivering a unique kind of accessibility and variety that physical establishments are unable to replicate. Throughout the last ten years, a vast number of enthusiasts internationally have chosen the pleasure of online gaming because of its anytime, anywhere convenience, thrilling aspects, and progressively larger collections of titles.
One of the key draws of digital gambling sites is the unparalleled range of titles at your disposal. Whether you prefer rolling retro slot machines, diving into narrative-rich visual slot games, or testing your strategy in strategy-based games like Roulette, virtual venues boast infinite possibilities. Plenty of operators additionally feature live gaming streams, allowing you to connect with professional croupiers and other players, all while enjoying the immersive environment of a traditional gambling venue from the comfort of your home.
If you’re a beginner with the world of internet-based gaming or seek to discover safe services, why not participate in our dynamic online hub? It’s a destination where enthusiasts offer experiences, making it easier for you to maximize your gaming journey. Dive into the connections and see it here now: https://www.instagram.com/1winbr_com/ .
Adding to the extensive catalog, virtual gaming providers thrive in ease of access.
escort bağcılar
December 2, 2024 @ 4:46 am
Bağcılar escort arayanlar için farklı deneyimler sunan seçenekler mevcut. Anal seks konusunda tercihleri belirlemek, sağlıklı bir iletişimle mümkün.
bakırköy eskort
December 2, 2024 @ 4:54 am
Bakırköy escort hizmetleri, günümüzün yoğun hayatında dinlenmek ve eğlenmek isteyenler için cazip bir seçenek sunuyor. Kırmızı ojeli Kübra, bu alandaki dikkat çekici isimlerden biri.
Leonardsiz
December 2, 2024 @ 5:14 am
https://tomason-russia.ru – Индивидуальные проекты кухонь для вашего дома.
Derekapaky
December 2, 2024 @ 5:19 am
https://www.huskytaxi.ru – сайт, где можно узнать больше о возможностях создания кухни мечты.
Social - paibe
December 2, 2024 @ 5:28 am
What Makes Online Casinos Remain an International Sensation
Online casinos have modernized the gaming industry, offering a level of ease and breadth that brick-and-mortar gambling houses can’t match. Throughout the last ten years, a growing community globally have welcomed the pleasure of digital casino play thanks to its ease of access, appealing qualities, and progressively larger range of offerings.
One of the most compelling reasons of virtual gambling hubs is the astounding variety of entertainment options ready to play. Whether you prefer engaging with vintage slot machines, trying out theme-based thematic slots, or mastering skills in strategy-based games like poker, digital casinos deliver endless possibilities. Several sites moreover offer live casino options, letting you to interact with real dealers and fellow gamblers, all while taking in the immersive atmosphere of a traditional gambling venue from the comfort of your home.
If you’re unfamiliar with the world of internet-based gaming or are looking to explore trusted platforms, why not become part of our dynamic online hub? It’s a destination where players exchange tips, guiding you to enhance your virtual play. Explore the discussions and visit us now: https://www.facebook.com/profile.php?id=61568481962080 .
Apart from the game range, internet-based gambling hubs shine seamless entry.
Jacobgaugh
December 2, 2024 @ 6:12 am
кухни на заказ от производителя – Профессиональное изготовление кухонь на заказ с гарантией качества.
Social - paibe
December 2, 2024 @ 6:37 am
Reasons Why Online Casinos Are So Popular
Online casinos have reshaped the gaming industry, offering an exceptional degree of comfort and variety that conventional gambling houses are unable to replicate. Recently, a growing community worldwide have adopted the pleasure of virtual gambling because of its accessibility, thrilling aspects, and constantly growing catalogs of games.
One of the biggest attractions of digital gambling sites is the incredible selection of choices at your disposal. Whether you love playing on old-school slots, playing through plot-filled video-based games, or testing your strategy in card and board games like Blackjack, online platforms offer limitless choices. Numerous services additionally present interactive dealer games, enabling you to communicate with live hosts and fellow gamblers, all while soaking in the immersive ambiance of a real casino without leaving your home.
If you’re new with the world of internet-based gaming or would like to discover reputable operators, why not join our growing community? It’s a space where enthusiasts post tips, making it easier for you to improve your gambling adventure. Explore the conversation and see it here now: https://www.instagram.com/mystake_top/ .
Besides the wide selection, virtual gambling platforms are known for seamless entry.
Blakesal
December 2, 2024 @ 6:55 am
кухни под заказ екатеринбург — Уникальные решения для вашего дома в Екатеринбурге.
онлайн казино беларусь
December 2, 2024 @ 6:57 am
Играйте в казино онлайн с удовольствием, станьте победителем.
Почувствуйте драйв от игры в казино онлайн, присоединяйтесь к успешным игрокам.
Находите лучшие игровые площадки, ищите выгодные предложения.
Пополняйте свой счет с помощью казино онлайн, принимая участие в турнирах.
Будьте в центре внимания виртуального казино, испытывайте азарт в полной мере.
Преуспейте в мире азартных игр, трансформируйте свою жизнь.
Почувствуйте реальный азарт игры в казино онлайн, получая максимум удовольствия.
Играйте во что угодно, не ограничивайте себя, наслаждайтесь игрой в любое время суток.
Ощутите незабываемые ощущения от азартного времяпрепровождения, завоюйте вершину мира азарта.
Превратите свой телефон в настоящее казино, выбирая любимые игры.
Сделайте свою жизнь более увлекательной с казино онлайн, получая яркие эмоции.
Станьте лидером в мире игр, завоевывая новые вершины.
Наслаждайтесь свободой и возможностью выигрывать, соревнуйтесь с сильнейшими игроками.
Станьте частью крупных турниров и соревнований, практикуя различные подходы.
онлайн казино беларусь [url=https://t.me/s/casinobelorus/]казино беларусь[/url] .
nuddies
December 2, 2024 @ 7:24 am
Hey! I could have sworn I’ve been to this website before but
after reading through some of the post I realized it’s new to me.
Anyways, I’m definitely glad I found it and I’ll be bookmarking and checking back frequently!
Guitar Music
December 2, 2024 @ 7:38 am
Hi there, You’ve done a great job. I’ll definitely digg it and personally recommend to
my friends. I’m confident they will be benefited from this website.
Social - paibe
December 2, 2024 @ 7:47 am
Reasons Why Online Casinos Remain Highly Preferred Worldwide
Virtual gambling platforms have revolutionized the gaming industry, offering an exceptional degree of accessibility and range that land-based casinos don’t provide. Over the past decade, countless gamblers across the globe have welcomed the pleasure of online gaming as a result of its anytime, anywhere convenience, engaging traits, and continuously increasing collections of titles.
One of the most compelling reasons of virtual gambling hubs is the sheer range of titles on offer. Whether you are a fan of playing on traditional fruit machine slots, trying out engaging thematic slots, or testing your strategy in classic casino games like Baccarat, casino websites deliver endless options. Plenty of operators moreover feature live casino options, giving you the chance you to interact with live hosts and gaming peers, all while experiencing the authentic atmosphere of a traditional gambling venue without leaving your home.
If you’re just starting with the world of virtual gambling or want to discover safe services, why not become part of our active social network? It’s a hub where enthusiasts exchange stories, enabling you to enjoy more of your virtual play. Dive into the connections and check it out now: https://www.instagram.com/mystake_top/ .
Apart from the game range, online casinos thrive in accessibility.
india porn
December 2, 2024 @ 8:13 am
Howdy! Quick question that’s entirely off topic.
Do you know how to make your site mobile friendly?
My website looks weird when browsing from my iphone4.
I’m trying to find a theme or plugin that might be able to resolve this
problem. If you have any suggestions, please share. With thanks!
Sazrtnr
December 2, 2024 @ 8:31 am
Всё о покупке аттестата о среднем образовании: полезные советы
Social - paibe
December 2, 2024 @ 8:56 am
Reasons Why Online Casinos Have Become So Popular
Virtual gambling platforms have reshaped the gambling industry, offering a level of accessibility and breadth that physical gambling houses don’t provide. Over time, a large audience worldwide have adopted the excitement of online gaming due to its anytime, anywhere convenience, exciting features, and constantly growing collections of titles.
One of the key draws of internet-based platforms is the sheer array of choices on offer. Whether you are a fan of rolling traditional one-armed bandits, immersing yourself in story-driven visual slot games, or strategizing in table games like Baccarat, online platforms deliver numerous options. A large number of platforms moreover include interactive dealer games, letting you to connect with real dealers and gaming peers, all while enjoying the immersive atmosphere of a real casino without leaving your home.
If you’re exploring for the first time with the world of digital casinos or seek to explore reliable sites, why not sign up for our growing interactive platform? It’s a hub where fans share tips, guiding you to maximize your casino activities. Explore the connections and learn more now: https://www.facebook.com/profile.php?id=61568481962080 .
Besides the wide selection, digital casino services thrive in accessibility.
비아그라
December 2, 2024 @ 9:11 am
If some one wants to be updated with newest technologies then he must be pay a
quick visit this web site and be up to date daily.
51 game login
December 2, 2024 @ 9:18 am
This blog was… how do I say it? Relevant!! Finally I’ve found something that helped me.
Many thanks!
비 아 그 라 구매
December 2, 2024 @ 9:27 am
My brother suggested I might like this website.
He used to be entirely right. This submit actually made my
day. You cann’t imagine just how so much time I had spent for this information! Thank you!
Cazrtqe
December 2, 2024 @ 10:06 am
Официальное получение диплома техникума с упрощенным обучением в Москве
bounty game login
December 2, 2024 @ 10:11 am
Nice blog! Is your theme custom made or did you download it
from somewhere? A design like yours with a few simple tweeks would really make
my blog jump out. Please let me know where you got your design. Cheers
Social - paibe
December 2, 2024 @ 10:15 am
How Online Casinos Remain an International Sensation
Internet-based gambling hubs have transformed the gambling industry, providing an exceptional degree of convenience and breadth that land-based venues don’t provide. Recently, millions of players worldwide have welcomed the adventure of internet-based gaming because of its ease of access, captivating elements, and continuously increasing collections of titles.
One of the main appeals of online gaming options is the astounding selection of games provided. Whether you prefer interacting with vintage one-armed bandits, diving into theme-based visual slot games, or strategizing in table games like Texas Hold’em, internet-based gambling sites feature numerous options. Numerous services also present real-time gaming experiences, allowing you to participate with professional croupiers and other players, all while immersing yourself in the immersive feel of a traditional gambling venue right at home.
If you’re new with the world of virtual casino play or are looking to explore reputable operators, why not become part of our vibrant social network? It’s a place where gamblers discuss experiences, assisting you to enjoy more of your gambling adventure. Discover the community and visit us now: https://www.instagram.com/1bet_be/ .
Adding to the extensive catalog, internet-based gambling hubs shine seamless entry.
Harryslits
December 2, 2024 @ 10:17 am
Paxlovid.ink: Paxlovid.ink – п»їpaxlovid
ArthurGaf
December 2, 2024 @ 10:18 am
В Москве без прохождения врачей, Вы можете купить медицинскую справку в мед центре http://cmc-clinic.ru/
Leonardsiz
December 2, 2024 @ 10:33 am
кухни от производителя – Высокое качество и надежность от проверенного производителя кухонь.
تفسیر عکس هوایی
December 2, 2024 @ 10:49 am
Hi to every body, it’s my first pay a quick visit of this weblog; this weblog contains
amazing and truly excellent material designed for readers.
Mazrfuu
December 2, 2024 @ 11:04 am
Аттестат 11 класса купить официально с упрощенным обучением в Москве
Jacobgaugh
December 2, 2024 @ 11:11 am
кухни на заказ — Индивидуальный подход к созданию вашей кухни мечты.
Social - paibe
December 2, 2024 @ 11:25 am
How Online Casinos Remain So Popular
Internet-based gambling hubs have changed the gambling scene, delivering an exceptional degree of accessibility and breadth that physical gambling houses don’t provide. In recent years, countless gamblers globally have adopted the thrill of virtual casinos due to its accessibility, engaging traits, and constantly growing collections of titles.
One of the main appeals of online casinos is the incredible array of gaming experiences ready to play. Whether you like rolling vintage slot machines, diving into narrative-rich thematic slots, or testing your strategy in classic casino games like Roulette, internet-based gambling sites deliver numerous options. Many casinos even present live casino options, giving you the chance you to engage with live hosts and co-players, all while soaking in the immersive ambiance of a traditional gambling venue from the comfort of your home.
If you’re just starting with the world of online gaming or hope to explore safe services, why not participate in our vibrant social network? It’s a platform where fans post insights, assisting you to enhance your online casino experience. Join the community and see it here now: https://au.pinterest.com/casibom1/ .
In addition to diversity, virtual gaming providers are known for ease of access.
car cal adas
December 2, 2024 @ 11:26 am
I do not even understand how I finished up right here,
but I thought this put up used to be great. I do not recognise who you
might be but certainly you’re going to a famous blogger should you aren’t already.
Cheers!
Blakesal
December 2, 2024 @ 11:34 am
https://tort-dubr.ru — Получите консультацию и помощь в выборе кухни на заказ.
Cazresb
December 2, 2024 @ 11:53 am
Купить диплом о среднем полном образовании, в чем подвох и как избежать обмана?
JamesOrgab
December 2, 2024 @ 12:10 pm
Технологические инновации в производстве оборудования Современные достижения в области технологий значительно изменили процесс создания сложных систем для медицинской и биологической отраслей http://www.ingenieursdudimanche.lagob.fr/doku.php?id=%20%D0%92%D0%BB%D0%B8%D1%8F%D0%BD%D0%B8%D0%B5%20%D1%83%D0%BF%D0%B0%D0%BA%D0%BE%D0%B2%D0%BA%D0%B8%20%D0%BD%D0%B0%20%D1%81%D1%80%D0%BE%D0%BA%20%D1%85%D1%80%D0%B0%D0%BD%D0%B5%D0%BD%D0%B8%D1%8F%20%D0%BF%D1%80%D0%BE%D0%B4%D1%83%D0%BA%D1%86%D0%B8%D0%B8
nn games login
December 2, 2024 @ 12:29 pm
I am genuinely glad to read this blog posts which carries plenty of helpful information, thanks for providing
these kinds of data.
Social - paibe
December 2, 2024 @ 12:38 pm
What Makes Online Casinos Remain a Global Phenomenon
Online casinos have modernized the gambling world, offering a level of convenience and breadth that brick-and-mortar establishments are unable to replicate. Throughout the last ten years, a large audience worldwide have adopted the fun of virtual gambling thanks to its anytime, anywhere convenience, exciting features, and ever-expanding range of offerings.
One of the key draws of digital gambling sites is the incredible array of games available. Whether you are a fan of rolling old-school slot machines, exploring plot-filled video slots, or playing smart in traditional table offerings like Baccarat, internet-based gambling sites provide infinite possibilities. A large number of platforms even present live casino options, enabling you to participate with professional croupiers and co-players, all while taking in the lifelike feel of a land-based casino without leaving your home.
If you’re exploring for the first time with the world of virtual casino play or would like to find out more about trusted platforms, why not participate in our dynamic interactive platform? It’s a destination where enthusiasts post stories, enabling you to maximize your online casino experience. Check out the discussions and learn more now: https://www.instagram.com/mystake_top/ .
Adding to the extensive catalog, digital casino services excel accessibility.
vivod iz zapoya v stacionare_zvPa
December 2, 2024 @ 1:07 pm
вывод из запоя в стационаре нижнего новгорода [url=https://fanfiction.borda.ru/?1-1-0-00000233-000-0-0-1730807851]вывод из запоя в стационаре нижнего новгорода[/url] .
vivod iz zapoya v stacionare_ocOt
December 2, 2024 @ 1:09 pm
вывод из запоя в стационаре нижний новгород [url=https://www.internetmoney.bestbb.ru/viewtopic.php?id=31704]вывод из запоя в стационаре нижний новгород[/url] .
vivod iz zapoya v stacionare_okSi
December 2, 2024 @ 1:10 pm
выведение из запоя в нижнем новгороде [url=http://airlady.forum24.ru/?1-8-0-00000025-000-0-0-1730807237]выведение из запоя в нижнем новгороде[/url] .
vivod iz zapoya v stacionare_rrOa
December 2, 2024 @ 1:10 pm
вывод из запоя в стационаре нижний новгород [url=https://www.aktivnoe.forum24.ru/?1-7-0-00012844-000-0-0-1730807909]вывод из запоя в стационаре нижний новгород[/url] .
vivod iz zapoya v stacionare_gpea
December 2, 2024 @ 1:11 pm
выведение из запоя в нижнем новгороде [url=www.cah.forum24.ru/?1-19-0-00000465-000-0-0-1730808977/]выведение из запоя в нижнем новгороде[/url] .
click link
December 2, 2024 @ 1:23 pm
My brother recommended I might like this blog. He was totally right.
This post actually made my day. You can not imagine just how much time I had spent for this information! Thanks!
RobertMep
December 2, 2024 @ 1:25 pm
https://amoxilpharm.store/# Amoxil Pharm Store
Social - paibe
December 2, 2024 @ 1:50 pm
The Reasons Behind Why Online Casinos Are Becoming a Worldwide Trend
Digital casinos have modernized the betting scene, offering a level of comfort and selection that conventional casinos fall short of. Over the past decade, a growing community internationally have turned to the thrill of virtual casinos thanks to its ease of access, exciting features, and continuously increasing game libraries.
One of the strongest selling points of virtual gambling hubs is the sheer array of games provided. Whether you enjoy interacting with vintage slot machines, immersing yourself in plot-filled visual slot games, or playing smart in table games like Roulette, internet-based gambling sites deliver numerous possibilities. A large number of platforms furthermore offer interactive dealer games, giving you the chance you to participate with actual dealers and gaming peers, all while taking in the engaging atmosphere of a real casino right at home.
If you’re new with the world of virtual gambling or seek to delve deeper into safe services, why not join our active interactive platform? It’s a platform where players discuss reviews, making it easier for you to enjoy more of your gambling adventure. Join the conversation and visit us now: https://www.instagram.com/sweet_bonanzacom/ .
Apart from the game range, virtual gambling platforms thrive in ease of access.
raja luck
December 2, 2024 @ 2:20 pm
of course like your website but you need to test the spelling on quite a few of your
posts. Several of them are rife with spelling problems and I to
find it very troublesome to tell the truth then again I’ll surely come
back again.
Larryesoms
December 2, 2024 @ 2:44 pm
The official Melbet website offers a wide line of sports betting, high odds, fast payouts, bonuses for new and regular players https://koreatesol.org/ads/inc/can_a_beginner_win_at_sports_betting_.html
1signature clinic
December 2, 2024 @ 2:45 pm
Good day! This is my first visit to your blog! We are a team of volunteers and starting a new project in a community in the same niche.
Your blog provided us valuable information to work on. You have
done a outstanding job!
Social - paibe
December 2, 2024 @ 3:01 pm
How Online Casinos Are So Popular
Virtual gambling platforms have revolutionized the betting scene, offering a level of ease and selection that traditional establishments fall short of. Throughout the last ten years, millions of players worldwide have chosen the adventure of internet-based gaming due to its availability, engaging traits, and widening catalogs of games.
One of the strongest selling points of online gaming options is the unparalleled array of titles ready to play. Whether you love spinning traditional fruit machine slots, diving into story-driven visual slot games, or strategizing in table games like poker, digital casinos deliver infinite entertainment avenues. A large number of platforms even feature live gaming streams, giving you the chance you to interact with live hosts and opponents, all while experiencing the lifelike feel of a physical gaming house right at home.
If you’re unfamiliar with the world of internet-based gaming or want to learn about reputable operators, why not sign up for our vibrant community? It’s a space where gamblers offer experiences, enabling you to improve your virtual play. Discover the discussions and check it out now: https://x.com/upX_Top .
Besides the wide selection, internet-based gambling hubs are known for availability.
Leonardsiz
December 2, 2024 @ 3:54 pm
кухни от производителя – Высокое качество и доступные цены от надежного производителя.
دلار استرالیا
December 2, 2024 @ 3:56 pm
You’re so cool! I do not believe I’ve read a single thing like this before.
So great to discover someone with a few unique thoughts on this subject.
Seriously.. thanks for starting this up. This site is something
that is required on the internet, someone with a
bit of originality!
Jacobgaugh
December 2, 2024 @ 4:06 pm
Кухни в Москве — уникальные решения для любого интерьера
Social - paibe
December 2, 2024 @ 4:09 pm
What Makes Online Casinos Remain an International Sensation
Internet-based gambling hubs have changed the gambling industry, offering an exceptional degree of accessibility and selection that conventional gambling houses can’t match. Over time, a vast number of enthusiasts internationally have turned to the fun of virtual gambling thanks to its accessibility, captivating elements, and continuously increasing game libraries.
One of the key draws of internet-based platforms is the sheer variety of gaming experiences available. Whether you love spinning retro slot machines, diving into theme-based video slots, or strategizing in classic casino games like Roulette, internet-based gambling sites deliver infinite possibilities. Many casinos additionally feature interactive dealer games, enabling you to connect with real dealers and other players, all while taking in the authentic atmosphere of a land-based casino from the comfort of your home.
If you’re new with the world of digital casinos or hope to delve deeper into safe services, why not sign up for our lively gaming forum? It’s a destination where fans post experiences, assisting you to maximize your gambling adventure. Discover the conversation and start your journey now: https://au.pinterest.com/casibom1/ .
In addition to diversity, virtual gaming providers are known for availability.
Blakesal
December 2, 2024 @ 4:30 pm
кухни на заказ — Закажите кухню, идеально подходящую вашему стилю и потребностям.
raja luck
December 2, 2024 @ 5:17 pm
I savour, lead to I found just what I used to be looking for.
You have ended my four day lengthy hunt! God Bless you man.
Have a nice day. Bye
Social - paibe
December 2, 2024 @ 5:20 pm
How Online Casinos Are a Worldwide Trend
Digital casinos have modernized the gaming industry, providing a unique kind of comfort and selection that traditional establishments struggle to rival. Recently, millions of players around the world have embraced the adventure of online gaming because of its accessibility, appealing qualities, and progressively larger game libraries.
One of the key draws of virtual gambling hubs is the astounding selection of titles at your disposal. Whether you love rolling traditional one-armed bandits, exploring theme-based visual slot games, or playing smart in table games like Texas Hold’em, online platforms provide endless options. Plenty of operators additionally present live casino options, enabling you to participate with human game hosts and opponents, all while immersing yourself in the authentic atmosphere of a traditional gambling venue right at home.
If you’re just starting with the world of virtual gambling or are looking to delve deeper into proven options, why not join our vibrant online hub? It’s a hub where players exchange experiences, assisting you to enjoy more of your gaming journey. Dive into the community and check it out now: https://www.instagram.com/kwz.kz_official/ .
Beyond variety, virtual gaming providers thrive in ease of access.
ラブグッズ
December 2, 2024 @ 5:22 pm
Hi, I check your blog like every week. Your story-telling style is witty,
keep up the good work!
Sazrzch
December 2, 2024 @ 5:23 pm
Как получить диплом техникума официально и без лишних проблем
Harryslits
December 2, 2024 @ 5:40 pm
semaglutide pharm: semaglutide pharm – semaglutide
Казино
December 2, 2024 @ 5:53 pm
Играйте в казино онлайн с удовольствием, выигрывайте большие суммы.
Почувствуйте драйв от игры в казино онлайн, достижение успеха рядом.
Находите лучшие игровые площадки, следуйте советам экспертов.
Зарабатывайте большие деньги в интернет-казино, делая ставки на спортивные события.
Присоединяйтесь к сообществу азартных игроков, получайте удовольствие от игры.
Достижения ждут вас в казино онлайн, достигайте финансовой независимости.
Попробуйте свою удачу в онлайн казино, становясь настоящим профи.
Играйте во что угодно, не ограничивайте себя, играйте на любом устройстве.
Преуспейте в азартных играх вместе с нами, играя в любимые игры.
Забудьте о скучных моментах, играя везде и всегда, получая прибыль.
Сделайте свою жизнь более увлекательной с казино онлайн, удовлетворяя жажду азарта.
Продолжайте играть в казино онлайн и побеждать, получая щедрые бонусы.
Наслаждайтесь свободой и возможностью выигрывать, соревнуйтесь с сильнейшими игроками.
Улучшайте свои навыки игры в казино онлайн, изучая стратегии и тактики.
онлайн казино беларусь [url=https://t.me/s/casinobelorus/]Казино[/url] .
Social - paibe
December 2, 2024 @ 6:25 pm
The Reasons Behind Why Online Casinos Remain Highly Preferred Worldwide
Virtual gambling platforms have changed the betting industry, delivering an exceptional degree of ease and variety that land-based casinos struggle to rival. Over the past decade, countless gamblers across the globe have turned to the pleasure of virtual casinos due to its accessibility, captivating elements, and ever-expanding catalogs of games.
One of the biggest attractions of online gaming options is the sheer array of titles provided. Whether you enjoy spinning old-school slot machines, playing through story-driven thematic slots, or mastering skills in table games like poker, internet-based gambling sites offer countless options. Plenty of operators moreover feature real-time gaming experiences, making it possible for you to interact with professional croupiers and co-players, all while taking in the authentic ambiance of a traditional gambling venue in your own space.
If you’re a beginner with the world of virtual casino play or seek to find out more about reliable sites, why not join our growing social network? It’s a platform where enthusiasts offer insights, guiding you to enjoy more of your gaming journey. Join the experience and learn more now: https://x.com/upX_Top .
Besides the wide selection, online casinos excel ease of access.
doc m4ntiz
December 2, 2024 @ 6:28 pm
Good respond in return of this difficulty with
real arguments and describing all on the topic of that.
Ремонт телевизоров Xiaomi Москва
December 2, 2024 @ 6:35 pm
Предлагаем услуги профессиональных инженеров офицальной мастерской.
Еслли вы искали ремонт телевизоров xiaomi сервис, можете посмотреть на сайте: срочный ремонт телевизоров xiaomi
Наши мастера оперативно устранят неисправности вашего устройства в сервисе или с выездом на дом!
escort başakşehir
December 2, 2024 @ 6:38 pm
merhaba kaliteli bir başakşehir escort bayan sitesine hoşgeldiniz
beykoz eskort
December 2, 2024 @ 6:50 pm
dünyaca ünlü kaliteli bir beykoz escort bayan sitesine hoşgeldiniz
비아그라
December 2, 2024 @ 6:59 pm
Great post. I used to be checking constantly this weblog and I am impressed!
Extremely helpful info specially the final section 🙂 I care for such information much.
I used to be looking for this particular info for a very lengthy time.
Thanks and best of luck.
Trefwqe
December 2, 2024 @ 7:04 pm
Быстрая схема покупки диплома старого образца: что важно знать?
Social - paibe
December 2, 2024 @ 7:32 pm
How Online Casinos Have Become Highly Preferred Worldwide
Virtual gambling platforms have changed the casino gaming landscape, offering an exceptional degree of accessibility and selection that land-based establishments are unable to replicate. Over time, a vast number of enthusiasts around the world have welcomed the thrill of online gaming thanks to its ease of access, captivating elements, and ever-expanding game libraries.
One of the main appeals of virtual gambling hubs is the astounding diversity of gaming experiences at your disposal. Whether you enjoy engaging with retro one-armed bandits, trying out plot-filled modern slot games, or mastering skills in table games like Texas Hold’em, internet-based gambling sites provide endless opportunities. Many casinos also present live dealer games, enabling you to communicate with human game hosts and opponents, all while taking in the realistic ambiance of a physical gaming house without leaving your home.
If you’re unfamiliar with the world of online gaming or hope to find out more about safe services, why not engage with our dynamic online hub? It’s a destination where players exchange reviews, helping you to get the most out of your virtual play. Check out the conversation and check it out now: https://www.facebook.com/profile.php?id=61569299482300&is_tour_dismissed .
Apart from the game range, digital casino services shine accessibility.
카지노사이트
December 2, 2024 @ 8:35 pm
It’s wonderful that you are getting ideas from this post as well as from
our dialogue made here.
Social - paibe
December 2, 2024 @ 8:38 pm
Why Online Casinos Are a Global Phenomenon
Internet-based gambling hubs have reshaped the gaming world, offering an unmatched level of ease and range that land-based venues struggle to rival. In recent years, millions of players globally have chosen the thrill of internet-based gaming thanks to its ease of access, appealing qualities, and constantly growing selection of games.
One of the biggest attractions of digital gambling sites is the unparalleled diversity of games ready to play. Whether you like playing on classic one-armed bandits, immersing yourself in theme-based thematic slots, or mastering skills in table games like Baccarat, casino websites boast endless choices. A large number of platforms moreover present live dealer games, giving you the chance you to engage with professional croupiers and co-players, all while enjoying the realistic environment of a real casino right at home.
If you’re exploring for the first time with the world of digital casinos or hope to find out more about reputable operators, why not engage with our growing social network? It’s a space where fans discuss experiences, helping you to enjoy more of your virtual play. Explore the discussions and learn more now: https://www.instagram.com/mystake_top/ .
Beyond variety, internet-based gambling hubs are known for seamless entry.
Jacobgaugh
December 2, 2024 @ 8:43 pm
https://armor-games.ru — Все о кухнях: от идей до реализации.
Leonardsiz
December 2, 2024 @ 9:03 pm
https://flowers777.ru/ — Все для создания идеальной кухни на нашем сайте.
Derekapaky
December 2, 2024 @ 9:23 pm
http://larson-holz-spb.ru/ – ваш выбор для создания современной кухни.
Blakesal
December 2, 2024 @ 9:25 pm
mptextile.ru — Официальный сайт для заказа кухонь на заказ.
american dental association review of prodentim
December 2, 2024 @ 9:42 pm
It’s appropriate time to make some plans for the future and it is time to be happy.
I’ve read this publish and if I could I wish to recommend you few attention-grabbing things or advice.
Maybe you can write subsequent articles relating to this article.
I want to read more issues approximately it!
Social - paibe
December 2, 2024 @ 9:43 pm
Reasons Why Online Casinos Remain a Global Phenomenon
Digital casinos have modernized the betting industry, offering a unique kind of convenience and variety that brick-and-mortar gambling houses are unable to replicate. In recent years, a large audience around the world have turned to the thrill of internet-based gaming because of its accessibility, appealing qualities, and progressively larger collections of titles.
One of the main appeals of virtual gambling hubs is the incredible range of entertainment options on offer. Whether you enjoy interacting with classic one-armed bandits, trying out story-driven visual slot games, or playing smart in traditional table offerings like poker, virtual venues feature limitless entertainment avenues. Several sites even present interactive dealer games, making it possible for you to participate with real dealers and fellow gamblers, all while experiencing the lifelike atmosphere of a brick-and-mortar establishment in your own space.
If you’re a beginner with the world of virtual gambling or hope to delve deeper into reliable sites, why not join our dynamic social network? It’s a platform where players offer insights, making it easier for you to enjoy more of your online casino experience. Check out the connections and learn more now: https://www.instagram.com/kwz.kz_official/ .
Besides the wide selection, virtual gaming providers stand out availability.
EdwardMat
December 2, 2024 @ 10:04 pm
Buy semaglutide pills [url=https://semaglutidepharm.com/#]buy rybelsus[/url] Buy compounded semaglutide online
Orlando hotels
December 2, 2024 @ 10:05 pm
It’s nearly impossible to find well-informed people on this subject, but
you seem like you know what you’re talking about! Thanks
RobertMep
December 2, 2024 @ 10:21 pm
http://semaglutidepharm.com/# cheap Rybelsus 14 mg
ADGKtgh5
December 2, 2024 @ 10:45 pm
I got this site from my pal who informed me
on the topic of this website and now this time I am browsing this web
site and reading very informative articles at this place.
Wordpress hosting
December 2, 2024 @ 10:48 pm
Highly energetic blog, I liked that a lot. Will there be
a part 2?
cegatmotor.com
December 2, 2024 @ 10:50 pm
I think this is among the most vital information for me.
And i’m glad reading your article. But should remark
on few general things, The site style is great, the articles is really excellent :
D. Good job, cheers
Social - paibe
December 2, 2024 @ 10:51 pm
What Makes Online Casinos Are Becoming a Worldwide Trend
Online casinos have changed the gambling market, delivering a unique kind of user-friendliness and diversity that brick-and-mortar venues fall short of. Throughout the last ten years, a growing community globally have adopted the thrill of internet-based gaming thanks to its availability, captivating elements, and constantly growing game libraries.
One of the strongest selling points of online gaming options is the incredible diversity of choices at your disposal. Whether you are a fan of interacting with traditional one-armed bandits, exploring story-driven visual slot games, or playing smart in table games like poker, online platforms offer numerous options. Plenty of operators additionally introduce real-time gaming experiences, giving you the chance you to communicate with actual dealers and fellow gamblers, all while soaking in the immersive feel of a land-based casino from anywhere you want.
If you’re a beginner with the world of virtual casino play or hope to discover proven options, why not become part of our vibrant interactive platform? It’s a place where enthusiasts share reviews, making it easier for you to enhance your casino activities. Join the connections and start your journey now: https://www.instagram.com/1winbr_com/ .
Adding to the extensive catalog, internet-based gambling hubs excel constant connectivity.
카지노사이트
December 2, 2024 @ 10:58 pm
Thanks for sharing your thoughts on 카지노사이트. Regards
Get Idea About Your Bit
December 2, 2024 @ 11:08 pm
you’re in reality a good webmaster. The web site loading pace is amazing.
It kind of feels that you are doing any distinctive trick.
Also, The contents are masterpiece. you’ve done a excellent activity
in this subject!
Social - paibe
December 2, 2024 @ 11:59 pm
How Online Casinos Are a Global Phenomenon
Internet-based gambling hubs have reshaped the casino gaming market, providing an exceptional degree of accessibility and variety that traditional casinos can’t match. Throughout the last ten years, millions of players internationally have turned to the fun of digital casino play thanks to its always-open nature, thrilling aspects, and widening collections of titles.
One of the most compelling reasons of digital gambling sites is the incredible selection of titles available. Whether you love playing on old-school slot machines, exploring theme-based modern slot games, or testing your strategy in card and board games like poker, digital casinos provide infinite entertainment avenues. Numerous services even feature live casino options, allowing you to participate with actual dealers and other players, all while taking in the immersive feel of a brick-and-mortar establishment from the comfort of your home.
If you’re a beginner with the world of digital casinos or seek to explore reliable sites, why not become part of our active community? It’s a space where players offer experiences, assisting you to enjoy more of your online casino experience. Dive into the experience and start your journey now: https://www.instagram.com/1bet_be/ .
Apart from the game range, internet-based gambling hubs stand out seamless entry.
bounty game
December 3, 2024 @ 12:36 am
Keep this going please, great job!
Harryslits
December 3, 2024 @ 12:45 am
Ivermectin Pharm: order minocycline 50mg – Ivermectin Pharm Store
51 game login
December 3, 2024 @ 1:06 am
I’m very happy to discover this site. I need to to thank you for your time for this
particularly wonderful read!! I definitely enjoyed every
little bit of it and I have you saved to fav to look at new stuff in your blog.
Social - paibe
December 3, 2024 @ 1:07 am
Reasons Why Online Casinos Remain an International Sensation
Digital casinos have modernized the casino gaming scene, offering an unmatched level of convenience and range that land-based gambling houses can’t match. In recent years, countless gamblers globally have chosen the fun of virtual casinos in light of its always-open nature, engaging traits, and widening catalogs of games.
One of the strongest selling points of online casinos is the vast diversity of entertainment options ready to play. Whether you prefer playing on retro slot machines, exploring narrative-rich video slots, or strategizing in table games like poker, online platforms deliver numerous opportunities. Many casinos even introduce live dealer games, making it possible for you to connect with actual dealers and opponents, all while immersing yourself in the immersive feel of a land-based casino from the comfort of your home.
If you’re just starting with the world of digital casinos or want to find out more about proven options, why not sign up for our vibrant online hub? It’s a platform where fans offer reviews, assisting you to get the most out of your gaming journey. Discover the conversation and visit us now: https://www.instagram.com/kwz.kz_official/ .
Adding to the extensive catalog, virtual gaming providers are known for ease of access.
Jacobgaugh
December 3, 2024 @ 1:25 am
кухни в москве — Большой выбор кухонь в Москве с индивидуальными проектами.
Eanrxii
December 3, 2024 @ 1:51 am
Официальная покупка диплома вуза с упрощенной программой обучения
Hervé Senni Music Streaming,
December 3, 2024 @ 1:59 am
I love what you guys are up too. This type of clever work and reporting!
Keep up the terrific works guys I’ve added you guys to our blogroll.
nexus for development
December 3, 2024 @ 2:10 am
Magnificent goods from you, man. I’ve take note
your stuff prior to and you are just too excellent. I
really like what you’ve received right here,
really like what you are saying and the best way by which you assert it.
You’re making it entertaining and you continue to care for to keep it sensible.
I can not wait to learn far more from you. This is really a great site.
Social - paibe
December 3, 2024 @ 2:11 am
Reasons Why Online Casinos Remain Highly Preferred Worldwide
Digital casinos have transformed the casino gaming world, providing an exceptional degree of user-friendliness and breadth that brick-and-mortar casinos struggle to rival. Over time, millions of players around the world have embraced the excitement of virtual gambling in light of its availability, captivating elements, and continuously increasing catalogs of games.
One of the biggest attractions of virtual gambling hubs is the incredible selection of gaming experiences on offer. Whether you like engaging with classic fruit machine slots, playing through narrative-rich video slots, or exercising tactics in table games like Texas Hold’em, online platforms offer numerous entertainment avenues. Numerous services furthermore feature live casino options, letting you to participate with professional croupiers and opponents, all while experiencing the realistic atmosphere of a land-based casino right at home.
If you’re a beginner with the world of virtual casino play or seek to explore reliable sites, why not become part of our growing online hub? It’s a place where fans offer experiences, helping you to enjoy more of your online casino experience. Dive into the experience and see it here now: https://www.instagram.com/1bet_be/ .
Apart from the game range, digital casino services excel seamless entry.
Visit here
December 3, 2024 @ 2:14 am
Hello! I could have sworn I’ve been to this site before but after browsing through a few
of the posts I realized it’s new to me. Anyways, I’m definitely happy I discovered it and I’ll be bookmarking it and checking back regularly!
Leonardsiz
December 3, 2024 @ 2:15 am
кухни от производителя – Надежность и качество прямо от производителя. Воплощаем ваши мечты о стильной кухне.
Blakesal
December 3, 2024 @ 2:17 am
кухни на заказ екатеринбург — Идеальные кухни на заказ, разработанные с учетом всех ваших пожеланий.
vivod iz zapoya v stacionare_wzOl
December 3, 2024 @ 3:11 am
выведение из запоя в стационаре [url=http://svarog.forum24.ru/?1-0-0-00000229-000-0-0-1730810647/]выведение из запоя в стационаре[/url] .
vivod iz zapoya v stacionare_nron
December 3, 2024 @ 3:12 am
выведение из запоя в стационаре [url=https://ya.7bb.ru/viewtopic.php?id=14622]выведение из запоя в стационаре[/url] .
Social - paibe
December 3, 2024 @ 3:20 am
What Makes Online Casinos Remain a Worldwide Trend
Virtual gambling platforms have transformed the casino gaming industry, providing a level of user-friendliness and range that land-based venues can’t match. Over the past decade, a vast number of enthusiasts worldwide have welcomed the excitement of digital casino play in light of its ease of access, thrilling aspects, and progressively larger game libraries.
One of the most compelling reasons of online gaming options is the astounding selection of gaming experiences provided. Whether you love spinning classic one-armed bandits, exploring plot-filled modern slot games, or testing your strategy in strategy-based games like Baccarat, casino websites provide countless choices. Many casinos moreover offer live gaming streams, making it possible for you to communicate with actual dealers and co-players, all while soaking in the realistic feel of a physical gaming house from anywhere you want.
If you’re a beginner with the world of virtual casino play or would like to explore reliable sites, why not sign up for our dynamic gaming forum? It’s a place where players share reviews, helping you to enjoy more of your gambling adventure. Dive into the community and visit us now: https://x.com/upX_Top .
Apart from the game range, virtual gambling platforms excel availability.
vivod iz zapoya v stacionare_rcMi
December 3, 2024 @ 3:20 am
вывод из запоя в стационаре [url=http://www.zarabotokdoma.creartuforo.com/viewtopic.php?id=11490#p23889]вывод из запоя в стационаре[/url] .
vivod iz zapoya v stacionare_qrma
December 3, 2024 @ 3:23 am
выведение из запоя нижний новгород [url=www.forumsilverstars.forum24.ru/?1-2-0-00000153-000-0-0-1730810963]выведение из запоя нижний новгород[/url] .
vivod iz zapoya v stacionare_aaOi
December 3, 2024 @ 3:24 am
выведение из запоя нижний новгород [url=http://rubiz.forum.cool/viewtopic.php?id=3754/]выведение из запоя нижний новгород[/url] .
Social - paibe
December 3, 2024 @ 4:26 am
Why Online Casinos Remain So Popular
Online casinos have reshaped the gaming market, providing an unmatched level of ease and variety that physical casinos struggle to rival. In recent years, millions of players around the world have embraced the fun of digital casino play because of its availability, thrilling aspects, and progressively larger range of offerings.
One of the biggest attractions of online gaming options is the unparalleled diversity of games available. Whether you love rolling old-school reel games, trying out theme-based visual slot games, or testing your strategy in card and board games like Texas Hold’em, online platforms offer limitless options. A large number of platforms even offer live casino options, making it possible for you to interact with human game hosts and gaming peers, all while experiencing the lifelike ambiance of a brick-and-mortar establishment right at home.
If you’re unfamiliar with the world of internet-based gaming or would like to find out more about reputable operators, why not join our vibrant gaming forum? It’s a space where players exchange stories, making it easier for you to maximize your virtual play. Discover the conversation and learn more now: https://www.facebook.com/profile.php?id=61568481962080 .
Besides the wide selection, online casinos thrive in ease of access.
Angelhob
December 3, 2024 @ 4:33 am
https://pilot-partner.ru – Простое решение для качественной фотосъемки вашего товара.
Michaelvok
December 3, 2024 @ 4:40 am
Интересная статья про мебель из массива дерева. Все, что нужно знать о мебели из сосны, березы, дуба или бука перед выбором и покупкой https://rempostroy.ru/arhitektura-i-dizayn/uhod-za-mebelyu-iz-massiva-sovety-i-pravila.html
Stevenrip
December 3, 2024 @ 4:42 am
Лабораторный компактный V-образный смеситель СФМ-11 подходит для смешивания различных материалов в областях фармацевтики, пищевой, химической, металлургии, керамики, электроники, аккумуляторов и т. д. и способен смешивать два или более сухих порошковых и/или гранулированных материалов вакуумные термоформеры
car insurance with sr22 in chicago
December 3, 2024 @ 5:02 am
If you’re unclear about the SR22 insurance demands in your condition, contact your
local area DMV for explanation. It is vital to totally comprehend
what is actually required to stay away from any type of lawful problems.
Harlequin Ichthyosis
December 3, 2024 @ 5:03 am
You really make it seem so easy with your presentation but I find
this matter to be really something that I think I would never understand.
It seems too complex and very broad for me. I am looking forward for your next post, I will try to get the
hang of it!
Social - paibe
December 3, 2024 @ 5:31 am
How Online Casinos Have Become an International Sensation
Virtual gambling platforms have transformed the gaming world, providing an unmatched level of user-friendliness and range that land-based establishments struggle to rival. Recently, a vast number of enthusiasts internationally have chosen the pleasure of virtual casinos due to its always-open nature, appealing qualities, and widening collections of titles.
One of the biggest attractions of digital gambling sites is the incredible variety of titles ready to play. Whether you are a fan of spinning classic one-armed bandits, trying out plot-filled thematic slots, or testing your strategy in classic casino games like Baccarat, casino websites offer limitless choices. Many casinos moreover feature live gaming streams, enabling you to connect with actual dealers and gaming peers, all while soaking in the engaging environment of a brick-and-mortar establishment in your own space.
If you’re new with the world of online gaming or are looking to delve deeper into reputable operators, why not participate in our growing online hub? It’s a space where enthusiasts discuss experiences, helping you to enjoy more of your online casino experience. Check out the connections and visit us now: https://www.facebook.com/profile.php?id=61568742522948 .
Beyond variety, digital casino services thrive in seamless entry.
Jacobgaugh
December 3, 2024 @ 6:18 am
http://goldcoon.ru – Удобный способ заказать стильную и функциональную кухню.
Social - paibe
December 3, 2024 @ 6:39 am
The Reasons Behind Why Online Casinos Are Becoming a Worldwide Trend
Virtual gambling platforms have transformed the gambling landscape, providing a level of comfort and variety that traditional gambling houses don’t provide. Over the past decade, a vast number of enthusiasts internationally have turned to the pleasure of internet-based gaming as a result of its ease of access, thrilling aspects, and progressively larger catalogs of games.
One of the main appeals of digital gambling sites is the astounding array of entertainment options on offer. Whether you love rolling traditional reel games, trying out plot-filled modern slot games, or exercising tactics in classic casino games like Blackjack, virtual venues feature endless options. A large number of platforms additionally feature interactive dealer games, allowing you to participate with actual dealers and other players, all while immersing yourself in the realistic atmosphere of a physical gaming house from anywhere you want.
If you’re just starting with the world of digital casinos or hope to learn about proven options, why not join our growing social network? It’s a destination where gamblers share insights, helping you to improve your gambling adventure. Dive into the community and check it out now: https://au.pinterest.com/casibom1/ .
In addition to diversity, internet-based gambling hubs thrive in ease of access.
RobertMep
December 3, 2024 @ 6:54 am
https://paxlovid.ink/# Paxlovid.ink
Blakesal
December 3, 2024 @ 7:17 am
кухни екатеринбург — Найдите идеальное решение для вашей кухни в Екатеринбурге.
Leonardsiz
December 3, 2024 @ 7:26 am
кухни на заказ спб – Создайте уникальную кухню, которая подчеркнет стиль вашего дома в Санкт-Петербурге.
Social - paibe
December 3, 2024 @ 7:44 am
What Makes Online Casinos Are Becoming So Popular
Internet-based gambling hubs have modernized the gaming industry, providing an exceptional degree of ease and variety that land-based gambling houses can’t match. Recently, a large audience across the globe have embraced the pleasure of internet-based gaming as a result of its ease of access, engaging traits, and continuously increasing game libraries.
One of the main appeals of online casinos is the vast diversity of gaming experiences provided. Whether you enjoy spinning traditional fruit machine slots, playing through theme-based modern slot games, or strategizing in table games like Blackjack, digital casinos offer numerous entertainment avenues. Plenty of operators additionally present live gaming streams, allowing you to interact with professional croupiers and opponents, all while immersing yourself in the realistic feel of a physical gaming house from anywhere you want.
If you’re a beginner with the world of virtual casino play or want to explore reliable sites, why not sign up for our vibrant interactive platform? It’s a hub where gamblers exchange experiences, making it easier for you to enhance your virtual play. Explore the experience and visit us now: https://www.instagram.com/kwz.kz_official/ .
Beyond variety, internet-based gambling hubs are known for availability.
real cash online casino
December 3, 2024 @ 7:53 am
Hi there! Quick question that’s totally off topic. Do you know how to make your site mobile friendly?
My web site looks weird when viewing from my
iphone 4. I’m trying to find a theme or plugin that might be able to correct this issue.
If you have any suggestions, please share.
With thanks!
Harryslits
December 3, 2024 @ 7:58 am
paxlovid price: Paxlovid.ink – Paxlovid.ink
카지노사이트
December 3, 2024 @ 8:24 am
I do not know whether it’s just me or if everyone else encountering problems with your website.
It appears like some of the written text in your posts
are running off the screen. Can someone else please provide feedback and let me know if this is
happening to them as well? This could be a issue with my browser because I’ve had this happen before.
Kudos
Xazrgdq
December 3, 2024 @ 8:41 am
Приобретение диплома ПТУ с сокращенной программой обучения в Москве
Social - paibe
December 3, 2024 @ 8:53 am
How Online Casinos Are Highly Preferred Worldwide
Digital casinos have modernized the gambling market, delivering a level of accessibility and selection that traditional establishments can’t match. Over time, countless gamblers worldwide have embraced the fun of digital casino play because of its anytime, anywhere convenience, engaging traits, and continuously increasing range of offerings.
One of the biggest attractions of virtual gambling hubs is the sheer selection of games provided. Whether you are a fan of engaging with classic one-armed bandits, exploring engaging visual slot games, or exercising tactics in strategy-based games like Roulette, virtual venues deliver limitless entertainment avenues. Numerous services additionally introduce live casino options, allowing you to engage with human game hosts and co-players, all while soaking in the realistic atmosphere of a land-based casino in your own space.
If you’re a beginner with the world of digital casinos or hope to delve deeper into safe services, why not join our lively interactive platform? It’s a space where gaming aficionados share experiences, assisting you to get the most out of your gambling adventure. Dive into the conversation and start your journey now: https://x.com/upX_Top .
Adding to the extensive catalog, virtual gambling platforms thrive in availability.
link mujurtoto2
December 3, 2024 @ 9:45 am
I used to be suggested this blog by means of my cousin. I’m no longer
positive whether this post is written by him as no one else know such
precise approximately my difficulty. You’re amazing!
Thanks!
Social - paibe
December 3, 2024 @ 10:01 am
Reasons Why Online Casinos Are Highly Preferred Worldwide
Internet-based gambling hubs have reshaped the gaming landscape, offering a unique kind of comfort and range that traditional venues fall short of. Recently, countless gamblers across the globe have embraced the excitement of virtual casinos because of its availability, captivating elements, and progressively larger game libraries.
One of the most compelling reasons of online gaming options is the unparalleled variety of choices provided. Whether you like playing on vintage reel games, immersing yourself in story-driven modern slot games, or mastering skills in card and board games like Blackjack, digital casinos provide infinite entertainment avenues. Plenty of operators also offer live gaming streams, enabling you to engage with human game hosts and gaming peers, all while immersing yourself in the engaging ambiance of a real casino from anywhere you want.
If you’re exploring for the first time with the world of internet-based gaming or hope to find out more about trusted platforms, why not engage with our dynamic interactive platform? It’s a place where enthusiasts share tips, enabling you to maximize your gambling adventure. Discover the community and see it here now: https://x.com/upX_Top .
Besides the wide selection, online casinos thrive in accessibility.
Techtra Automotive Academy
December 3, 2024 @ 10:48 am
Aw, this was a very good post. Taking a few
minutes and actual effort to generate a great article… but what can I say… I procrastinate a lot and
never manage to get anything done.
PUB 189
December 3, 2024 @ 11:03 am
Hi there are using WordPress for your blog platform?
I’m new to the blog world but I’m trying to get started and set up my own. Do you need any coding expertise to make your own blog?
Any help would be really appreciated!
Jacobgaugh
December 3, 2024 @ 11:05 am
кухни на заказ — Индивидуальный подход к созданию вашей кухни мечты.
Derekapaky
December 3, 2024 @ 11:08 am
кухни на заказ спб недорого – оптимальный вариант для тех, кто ищет качественную мебель по доступной цене.
Social - paibe
December 3, 2024 @ 11:09 am
The Reasons Behind Why Online Casinos Are Highly Preferred Worldwide
Internet-based gambling hubs have reshaped the betting market, offering a unique kind of user-friendliness and range that traditional establishments fall short of. Over the past decade, countless gamblers worldwide have embraced the excitement of digital casino play because of its ease of access, appealing qualities, and ever-expanding game libraries.
One of the most compelling reasons of online casinos is the incredible array of titles ready to play. Whether you prefer engaging with classic fruit machine slots, immersing yourself in story-driven video slots, or playing smart in traditional table offerings like Roulette, internet-based gambling sites deliver countless options. A large number of platforms furthermore offer interactive dealer games, enabling you to participate with human game hosts and other players, all while soaking in the immersive feel of a brick-and-mortar establishment from anywhere you want.
If you’re exploring for the first time with the world of virtual casino play or seek to learn about reliable sites, why not join our active gaming forum? It’s a place where enthusiasts share insights, making it easier for you to improve your gaming journey. Join the discussions and learn more now: https://www.facebook.com/profile.php?id=61568958127704 .
Beyond variety, digital casino services shine availability.
Mazrxzu
December 3, 2024 @ 11:35 am
Купить диплом ВУЗа России
Cazrore
December 3, 2024 @ 11:51 am
Можно ли быстро купить диплом старого образца и в чем подвох?
Blakesal
December 3, 2024 @ 12:09 pm
изготовление кухни на заказ — Качественное и профессиональное изготовление кухонь по индивидуальным проектам.
dondego
December 3, 2024 @ 12:19 pm
You really make it seem so easy with your presentation but I find
this topic to be actually something that I think
I would never understand. It seems too complicated and
extremely broad for me. I’m looking forward for your next post, I will try to get the hang of it!
Leonardsiz
December 3, 2024 @ 12:38 pm
https://tsmk-altai.ru – Индивидуальный подход и качественное исполнение вашей кухни.
fzxyr839
December 3, 2024 @ 1:11 pm
b138752
click this m4093533
beylikdüzü escort bayan
December 3, 2024 @ 1:18 pm
Beylikdüzü escort dünyası, estetik ve zarafetin buluştuğu bir yer. Bu yazıda Kainat Güzeli Asena’nın hikayesini keşfedeceksiniz.
dzieci porno
December 3, 2024 @ 1:34 pm
If some one wishes to be updated with hottest technologies after that he must be pay a quick visit this web page and be up to date every day.
honda ct125 dax
December 3, 2024 @ 1:35 pm
A motivating discussion is definitely worth comment.
I do think that you should write more about this
subject, it may not be a taboo matter but generally folks don’t discuss these topics.
To the next! Cheers!!
büyükçekmece escort
December 3, 2024 @ 1:59 pm
Dünyaca ünlü büyükçekmece escort sitesine hoşgeldiniz burda kendinize uygun bir kaliteli eş adayı bulabilirsiniz
DonaldIncex
December 3, 2024 @ 2:29 pm
Термоформер Multivac R085 предназначен для упаковки непищевых, пищевых и медицинских продуктов в мягкую и в жёсткую плёнку и вакуумирования смеситель бинов
Michaelvok
December 3, 2024 @ 2:35 pm
Блистерная упаковка (контурные ячейки) – вид пластиковой упаковки для фармацевтической продукции. Данный тип упаковки имеет множество преимущество: гигиеничность, компактность, защищенность от подделок и механических повреждений, удобство использования и пр вакуумная термоформовочная машина
FrankPubre
December 3, 2024 @ 2:57 pm
Ivermectin Pharm: ivermectin 0.5 – Ivermectin Pharm Store
brlwm229
December 3, 2024 @ 3:00 pm
h682952
additional resources d4279479
Angelhob
December 3, 2024 @ 3:02 pm
http://mirdolmenov.ru – Все подробности о фотосъемке для маркетплейсов и магазинов.
Harryslits
December 3, 2024 @ 3:03 pm
neurontin cost: neurontin online – Gabapentin Pharm
situs phising
December 3, 2024 @ 3:08 pm
Thanks for sharing your thoughts on situs phising.
Regards
네이버 아이디 구매
December 3, 2024 @ 3:09 pm
https://medium.com/@carlfrancoh38793/%EB%84%A4%EC%9D%B4%EB%B2%84-%EC%95%84%EC%9D%B4%EB%94%94-%EB%B9%84%EB%B0%80%EB%B2%88%ED%98%B8-%EB%B3%B5%EA%B5%AC%ED%95%98%EB%8A%94-5%EA%B0%80%EC%A7%80-%EB%B0%A9%EB%B2%95-216aafd6cf7c
www.gz-jj.com
December 3, 2024 @ 3:12 pm
Seriously tons of fantastic information.
Feel free to visit my web-site – クロムハーツ スーパーコピー 金 銀 (http://www.gz-jj.com/comment/html/?84816.html)
RobertMep
December 3, 2024 @ 3:22 pm
https://gabapentinpharm.com/# neurontin coupon
スーパーコピー わかる ファッ
December 3, 2024 @ 3:35 pm
Thanks a lot! Excellent stuff!
Here is my web-site; http://www.cqcici.com/comment/html/?170641.html
Jacobgaugh
December 3, 2024 @ 3:49 pm
http://www.taxipuma.ru — удобный интерфейс и полный каталог кухонь
Lazrjnx
December 3, 2024 @ 4:25 pm
Как получить диплом техникума с упрощенным обучением в Москве официально
카지노사이트
December 3, 2024 @ 4:35 pm
Hi all, here every one is sharing these kinds of familiarity,
so it’s nice to read this weblog, and I used to pay a quick visit
this blog all the time.
Blakesal
December 3, 2024 @ 5:02 pm
кухни под заказ екатеринбург — Широкий выбор кухонь под заказ с доставкой по Екатеринбургу.
Leonardsiz
December 3, 2024 @ 5:32 pm
https://viktorova-ts.ru – Удобный сервис для выбора и заказа кухни.
Harlequin Ichthyosis
December 3, 2024 @ 6:33 pm
Everything is very open with a really clear explanation of the issues.
It was truly informative. Your website is very useful.
Thanks for sharing!
check out this site
December 3, 2024 @ 6:52 pm
Heya i’m for the primary time here. I found this board and I in finding It really helpful & it helped
me out a lot. I’m hoping to present one thing back and aid others such as you aided
me.
Harlequin Ichthyosis
December 3, 2024 @ 6:54 pm
As the admin of this web site is working, no question very quickly it
will be well-known, due to its quality contents.
Derekapaky
December 3, 2024 @ 7:12 pm
https://tatarstan-heroes.ru – ресурс для выбора и заказа мебели на кухню.
Going Here
December 3, 2024 @ 7:19 pm
It’s going to be end of mine day, but before end I am reading this fantastic
article to improve my know-how.
vivod iz zapoya v stacionare_ipOr
December 3, 2024 @ 7:22 pm
вывод из запоя стационарно [url=https://pandora.ukrbb.net/viewtopic.php?f=2&t=12320]вывод из запоя стационарно[/url] .
vivod iz zapoya v stacionare_dxen
December 3, 2024 @ 7:24 pm
вывод из запоя в нижнем новгороде [url=dexanet.ukrbb.net/viewtopic.php?f=14&t=20414]вывод из запоя в нижнем новгороде[/url] .
vivod iz zapoya v stacionare_adpn
December 3, 2024 @ 7:27 pm
выведение из запоя в стационаре [url=www.golosa.ukrbb.net/viewtopic.php?f=3&t=7479/]выведение из запоя в стационаре[/url] .
vivod iz zapoya v stacionare_ywsa
December 3, 2024 @ 7:27 pm
наркология вывод из запоя в стационаре [url=https://kryto.ukrbb.net/viewtopic.php?f=3&t=1175/]наркология вывод из запоя в стационаре[/url] .
kyhni na zakaz_koOn
December 3, 2024 @ 7:28 pm
кухни на заказ [url=http://mebeldinastiya.ru/]http://mebeldinastiya.ru/[/url] .
Jacobgaugh
December 3, 2024 @ 7:57 pm
кухни москва — Большой выбор современных кухонь в Москве.
freespins
December 3, 2024 @ 8:02 pm
I am really thankful to the holder of this site who has shared this fantastic article at here.
LOPIUTV04
December 3, 2024 @ 8:15 pm
What’s up Dear, are you actually visiting this web site on a regular basis, if so then you will definitely get good experience.
casino extra mon compte
December 3, 2024 @ 8:23 pm
Lors du retrait de vos gains, les sites de casino en ligne ont pour habitude de confirmer vos documents de vérification.
DRTEYT783
December 3, 2024 @ 8:40 pm
Please let me know if you’re looking for a writer for your blog.
You have some really great posts and I believe I would be
a good asset. If you ever want to take some of the load off, I’d absolutely love to write some
articles for your blog in exchange for a link back to mine.
Please shoot me an email if interested. Kudos!
Yakima Web Design
December 3, 2024 @ 8:40 pm
Incredible! This blog looks exactly like my old
one! It’s on a totally different topic but it has pretty much the same layout
and design. Wonderful choice of colors!
Blakesal
December 3, 2024 @ 9:26 pm
mptextile.ru/ — Качественные кухни на заказ от профессионалов.
Angelhob
December 3, 2024 @ 10:01 pm
5hat.ru – Перейдите на сайт для получения подробной информации о наших услугах фотосъемки.
tyre shop near me open now
December 3, 2024 @ 10:04 pm
I visited several blogs but the audio quality for audio songs existing at this web page is truly superb.
Leonardsiz
December 3, 2024 @ 10:07 pm
кухни на заказ в спб — Кухни на заказ в Санкт-Петербурге: от классики до современного стиля.
Richardvak
December 3, 2024 @ 10:26 pm
[url=][/url]
Временная регистрация в СПб: Быстро и Легально!
Ищете, где оформить временную регистрацию в Санкт-Петербурге?
Мы гарантируем быстрое и легальное оформление без очередей и лишних документов.
Ваше спокойствие – наша забота!
Минимум усилий • Максимум удобства • Полная легальность
Свяжитесь с нами прямо сейчас!
Временная регистрация
[url=][/url]
Robertvox
December 3, 2024 @ 10:36 pm
how to buy clomid price: buy cheap clomid no prescription – can i buy clomid without rx
Jacobgaugh
December 3, 2024 @ 11:58 pm
https://www.gft-leasing.ru/ — Начните свой путь к идеальной кухне с нами.
Dnrtpvo
December 4, 2024 @ 12:23 am
Как приобрести аттестат о среднем образовании в Москве и других городах
whatsapp网页版登录入口
December 4, 2024 @ 12:39 am
Hello, i think that i saw you visited my weblog thus i came to “return the favor”.I’m attempting
to find things to improve my site!I suppose its ok to use some of your ideas!!
Sazrygy
December 4, 2024 @ 12:57 am
Легальные способы покупки диплома о среднем полном образовании
Derekapaky
December 4, 2024 @ 1:20 am
http://uristhmao.ru – удобный ресурс для создания уникальной кухни.
카지노사이트
December 4, 2024 @ 1:31 am
Undeniably consider that which you stated. Your favourite justification seemed to be on the web the easiest factor
to bear in mind of. I say to you, I definitely get annoyed while other people think about issues that
they just do not recognise about. You controlled to hit the nail upon the
highest as well as outlined out the whole thing
without having side-effects , other people could take a
signal. Will probably be back to get more. Thanks
GamaCasino
December 4, 2024 @ 1:32 am
[url=https://gama-casino-apk.ru]http://www.gama-casino-apk.ru[/url]
Установить последнюю версию приложения казино онлайн gama – выигрывай сегодня!
http://gama-casino-apk.ru
Blakesal
December 4, 2024 @ 1:45 am
https://www.north-web.ru — Получите подробности об услугах на нашем сайте.
blood sugar supplements
December 4, 2024 @ 2:14 am
Hello, i believe that i noticed you visited my weblog
so i got here to go back the choose?.I am attempting to
in finding things to improve my web site!I guess its ok to
use a few of your concepts!!
Leonardsiz
December 4, 2024 @ 2:36 am
https://tir92.ru/ – Мы знаем, как сделать вашу кухню комфортной и стильной.
GHTOPHTGF
December 4, 2024 @ 2:52 am
I really like your blog.. very nice colors &
theme. Did you create this website yourself or did you hire someone to do it for you?
Plz answer back as I’m looking to construct my own blog and would like
to find out where u got this from. many thanks
Buy Google Reviews Cheap
December 4, 2024 @ 2:55 am
Thanks for a marvelous posting! I genuinely enjoyed reading it, you’re a great author.I will
make certain to bookmark your blog and will come back at some point.
I want to encourage yourself to continue your great work, have a nice day!
natural clay toothpaste
December 4, 2024 @ 3:18 am
What’s up mates, how is everything, and what you would like to say concerning this article, in my view its in fact remarkable designed for me.
Howardteasy
December 4, 2024 @ 3:25 am
Компания Ист-Пак приняла участие в выставке Pharmtech & Ingredients 2024 — крупнейшей в России и странах ближнего зарубежья международной выставке оборудования, сырья и технологий для фармацевтического производства вакуумные термоформеры
Jimmieclict
December 4, 2024 @ 3:26 am
https://azithromycinus.com/# zithromax cost canada
ReggieBeaum
December 4, 2024 @ 3:28 am
Pin up login to the official website. Play online casino Pin Up for real money. Register on the Pin Up Casino website and get bonuses pin up casino
Sazrbfi
December 4, 2024 @ 3:38 am
Всё о покупке аттестата о среднем образовании: полезные советы
Harlequin Ichthyosis
December 4, 2024 @ 3:59 am
obviously like your website however you need to test the spelling on several
of your posts. Many of them are rife with spelling problems and I to find it very troublesome to inform the truth then again I will
surely come again again.
Rileypaf
December 4, 2024 @ 4:23 am
Viagra * Cialis * Levitra
All the products you are looking an eye to are currently available for the duration of 1+1.
4 more tablets of one of the following services: Viagra * Cialis * Levitra
https://xn--2i0bm4p0sf2wh7vdmsy.net
Angelhob
December 4, 2024 @ 4:27 am
https://www.bagroo.ru/ – Посетите наш сайт для консультации и заказа съемки товаров.
Jacobgaugh
December 4, 2024 @ 4:31 am
кухни в москве – Подбор кухонь с установкой в Москве и области.
Jaimeget
December 4, 2024 @ 4:53 am
zithromax z-pak [url=https://azithromycinus.com/#]zithromax 250 mg australia[/url] zithromax over the counter
iwinclub6688
December 4, 2024 @ 5:07 am
Yesterday, while I was at work, my sister stole my iPad and tested to see if it can survive a 30 foot drop,
just so she can be a youtube sensation. My apple ipad is
now broken and she has 83 views. I know this is totally off topic but I had to share it with someone!
naklejki z kosmonautami
December 4, 2024 @ 5:53 am
I enjoy what you guys tend too be up too. This kind of clever wwork
and coverage! Keep up the amazing works guys I’ve indluded you guys to my own blogroll. https://storage.googleapis.com/naklejkinasciane/naklejki-dzieciece-na-sciane/Naklejka-na-cian-Geisha.html
ADGKLIYT7
December 4, 2024 @ 5:54 am
What a data of un-ambiguity and preserveness of precious knowledge about unpredicted feelings.
Social - paibe
December 4, 2024 @ 6:05 am
The Reasons Behind Why Online Casinos Are a Worldwide Trend
Internet-based gambling hubs have modernized the gaming market, delivering a unique kind of convenience and selection that traditional casinos struggle to rival. Recently, countless gamblers around the world have chosen the adventure of online gaming due to its anytime, anywhere convenience, appealing qualities, and ever-expanding game libraries.
One of the biggest attractions of digital gambling sites is the vast range of entertainment options ready to play. Whether you like engaging with vintage one-armed bandits, playing through plot-filled video slots, or testing your strategy in traditional table offerings like Blackjack, digital casinos deliver infinite possibilities. Numerous services additionally introduce live casino options, enabling you to interact with actual dealers and fellow gamblers, all while experiencing the engaging ambiance of a brick-and-mortar establishment from the comfort of your home.
If you’re just starting with the world of online gaming or are looking to explore reliable sites, why not sign up for our dynamic social network? It’s a platform where players discuss stories, helping you to enjoy more of your gambling adventure. Explore the connections and check it out now: https://www.instagram.com/kwz.kz_official/ .
In addition to diversity, internet-based gambling hubs excel seamless entry.
Nevada website builders
December 4, 2024 @ 6:19 am
Pretty great post. I simply stumbled upon your weblog and wanted to mention that I’ve really loved surfing around
your weblog posts. In any case I’ll be subscribing to your rss feed
and I am hoping you write again very soon!
Blakesal
December 4, 2024 @ 6:37 am
кухни на заказ екатеринбург — Изготовление стильных и функциональных кухонь на заказ в Екатеринбурге.
xxx
December 4, 2024 @ 6:40 am
Pretty element of content. I just stumbled upon your blog and in accession capital to say that I get actually loved account your weblog posts.
Anyway I will be subscribing for your augment or even I success you get
entry to persistently quickly.
WesleyExele
December 4, 2024 @ 6:41 am
У нас есть всё для охоты, рыбалки и туризма. От рыболовных снастей и прикормки, одежды и обуви, до лодок и моторов, ножей и ружей, а также туристического рыбалка и охота
Social - paibe
December 4, 2024 @ 7:11 am
How Online Casinos Have Become a Global Phenomenon
Digital casinos have transformed the betting scene, delivering a unique kind of user-friendliness and variety that traditional venues fall short of. In recent years, countless gamblers internationally have chosen the thrill of online gaming due to its ease of access, thrilling aspects, and progressively larger catalogs of games.
One of the biggest attractions of internet-based platforms is the astounding range of gaming experiences at your disposal. Whether you prefer interacting with old-school slots, playing through story-driven modern slot games, or exercising tactics in card and board games like Baccarat, internet-based gambling sites provide countless opportunities. Numerous services additionally introduce live gaming streams, giving you the chance you to engage with real dealers and opponents, all while experiencing the immersive ambiance of a brick-and-mortar establishment from anywhere you want.
If you’re new with the world of digital casinos or want to explore reliable sites, why not become part of our active interactive platform? It’s a hub where gaming aficionados exchange tips, making it easier for you to enjoy more of your gaming journey. Join the conversation and start your journey now: https://www.facebook.com/profile.php?id=61568742522948 .
Beyond variety, digital casino services excel constant connectivity.
Derekapaky
December 4, 2024 @ 7:28 am
http://huskytaxi.ru/ – ваше решение для уникального дизайна кухни.
SGTRYIOP3
December 4, 2024 @ 7:28 am
Wonderful goods from you, man. I’ve remember your stuff prior to and you
are just too wonderful. I actually like what you’ve bought here, really like what you are stating and the best way during which you are saying it.
You are making it enjoyable and you still care for to keep it sensible.
I can’t wait to learn much more from you. That is actually a tremendous site.
Leonardsiz
December 4, 2024 @ 7:36 am
кухни под заказ – Уникальные и практичные решения для вашей кухни.
Harlequin Ichthyosis
December 4, 2024 @ 7:41 am
Good day! I could have sworn I’ve been to this site before but
after reading through some of the post I realized it’s new to me.
Nonetheless, I’m definitely happy I found it and I’ll be book-marking and checking back often!
Social - paibe
December 4, 2024 @ 8:17 am
Why Online Casinos Are Becoming a Global Phenomenon
Internet-based gambling hubs have transformed the casino gaming world, offering an unmatched level of user-friendliness and diversity that physical casinos can’t match. In recent years, a large audience internationally have turned to the fun of virtual gambling because of its accessibility, captivating elements, and ever-expanding game libraries.
One of the strongest selling points of virtual gambling hubs is the vast variety of titles on offer. Whether you enjoy engaging with vintage reel games, playing through narrative-rich visual slot games, or testing your strategy in strategy-based games like Texas Hold’em, online platforms provide numerous entertainment avenues. Several sites additionally introduce live casino options, making it possible for you to connect with real dealers and opponents, all while taking in the realistic environment of a real casino without leaving your home.
If you’re exploring for the first time with the world of virtual casino play or would like to explore safe services, why not participate in our lively interactive platform? It’s a platform where gaming aficionados share tips, making it easier for you to improve your virtual play. Dive into the connections and check it out now: https://www.instagram.com/1bet_be/ .
Besides the wide selection, digital casino services stand out accessibility.
Harlequin Ichthyosis
December 4, 2024 @ 8:44 am
Marvelous, what a web site it is! This web site presents helpful information to
us, keep it up.
halkalı escort
December 4, 2024 @ 9:11 am
dünyaca ünlü kalitenin tek adresi halkalı escort atakent escort sitesine hoşgeldiniz
Jacobgaugh
December 4, 2024 @ 9:21 am
кухни на заказ москва — Уникальные кухонные решения для жителей Москвы.
Social - paibe
December 4, 2024 @ 9:23 am
The Reasons Behind Why Online Casinos Have Become So Popular
Digital casinos have transformed the gaming scene, providing a level of comfort and range that conventional establishments fall short of. Throughout the last ten years, a vast number of enthusiasts worldwide have turned to the pleasure of digital casino play because of its anytime, anywhere convenience, exciting features, and continuously increasing catalogs of games.
One of the most compelling reasons of online gaming options is the vast diversity of games provided. Whether you like engaging with traditional slots, immersing yourself in story-driven thematic slots, or mastering skills in traditional table offerings like Texas Hold’em, casino websites feature countless possibilities. Numerous services even introduce live gaming streams, making it possible for you to participate with live hosts and co-players, all while immersing yourself in the engaging feel of a traditional gambling venue in your own space.
If you’re just starting with the world of internet-based gaming or want to discover proven options, why not sign up for our vibrant gaming forum? It’s a hub where players share experiences, guiding you to improve your online casino experience. Discover the connections and start your journey now: https://www.instagram.com/mystake_top/ .
Apart from the game range, virtual gaming providers are known for ease of access.
kurtköy eskort
December 4, 2024 @ 9:37 am
Pendik Escort Kurtköy Escort keyifli anlar yaşamak isteyenler için ideal seçenekler sunuyor. Pendik escort bayanlar arasından seçim yaparak, farklı deneyimler keşfedebilirsiniz.
WilliamMed
December 4, 2024 @ 9:51 am
Crazy Time is a popular live game show from Evolution Gaming in real time with live dealer. List of the best casinos to play Crazy Time Live Wheel https://crazytimelive-bn.tumblr.com/
Angelhob
December 4, 2024 @ 9:55 am
предметная фотосъемка москва – Профессиональная фотосъемка предметов для рекламы, каталогов и сайтов.
Harlequin Ichthyosis
December 4, 2024 @ 10:08 am
Hey! I’m at work browsing your blog from my new iphone 3gs!
Just wanted to say I love reading your blog and look forward to all your posts!
Keep up the fantastic work!
Social - paibe
December 4, 2024 @ 10:30 am
Why Online Casinos Are Becoming Highly Preferred Worldwide
Online casinos have transformed the casino gaming world, offering a unique kind of accessibility and selection that physical venues fall short of. Over the past decade, a vast number of enthusiasts around the world have adopted the fun of online gaming because of its always-open nature, exciting features, and constantly growing collections of titles.
One of the key draws of internet-based platforms is the unparalleled diversity of gaming experiences on offer. Whether you like playing on old-school one-armed bandits, exploring narrative-rich modern slot games, or playing smart in classic casino games like poker, casino websites provide endless options. Numerous services also offer real-time gaming experiences, making it possible for you to communicate with human game hosts and other players, all while experiencing the authentic feel of a brick-and-mortar establishment from the comfort of your home.
If you’re new with the world of digital casinos or want to explore trusted platforms, why not engage with our active online hub? It’s a hub where gamblers post insights, enabling you to enhance your gambling adventure. Join the conversation and learn more now: https://ru.pinterest.com/UpX_Official/ .
Adding to the extensive catalog, internet-based gambling hubs excel seamless entry.
Sazrasv
December 4, 2024 @ 10:39 am
Приобретение школьного аттестата с официальным упрощенным обучением в Москве
Ismaelaveds
December 4, 2024 @ 10:56 am
https://renvills-hotel.ru/rooms/
maps.app.goo.gl
December 4, 2024 @ 11:26 am
Hi, I do believe this is an excellent website. I stumbledupon it 😉 I’m going to revisit yet again since i have book-marked it.
Money and freedom is the best way to change, may you be rich and continue to help other people.
Blakesal
December 4, 2024 @ 11:31 am
кухни под заказ екатеринбург — Закажите кухню на заказ с доставкой по Екатеринбургу.
Social - paibe
December 4, 2024 @ 11:38 am
Reasons Why Online Casinos Have Become a Global Phenomenon
Virtual gambling platforms have revolutionized the casino gaming landscape, providing an exceptional degree of comfort and range that brick-and-mortar venues don’t provide. In recent years, millions of players across the globe have embraced the fun of internet-based gaming due to its anytime, anywhere convenience, appealing qualities, and continuously increasing selection of games.
One of the key draws of online casinos is the vast selection of titles provided. Whether you are a fan of interacting with retro fruit machine slots, exploring story-driven video-based games, or exercising tactics in card and board games like poker, casino websites offer countless entertainment avenues. Plenty of operators even offer interactive dealer games, making it possible for you to participate with real dealers and co-players, all while soaking in the realistic vibes of a land-based casino right at home.
If you’re unfamiliar with the world of virtual gambling or want to learn about reputable operators, why not become part of our vibrant social network? It’s a space where players discuss insights, making it easier for you to maximize your online casino experience. Join the discussions and see it here now: https://www.instagram.com/sweet_bonanzacom/ .
Apart from the game range, digital casino services stand out ease of access.
WaltonSer
December 4, 2024 @ 11:53 am
[url=][/url]
Временная регистрация в Санкт-Петербурге: Быстро и Легально!
Ищете, где оформить временную регистрацию в СПб?
Мы гарантируем быстрое и легальное оформление без очередей и лишних документов.
Ваше спокойствие – наша забота!
Минимум усилий • Максимум удобства • Полная легальность
Свяжитесь с нами прямо сейчас!
Временная регистрация в СПб
[url=][/url]
AI Low code Platform
December 4, 2024 @ 12:10 pm
Ahaa, its pleasant discussion concerning this piece of
writing here at this website, I have read all that, so at this time me also commenting here.
wing888
December 4, 2024 @ 12:31 pm
Your style is very unique in comparison to other people I’ve read stuff
from. Many thanks for posting when you’ve got the opportunity,
Guess I’ll just book mark this web site.
Leonardsiz
December 4, 2024 @ 12:38 pm
https://www.tsmk-altai.ru/ – Сделайте первый шаг к кухне вашей мечты.
Social - paibe
December 4, 2024 @ 12:44 pm
Reasons Why Online Casinos Have Become an International Sensation
Online casinos have modernized the gaming scene, offering a unique kind of user-friendliness and selection that conventional establishments struggle to rival. Over the past decade, a growing community globally have chosen the adventure of virtual gambling as a result of its always-open nature, appealing qualities, and continuously increasing game libraries.
One of the strongest selling points of digital gambling sites is the unparalleled array of titles available. Whether you enjoy spinning old-school one-armed bandits, immersing yourself in theme-based visual slot games, or strategizing in table games like Roulette, casino websites offer infinite opportunities. Several sites even offer live gaming streams, enabling you to interact with professional croupiers and other players, all while soaking in the immersive environment of a land-based casino right at home.
If you’re a beginner with the world of online gaming or seek to delve deeper into safe services, why not join our growing interactive platform? It’s a destination where gamblers discuss tips, enabling you to enhance your gaming journey. Join the experience and check it out now: https://www.facebook.com/profile.php?id=61568742522948 .
Adding to the extensive catalog, online casinos excel seamless entry.
Lazreax
December 4, 2024 @ 12:45 pm
Пошаговая инструкция по безопасной покупке диплома о высшем образовании
JeffreyZex
December 4, 2024 @ 12:51 pm
https://hoteltukan.ru/nomera-i-tseny/
Michaelabach
December 4, 2024 @ 1:05 pm
Портал строительства https://mbmedicall.com ресурс для поиска информации о ремонте и строительстве. Полезные советы, технологии, инструкции и рекомендации для профессионалов и новичков.
snyatie zapoya na domy_iypn
December 4, 2024 @ 1:38 pm
вывод из запоя на дому нижний [url=http://www.gaslo.ukrbb.net/viewtopic.php?f=13&t=3419]вывод из запоя на дому нижний[/url] .
snyatie zapoya na domy_drsl
December 4, 2024 @ 1:42 pm
капельница от запоя на дому нижний новгород [url=http://mymoscow.forum24.ru/?1-1-0-00001723-000-0-0-1730813000/]капельница от запоя на дому нижний новгород[/url] .
snyatie zapoya na domy_jkma
December 4, 2024 @ 1:42 pm
нижний новгород вывод из запоя на дому [url=http://www.www.rolandus.org/forum/viewtopic.php?p=106474]нижний новгород вывод из запоя на дому[/url] .
snyatie zapoya na domy_vdPr
December 4, 2024 @ 1:44 pm
капельница от запоя вызов на дом [url=http://uaforum.ukrbb.net/viewtopic.php?f=13&t=3249]капельница от запоя вызов на дом[/url] .
snyatie zapoya na domy_tlOi
December 4, 2024 @ 1:44 pm
вывод. из. запоя. на. дому. [url=https://www.astana.forum24.ru/?1-1-0-00000090-000-0-0-1730813261]вывод. из. запоя. на. дому.[/url] .
Social - paibe
December 4, 2024 @ 1:51 pm
The Reasons Behind Why Online Casinos Are Becoming a Global Phenomenon
Virtual gambling platforms have reshaped the betting industry, offering an unmatched level of comfort and diversity that traditional gambling houses fall short of. Recently, a vast number of enthusiasts internationally have embraced the fun of internet-based gaming thanks to its always-open nature, engaging traits, and widening selection of games.
One of the strongest selling points of virtual gambling hubs is the astounding variety of choices ready to play. Whether you prefer playing on traditional slot machines, diving into engaging modern slot games, or exercising tactics in table games like poker, online platforms deliver endless opportunities. Many casinos furthermore feature live gaming streams, giving you the chance you to communicate with live hosts and co-players, all while immersing yourself in the authentic atmosphere of a real casino right at home.
If you’re just starting with the world of virtual casino play or are looking to explore reliable sites, why not become part of our vibrant online hub? It’s a platform where gamblers share tips, assisting you to enhance your gaming journey. Join the connections and start your journey now: https://t.me/casibomgiris_net .
Apart from the game range, internet-based gambling hubs shine constant connectivity.
Jaimeget
December 4, 2024 @ 2:05 pm
where buy generic clomid online [url=https://clomid.store/#]can i get clomid without dr prescription[/url] where to buy generic clomid no prescription
JulioSnash
December 4, 2024 @ 2:06 pm
В зависимости от размера, толщины листового металла, из которого изготовлены трубы, меняется степень выдерживаемой нагрузки. Больше информации о каждом виде изделия, включая технические характеристики, вы получите, перейдя в карточку товара продукция металлопроката
Willieplada
December 4, 2024 @ 2:10 pm
Труба профильная, выполненная таким методом, также отличается повышенной пластичностью. Поэтому она подходит для изготовления различных заготовок, применяемых в каркасном строительстве и мебельном производстве метал купить
Jacobgaugh
December 4, 2024 @ 2:21 pm
Кухни в Москве — уникальные решения для любого интерьера
Social - paibe
December 4, 2024 @ 3:01 pm
Why Online Casinos Have Become So Popular
Online casinos have changed the betting market, delivering an exceptional degree of convenience and variety that brick-and-mortar venues fall short of. Over the past decade, millions of players across the globe have adopted the fun of virtual gambling due to its ease of access, appealing qualities, and continuously increasing range of offerings.
One of the strongest selling points of internet-based platforms is the unparalleled selection of entertainment options provided. Whether you enjoy rolling old-school fruit machine slots, playing through narrative-rich modern slot games, or exercising tactics in card and board games like Roulette, casino websites boast infinite choices. Numerous services also feature live gaming streams, giving you the chance you to interact with human game hosts and other players, all while enjoying the lifelike ambiance of a land-based casino right at home.
If you’re a beginner with the world of online gaming or seek to learn about proven options, why not join our growing community? It’s a place where enthusiasts offer experiences, assisting you to get the most out of your virtual play. Explore the experience and visit us now: https://www.instagram.com/sweet_bonanzacom/ .
Adding to the extensive catalog, digital casino services shine ease of access.
Lazrcpt
December 4, 2024 @ 3:10 pm
Всё, что нужно знать о покупке аттестата о среднем образовании без рисков
Mazrgqx
December 4, 2024 @ 3:55 pm
Купить диплом судоводителя
Investigate This Site
December 4, 2024 @ 3:57 pm
Do not ignore mouthwatering choices, either. Green spinach and feta pancakes make a delightful enhancement to a brunch snack bar, while packed white potato pancakes can be actually a favorite at a laid-back party. Along with simple preparation and never-ending variants, pancakes are actually the utmost dish that delivers folks together. Therefore, the upcoming time you’re intending an occasion, look at whipping up a batch of hot cakes to create tasty minds that every person will certainly value, https://www.exoltech.net/blogs/181793/D%C5%AFvod-Pancake-Recepty-Mo%C5%BEn%C3%A1-B%C3%BDt-Vlastn%C4%9B-v-Ka%C5%BEd%C3%BD-Doma-Kucha%C5%99ka.
Social - paibe
December 4, 2024 @ 4:09 pm
Why Online Casinos Remain a Global Phenomenon
Online casinos have changed the casino gaming scene, delivering an unmatched level of convenience and selection that conventional venues are unable to replicate. In recent years, a vast number of enthusiasts worldwide have embraced the thrill of virtual gambling due to its anytime, anywhere convenience, engaging traits, and ever-expanding collections of titles.
One of the most compelling reasons of internet-based platforms is the unparalleled variety of games available. Whether you love interacting with old-school slots, immersing yourself in engaging visual slot games, or testing your strategy in strategy-based games like Baccarat, casino websites offer countless possibilities. A large number of platforms moreover present live dealer games, making it possible for you to communicate with actual dealers and opponents, all while soaking in the immersive ambiance of a traditional gambling venue from anywhere you want.
If you’re unfamiliar with the world of virtual gambling or would like to explore trusted platforms, why not participate in our dynamic interactive platform? It’s a destination where gaming aficionados post experiences, helping you to get the most out of your gaming journey. Check out the experience and start your journey now: https://www.instagram.com/1bet_be/ .
In addition to diversity, virtual gambling platforms excel availability.
Get More Info
December 4, 2024 @ 5:04 pm
If you are actually seeking a tutoring company in Prague, it’s because you value quality learning and individual focus for your kid. Expert instructors center on enriching skill-sets, constructing confidence, and readying pupils for scholarly problems. Along with tailored approaches and knowledgeable teachers, tutoring services encourage pupils to reach their complete possibility and prosper in institution and past, https://start.me/w/RglXde.
비아그라
December 4, 2024 @ 5:06 pm
Wow, incredible blog layout! How long have you been blogging
for? you made blogging look easy. The overall look of your
site is fantastic, let alone the content!
Social - paibe
December 4, 2024 @ 5:15 pm
How Online Casinos Have Become a Worldwide Trend
Digital casinos have changed the casino gaming world, offering an unmatched level of accessibility and diversity that brick-and-mortar gambling houses can’t match. Over time, countless gamblers around the world have adopted the adventure of online gaming due to its ease of access, captivating elements, and constantly growing game libraries.
One of the most compelling reasons of online casinos is the astounding diversity of gaming experiences available. Whether you are a fan of interacting with vintage one-armed bandits, playing through narrative-rich thematic slots, or strategizing in card and board games like Baccarat, internet-based gambling sites offer infinite possibilities. Many casinos moreover offer live casino options, giving you the chance you to communicate with professional croupiers and co-players, all while enjoying the lifelike atmosphere of a brick-and-mortar establishment without leaving your home.
If you’re unfamiliar with the world of digital casinos or are looking to find out more about trusted platforms, why not join our dynamic social network? It’s a destination where enthusiasts exchange experiences, assisting you to enhance your virtual play. Join the experience and learn more now: https://www.instagram.com/kwz.kz_official/ .
Beyond variety, online casinos stand out accessibility.
Stephany
December 4, 2024 @ 5:17 pm
Hello! I just wanted to ask if you ever have any issues with
hackers? My last blog (wordpress) was hacked and I ended up losing many months
of hard work due to no back up. Do you have any solutions
to stop hackers?
Angelhob
December 4, 2024 @ 5:35 pm
https://www.pilot-partner.ru – Перейдите на сайт, чтобы ознакомиться с услугами и примерами работ.
luxury car rental kl
December 4, 2024 @ 6:02 pm
This piece of writing will help the internet people for building up new webpage or even a blog
from start to end.
click link
December 4, 2024 @ 6:02 pm
Very quickly this site will be famous among all blogging viewers,
due to it’s good articles or reviews
Social - paibe
December 4, 2024 @ 6:20 pm
Why Online Casinos Have Become an International Sensation
Virtual gambling platforms have modernized the casino gaming industry, delivering a unique kind of user-friendliness and breadth that conventional establishments are unable to replicate. In recent years, a vast number of enthusiasts worldwide have adopted the fun of virtual casinos in light of its always-open nature, appealing qualities, and constantly growing range of offerings.
One of the biggest attractions of online casinos is the vast variety of gaming experiences at your disposal. Whether you are a fan of playing on old-school slots, immersing yourself in engaging visual slot games, or exercising tactics in strategy-based games like Blackjack, internet-based gambling sites offer countless possibilities. Plenty of operators also present live dealer games, allowing you to participate with real dealers and gaming peers, all while immersing yourself in the engaging feel of a land-based casino right at home.
If you’re exploring for the first time with the world of internet-based gaming or are looking to find out more about trusted platforms, why not join our lively social network? It’s a space where gaming aficionados share experiences, making it easier for you to enjoy more of your gaming journey. Discover the community and check it out now: https://www.instagram.com/1winbr_com/ .
Besides the wide selection, virtual gambling platforms shine ease of access.
i爱思助手
December 4, 2024 @ 6:39 pm
Tremendous things here. I am very happy to look your article.
Thank you a lot and I am having a look forward to touch you.
Will you kindly drop me a mail?
mamiqq co
December 4, 2024 @ 6:49 pm
I like it whenever people get together and share ideas.
Great website, continue the good work!
Social - paibe
December 4, 2024 @ 7:27 pm
What Makes Online Casinos Have Become a Worldwide Trend
Digital casinos have transformed the casino gaming scene, providing an exceptional degree of user-friendliness and diversity that traditional casinos struggle to rival. Throughout the last ten years, a vast number of enthusiasts across the globe have adopted the fun of digital casino play as a result of its accessibility, captivating elements, and ever-expanding collections of titles.
One of the main appeals of online gaming options is the astounding array of entertainment options on offer. Whether you enjoy spinning classic slots, playing through narrative-rich video-based games, or playing smart in traditional table offerings like poker, digital casinos feature limitless options. Many casinos additionally introduce live gaming streams, enabling you to connect with human game hosts and other players, all while immersing yourself in the authentic vibes of a real casino from anywhere you want.
If you’re a beginner with the world of digital casinos or would like to learn about safe services, why not join our lively social network? It’s a place where gamblers post reviews, making it easier for you to enhance your casino activities. Join the experience and see it here now: https://www.instagram.com/mystake_top/ .
Apart from the game range, digital casino services shine ease of access.
Social - paibe
December 4, 2024 @ 8:32 pm
What Makes Online Casinos Are an International Sensation
Online casinos have modernized the gaming market, providing a unique kind of convenience and breadth that physical casinos don’t provide. Recently, millions of players worldwide have chosen the fun of digital casino play thanks to its availability, appealing qualities, and ever-expanding game libraries.
One of the biggest attractions of digital gambling sites is the incredible diversity of titles available. Whether you prefer rolling retro fruit machine slots, playing through story-driven thematic slots, or playing smart in traditional table offerings like poker, online platforms offer endless options. Plenty of operators additionally feature interactive dealer games, making it possible for you to interact with real dealers and gaming peers, all while soaking in the engaging environment of a brick-and-mortar establishment without leaving your home.
If you’re just starting with the world of online gaming or seek to explore reliable sites, why not participate in our lively community? It’s a hub where fans discuss insights, making it easier for you to get the most out of your virtual play. Dive into the community and visit us now: https://www.facebook.com/profile.php?id=61569299482300&is_tour_dismissed .
Besides the wide selection, virtual gaming providers thrive in ease of access.
vietyouthvn
December 4, 2024 @ 8:40 pm
Wow, marvelous weblog structure! How lengthy have you ever been blogging for?
you make running a blog glance easy. The whole glance of your website is great, as well as the content material!
Derekapaky
December 4, 2024 @ 8:47 pm
кухня на заказ – идеальное решение для реализации индивидуальных дизайнерских идей.
beo777
December 4, 2024 @ 9:04 pm
Hi there! I’m at work surfing around your blog from my new iphone 4!
Just wanted to say I love reading through your blog and look forward to all
your posts! Carry on the excellent work!
Social - paibe
December 4, 2024 @ 9:35 pm
What Makes Online Casinos Remain an International Sensation
Internet-based gambling hubs have reshaped the betting scene, delivering an unmatched level of ease and variety that physical establishments struggle to rival. Over the past decade, a growing community across the globe have welcomed the excitement of online gaming because of its always-open nature, exciting features, and constantly growing selection of games.
One of the key draws of internet-based platforms is the incredible diversity of titles ready to play. Whether you enjoy playing on classic reel games, trying out engaging thematic slots, or playing smart in classic casino games like Texas Hold’em, virtual venues boast countless choices. Numerous services moreover feature live gaming streams, allowing you to interact with human game hosts and opponents, all while enjoying the engaging ambiance of a real casino from anywhere you want.
If you’re just starting with the world of online gaming or hope to find out more about safe services, why not join our dynamic community? It’s a place where gamblers discuss insights, helping you to enjoy more of your virtual play. Join the community and learn more now: https://ru.pinterest.com/UpX_Official/ .
In addition to diversity, internet-based gambling hubs thrive in accessibility.
Social - paibe
December 4, 2024 @ 10:39 pm
How Online Casinos Have Become a Worldwide Trend
Virtual gambling platforms have transformed the gambling scene, delivering a unique kind of convenience and range that conventional gambling houses are unable to replicate. Recently, a large audience around the world have welcomed the fun of digital casino play due to its availability, appealing qualities, and continuously increasing game libraries.
One of the most compelling reasons of online casinos is the incredible diversity of entertainment options available. Whether you prefer spinning vintage reel games, diving into engaging visual slot games, or playing smart in classic casino games like Blackjack, virtual venues deliver endless choices. Numerous services additionally introduce real-time gaming experiences, making it possible for you to interact with human game hosts and other players, all while enjoying the authentic environment of a brick-and-mortar establishment right at home.
If you’re a beginner with the world of virtual gambling or hope to discover trusted platforms, why not participate in our vibrant social network? It’s a hub where fans share stories, making it easier for you to maximize your gambling adventure. Explore the conversation and visit us now: https://ru.pinterest.com/UpX_Official/ .
Adding to the extensive catalog, online casinos shine ease of access.
267908
December 4, 2024 @ 10:48 pm
Hello very nice web site!! Guy .. Excellent .. Wonderful ..
I will bookmark your website and take the feeds also?
I am satisfied to seek out numerous useful info right here within the post, we’d like work out more strategies in this regard,
thanks for sharing. . . . . .
raja luck login
December 4, 2024 @ 11:00 pm
Your style is very unique in comparison to other folks I have
read stuff from. Thank you for posting when you have the opportunity, Guess I’ll just book mark this web site.
ArmandoWal
December 4, 2024 @ 11:06 pm
how to get clomid for sale: get cheap clomid – where to get generic clomid
Matthewpok
December 4, 2024 @ 11:08 pm
https://cytotec.top/# Misoprostol 200 mg buy online
ciprofloxacin generic price [url=https://ciprofloxacin.cheap/#]ciprofloxacin order online[/url] buy cipro online canada
Cazrbsg
December 4, 2024 @ 11:37 pm
Как официально купить аттестат 11 класса с упрощенным обучением в Москве
Social - paibe
December 4, 2024 @ 11:43 pm
The Reasons Behind Why Online Casinos Are Becoming a Global Phenomenon
Internet-based gambling hubs have changed the gaming world, delivering an unmatched level of user-friendliness and diversity that brick-and-mortar gambling houses don’t provide. Recently, a growing community globally have turned to the pleasure of internet-based gaming as a result of its availability, engaging traits, and progressively larger range of offerings.
One of the strongest selling points of internet-based platforms is the unparalleled selection of choices provided. Whether you are a fan of playing on retro fruit machine slots, immersing yourself in plot-filled modern slot games, or strategizing in table games like Baccarat, casino websites feature countless opportunities. Numerous services even include real-time gaming experiences, enabling you to connect with real dealers and fellow gamblers, all while enjoying the lifelike ambiance of a brick-and-mortar establishment from the comfort of your home.
If you’re unfamiliar with the world of virtual gambling or are looking to discover proven options, why not participate in our lively social network? It’s a platform where gaming aficionados offer experiences, enabling you to enhance your casino activities. Discover the connections and visit us now: https://www.facebook.com/profile.php?id=61569299482300&is_tour_dismissed .
In addition to diversity, virtual gaming providers shine accessibility.
Angelhob
December 4, 2024 @ 11:57 pm
съемка ювелирных изделий – Изящные фотографии, подчеркивающие красоту ваших украшений.
Social - paibe
December 5, 2024 @ 12:49 am
Reasons Why Online Casinos Are Highly Preferred Worldwide
Digital casinos have reshaped the gambling world, delivering a level of accessibility and diversity that traditional venues struggle to rival. Recently, a vast number of enthusiasts around the world have turned to the adventure of virtual casinos as a result of its accessibility, engaging traits, and ever-expanding selection of games.
One of the main appeals of virtual gambling hubs is the sheer range of titles at your disposal. Whether you love spinning classic slots, exploring engaging thematic slots, or playing smart in classic casino games like Roulette, casino websites offer infinite options. Numerous services also introduce live dealer games, letting you to participate with professional croupiers and fellow gamblers, all while immersing yourself in the immersive atmosphere of a brick-and-mortar establishment from the comfort of your home.
If you’re new with the world of internet-based gaming or want to discover reputable operators, why not join our active interactive platform? It’s a hub where players post insights, guiding you to enhance your virtual play. Join the conversation and check it out now: https://x.com/upX_Top .
Apart from the game range, internet-based gambling hubs thrive in availability.
Mazrcjq
December 5, 2024 @ 1:04 am
Как получить диплом техникума официально и без лишних проблем
Cazrunh
December 5, 2024 @ 1:50 am
Официальная покупка диплома вуза с сокращенной программой в Москве
Social - paibe
December 5, 2024 @ 1:54 am
Reasons Why Online Casinos Have Become a Worldwide Trend
Online casinos have reshaped the casino gaming scene, delivering an exceptional degree of user-friendliness and variety that land-based casinos don’t provide. Throughout the last ten years, a growing community across the globe have welcomed the excitement of virtual gambling as a result of its ease of access, engaging traits, and progressively larger collections of titles.
One of the biggest attractions of internet-based platforms is the astounding variety of games provided. Whether you enjoy playing on traditional reel games, immersing yourself in plot-filled visual slot games, or exercising tactics in traditional table offerings like Texas Hold’em, internet-based gambling sites boast limitless possibilities. Plenty of operators even present live gaming streams, enabling you to engage with live hosts and opponents, all while taking in the authentic atmosphere of a brick-and-mortar establishment from anywhere you want.
If you’re unfamiliar with the world of internet-based gaming or would like to discover trusted platforms, why not join our dynamic social network? It’s a platform where gaming aficionados discuss insights, helping you to improve your gambling adventure. Join the community and start your journey now: https://www.facebook.com/profile.php?id=61568692273712 .
Beyond variety, internet-based gambling hubs stand out availability.
Robertvox
December 5, 2024 @ 2:00 am
lisinopril 20 mg tab price: zestoretic tabs – zestril 20 mg
JulioSnash
December 5, 2024 @ 2:40 am
Здесь есть статьи о любимых блюдах, кулинарные новости, кухонные лайфхаки и инструкции от лучших шеф-поваров как приготовить
Social - paibe
December 5, 2024 @ 2:58 am
Why Online Casinos Remain a Worldwide Trend
Internet-based gambling hubs have reshaped the gambling world, delivering an exceptional degree of comfort and variety that brick-and-mortar establishments struggle to rival. Recently, a growing community internationally have welcomed the pleasure of virtual gambling as a result of its availability, thrilling aspects, and ever-expanding game libraries.
One of the key draws of online gaming options is the vast range of games provided. Whether you enjoy spinning retro fruit machine slots, diving into story-driven thematic slots, or playing smart in strategy-based games like Baccarat, internet-based gambling sites feature numerous possibilities. Many casinos also feature real-time gaming experiences, giving you the chance you to engage with professional croupiers and co-players, all while soaking in the engaging atmosphere of a traditional gambling venue from anywhere you want.
If you’re unfamiliar with the world of virtual casino play or would like to find out more about reliable sites, why not engage with our dynamic community? It’s a space where enthusiasts offer insights, making it easier for you to improve your gambling adventure. Join the experience and visit us now: https://au.pinterest.com/casibom1/ .
Apart from the game range, virtual gaming providers excel seamless entry.
Harlequin Ichthyosis
December 5, 2024 @ 3:08 am
Very good information. Lucky me I found your site by chance (stumbleupon).
I’ve saved it for later!
Derekapaky
December 5, 2024 @ 3:27 am
http://www.tatarstan-heroes.ru – удобный сервис для выбора и заказа кухонь.
Social - paibe
December 5, 2024 @ 4:03 am
Reasons Why Online Casinos Are Highly Preferred Worldwide
Virtual gambling platforms have modernized the gambling landscape, delivering a level of comfort and breadth that traditional venues struggle to rival. Over the past decade, a growing community across the globe have welcomed the excitement of internet-based gaming in light of its accessibility, captivating elements, and continuously increasing catalogs of games.
One of the key draws of digital gambling sites is the vast range of choices available. Whether you prefer rolling old-school fruit machine slots, playing through plot-filled video slots, or mastering skills in card and board games like Texas Hold’em, digital casinos offer limitless options. Numerous services furthermore present live casino options, making it possible for you to participate with human game hosts and fellow gamblers, all while experiencing the authentic vibes of a real casino right at home.
If you’re unfamiliar with the world of virtual gambling or are looking to explore reliable sites, why not sign up for our vibrant interactive platform? It’s a platform where fans offer insights, assisting you to enhance your online casino experience. Dive into the experience and visit us now: https://www.facebook.com/profile.php?id=61568742522948 .
Beyond variety, virtual gambling platforms stand out availability.
https://Storage.Googleapis.com/naklejkinasciane/Naklejki-na-sciane-/Seamless-ethnic-pattern-background.html
December 5, 2024 @ 4:13 am
Great post. I am facing many of these issues as well.. https://Storage.Googleapis.com/naklejkinasciane/Naklejki-na-sciane-/Seamless-ethnic-pattern-background.html
Social - paibe
December 5, 2024 @ 5:09 am
Reasons Why Online Casinos Have Become a Global Phenomenon
Online casinos have reshaped the casino gaming scene, offering a level of user-friendliness and range that brick-and-mortar establishments don’t provide. In recent years, countless gamblers internationally have welcomed the fun of virtual gambling in light of its accessibility, exciting features, and widening selection of games.
One of the biggest attractions of digital gambling sites is the sheer diversity of gaming experiences on offer. Whether you prefer rolling retro slot machines, playing through narrative-rich video-based games, or mastering skills in traditional table offerings like Baccarat, virtual venues boast endless opportunities. Plenty of operators moreover include live dealer games, letting you to engage with real dealers and opponents, all while experiencing the immersive ambiance of a real casino from the comfort of your home.
If you’re new with the world of online gaming or would like to delve deeper into proven options, why not participate in our active online hub? It’s a place where players offer experiences, helping you to maximize your casino activities. Join the conversation and learn more now: https://www.instagram.com/mystake_top/ .
In addition to diversity, online casinos thrive in accessibility.
Rileypaf
December 5, 2024 @ 5:09 am
Viagra * Cialis * Levitra
All the products you are looking suitable are currently convenient in support of 1+1.
4 more tablets of one of the following services: Viagra * Cialis * Levitra
https://hp9.kr
Jimmieclict
December 5, 2024 @ 5:49 am
https://azithromycinus.com/# can you buy zithromax over the counter in canada
Social - paibe
December 5, 2024 @ 6:15 am
How Online Casinos Have Become an International Sensation
Internet-based gambling hubs have reshaped the casino gaming landscape, offering a unique kind of ease and diversity that physical venues fall short of. Throughout the last ten years, a vast number of enthusiasts around the world have adopted the adventure of online gaming in light of its availability, exciting features, and constantly growing game libraries.
One of the strongest selling points of internet-based platforms is the incredible diversity of choices provided. Whether you are a fan of interacting with old-school slots, trying out narrative-rich video slots, or playing smart in card and board games like Blackjack, online platforms offer infinite possibilities. Several sites additionally present real-time gaming experiences, making it possible for you to communicate with real dealers and co-players, all while soaking in the realistic environment of a brick-and-mortar establishment right at home.
If you’re unfamiliar with the world of digital casinos or are looking to learn about reputable operators, why not engage with our growing social network? It’s a space where gamblers share stories, guiding you to improve your virtual play. Discover the experience and start your journey now: https://www.facebook.com/profile.php?id=61568742522948 .
Adding to the extensive catalog, virtual gaming providers are known for seamless entry.
Artificial intelligence
December 5, 2024 @ 6:22 am
What’s up, of course this article is truly fastidious and I have
learned lot of things from it about blogging. thanks.
Leonardsiz
December 5, 2024 @ 6:25 am
кухни на заказ спб – Закажите кухню в Санкт-Петербурге, полностью адаптированную под ваши нужды.
Jaimeget
December 5, 2024 @ 6:34 am
cost of cheap clomid pills [url=https://clomid.store/#]how can i get cheap clomid without dr prescription[/url] can you buy cheap clomid without rx
MarvinMed
December 5, 2024 @ 6:59 am
В каждой игре существуют предметы, которые считаются особенно ценными и редкими. Эти артефакты зачастую становятся основным объектом стремлений игроков, так как могут существенно повлиять на их успех в игровом процессе. Их получение требует значительных усилий, времени или даже удачи, а их использование открывает новые возможности для развития персонажа http://wiki.why42.ru/index.php?title=%20%D0%A2%D0%BE%D0%BD%D0%BA%D0%BE%D1%81%D1%82%D0%B8%20%D0%B2%D1%8B%D0%B1%D0%BE%D1%80%D0%B0%20%D0%BA%D0%BE%D0%BB%D0%B5%D1%81%D0%BD%D1%8B%D1%85%20%D0%B4%D0%B8%D1%81%D0%BA%D0%BE%D0%B2
Cazrbyw
December 5, 2024 @ 7:05 am
Как приобрести диплом техникума с минимальными рисками
Social - paibe
December 5, 2024 @ 7:23 am
What Makes Online Casinos Have Become So Popular
Internet-based gambling hubs have revolutionized the gaming industry, delivering an exceptional degree of comfort and range that physical establishments don’t provide. Throughout the last ten years, millions of players around the world have chosen the adventure of virtual gambling in light of its anytime, anywhere convenience, engaging traits, and ever-expanding range of offerings.
One of the key draws of digital gambling sites is the sheer selection of choices ready to play. Whether you enjoy playing on old-school one-armed bandits, immersing yourself in story-driven video slots, or testing your strategy in card and board games like Texas Hold’em, virtual venues feature infinite possibilities. Several sites furthermore offer live dealer games, enabling you to participate with live hosts and opponents, all while enjoying the engaging feel of a land-based casino in your own space.
If you’re a beginner with the world of virtual casino play or hope to explore reputable operators, why not become part of our dynamic gaming forum? It’s a platform where gamblers offer experiences, helping you to improve your online casino experience. Dive into the community and see it here now: https://ru.pinterest.com/UpX_Official/ .
Apart from the game range, online casinos excel ease of access.
Angelhob
December 5, 2024 @ 7:33 am
фотосъемка одежды на моделях – Профессиональная съемка одежды на моделях для онлайн-магазинов и брендов.
JasonGOP
December 5, 2024 @ 7:33 am
Закончил универ и начал искать работу: сложности начались уже на этапе создания резюме. Денег на профессиональную консультацию HR на хедхантере было откровенно жалко, поэтому искал другие способы подготовки хорошего CV. Суммировал весь свой опыт и сделал неожиданные для себя выводы Резюме для интервью
binance
December 5, 2024 @ 7:35 am
Thanks for sharing. I read many of your blog posts, cool, your blog is very good.
JosephMossy
December 5, 2024 @ 7:36 am
[url=][/url]
Временная регистрация в Санкт-Петербурге: Быстро и Легально!
Ищете, где оформить временную регистрацию в СПБ?
Мы гарантируем быстрое и легальное оформление без очередей и лишних документов.
Ваше спокойствие – наша забота!
Минимум усилий • Максимум удобства • Полная легальность
Свяжитесь с нами прямо сейчас!
Временная регистрация в СПБ
[url=][/url]
Sazrqgv
December 5, 2024 @ 7:53 am
Официальная покупка диплома ПТУ с упрощенной программой обучения
24 hour electrician near me
December 5, 2024 @ 8:03 am
Thank you for the good writeup. It in fact was a amusement
account it. Look advanced to more added agreeable from you!
However, how can we communicate?
Jacobgaugh
December 5, 2024 @ 8:07 am
кухни под заказ — Закажите кухню с учетом ваших потребностей и размеров помещения.
snyatie zapoya na domy_ixpn
December 5, 2024 @ 8:21 am
вывод запой нижний [url=www.gonochki.forum24.ru/?1-10-0-00001142-000-0-0-1730815393/]вывод запой нижний[/url] .
snyatie zapoya na domy_niOa
December 5, 2024 @ 8:24 am
капельница от запоя на дому нижний новгород [url=https://www.fanfiction.borda.ru/?1-1-0-00000234-000-0-0-1730813920]капельница от запоя на дому нижний новгород[/url] .
snyatie zapoya na domy_joEn
December 5, 2024 @ 8:25 am
вывод. из. запоя. на. дому. [url=www.cah.forum24.ru/?1-19-0-00000466-000-0-0-1730814396/]вывод. из. запоя. на. дому.[/url] .
Social - paibe
December 5, 2024 @ 8:29 am
What Makes Online Casinos Are Becoming a Worldwide Trend
Internet-based gambling hubs have transformed the betting industry, delivering a unique kind of comfort and variety that land-based venues struggle to rival. Recently, a vast number of enthusiasts worldwide have welcomed the pleasure of virtual casinos because of its accessibility, exciting features, and progressively larger range of offerings.
One of the strongest selling points of virtual gambling hubs is the unparalleled array of choices at your disposal. Whether you enjoy engaging with old-school one-armed bandits, trying out theme-based modern slot games, or testing your strategy in card and board games like Baccarat, digital casinos feature limitless entertainment avenues. A large number of platforms also include live gaming streams, making it possible for you to interact with professional croupiers and fellow gamblers, all while taking in the engaging ambiance of a land-based casino in your own space.
If you’re new with the world of virtual casino play or want to learn about trusted platforms, why not sign up for our lively online hub? It’s a place where enthusiasts discuss stories, enabling you to get the most out of your gaming journey. Discover the experience and check it out now: https://x.com/upX_Top .
Beyond variety, virtual gaming providers shine constant connectivity.
snyatie zapoya na domy_leMt
December 5, 2024 @ 8:36 am
капельница при алкогольной интоксикации на дому цена [url=http://automobilist.forum24.ru/?1-19-0-00000144-000-0-0-1730815035]капельница при алкогольной интоксикации на дому цена[/url] .
snyatie zapoya na domy_vckl
December 5, 2024 @ 8:50 am
нарколог на дом вывод из запоя цена [url=dubna.myqip.ru/?1-5-0-00000288-000-0-0-1730814278]нарколог на дом вывод из запоя цена[/url] .
mzplay
December 5, 2024 @ 9:22 am
MZPlay is the best money making platform in Malaysia right now
Social - paibe
December 5, 2024 @ 9:36 am
The Reasons Behind Why Online Casinos Have Become a Global Phenomenon
Virtual gambling platforms have transformed the betting market, offering a unique kind of convenience and selection that conventional casinos don’t provide. Throughout the last ten years, millions of players globally have welcomed the fun of virtual gambling in light of its always-open nature, engaging traits, and widening game libraries.
One of the biggest attractions of digital gambling sites is the unparalleled selection of choices ready to play. Whether you prefer spinning vintage one-armed bandits, immersing yourself in plot-filled thematic slots, or playing smart in table games like Baccarat, online platforms deliver limitless possibilities. Several sites even introduce live casino options, letting you to communicate with real dealers and other players, all while taking in the lifelike feel of a brick-and-mortar establishment right at home.
If you’re new with the world of online gaming or hope to find out more about proven options, why not engage with our growing online hub? It’s a hub where gaming aficionados exchange experiences, enabling you to improve your online casino experience. Check out the conversation and see it here now: https://www.linkedin.com/in/my-stake-a096b427a/ .
Apart from the game range, online casinos shine availability.
here
December 5, 2024 @ 9:43 am
Do you have a spam problem on this website; I also am a blogger, and
I was wondering your situation; we have developed some nice procedures and we
are looking to exchange methods with other folks, why not
shoot me an e-mail if interested.
bokep
December 5, 2024 @ 10:32 am
whoah this weblog is wonderful i love studying your articles.
Stay up the good work! You already know, a lot of persons
are searching around for this information, you can aid them greatly.
mamiqq pkv
December 5, 2024 @ 10:44 am
Hiya! I know this is kinda off topic nevertheless I’d figured I’d ask.
Would you be interested in trading links or maybe guest writing a blog article or vice-versa?
My blog covers a lot of the same subjects as yours and I think we could greatly benefit from
each other. If you are interested feel free to send me an email.
I look forward to hearing from you! Superb blog by the
way!
Social - paibe
December 5, 2024 @ 10:45 am
Why Online Casinos Are So Popular
Virtual gambling platforms have reshaped the gaming industry, delivering an unmatched level of comfort and breadth that brick-and-mortar gambling houses fall short of. Throughout the last ten years, countless gamblers globally have turned to the adventure of virtual gambling thanks to its availability, appealing qualities, and ever-expanding game libraries.
One of the key draws of online gaming options is the incredible variety of choices at your disposal. Whether you enjoy engaging with old-school one-armed bandits, trying out theme-based visual slot games, or exercising tactics in classic casino games like Baccarat, virtual venues deliver countless choices. Several sites moreover offer real-time gaming experiences, letting you to interact with human game hosts and fellow gamblers, all while enjoying the authentic environment of a physical gaming house right at home.
If you’re unfamiliar with the world of digital casinos or are looking to learn about reliable sites, why not join our dynamic social network? It’s a destination where gamblers discuss experiences, assisting you to maximize your gaming journey. Join the community and check it out now: https://x.com/upX_Top .
Besides the wide selection, virtual gambling platforms thrive in seamless entry.
automotive photo retouching
December 5, 2024 @ 10:54 am
Thanks for your personal marvelous posting!
I genuinely enjoyed reading it, you are a great author. I
will always bookmark your blog and will come back someday.
I want to encourage you to ultimately continue your great work, have a nice
weekend!
Leonardsiz
December 5, 2024 @ 11:00 am
кухни на заказ в спб — Кухни на заказ в Санкт-Петербурге: от классики до современного стиля.
Derekapaky
December 5, 2024 @ 11:11 am
кухни на заказ в спб – это функциональность и стиль, созданные специально для вашего дома.
Trefbkh
December 5, 2024 @ 11:26 am
Приобретение диплома ПТУ с сокращенной программой обучения в Москве
Social - paibe
December 5, 2024 @ 11:52 am
Why Online Casinos Remain a Global Phenomenon
Internet-based gambling hubs have modernized the casino gaming market, offering a unique kind of ease and breadth that land-based gambling houses struggle to rival. Recently, a vast number of enthusiasts globally have turned to the adventure of online gaming as a result of its accessibility, thrilling aspects, and progressively larger game libraries.
One of the biggest attractions of digital gambling sites is the vast array of choices provided. Whether you are a fan of rolling classic reel games, exploring engaging thematic slots, or playing smart in strategy-based games like Texas Hold’em, virtual venues boast limitless options. Plenty of operators even feature interactive dealer games, allowing you to engage with professional croupiers and gaming peers, all while experiencing the authentic ambiance of a land-based casino without leaving your home.
If you’re unfamiliar with the world of virtual gambling or want to learn about reputable operators, why not sign up for our active interactive platform? It’s a platform where gaming aficionados share reviews, assisting you to enjoy more of your gambling adventure. Join the connections and learn more now: https://www.linkedin.com/in/my-stake-a096b427a/ .
Adding to the extensive catalog, online casinos are known for availability.
PatrickSpeld
December 5, 2024 @ 11:55 am
Шины являются важной частью любой специальной техники, обеспечивая её проходимость, устойчивость и безопасность на различных типах покрытия. Правильный выбор покрышек влияет на эффективность работы машины и её долговечность, а также помогает избежать возможных поломок и аварийных ситуаций http://wiki.catfolks.net/doku.php?id=%D0%9A%D0%B0%D0%BA%20%D0%B2%D1%8B%D0%B1%D1%80%D0%B0%D1%82%D1%8C%20%D1%88%D0%B8%D0%BD%D1%8B%20%D0%B4%D0%BB%D1%8F%20%D1%81%D0%BF%D0%B5%D1%86%D1%82%D0%B5%D1%85%D0%BD%D0%B8%D0%BA%D0%B8
Slottica Casino
December 5, 2024 @ 12:05 pm
Hi there to all, how is everything, I think every one is getting more from this website,
and your views are nice for new users.
Robertvox
December 5, 2024 @ 12:11 pm
medicine lisinopril 10 mg: lisinopril 10 best price – lisinopril online canadian pharmacy
Jacobgaugh
December 5, 2024 @ 12:48 pm
кухни под заказ — Разнообразные варианты кухонь, выполненные на заказ.
Matthewpok
December 5, 2024 @ 12:59 pm
http://azithromycinus.com/# zithromax canadian pharmacy
where can i get clomid no prescription [url=https://clomid.store/#]how can i get cheap clomid no prescription[/url] where can i get generic clomid pills
Social - paibe
December 5, 2024 @ 1:01 pm
The Reasons Behind Why Online Casinos Are a Worldwide Trend
Virtual gambling platforms have revolutionized the casino gaming world, delivering an unmatched level of comfort and variety that traditional venues fall short of. Over time, countless gamblers worldwide have embraced the thrill of virtual gambling due to its ease of access, exciting features, and constantly growing selection of games.
One of the key draws of online casinos is the sheer array of titles available. Whether you love engaging with vintage slots, trying out theme-based thematic slots, or exercising tactics in card and board games like Blackjack, casino websites feature limitless entertainment avenues. Several sites also introduce live gaming streams, making it possible for you to engage with professional croupiers and opponents, all while soaking in the authentic feel of a traditional gambling venue right at home.
If you’re unfamiliar with the world of digital casinos or seek to find out more about reliable sites, why not sign up for our lively social network? It’s a space where gaming aficionados offer stories, guiding you to enhance your casino activities. Explore the discussions and learn more now: https://t.me/casibomgiris_net .
In addition to diversity, virtual gambling platforms excel seamless entry.
Imbaslot Login
December 5, 2024 @ 1:03 pm
What’s up, I would like to subscribe for this weblog to take most up-to-date updates, therefore where can i do it
please assist.
autospin777
December 5, 2024 @ 1:12 pm
It’s very straightforward to find out any matter on web
as compared to textbooks, as I found this article at this site.
taktichn__zhKr
December 5, 2024 @ 1:30 pm
Неотразимый стиль современных тактичных штанов, как сочетать их с другой одеждой.
Неотъемлемая часть гардероба – тактичные штаны, которые подчеркнут ваш стиль и индивидуальность.
Идеальные тактичные штаны: находка для занятых людей, который подчеркнет вашу уверенность и статус.
Тактичные штаны для активного отдыха: важный элемент гардероба, которые подчеркнут вашу спортивную натуру.
Как выбрать тактичные штаны под свой стиль?, чтобы подчеркнуть свою уникальность и индивидуальность.
Секрет стильных мужчин: тактичные штаны, которые подчеркнут ваш вкус и качество вашей одежды.
Тактичные штаны: универсальный выбор для различных ситуаций, которые подчеркнут ваш профессионализм и серьезность.
тактичні штани [url=https://dffrgrgrgdhajshf.com.ua/]https://dffrgrgrgdhajshf.com.ua/[/url] .
Social - paibe
December 5, 2024 @ 2:08 pm
What Makes Online Casinos Have Become So Popular
Internet-based gambling hubs have transformed the gambling world, offering a unique kind of ease and range that land-based casinos struggle to rival. Recently, millions of players worldwide have embraced the thrill of internet-based gaming due to its always-open nature, captivating elements, and progressively larger game libraries.
One of the main appeals of digital gambling sites is the sheer selection of games provided. Whether you enjoy engaging with traditional one-armed bandits, playing through theme-based thematic slots, or playing smart in traditional table offerings like Roulette, digital casinos boast infinite options. Plenty of operators additionally introduce real-time gaming experiences, allowing you to interact with professional croupiers and fellow gamblers, all while immersing yourself in the engaging vibes of a traditional gambling venue right at home.
If you’re exploring for the first time with the world of digital casinos or seek to delve deeper into safe services, why not join our active gaming forum? It’s a destination where enthusiasts share tips, making it easier for you to maximize your virtual play. Join the discussions and check it out now: https://www.linkedin.com/in/my-stake-a096b427a/ .
In addition to diversity, virtual gambling platforms excel availability.
esim europe
December 5, 2024 @ 2:31 pm
Hey there, You’ve done a great job. I’ll certainly digg it
and personally suggest to my friends. I am confident they will be benefited from this website.
telegram 中文 版 下载
December 5, 2024 @ 2:50 pm
At this time I am going away to do my breakfast, later than having my breakfast
coming over again to read further news.
Jimmieclict
December 5, 2024 @ 2:58 pm
http://lisinoprilus.com/# zestril 20 mg tab
natural ED supplements
December 5, 2024 @ 3:02 pm
Wonderful article! This is the kind of information that
should be shared around the net. Disgrace on the search engines for no longer
positioning this submit upper! Come on over and consult with my website .
Thanks =)
Angelhob
December 5, 2024 @ 3:03 pm
предметная фотосъемка москва – Высококачественная фотосъемка различных предметов для каталогов и онлайн-платформ.
Social - paibe
December 5, 2024 @ 3:15 pm
Why Online Casinos Have Become So Popular
Digital casinos have changed the gaming scene, offering a unique kind of user-friendliness and range that conventional establishments fall short of. Over the past decade, a large audience across the globe have welcomed the thrill of digital casino play in light of its accessibility, exciting features, and continuously increasing range of offerings.
One of the most compelling reasons of online gaming options is the unparalleled range of titles available. Whether you enjoy interacting with old-school reel games, exploring theme-based visual slot games, or playing smart in traditional table offerings like Roulette, online platforms feature endless choices. Numerous services even present real-time gaming experiences, allowing you to participate with live hosts and opponents, all while experiencing the realistic feel of a real casino from anywhere you want.
If you’re just starting with the world of online gaming or want to find out more about safe services, why not engage with our dynamic gaming forum? It’s a place where fans discuss stories, enabling you to get the most out of your virtual play. Join the community and see it here now: https://ru.pinterest.com/UpX_Official/ .
In addition to diversity, internet-based gambling hubs stand out availability.
Mazrnlp
December 5, 2024 @ 3:30 pm
Купить диплом экономиста – оптимальное решение
Professional Year
December 5, 2024 @ 3:55 pm
This article is truly a fastidious one it assists new the web
viewers, who are wishing in favor of blogging.
Leonardsiz
December 5, 2024 @ 4:09 pm
https://www.tir92.ru – Посетите наш сайт и узнайте больше о том, как мы создаем кухни для вас.
Jaimeget
December 5, 2024 @ 4:11 pm
zithromax 500 without prescription [url=https://azithromycinus.com/#]buy zithromax 1000mg online[/url] zithromax order online uk
Social - paibe
December 5, 2024 @ 4:21 pm
What Makes Online Casinos Have Become a Worldwide Trend
Online casinos have reshaped the gaming scene, delivering an unmatched level of user-friendliness and variety that physical establishments struggle to rival. Over time, a vast number of enthusiasts across the globe have turned to the pleasure of virtual casinos in light of its anytime, anywhere convenience, exciting features, and ever-expanding game libraries.
One of the key draws of online gaming options is the astounding variety of choices available. Whether you are a fan of rolling old-school slot machines, trying out theme-based video slots, or testing your strategy in table games like poker, online platforms boast numerous opportunities. Plenty of operators also include real-time gaming experiences, making it possible for you to connect with professional croupiers and gaming peers, all while enjoying the realistic ambiance of a traditional gambling venue from anywhere you want.
If you’re new with the world of virtual casino play or hope to find out more about reliable sites, why not engage with our lively online hub? It’s a hub where fans discuss stories, making it easier for you to get the most out of your online casino experience. Discover the conversation and visit us now: https://www.facebook.com/profile.php?id=61568958127704 .
Beyond variety, internet-based gambling hubs excel seamless entry.
CBD Oil UK Online
December 5, 2024 @ 4:36 pm
Hey! I just wanted to ask if you ever have any issues with hackers?
My last blog (wordpress) was hacked and I ended up
losing a few months of hard work due to no back up.
Do you have any solutions to stop hackers?
Social - paibe
December 5, 2024 @ 5:23 pm
The Reasons Behind Why Online Casinos Are Becoming an International Sensation
Internet-based gambling hubs have changed the betting industry, offering an exceptional degree of user-friendliness and variety that physical gambling houses are unable to replicate. In recent years, a growing community across the globe have embraced the excitement of digital casino play due to its always-open nature, captivating elements, and continuously increasing selection of games.
One of the main appeals of internet-based platforms is the sheer selection of entertainment options provided. Whether you prefer playing on traditional one-armed bandits, trying out theme-based modern slot games, or strategizing in strategy-based games like Texas Hold’em, online platforms feature infinite entertainment avenues. Plenty of operators additionally introduce real-time gaming experiences, enabling you to communicate with actual dealers and opponents, all while experiencing the realistic vibes of a traditional gambling venue right at home.
If you’re exploring for the first time with the world of virtual gambling or would like to explore reputable operators, why not engage with our lively interactive platform? It’s a platform where fans discuss stories, helping you to improve your virtual play. Explore the connections and visit us now: https://au.pinterest.com/casibom1/ .
In addition to diversity, digital casino services stand out accessibility.
Read My Reviews Here
December 5, 2024 @ 5:40 pm
Hello, I would like to subscribe for this website to take newest updates, thus
where can i do it please help.
Kup Viagrę Online
December 5, 2024 @ 5:42 pm
My partner and I stumbled over here coming from a different page and thought I might
as well check things out. I like what I see so now i am following you.
Look forward to looking into your web page yet again.
Find More About This
December 5, 2024 @ 5:48 pm
Pretty nice post. I just stumbled upon your weblog
and wished to say that I have really enjoyed
browsing your blog posts. After all I’ll be subscribing to your feed and I hope you write again soon!
Jacobgaugh
December 5, 2024 @ 5:49 pm
кухни в москве – Подбор кухонь с установкой в Москве и области.
검색 엔진 최적화
December 5, 2024 @ 5:58 pm
백링크는 웹사이트의 신뢰성을 높이고 검색 엔진 최적화(SEO)에 중요한 역할을 합니다.
유용한 콘텐츠를 제공함으로써 다른 사이트로부터
백링크를 유도하면 자연스럽게 트래픽이 증가하게 됩니다.
Derekapaky
December 5, 2024 @ 6:02 pm
кухня на заказ в спб – отличный способ получить интерьер вашей мечты с учетом всех пожеланий.
Навесы на заказ в Ленинградской области
December 5, 2024 @ 6:05 pm
Собственное производство металлоконструкций. Если вас интересует Навесы из профильной трубы мы предлогаем изготовление под ключ навес из металлопрофиля цена
Social - paibe
December 5, 2024 @ 6:26 pm
Why Online Casinos Have Become So Popular
Digital casinos have transformed the casino gaming landscape, offering an unmatched level of accessibility and diversity that traditional venues don’t provide. Over the past decade, countless gamblers globally have embraced the adventure of online gaming thanks to its always-open nature, exciting features, and widening selection of games.
One of the most compelling reasons of virtual gambling hubs is the unparalleled diversity of games ready to play. Whether you love engaging with classic slot machines, immersing yourself in narrative-rich video-based games, or mastering skills in card and board games like Blackjack, internet-based gambling sites offer endless choices. Numerous services additionally present live casino options, allowing you to participate with live hosts and fellow gamblers, all while experiencing the engaging atmosphere of a brick-and-mortar establishment from the comfort of your home.
If you’re exploring for the first time with the world of internet-based gaming or would like to discover proven options, why not join our active interactive platform? It’s a place where gamblers post reviews, assisting you to improve your virtual play. Dive into the conversation and start your journey now: https://www.instagram.com/1winbr_com/ .
Adding to the extensive catalog, online casinos excel accessibility.
ovarian cancer symptoms after menopause
December 5, 2024 @ 6:28 pm
you’re actually a just right webmaster. The web site
loading pace is amazing. It seems that you’re
doing any distinctive trick. Moreover, The contents are masterpiece.
you have performed a magnificent task in this
subject!
Where is American Quality Ammunition Manufactured
December 5, 2024 @ 6:40 pm
Hi i am kavin, its my first occasion to commenting anyplace, when i read this piece of writing i thought i could also create comment
due to this sensible paragraph.
Social - paibe
December 5, 2024 @ 7:29 pm
Why Online Casinos Have Become an International Sensation
Virtual gambling platforms have modernized the gambling landscape, offering an unmatched level of convenience and breadth that physical establishments don’t provide. Throughout the last ten years, millions of players internationally have chosen the pleasure of online gaming in light of its accessibility, captivating elements, and progressively larger catalogs of games.
One of the most compelling reasons of virtual gambling hubs is the vast variety of gaming experiences on offer. Whether you like interacting with traditional fruit machine slots, exploring story-driven thematic slots, or mastering skills in traditional table offerings like Texas Hold’em, casino websites offer endless choices. A large number of platforms moreover feature live dealer games, making it possible for you to interact with human game hosts and gaming peers, all while soaking in the lifelike vibes of a physical gaming house in your own space.
If you’re just starting with the world of internet-based gaming or would like to find out more about safe services, why not sign up for our dynamic online hub? It’s a destination where enthusiasts discuss experiences, making it easier for you to enjoy more of your gambling adventure. Check out the conversation and visit us now: https://www.instagram.com/mystake_top/ .
In addition to diversity, online casinos shine ease of access.
Dnrtqdd
December 5, 2024 @ 8:05 pm
Как безопасно купить диплом колледжа или ПТУ в России, что важно знать
wilkinson laundry chute parts
December 5, 2024 @ 8:31 pm
I am regular reader, how are you everybody? This paragraph posted at
this web site is in fact pleasant.
Review my web-site wilkinson laundry chute parts
Social - paibe
December 5, 2024 @ 8:36 pm
How Online Casinos Are Becoming a Worldwide Trend
Online casinos have reshaped the gaming industry, delivering a unique kind of ease and range that brick-and-mortar casinos struggle to rival. Recently, millions of players worldwide have adopted the adventure of internet-based gaming due to its accessibility, thrilling aspects, and widening collections of titles.
One of the strongest selling points of internet-based platforms is the vast array of entertainment options at your disposal. Whether you prefer playing on traditional reel games, trying out plot-filled video slots, or exercising tactics in table games like poker, digital casinos deliver endless possibilities. Several sites also include live dealer games, letting you to participate with actual dealers and co-players, all while taking in the immersive atmosphere of a physical gaming house without leaving your home.
If you’re exploring for the first time with the world of virtual gambling or seek to discover trusted platforms, why not join our vibrant social network? It’s a hub where fans post reviews, enabling you to maximize your gambling adventure. Discover the community and learn more now: https://x.com/upX_Top .
In addition to diversity, internet-based gambling hubs shine constant connectivity.
tội phạm tính dục
December 5, 2024 @ 8:47 pm
Hi Dear, are you truly visiting this site daily, if so
afterward you will definitely obtain fastidious know-how.
Leonardsiz
December 5, 2024 @ 8:50 pm
https://tomason-russia.ru/ – Высокое качество и стиль в каждой детали вашей кухни.
sell whatsapp channels
December 5, 2024 @ 9:00 pm
Spot on with this write-up, I actually think this site needs a lot more attention. I’ll
probably be back again to read through more, thanks for the advice!
Angelhob
December 5, 2024 @ 9:10 pm
предметная фотосъемка товаров – Эффективная съемка товаров для привлечения покупателей и увеличения продаж.
Visit Here
December 5, 2024 @ 9:10 pm
The glamorous environments allow you to breathe greatly and appreciate the minute, all while sipping on a fascinating natural tea. You’ll leave behind emotion rejuvenated and all set to look into the captivating streets of Prague. Staying at this luxury accommodation suggests prioritizing your welfare, ensuring you come back home not merely kicked back, yet absolutely invigorated. Welcome this chance for spectacular leisure and wellness in the course of your visit, https://www.tumblr.com/jocelynsfairbank/768824528657874944/nejlep%C5%A1%C3%AD-funkce-o%C4%8Dek%C3%A1vat-v-luxusn%C3%AD-resort-core?source=share.
Social - paibe
December 5, 2024 @ 9:42 pm
How Online Casinos Are Becoming Highly Preferred Worldwide
Internet-based gambling hubs have reshaped the gambling landscape, offering an unmatched level of convenience and variety that traditional casinos can’t match. Over time, millions of players across the globe have adopted the excitement of internet-based gaming due to its accessibility, appealing qualities, and widening selection of games.
One of the key draws of virtual gambling hubs is the astounding diversity of choices available. Whether you like engaging with retro slots, diving into story-driven thematic slots, or mastering skills in traditional table offerings like Blackjack, virtual venues offer limitless options. Many casinos furthermore introduce interactive dealer games, letting you to connect with professional croupiers and opponents, all while enjoying the authentic environment of a brick-and-mortar establishment from the comfort of your home.
If you’re a beginner with the world of virtual gambling or seek to discover reliable sites, why not become part of our lively gaming forum? It’s a space where gaming aficionados discuss stories, making it easier for you to get the most out of your gaming journey. Discover the discussions and learn more now: https://www.linkedin.com/in/my-stake-a096b427a/ .
In addition to diversity, online casinos stand out ease of access.
Robertvox
December 5, 2024 @ 9:57 pm
п»їcytotec pills online: Cytotec 200mcg price – purchase cytotec
Pag-sign Up
December 5, 2024 @ 9:59 pm
I don’t think the title of your article matches the content lol. Just kidding, mainly because I had some doubts after reading the article.
Jacobgaugh
December 5, 2024 @ 10:21 pm
кухни от производителя — Прямые поставки качественных кухонь от проверенных фабрик.
Empresa de tratamiento de humedades
December 5, 2024 @ 10:32 pm
Spot on with this write-up, I really think this website needs
much more attention. I’ll probably be back again to
see more, thanks for the advice!
Singapore confinement nanny
December 5, 2024 @ 10:36 pm
Having read this I believed it was extremely enlightening.
I appreciate you taking the time and energy to put this informative article together.
I once again find myself personally spending a lot of time both reading and posting comments.
But so what, it was still worthwhile!
JosephMossy
December 5, 2024 @ 10:40 pm
[url=][/url]
Временная регистрация в Санкт-Петербурге: Быстро и Легально!
Ищете, где оформить временную регистрацию в СПБ?
Мы гарантируем быстрое и легальное оформление без очередей и лишних документов.
Ваше спокойствие – наша забота!
Минимум усилий • Максимум удобства • Полная легальность
Свяжитесь с нами прямо сейчас!
Временная регистрация в СПБ
[url=][/url]
Social - paibe
December 5, 2024 @ 10:46 pm
What Makes Online Casinos Are Highly Preferred Worldwide
Online casinos have modernized the gambling scene, providing a unique kind of comfort and variety that land-based venues fall short of. Over the past decade, a growing community internationally have chosen the pleasure of virtual gambling thanks to its always-open nature, captivating elements, and ever-expanding game libraries.
One of the most compelling reasons of virtual gambling hubs is the unparalleled array of gaming experiences on offer. Whether you enjoy playing on classic slots, immersing yourself in plot-filled visual slot games, or exercising tactics in strategy-based games like Roulette, casino websites offer limitless possibilities. A large number of platforms even present live gaming streams, allowing you to engage with actual dealers and opponents, all while enjoying the immersive feel of a land-based casino in your own space.
If you’re new with the world of internet-based gaming or would like to learn about proven options, why not join our growing interactive platform? It’s a destination where gamblers discuss insights, helping you to enhance your online casino experience. Join the conversation and start your journey now: https://www.instagram.com/1winbr_com/ .
Beyond variety, digital casino services excel seamless entry.
토토사이트
December 5, 2024 @ 11:04 pm
안전한 토토사이트와 메이저사이트를 선택하면, 믿을 수 있는 환경에서 메이저놀이터의 다양한 게임을 즐길 수 있습니다.
이러한 사이트들은 사용자에게 안전하고 즐거운
경험을 제공하여, 게임에 대한 신뢰성을 높이는 데
큰 역할을 합니다.
snyatie zapoya na domy_rber
December 5, 2024 @ 11:29 pm
сделать капельницу на дому от алкоголя [url=https://ideya.forums.party/viewtopic.php?id=668/]сделать капельницу на дому от алкоголя[/url] .
apkportalnik
December 5, 2024 @ 11:30 pm
закачать приложения онлайн казино https://funke-schluesseldienst.de/luchshie-onlajn-kazino-igrat-online-casino-na-7/
snyatie zapoya na domy_bbpr
December 5, 2024 @ 11:31 pm
сколько стоит капельница на дому от запоя [url=www.svarog.forum24.ru/?1-0-0-00000230-000-0-0-1730815368]сколько стоит капельница на дому от запоя[/url] .
snyatie zapoya na domy_ftSa
December 5, 2024 @ 11:32 pm
капельница от запоя нижний новгород [url=forumsilverstars.forum24.ru/?1-2-0-00000154-000-0-0-1730815566]капельница от запоя нижний новгород [/url] .
Derekapaky
December 5, 2024 @ 11:48 pm
кухня на заказ в спб – идеальный вариант для тех, кто хочет создать уютный и современный интерьер.
Social - paibe
December 5, 2024 @ 11:50 pm
How Online Casinos Remain So Popular
Virtual gambling platforms have modernized the gambling scene, offering a level of user-friendliness and selection that physical casinos fall short of. Over time, countless gamblers globally have welcomed the excitement of virtual gambling due to its availability, engaging traits, and continuously increasing collections of titles.
One of the biggest attractions of online gaming options is the astounding selection of gaming experiences provided. Whether you like engaging with traditional slots, diving into narrative-rich video-based games, or strategizing in card and board games like Roulette, internet-based gambling sites feature countless possibilities. Several sites furthermore present real-time gaming experiences, making it possible for you to connect with live hosts and co-players, all while immersing yourself in the realistic atmosphere of a land-based casino from the comfort of your home.
If you’re a beginner with the world of internet-based gaming or want to discover safe services, why not engage with our growing online hub? It’s a space where players offer tips, helping you to enhance your gambling adventure. Check out the connections and see it here now: https://t.me/casibomgiris_net .
Besides the wide selection, online casinos shine availability.
สล็อต pp
December 5, 2024 @ 11:54 pm
You suggested that terrifically! https://maps.google.com.ai/url?q=https://ppslot-th.me
Juragan4d
December 6, 2024 @ 12:06 am
Magnificent beat ! I would like to apprentice while you amend your
web site, how could i subscribe for a blog site?
The account helped me a acceptable deal. I had been a little bit acquainted
of this your broadcast offered bright clear concept
My web
December 6, 2024 @ 12:26 am
Hello, yup this piece of writing is really good and I
have learned lot of things from it regarding blogging.
thanks.
Crypto asset recovery results
December 6, 2024 @ 12:26 am
Hello mates, how is everything, and what you desire to say about this paragraph, in my view its truly remarkable for me.
Social - paibe
December 6, 2024 @ 12:57 am
The Reasons Behind Why Online Casinos Are So Popular
Virtual gambling platforms have transformed the gaming scene, delivering an exceptional degree of accessibility and range that traditional gambling houses are unable to replicate. In recent years, a growing community worldwide have embraced the adventure of internet-based gaming as a result of its anytime, anywhere convenience, engaging traits, and progressively larger collections of titles.
One of the biggest attractions of digital gambling sites is the vast selection of entertainment options ready to play. Whether you enjoy interacting with vintage fruit machine slots, trying out engaging video slots, or testing your strategy in table games like Texas Hold’em, online platforms feature endless opportunities. Many casinos furthermore feature real-time gaming experiences, giving you the chance you to engage with professional croupiers and other players, all while taking in the immersive atmosphere of a traditional gambling venue from anywhere you want.
If you’re a beginner with the world of virtual casino play or seek to find out more about reliable sites, why not sign up for our vibrant interactive platform? It’s a platform where fans discuss insights, making it easier for you to get the most out of your casino activities. Join the connections and learn more now: https://www.instagram.com/mystake_top/ .
Beyond variety, internet-based gambling hubs excel ease of access.
Leonardsiz
December 6, 2024 @ 1:22 am
кухни от производителя – Высокое качество и надежность от проверенного производителя кухонь.
Jaimeget
December 6, 2024 @ 1:34 am
buy zithromax online cheap [url=https://azithromycinus.com/#]zithromax buy online no prescription[/url] average cost of generic zithromax
Fałszywe opinie o Slottica
December 6, 2024 @ 1:52 am
Oh my goodness! Amazing article dude! Thank you, However I am going through troubles with your RSS.
I don’t know the reason why I can’t subscribe to
it. Is there anyone else getting identical RSS issues? Anyone who knows the answer will you kindly respond?
Thanx!!
Social - paibe
December 6, 2024 @ 2:03 am
How Online Casinos Remain a Worldwide Trend
Online casinos have revolutionized the betting market, offering an unmatched level of ease and selection that traditional casinos are unable to replicate. Over time, countless gamblers across the globe have welcomed the pleasure of virtual casinos thanks to its always-open nature, engaging traits, and constantly growing range of offerings.
One of the key draws of online casinos is the vast diversity of games at your disposal. Whether you like spinning vintage reel games, immersing yourself in engaging video slots, or testing your strategy in strategy-based games like Blackjack, digital casinos deliver countless opportunities. Several sites also introduce live dealer games, enabling you to communicate with actual dealers and co-players, all while taking in the engaging vibes of a land-based casino from anywhere you want.
If you’re new with the world of internet-based gaming or seek to explore trusted platforms, why not engage with our dynamic community? It’s a platform where fans post experiences, assisting you to maximize your online casino experience. Join the conversation and check it out now: https://www.facebook.com/profile.php?id=61568692273712 .
Apart from the game range, online casinos are known for seamless entry.
QWERTYU7
December 6, 2024 @ 2:27 am
Hello there, You’ve done a great job. I’ll certainly digg it
and personally suggest to my friends. I’m
sure they’ll be benefited from this site.
Matthewpok
December 6, 2024 @ 2:38 am
http://azithromycinus.com/# where to buy zithromax in canada
zithromax 250mg [url=https://azithromycinus.com/#]generic zithromax 500mg india[/url] buy zithromax 1000 mg online
snyatie zapoya na domy_kwml
December 6, 2024 @ 2:48 am
капельница от похмелья на дому [url=www.ximki.ukrbb.net/viewtopic.php?f=12&t=3712/]капельница от похмелья на дому[/url] .
outdoor trash can cleaner
December 6, 2024 @ 2:55 am
It’s an remarkable paragraph for all the internet users; they will take
benefit from it I am sure.
Jacobgaugh
December 6, 2024 @ 2:57 am
https://www.taxipuma.ru — посетите наш сайт, чтобы узнать больше о наших услугах
Angelhob
December 6, 2024 @ 3:05 am
фотограф предметная съемка – Профессиональная помощь в создании коммерческих фотографий.
Social - paibe
December 6, 2024 @ 3:08 am
The Reasons Behind Why Online Casinos Have Become an International Sensation
Internet-based gambling hubs have changed the gambling scene, delivering an unmatched level of convenience and selection that conventional venues can’t match. Over the past decade, millions of players worldwide have turned to the adventure of online gaming due to its anytime, anywhere convenience, appealing qualities, and ever-expanding catalogs of games.
One of the strongest selling points of online gaming options is the unparalleled array of gaming experiences ready to play. Whether you prefer interacting with traditional fruit machine slots, trying out engaging video-based games, or testing your strategy in strategy-based games like poker, online platforms deliver infinite choices. Many casinos furthermore introduce live dealer games, making it possible for you to engage with live hosts and gaming peers, all while soaking in the immersive atmosphere of a land-based casino in your own space.
If you’re unfamiliar with the world of virtual gambling or hope to find out more about reputable operators, why not engage with our active gaming forum? It’s a place where fans post reviews, assisting you to maximize your online casino experience. Join the community and check it out now: https://ru.pinterest.com/UpX_Official/ .
Adding to the extensive catalog, digital casino services stand out accessibility.
garbage chute handle
December 6, 2024 @ 3:27 am
Why users still make use of to read news papers when in this
technological world all is available on net?
Check out my web page :: garbage chute handle
Barnsley escorts
December 6, 2024 @ 3:32 am
fantastic post, very informative. I ponder why the other experts of this
sector don’t understand this. You should proceed your writing.
I’m sure, you’ve a huge readers’ base already!
trash chute hydraulic closer
December 6, 2024 @ 3:39 am
whoah this weblog is wonderful i really like reading your
articles. Keep up the great work! You know, lots of
persons are looking around for this information, you can help them greatly.
Feel free to surf to my blog: trash chute hydraulic closer
Leeds Escorts Agency
December 6, 2024 @ 3:59 am
Hey there just wanted to give you a quick heads up. The text in your article seem
to be running off the screen in Firefox. I’m not sure if
this is a formatting issue or something to do with internet browser compatibility but I figured I’d post to let you know.
The design and style look great though! Hope you get the problem fixed soon. Thanks
Social - paibe
December 6, 2024 @ 4:15 am
What Makes Online Casinos Remain Highly Preferred Worldwide
Virtual gambling platforms have transformed the gaming landscape, offering an exceptional degree of convenience and selection that land-based gambling houses fall short of. Over the past decade, a growing community across the globe have turned to the pleasure of internet-based gaming due to its ease of access, exciting features, and progressively larger game libraries.
One of the main appeals of online casinos is the astounding array of entertainment options available. Whether you like playing on old-school slots, playing through story-driven thematic slots, or mastering skills in traditional table offerings like Texas Hold’em, online platforms feature infinite entertainment avenues. Numerous services also present live casino options, letting you to participate with human game hosts and co-players, all while immersing yourself in the authentic ambiance of a traditional gambling venue from the comfort of your home.
If you’re a beginner with the world of virtual gambling or would like to find out more about reliable sites, why not participate in our vibrant online hub? It’s a hub where enthusiasts discuss tips, assisting you to enjoy more of your casino activities. Discover the community and start your journey now: https://www.instagram.com/1bet_be/ .
Besides the wide selection, virtual gaming providers are known for accessibility.
houston texas best driveway repairs
December 6, 2024 @ 4:22 am
Oh my goodness! Amazing article dude! Thanks, However I am experiencing difficulties
with your RSS. I don’t understand the reason why
I can’t subscribe to it. Is there anyone else having similar RSS problems?
Anyone that knows the solution can you kindly respond? Thanks!!
جادوی اینترنت
December 6, 2024 @ 4:24 am
What’s up, all the time i used to check web site posts here in the early hours in the
break of day, because i enjoy to gain knowledge
of more and more.
View more
December 6, 2024 @ 4:25 am
Greetings from Florida! I’m bored to death at work so I decided to browse your website on my iphone during lunch break.
I love the information you provide here and can’t
wait to take a look when I get home. I’m surprised at how fast your
blog loaded on my cell phone .. I’m not even using WIFI,
just 3G .. Anyways, good blog!
Francisshece
December 6, 2024 @ 4:40 am
не упустите возможность! vavada официальный сайт — ваш быстрый старт в мире онлайн-развлечений. переходите на зеркало vavada и начните играть прямо сейчас. доступ к лучшим бонусам станет вашим в несколько кликов. удобно на любом устройстве!
Matthewsic
December 6, 2024 @ 4:47 am
не упустите возможность! вавада телеграм — ваш быстрый старт в мире онлайн-развлечений. переходите на зеркало vavada и начните играть прямо сейчас. доступ к казино станет вашим в несколько кликов. удобно на любом устройстве!
WEGKLLIYG2
December 6, 2024 @ 5:04 am
Informative article, totally what I wanted to find.
สล็อต pp
December 6, 2024 @ 5:13 am
With thanks. Terrific stuff. https://images.google.com.bd/url?q=https://pp-slot.vip
honeymoney online casino
December 6, 2024 @ 5:22 am
honeymoney online casino
Social - paibe
December 6, 2024 @ 5:22 am
The Reasons Behind Why Online Casinos Remain So Popular
Online casinos have modernized the gaming world, offering a level of convenience and range that brick-and-mortar venues struggle to rival. Recently, millions of players worldwide have chosen the fun of virtual casinos because of its anytime, anywhere convenience, captivating elements, and continuously increasing catalogs of games.
One of the strongest selling points of internet-based platforms is the unparalleled variety of gaming experiences provided. Whether you prefer interacting with vintage one-armed bandits, diving into story-driven visual slot games, or strategizing in traditional table offerings like poker, casino websites deliver numerous options. A large number of platforms even include live dealer games, letting you to interact with human game hosts and gaming peers, all while soaking in the lifelike atmosphere of a physical gaming house from the comfort of your home.
If you’re just starting with the world of digital casinos or hope to discover reliable sites, why not sign up for our vibrant interactive platform? It’s a hub where fans post stories, guiding you to maximize your online casino experience. Dive into the community and see it here now: https://www.facebook.com/profile.php?id=61568481962080 .
Beyond variety, internet-based gambling hubs excel ease of access.
Derekapaky
December 6, 2024 @ 5:42 am
seo-wolf.ru/ – платформа для выбора стильной и функциональной мебели.
Leonardsiz
December 6, 2024 @ 5:53 am
кухни санкт петербург – Лучшие решения для кухни от профессионалов в Санкт-Петербурге.
Lazrgaw
December 6, 2024 @ 6:12 am
Быстрое обучение и получение диплома магистра – возможно ли это?
honeymoney зеркало
December 6, 2024 @ 6:43 am
бездепозитные фриспины хани мани
Social - paibe
December 6, 2024 @ 6:45 am
Why Online Casinos Are Becoming So Popular
Virtual gambling platforms have changed the gambling industry, delivering a level of accessibility and breadth that land-based gambling houses are unable to replicate. Over the past decade, a large audience around the world have embraced the fun of digital casino play due to its availability, exciting features, and ever-expanding selection of games.
One of the key draws of online gaming options is the incredible array of choices on offer. Whether you love playing on vintage slots, exploring engaging modern slot games, or playing smart in strategy-based games like Texas Hold’em, internet-based gambling sites offer limitless opportunities. Plenty of operators furthermore introduce live dealer games, letting you to connect with professional croupiers and fellow gamblers, all while enjoying the immersive atmosphere of a traditional gambling venue right at home.
If you’re unfamiliar with the world of virtual casino play or seek to discover trusted platforms, why not engage with our growing online hub? It’s a place where enthusiasts offer stories, enabling you to enjoy more of your gambling adventure. Discover the experience and visit us now: https://www.instagram.com/kwz.kz_official/ .
Apart from the game range, virtual gambling platforms stand out constant connectivity.
Jeremymup
December 6, 2024 @ 7:18 am
сеть магазинов на Дальнем Востоке России, успешно функционирующая с 2016 года. Наше присутствие в регионе обеспечивает удобство покупок для жителей Уссурийска, Владивостока и Хабаровска https://mirotvorec-dv.ru/catalog/snaryazhenie/
Jacobgaugh
December 6, 2024 @ 7:19 am
фабрика кухня — Прямые поставки от производителя: кухни высокого качества.
end dump trailer haul out service
December 6, 2024 @ 7:24 am
If you wish for to get a great deal from this article then you have to apply such strategies to your won blog.
RobertPiela
December 6, 2024 @ 7:34 am
Lightning Storm — это захватывающий видеослот, созданный компанией Aristocrat Technologies, известной своими высококачественными играми для наземных и онлайн-казино. Lightning Storm выделяется своим уникальным дизайном, интересными механиками и высокими потенциальными выигрышами. Давайте рассмотрим основные аспекты этой игры https://teip-irk.ru/media/pgs/crazy_pachinko_.html
Social - paibe
December 6, 2024 @ 7:50 am
Reasons Why Online Casinos Have Become a Global Phenomenon
Virtual gambling platforms have transformed the gambling landscape, providing an unmatched level of accessibility and selection that brick-and-mortar venues don’t provide. Throughout the last ten years, a growing community worldwide have chosen the fun of internet-based gaming due to its accessibility, exciting features, and constantly growing range of offerings.
One of the key draws of digital gambling sites is the sheer variety of titles ready to play. Whether you enjoy rolling old-school slots, diving into plot-filled modern slot games, or exercising tactics in table games like poker, online platforms feature numerous choices. Numerous services moreover introduce interactive dealer games, making it possible for you to communicate with human game hosts and opponents, all while taking in the lifelike vibes of a physical gaming house without leaving your home.
If you’re new with the world of virtual gambling or would like to delve deeper into safe services, why not participate in our active community? It’s a space where players exchange experiences, helping you to enjoy more of your gambling adventure. Dive into the conversation and check it out now: https://www.instagram.com/1winbr_com/ .
Adding to the extensive catalog, digital casino services excel accessibility.
anti aging serum
December 6, 2024 @ 8:05 am
Howdy! I realize this is sort of off-topic however I had to ask.
Does building a well-established blog such as yours take a large amount
of work? I’m brand new to operating a blog however I do write in my diary everyday.
I’d like to start a blog so I will be able to share my experience
and views online. Please let me know if you have any ideas or tips for brand
new aspiring bloggers. Appreciate it!
ArmandoWal
December 6, 2024 @ 8:05 am
lisinopril 420: cost of lisinopril 40 mg – lisinopril 10 mg
Jimmieclict
December 6, 2024 @ 8:30 am
https://ciprofloxacin.cheap/# cipro pharmacy
Social - paibe
December 6, 2024 @ 8:59 am
The Reasons Behind Why Online Casinos Have Become an International Sensation
Internet-based gambling hubs have transformed the gambling world, providing an unmatched level of convenience and diversity that traditional venues can’t match. In recent years, a growing community across the globe have chosen the pleasure of digital casino play because of its anytime, anywhere convenience, exciting features, and continuously increasing catalogs of games.
One of the strongest selling points of virtual gambling hubs is the astounding range of titles ready to play. Whether you like playing on retro slot machines, playing through engaging modern slot games, or mastering skills in strategy-based games like Baccarat, casino websites offer countless entertainment avenues. A large number of platforms also introduce live dealer games, making it possible for you to connect with professional croupiers and gaming peers, all while immersing yourself in the realistic vibes of a real casino from anywhere you want.
If you’re a beginner with the world of virtual casino play or want to explore reputable operators, why not become part of our dynamic gaming forum? It’s a hub where enthusiasts share experiences, helping you to maximize your virtual play. Check out the experience and start your journey now: https://www.instagram.com/1bet_be/ .
Besides the wide selection, online casinos stand out ease of access.
Angelhob
December 6, 2024 @ 9:04 am
https://mirdolmenov.ru – Примеры успешных проектов и отзывы клиентов.
JosephMossy
December 6, 2024 @ 9:17 am
[url=][/url]
Временная регистрация в Санкт-Петербурге: Быстро и Легально!
Ищете, где оформить временную регистрацию в СПБ?
Мы гарантируем быстрое и легальное оформление без очередей и лишних документов.
Ваше спокойствие – наша забота!
Минимум усилий • Максимум удобства • Полная легальность
Свяжитесь с нами прямо сейчас!
Временная регистрация в СПБ
[url=][/url]
snyatie zapoya na domy_geMt
December 6, 2024 @ 10:06 am
капельница от запоя вызов [url=www.kyevlyn.ukrbb.net/viewtopic.php?f=2&t=13678]капельница от запоя вызов[/url] .
Social - paibe
December 6, 2024 @ 10:08 am
Why Online Casinos Have Become an International Sensation
Digital casinos have transformed the casino gaming market, providing an exceptional degree of user-friendliness and diversity that conventional venues don’t provide. Over the past decade, a large audience globally have turned to the thrill of digital casino play due to its accessibility, exciting features, and ever-expanding selection of games.
One of the biggest attractions of virtual gambling hubs is the vast diversity of entertainment options at your disposal. Whether you prefer spinning old-school fruit machine slots, diving into theme-based video slots, or mastering skills in table games like Roulette, internet-based gambling sites offer endless possibilities. Plenty of operators also include real-time gaming experiences, making it possible for you to communicate with actual dealers and co-players, all while immersing yourself in the realistic ambiance of a physical gaming house right at home.
If you’re new with the world of internet-based gaming or would like to discover reputable operators, why not join our growing community? It’s a destination where players exchange tips, guiding you to maximize your virtual play. Explore the connections and start your journey now: https://www.instagram.com/sweet_bonanzacom/ .
Adding to the extensive catalog, virtual gaming providers stand out constant connectivity.
این سایت تایید شده را بسیار مفید یافتم
December 6, 2024 @ 10:10 am
عرض ادب! این سایت برای من یه منبع عالی شده.
امیدوارم همیشه موفق باشید و مقالات خوبی بنویسید.
دمتون گرم!
Leonardsiz
December 6, 2024 @ 10:25 am
кухни от производителя — Кухни от производителя: качество и доступная цена.
Jaimeget
December 6, 2024 @ 10:50 am
cipro [url=http://ciprofloxacin.cheap/#]buy cipro cheap[/url] ciprofloxacin 500mg buy online
Social - paibe
December 6, 2024 @ 11:12 am
The Reasons Behind Why Online Casinos Are Becoming So Popular
Digital casinos have reshaped the gambling market, delivering an unmatched level of accessibility and variety that brick-and-mortar gambling houses don’t provide. Over the past decade, a large audience internationally have turned to the pleasure of virtual gambling because of its ease of access, thrilling aspects, and ever-expanding catalogs of games.
One of the key draws of virtual gambling hubs is the astounding range of choices on offer. Whether you enjoy interacting with retro slot machines, immersing yourself in theme-based thematic slots, or testing your strategy in classic casino games like poker, internet-based gambling sites deliver endless entertainment avenues. A large number of platforms also introduce live dealer games, making it possible for you to connect with human game hosts and other players, all while taking in the authentic vibes of a physical gaming house in your own space.
If you’re a beginner with the world of virtual gambling or are looking to discover safe services, why not join our growing gaming forum? It’s a place where enthusiasts offer reviews, enabling you to enjoy more of your casino activities. Join the experience and learn more now: https://au.pinterest.com/casibom1/ .
Adding to the extensive catalog, internet-based gambling hubs excel ease of access.
スーパーコピー ブルガリ ピアス fx
December 6, 2024 @ 11:12 am
Reliable information With thanks.
my blog post … http://www.zgcksxy.com/
Betchain
December 6, 2024 @ 11:23 am
Wow that was strange. I just wrote an incredibly long
comment but after I clicked submit my comment didn’t appear.
Grrrr… well I’m not writing all that over again. Anyways, just wanted to say superb blog!
ThomasLum
December 6, 2024 @ 11:25 am
Обратите внимание на гинеколога в Варшаве, Анастасию Юрьевну. Это врач с огромным опытом, которая помогает пациенткам в самых разных ситуациях. Она занимается гинекологическими осмотрами, ультразвуковой диагностикой, лечением воспалительных процессов и консультациями по вопросам планирования семьи http://melanz.phorum.pl/viewtopic.php?f=4&t=93426
BLPOYTUIG
December 6, 2024 @ 11:33 am
Hi, i feel that i saw you visited my web site so i got here to return the favor?.I’m trying
to find issues to improve my web site!I guess its adequate to use a few of
your concepts!!
Jacobgaugh
December 6, 2024 @ 11:37 am
http://gft-leasing.ru/ — Ознакомьтесь с нашими проектами и найдите свое решение.
betflik
December 6, 2024 @ 11:41 am
Many thanks. Plenty of information.
https://clients1.google.co.tz/url?q=https://betflik28.asia
сейф взломостойкий
December 6, 2024 @ 11:58 am
Тут можно преобрести сейфы офисные взломостойкие сейф пожаровзломостойкие купить
Derekapaky
December 6, 2024 @ 12:08 pm
кухни на заказ – это идеальное решение для реализации любых дизайнерских идей.
Social - paibe
December 6, 2024 @ 12:20 pm
Reasons Why Online Casinos Are Highly Preferred Worldwide
Internet-based gambling hubs have revolutionized the gaming scene, providing an unmatched level of accessibility and diversity that land-based establishments don’t provide. Over the past decade, a growing community worldwide have turned to the fun of virtual casinos in light of its anytime, anywhere convenience, appealing qualities, and continuously increasing catalogs of games.
One of the most compelling reasons of digital gambling sites is the incredible selection of entertainment options available. Whether you enjoy interacting with traditional slots, diving into narrative-rich video slots, or testing your strategy in strategy-based games like Texas Hold’em, online platforms deliver endless opportunities. Many casinos also include interactive dealer games, enabling you to interact with live hosts and other players, all while immersing yourself in the engaging environment of a real casino without leaving your home.
If you’re unfamiliar with the world of virtual gambling or seek to delve deeper into reliable sites, why not participate in our dynamic gaming forum? It’s a platform where fans offer reviews, helping you to maximize your virtual play. Join the community and start your journey now: https://www.instagram.com/1winbr_com/ .
Besides the wide selection, digital casino services shine availability.
dallas piano teachers
December 6, 2024 @ 12:26 pm
Magnificent goods from you, man. I have be aware your stuff previous to and you are
simply too great. I actually like what you have obtained
right here, really like what you’re saying and the best way during which you say it.
You are making it enjoyable and you continue to care for to stay
it sensible. I cant wait to read much more from you.
That is actually a terrific web site.
Cazruod
December 6, 2024 @ 1:08 pm
Реально ли приобрести диплом стоматолога? Основные шаги
Click here
December 6, 2024 @ 1:18 pm
Hi to all, it’s genuinely a fastidious for me to visit this
website, it includes important Information.
Social - paibe
December 6, 2024 @ 1:26 pm
How Online Casinos Are Becoming a Worldwide Trend
Digital casinos have transformed the gaming market, offering an unmatched level of user-friendliness and variety that land-based venues struggle to rival. Over the past decade, a large audience globally have adopted the thrill of digital casino play as a result of its anytime, anywhere convenience, appealing qualities, and progressively larger selection of games.
One of the strongest selling points of online casinos is the incredible range of choices provided. Whether you are a fan of spinning traditional fruit machine slots, exploring engaging video slots, or exercising tactics in strategy-based games like poker, virtual venues offer limitless possibilities. Many casinos even introduce live casino options, enabling you to connect with real dealers and fellow gamblers, all while enjoying the authentic atmosphere of a brick-and-mortar establishment from the comfort of your home.
If you’re just starting with the world of digital casinos or would like to explore proven options, why not join our vibrant interactive platform? It’s a space where gamblers post tips, making it easier for you to improve your online casino experience. Check out the conversation and check it out now: https://www.facebook.com/profile.php?id=61568958127704 .
In addition to diversity, online casinos thrive in constant connectivity.
Jimmybiown
December 6, 2024 @ 1:39 pm
Mostbet, Turkiye’deki kullan?c?lar icin genis bir bahis ve casino oyunlar? secenegi sunan populer bir platformdur https://hurifo.org/
Social - paibe
December 6, 2024 @ 2:32 pm
Why Online Casinos Are Becoming So Popular
Online casinos have changed the betting landscape, offering a level of accessibility and range that physical casinos fall short of. Throughout the last ten years, a vast number of enthusiasts globally have welcomed the thrill of virtual gambling because of its availability, exciting features, and widening range of offerings.
One of the most compelling reasons of internet-based platforms is the vast range of choices ready to play. Whether you enjoy rolling vintage one-armed bandits, trying out narrative-rich modern slot games, or playing smart in card and board games like Roulette, casino websites deliver countless opportunities. Numerous services additionally introduce live gaming streams, giving you the chance you to connect with professional croupiers and fellow gamblers, all while immersing yourself in the engaging environment of a traditional gambling venue right at home.
If you’re a beginner with the world of internet-based gaming or are looking to learn about safe services, why not engage with our growing online hub? It’s a hub where gaming aficionados post experiences, making it easier for you to improve your gambling adventure. Dive into the connections and start your journey now: https://www.instagram.com/kwz.kz_official/ .
Besides the wide selection, virtual gaming providers thrive in ease of access.
ArmandoWal
December 6, 2024 @ 2:49 pm
lisinopril 20mg tablets cost: lisinopril 1 mg tablet – lisinopril 10 mg pill
Leonardsiz
December 6, 2024 @ 3:14 pm
кухни санкт петербург – Выбирайте кухню у профессионалов в Санкт-Петербурге. Гарантируем качество и стиль.
GamaCasino
December 6, 2024 @ 3:22 pm
[url=https://gama-casino-apk.ru]http://gama-casino-apk.ru[/url]
Установить приложение онлайн казино gama – выигрывай сейчас!
https://www.gama-casino-apk.ru
Social - paibe
December 6, 2024 @ 3:38 pm
Reasons Why Online Casinos Are So Popular
Internet-based gambling hubs have reshaped the gambling world, providing an unmatched level of convenience and diversity that land-based establishments fall short of. In recent years, a vast number of enthusiasts globally have embraced the fun of internet-based gaming thanks to its anytime, anywhere convenience, captivating elements, and progressively larger game libraries.
One of the most compelling reasons of online casinos is the sheer array of gaming experiences available. Whether you like interacting with retro one-armed bandits, playing through theme-based thematic slots, or playing smart in card and board games like poker, casino websites provide numerous possibilities. Several sites even present live gaming streams, making it possible for you to interact with human game hosts and fellow gamblers, all while soaking in the immersive feel of a real casino right at home.
If you’re new with the world of virtual casino play or would like to find out more about reliable sites, why not sign up for our growing online hub? It’s a platform where gaming aficionados post stories, enabling you to maximize your online casino experience. Explore the conversation and learn more now: https://t.me/casibomgiris_net .
Apart from the game range, online casinos shine availability.
Casino
December 6, 2024 @ 3:41 pm
Saved as a favorite, I love your web site!
Jacobgaugh
December 6, 2024 @ 3:58 pm
кухня на заказ недорого москва – Кухни на заказ по доступной цене для жителей Москвы.
Matthewpok
December 6, 2024 @ 4:04 pm
http://cytotec.top/# buy cytotec over the counter
zithromax 1000 mg online [url=https://azithromycinus.com/#]zithromax 500 tablet[/url] azithromycin zithromax
rgo 303
December 6, 2024 @ 4:14 pm
Its like you read my mind! You seem to know a lot about this, like you
wrote the book in it or something. I think that you can do with some pics to
drive the message home a little bit, but instead of that, this is wonderful blog.
An excellent read. I will definitely be back.
Gk88
December 6, 2024 @ 4:29 pm
Thanks for sharing your thoughts on Gk88. Regards
Social - paibe
December 6, 2024 @ 4:41 pm
Reasons Why Online Casinos Remain Highly Preferred Worldwide
Online casinos have modernized the gaming market, delivering an unmatched level of ease and breadth that conventional casinos can’t match. In recent years, a growing community internationally have turned to the pleasure of virtual gambling in light of its ease of access, exciting features, and ever-expanding catalogs of games.
One of the biggest attractions of digital gambling sites is the astounding range of games at your disposal. Whether you like spinning old-school slots, exploring engaging visual slot games, or exercising tactics in table games like Blackjack, digital casinos feature endless choices. Numerous services moreover include live casino options, letting you to connect with real dealers and fellow gamblers, all while immersing yourself in the lifelike feel of a physical gaming house without leaving your home.
If you’re new with the world of virtual gambling or would like to discover reliable sites, why not participate in our growing gaming forum? It’s a space where gamblers exchange stories, enabling you to enjoy more of your gaming journey. Explore the connections and start your journey now: https://www.facebook.com/profile.php?id=61568742522948 .
Besides the wide selection, virtual gaming providers stand out availability.
Angelhob
December 6, 2024 @ 5:04 pm
предметная съемка косметики – Съемка косметических товаров с акцентом на их текстуру и привлекательный внешний вид.
Robertvox
December 6, 2024 @ 5:07 pm
generic zithromax medicine: can i buy zithromax online – zithromax canadian pharmacy
Casino
December 6, 2024 @ 5:20 pm
Thank you a lot for sharing this with all people you
actually understand what you’re talking about!
Bookmarked. Kindly also discuss with my web site =).
We will have a hyperlink exchange arrangement
among us
Sazrbfz
December 6, 2024 @ 5:31 pm
Диплом вуза купить официально с упрощенным обучением в Москве
Social - paibe
December 6, 2024 @ 5:43 pm
The Reasons Behind Why Online Casinos Have Become an International Sensation
Internet-based gambling hubs have changed the gambling market, delivering a unique kind of accessibility and selection that conventional casinos struggle to rival. In recent years, a vast number of enthusiasts across the globe have chosen the fun of virtual casinos as a result of its ease of access, captivating elements, and ever-expanding selection of games.
One of the most compelling reasons of internet-based platforms is the vast selection of games ready to play. Whether you enjoy interacting with traditional reel games, diving into story-driven thematic slots, or testing your strategy in classic casino games like Baccarat, online platforms feature infinite entertainment avenues. Plenty of operators also introduce live dealer games, giving you the chance you to interact with live hosts and gaming peers, all while experiencing the authentic atmosphere of a traditional gambling venue right at home.
If you’re a beginner with the world of digital casinos or want to discover reputable operators, why not join our growing interactive platform? It’s a place where gamblers share reviews, making it easier for you to maximize your gaming journey. Check out the community and learn more now: https://www.facebook.com/profile.php?id=61568692273712 .
In addition to diversity, virtual gaming providers are known for constant connectivity.
Jacobgaugh
December 6, 2024 @ 6:25 pm
кухни на заказ от производителя — Эксклюзивные кухни на заказ от производителя с гарантией качества.
Jacobgaugh
December 6, 2024 @ 6:28 pm
http://www.pitaniedetey.ru – Официальный сайт компании, специализирующейся на кухнях.
スーパーコピー ジョーダン ltd
December 6, 2024 @ 6:31 pm
You actually explained that perfectly.
Feel free to visit my web page :: http://www.buy-Aeds.com/comment/html/?167833.html
Social - paibe
December 6, 2024 @ 6:43 pm
Why Online Casinos Are Becoming a Global Phenomenon
Virtual gambling platforms have reshaped the gaming landscape, delivering an exceptional degree of comfort and breadth that land-based gambling houses don’t provide. Over the past decade, a vast number of enthusiasts internationally have turned to the excitement of virtual casinos because of its anytime, anywhere convenience, engaging traits, and constantly growing range of offerings.
One of the key draws of online gaming options is the incredible diversity of choices ready to play. Whether you are a fan of rolling old-school slot machines, exploring narrative-rich visual slot games, or playing smart in classic casino games like Roulette, virtual venues feature infinite choices. Several sites even introduce live dealer games, giving you the chance you to interact with human game hosts and other players, all while experiencing the immersive ambiance of a physical gaming house from anywhere you want.
If you’re exploring for the first time with the world of digital casinos or seek to delve deeper into proven options, why not participate in our lively online hub? It’s a place where gamblers discuss experiences, making it easier for you to enjoy more of your online casino experience. Explore the experience and learn more now: https://t.me/casibomgiris_net .
In addition to diversity, online casinos are known for seamless entry.
vivod iz zapoya v stacionare_syKa
December 6, 2024 @ 7:27 pm
вывод из запоя в наркологическом стационаре Самары [url=http://honey.ukrbb.net/viewtopic.php?f=45&t=16727]вывод из запоя в наркологическом стационаре Самары[/url] .
vivod iz zapoya v stacionare_wist
December 6, 2024 @ 7:37 pm
быстрый вывод из запоя в стационаре [url=http://superstar.ukrbb.net/viewtopic.php?f=3&t=688/]быстрый вывод из запоя в стационаре[/url] .
vivod iz zapoya v stacionare_vkpt
December 6, 2024 @ 7:37 pm
лечению наркозависимости в стационаре [url=www.eisberg.forum24.ru/?1-1-0-00002019-000-0-0-1730816715/]лечению наркозависимости в стационаре[/url] .
pozdravleniya na svadby _wxMn
December 6, 2024 @ 7:44 pm
поздравления с днем рождения маме [url=pro-happy-birthday.ru]поздравления с днем рождения маме[/url] .
Social - paibe
December 6, 2024 @ 7:44 pm
What Makes Online Casinos Are So Popular
Virtual gambling platforms have revolutionized the casino gaming scene, providing an unmatched level of user-friendliness and range that conventional venues fall short of. Over the past decade, millions of players internationally have chosen the fun of online gaming as a result of its availability, thrilling aspects, and widening selection of games.
One of the strongest selling points of internet-based platforms is the vast variety of games on offer. Whether you love rolling retro fruit machine slots, trying out narrative-rich modern slot games, or playing smart in strategy-based games like Baccarat, virtual venues offer numerous entertainment avenues. Plenty of operators moreover offer live casino options, giving you the chance you to communicate with professional croupiers and gaming peers, all while experiencing the realistic vibes of a brick-and-mortar establishment from the comfort of your home.
If you’re a beginner with the world of digital casinos or seek to learn about proven options, why not become part of our lively community? It’s a destination where players share reviews, assisting you to get the most out of your casino activities. Join the discussions and visit us now: https://au.pinterest.com/casibom1/ .
Besides the wide selection, internet-based gambling hubs excel constant connectivity.
Jaimeget
December 6, 2024 @ 8:01 pm
get cheap clomid without a prescription [url=http://clomid.store/#]how can i get clomid no prescription[/url] where to get clomid without dr prescription
Leonardsiz
December 6, 2024 @ 8:02 pm
https://www.tomason-russia.ru/ – Сделайте шаг к созданию кухни вашей мечты.
Derekapaky
December 6, 2024 @ 8:15 pm
кухни на заказ – сочетание оригинального дизайна и практичности.
website
December 6, 2024 @ 8:18 pm
WOW just what I was looking for. Came here
by searching for website
cooking oil recycling dallas
December 6, 2024 @ 8:26 pm
Simply wish to say your article is as surprising.
The clearness to your publish is simply cool and i could assume you
are knowledgeable in this subject. Fine with your permission let me to seize
your feed to keep up to date with drawing close post.
Thanks 1,000,000 and please keep up the rewarding work.
Social - paibe
December 6, 2024 @ 8:45 pm
Why Online Casinos Have Become a Global Phenomenon
Online casinos have reshaped the betting scene, providing an exceptional degree of ease and variety that conventional gambling houses don’t provide. Over time, a growing community across the globe have chosen the thrill of internet-based gaming as a result of its ease of access, exciting features, and continuously increasing collections of titles.
One of the main appeals of online gaming options is the unparalleled diversity of games on offer. Whether you are a fan of playing on retro slots, playing through narrative-rich modern slot games, or exercising tactics in strategy-based games like Roulette, digital casinos deliver countless options. A large number of platforms even offer live casino options, allowing you to communicate with live hosts and opponents, all while experiencing the authentic atmosphere of a land-based casino from the comfort of your home.
If you’re new with the world of online gaming or want to delve deeper into trusted platforms, why not sign up for our vibrant social network? It’s a platform where fans share insights, guiding you to get the most out of your gaming journey. Dive into the conversation and visit us now: https://www.facebook.com/profile.php?id=61568481962080 .
Beyond variety, digital casino services thrive in seamless entry.
ArmandoWal
December 6, 2024 @ 9:36 pm
Cytotec 200mcg price: buy cytotec – cytotec online
Social - paibe
December 6, 2024 @ 9:45 pm
How Online Casinos Have Become Highly Preferred Worldwide
Digital casinos have modernized the gaming market, delivering a level of accessibility and breadth that brick-and-mortar gambling houses fall short of. Over the past decade, a growing community across the globe have embraced the excitement of virtual gambling thanks to its ease of access, engaging traits, and ever-expanding game libraries.
One of the main appeals of online casinos is the unparalleled range of games at your disposal. Whether you are a fan of playing on traditional fruit machine slots, playing through plot-filled visual slot games, or playing smart in classic casino games like Texas Hold’em, casino websites deliver limitless entertainment avenues. Plenty of operators moreover offer interactive dealer games, allowing you to engage with live hosts and fellow gamblers, all while experiencing the engaging environment of a traditional gambling venue from anywhere you want.
If you’re a beginner with the world of digital casinos or would like to discover proven options, why not engage with our active interactive platform? It’s a platform where players post experiences, helping you to maximize your online casino experience. Join the conversation and visit us now: https://x.com/upX_Top .
Adding to the extensive catalog, digital casino services shine constant connectivity.
Lazrxgr
December 6, 2024 @ 10:08 pm
Вопросы и ответы: можно ли быстро купить диплом старого образца?
Start here
December 6, 2024 @ 10:13 pm
I believe everything said was very reasonable. But, consider this, suppose
you were to write a killer headline? I mean, I don’t want to tell you how to run your
website, however what if you added a post title that makes people desire more?
I mean Filling in the gaps of HIV is kinda boring.
You might look at Yahoo’s home page and see how they create news titles to grab people interested.
You might add a related video or a picture or two
to get readers excited about everything’ve got to say.
Just my opinion, it could make your website a little livelier.
balinese massage near me
December 6, 2024 @ 10:37 pm
Thanks for some other wonderful article.
The place else may anybody get that type of info in such an ideal means of writing?
I’ve a presentation next week, and I am at the search for such information.
Social - paibe
December 6, 2024 @ 10:47 pm
Why Online Casinos Are Becoming So Popular
Online casinos have modernized the gaming scene, providing a unique kind of convenience and range that traditional establishments struggle to rival. Over time, a vast number of enthusiasts across the globe have embraced the fun of online gaming because of its accessibility, appealing qualities, and continuously increasing selection of games.
One of the main appeals of online gaming options is the sheer diversity of choices at your disposal. Whether you like spinning retro slot machines, exploring engaging video-based games, or strategizing in card and board games like Blackjack, casino websites boast endless options. A large number of platforms furthermore introduce real-time gaming experiences, giving you the chance you to connect with professional croupiers and fellow gamblers, all while soaking in the engaging ambiance of a real casino without leaving your home.
If you’re new with the world of digital casinos or would like to find out more about reputable operators, why not join our growing interactive platform? It’s a space where enthusiasts share reviews, guiding you to get the most out of your virtual play. Dive into the community and check it out now: https://www.facebook.com/profile.php?id=61568958127704 .
Adding to the extensive catalog, online casinos are known for ease of access.
Review Casino. Casino
December 6, 2024 @ 10:51 pm
I’m really impressed with your writing skills and also with the layout
on your blog. Is this a paid theme or did you customize
it yourself? Anyway keep up the nice quality writing, it
is rare to see a nice blog like this one nowadays.
escort kadıköy
December 6, 2024 @ 10:58 pm
bomba gibi kadıköy escort sitesine hoşgeldiniz https://kadikoyescortbul.com/
Jacobgaugh
December 6, 2024 @ 11:16 pm
mtucizone.ru/ — Полный ассортимент кухонь для вашего удобства.
Sazrivo
December 6, 2024 @ 11:20 pm
Купить диплом старого образца, можно ли это сделать по быстрой схеме?
Jacobgaugh
December 6, 2024 @ 11:23 pm
фабрика кухня – Прямая работа с фабрикой для создания качественной и стильной кухни.
bitcoin cash MLM business plan
December 6, 2024 @ 11:39 pm
My brother suggested I may like this web site. He used
to be entirely right. This publish truly made my day.
You can not consider just how so much time I had spent for this information! Thanks!
Sazrfqn
December 6, 2024 @ 11:41 pm
Легальная покупка школьного аттестата с упрощенной программой обучения
Social - paibe
December 6, 2024 @ 11:49 pm
Reasons Why Online Casinos Remain So Popular
Internet-based gambling hubs have transformed the betting scene, offering an exceptional degree of convenience and range that conventional casinos are unable to replicate. In recent years, a growing community internationally have embraced the excitement of virtual gambling as a result of its availability, captivating elements, and widening range of offerings.
One of the biggest attractions of online casinos is the vast diversity of gaming experiences ready to play. Whether you love rolling retro reel games, exploring plot-filled visual slot games, or mastering skills in card and board games like Roulette, internet-based gambling sites offer infinite choices. Several sites furthermore offer live casino options, giving you the chance you to interact with human game hosts and co-players, all while experiencing the engaging atmosphere of a brick-and-mortar establishment right at home.
If you’re exploring for the first time with the world of digital casinos or are looking to find out more about proven options, why not engage with our lively interactive platform? It’s a place where players share reviews, assisting you to maximize your gaming journey. Dive into the conversation and see it here now: https://www.facebook.com/profile.php?id=61568742522948 .
Beyond variety, internet-based gambling hubs thrive in constant connectivity.
Angelhob
December 7, 2024 @ 12:02 am
предметная съемка для маркетплейсов – Услуги фотосъемки товаров, соответствующие требованиям крупных маркетплейсов.
Social - paibe
December 7, 2024 @ 12:50 am
How Online Casinos Remain a Worldwide Trend
Internet-based gambling hubs have transformed the gambling industry, delivering an exceptional degree of user-friendliness and range that conventional establishments can’t match. Recently, a vast number of enthusiasts globally have adopted the adventure of online gaming due to its anytime, anywhere convenience, appealing qualities, and constantly growing game libraries.
One of the main appeals of internet-based platforms is the sheer variety of entertainment options at your disposal. Whether you love engaging with retro fruit machine slots, trying out engaging video-based games, or mastering skills in traditional table offerings like Roulette, casino websites deliver limitless choices. A large number of platforms furthermore offer live casino options, making it possible for you to connect with actual dealers and co-players, all while enjoying the realistic atmosphere of a brick-and-mortar establishment from the comfort of your home.
If you’re new with the world of virtual gambling or would like to learn about reputable operators, why not become part of our growing gaming forum? It’s a place where enthusiasts exchange tips, enabling you to get the most out of your casino activities. Discover the conversation and check it out now: https://www.instagram.com/sweet_bonanzacom/ .
Adding to the extensive catalog, virtual gambling platforms excel availability.
Leonardsiz
December 7, 2024 @ 12:59 am
https://www.tsmk-altai.ru – Официальный сайт с полным спектром услуг по созданию кухни.
DHJJGFRHHG5
December 7, 2024 @ 1:27 am
My partner and I stumbled over here different page
and thought I should check things out. I like what I see so now i’m following you.
Look forward to going over your web page repeatedly.
Go To This Web-Site
December 7, 2024 @ 1:49 am
I really like what you guys are usually up too.
This sort of clever work and reporting! Keep up the excellent works guys I’ve incorporated you guys to my personal
blogroll.
Social - paibe
December 7, 2024 @ 1:51 am
The Reasons Behind Why Online Casinos Are So Popular
Digital casinos have transformed the casino gaming landscape, offering an unmatched level of accessibility and range that land-based casinos don’t provide. Throughout the last ten years, millions of players across the globe have turned to the adventure of internet-based gaming in light of its ease of access, appealing qualities, and progressively larger catalogs of games.
One of the biggest attractions of online gaming options is the incredible diversity of titles ready to play. Whether you enjoy interacting with retro one-armed bandits, trying out narrative-rich video-based games, or exercising tactics in classic casino games like Blackjack, digital casinos boast numerous opportunities. Numerous services even feature real-time gaming experiences, giving you the chance you to engage with real dealers and other players, all while immersing yourself in the realistic atmosphere of a land-based casino right at home.
If you’re just starting with the world of virtual gambling or want to delve deeper into trusted platforms, why not become part of our dynamic online hub? It’s a space where fans offer stories, making it easier for you to enhance your online casino experience. Discover the connections and start your journey now: https://x.com/upX_Top .
Beyond variety, virtual gaming providers are known for availability.
soikeo
December 7, 2024 @ 2:29 am
Whats up are using WordPress for your blog platform? I’m
new to the blog world but I’m trying to get started and set up my own.
Do you need any coding expertise to make your own blog? Any help would
be really appreciated!
Robertvox
December 7, 2024 @ 2:33 am
purchase cipro: ciprofloxacin 500mg buy online – where can i buy cipro online
Casino
December 7, 2024 @ 2:50 am
Hi there, I enjoy reading all of your article. I wanted to write a little comment to support
you.
Social - paibe
December 7, 2024 @ 2:53 am
Why Online Casinos Are Highly Preferred Worldwide
Online casinos have changed the casino gaming landscape, offering an exceptional degree of user-friendliness and selection that land-based venues can’t match. Over time, countless gamblers across the globe have adopted the pleasure of digital casino play due to its accessibility, engaging traits, and continuously increasing catalogs of games.
One of the key draws of online casinos is the incredible diversity of gaming experiences on offer. Whether you enjoy interacting with old-school reel games, diving into plot-filled video slots, or testing your strategy in strategy-based games like Texas Hold’em, internet-based gambling sites provide infinite opportunities. Plenty of operators moreover present live casino options, allowing you to connect with live hosts and other players, all while immersing yourself in the lifelike ambiance of a traditional gambling venue in your own space.
If you’re a beginner with the world of virtual gambling or are looking to find out more about proven options, why not participate in our vibrant interactive platform? It’s a hub where gaming aficionados share stories, assisting you to improve your virtual play. Join the experience and learn more now: https://www.instagram.com/1bet_be/ .
Besides the wide selection, internet-based gambling hubs shine ease of access.
این سایت دانشگاهی را پیشنهاد میدهم
December 7, 2024 @ 2:53 am
عرض ادب! محتوای سایت شما واقعاً خاص و
عالیه. امیدوارم همیشه موفق باشید و این کیفیت بالا رو
حفظ کنید. دمتون گرم!
Manchester escorts
December 7, 2024 @ 3:08 am
Excellent article. Keep writing such kind of information on your blog.
Im really impressed by your site.
Hi there, You’ve done an excellent job. I’ll certainly digg it and
for my part suggest to my friends. I’m confident they will be benefited from
this website.
Derekapaky
December 7, 2024 @ 3:08 am
http://www.huskytaxi.ru – удобный сервис для тех, кто ценит комфорт и качество.
CurtisDogue
December 7, 2024 @ 3:21 am
https://mayaschool.ru
EdwardLoarl
December 7, 2024 @ 3:51 am
Оптимизируйте работу своего бизнеса с надежным и мощным VPS-хостингом. Аренда выделенных виртуальных серверов с гарантированной производительностью и надежностью сервер арендовать
Social - paibe
December 7, 2024 @ 3:55 am
How Online Casinos Are Becoming a Global Phenomenon
Virtual gambling platforms have revolutionized the gaming world, offering an exceptional degree of comfort and selection that conventional gambling houses can’t match. Over the past decade, a growing community around the world have welcomed the excitement of virtual casinos thanks to its ease of access, exciting features, and continuously increasing range of offerings.
One of the strongest selling points of online gaming options is the vast array of choices ready to play. Whether you like interacting with traditional slots, trying out narrative-rich video slots, or mastering skills in traditional table offerings like poker, digital casinos offer limitless possibilities. Several sites additionally present live dealer games, enabling you to engage with professional croupiers and gaming peers, all while taking in the lifelike ambiance of a land-based casino without leaving your home.
If you’re new with the world of online gaming or are looking to find out more about reliable sites, why not become part of our dynamic interactive platform? It’s a place where fans offer stories, assisting you to enhance your virtual play. Explore the community and see it here now: https://www.facebook.com/profile.php?id=61569299482300&is_tour_dismissed .
Beyond variety, digital casino services are known for constant connectivity.
Top-Porno-Casino
December 7, 2024 @ 3:55 am
I like reading through an article that will make
people think. Also, thanks for permitting me to comment!
ArmandoWal
December 7, 2024 @ 3:59 am
Cytotec 200mcg price: cytotec pills buy online – п»їcytotec pills online
Jacobgaugh
December 7, 2024 @ 4:07 am
кухни москва — Большой выбор современных кухонь в Москве.
Jacobgaugh
December 7, 2024 @ 4:24 am
кухни на заказ москва – Изготовление кухонь на заказ в Москве с учетом всех ваших пожеланий.
Social - paibe
December 7, 2024 @ 4:56 am
The Reasons Behind Why Online Casinos Are a Worldwide Trend
Digital casinos have reshaped the gaming industry, providing a unique kind of comfort and selection that brick-and-mortar venues don’t provide. In recent years, a vast number of enthusiasts across the globe have adopted the pleasure of internet-based gaming because of its anytime, anywhere convenience, exciting features, and widening range of offerings.
One of the strongest selling points of internet-based platforms is the astounding diversity of choices ready to play. Whether you like engaging with classic fruit machine slots, playing through story-driven video-based games, or exercising tactics in table games like poker, virtual venues feature endless options. Numerous services moreover introduce interactive dealer games, allowing you to participate with actual dealers and gaming peers, all while immersing yourself in the immersive environment of a brick-and-mortar establishment in your own space.
If you’re just starting with the world of online gaming or would like to find out more about trusted platforms, why not participate in our active social network? It’s a platform where enthusiasts exchange experiences, helping you to improve your gaming journey. Discover the discussions and start your journey now: https://ru.pinterest.com/UpX_Official/ .
Beyond variety, online casinos are known for ease of access.
Maya
December 7, 2024 @ 4:58 am
You said it very well..
Feel free to surf to my web-site スーパーコピー ルイヴィトン マフラー 中古 (https://www.redly.vip/2245815)
Continue
December 7, 2024 @ 5:11 am
Hi every one, here every one is sharing these kinds of familiarity, thus it’s fastidious to read this web site, and I used to pay a quick visit this web site all the
time.
101 game login
December 7, 2024 @ 5:22 am
It is in point of fact a great and helpful piece of information.
I am glad that you shared this useful info with us. Please stay
us up to date like this. Thank you for sharing.
Social - paibe
December 7, 2024 @ 5:58 am
The Reasons Behind Why Online Casinos Remain a Global Phenomenon
Online casinos have transformed the betting scene, providing an exceptional degree of convenience and variety that traditional casinos struggle to rival. Recently, a vast number of enthusiasts internationally have chosen the pleasure of internet-based gaming as a result of its availability, appealing qualities, and ever-expanding collections of titles.
One of the strongest selling points of internet-based platforms is the sheer range of choices available. Whether you like playing on old-school one-armed bandits, immersing yourself in engaging video-based games, or exercising tactics in card and board games like poker, casino websites boast infinite possibilities. Several sites furthermore present real-time gaming experiences, making it possible for you to communicate with human game hosts and other players, all while soaking in the engaging vibes of a brick-and-mortar establishment in your own space.
If you’re a beginner with the world of online gaming or are looking to explore reputable operators, why not sign up for our active interactive platform? It’s a space where players post experiences, enabling you to maximize your gaming journey. Discover the community and see it here now: https://www.instagram.com/sweet_bonanzacom/ .
Besides the wide selection, virtual gaming providers are known for constant connectivity.
Go88
December 7, 2024 @ 6:46 am
You made some decent points there. I looked on the web
for more information about the issue and found most people
will go along with your views on this web site.
Social - paibe
December 7, 2024 @ 7:00 am
Why Online Casinos Are Becoming a Worldwide Trend
Virtual gambling platforms have changed the gambling industry, offering an unmatched level of convenience and range that traditional casinos don’t provide. Throughout the last ten years, millions of players internationally have welcomed the thrill of digital casino play in light of its ease of access, appealing qualities, and constantly growing collections of titles.
One of the biggest attractions of internet-based platforms is the sheer variety of entertainment options available. Whether you like interacting with old-school slot machines, diving into engaging video slots, or exercising tactics in strategy-based games like Baccarat, internet-based gambling sites feature countless choices. Plenty of operators furthermore introduce live casino options, letting you to interact with real dealers and other players, all while enjoying the realistic atmosphere of a traditional gambling venue without leaving your home.
If you’re a beginner with the world of internet-based gaming or seek to explore safe services, why not join our active community? It’s a destination where fans exchange reviews, making it easier for you to enhance your gaming journey. Check out the experience and visit us now: https://www.instagram.com/mystake_top/ .
In addition to diversity, internet-based gambling hubs shine availability.
Angelhob
December 7, 2024 @ 7:34 am
предметная съемка для маркетплейсов – Фотосъемка товаров, соответствующая требованиям крупнейших маркетплейсов.
betflik88
December 7, 2024 @ 7:40 am
You mentioned this wonderfully. https://cse.google.ne/url?q=https://betflix-88.one
Social - paibe
December 7, 2024 @ 8:02 am
How Online Casinos Are Highly Preferred Worldwide
Internet-based gambling hubs have revolutionized the betting scene, offering a level of comfort and variety that land-based casinos are unable to replicate. Over the past decade, millions of players across the globe have embraced the excitement of digital casino play because of its ease of access, thrilling aspects, and constantly growing selection of games.
One of the biggest attractions of virtual gambling hubs is the astounding range of entertainment options on offer. Whether you love rolling traditional reel games, diving into story-driven visual slot games, or exercising tactics in traditional table offerings like Blackjack, digital casinos provide infinite possibilities. Plenty of operators also present real-time gaming experiences, allowing you to interact with professional croupiers and fellow gamblers, all while immersing yourself in the realistic ambiance of a traditional gambling venue right at home.
If you’re just starting with the world of digital casinos or hope to learn about trusted platforms, why not participate in our vibrant online hub? It’s a destination where gamblers offer reviews, enabling you to get the most out of your online casino experience. Explore the community and learn more now: https://www.facebook.com/profile.php?id=61569299482300&is_tour_dismissed .
Besides the wide selection, online casinos thrive in seamless entry.
rooftop solutions
December 7, 2024 @ 8:09 am
A person necessarily help to make significantly articles I would state.
This is the very first time I frequented your web page and
thus far? I amazed with the analysis you made to create this particular submit incredible.
Excellent task!
ارزان ترین فالوور فیک
December 7, 2024 @ 8:41 am
Everyone loves it when folks come together and
share thoughts. Great blog, continue the good work!
Jacobgaugh
December 7, 2024 @ 8:52 am
http://gft-leasing.ru/ — Ознакомьтесь с нашими проектами и найдите свое решение.
Social - paibe
December 7, 2024 @ 9:07 am
Why Online Casinos Are So Popular
Internet-based gambling hubs have revolutionized the gambling landscape, providing a unique kind of ease and variety that brick-and-mortar establishments are unable to replicate. Over the past decade, countless gamblers around the world have chosen the pleasure of virtual gambling because of its always-open nature, appealing qualities, and widening game libraries.
One of the biggest attractions of online casinos is the astounding variety of choices at your disposal. Whether you are a fan of rolling traditional reel games, playing through plot-filled video slots, or testing your strategy in traditional table offerings like Texas Hold’em, casino websites offer numerous options. Numerous services also introduce live casino options, letting you to communicate with human game hosts and other players, all while enjoying the authentic feel of a physical gaming house in your own space.
If you’re just starting with the world of virtual casino play or seek to discover safe services, why not participate in our active interactive platform? It’s a platform where enthusiasts offer insights, assisting you to maximize your virtual play. Join the community and start your journey now: https://www.facebook.com/profile.php?id=61568481962080 .
In addition to diversity, digital casino services thrive in ease of access.
Jimmieclict
December 7, 2024 @ 9:29 am
https://lisinoprilus.com/# zestril 20 mg tab
giftcardmall mygift
December 7, 2024 @ 9:58 am
What’s up, all is going well here and ofcourse every one is sharing information, that’s in fact excellent, keep up writing.
WaltonSer
December 7, 2024 @ 10:12 am
[url=][/url]
Временная регистрация в Санкт-Петербурге: Быстро и Легально!
Ищете, где оформить временную регистрацию в СПб?
Мы гарантируем быстрое и легальное оформление без очередей и лишних документов.
Ваше спокойствие – наша забота!
Минимум усилий • Максимум удобства • Полная легальность
Свяжитесь с нами прямо сейчас!
Временная регистрация в СПб
[url=][/url]
Social - paibe
December 7, 2024 @ 10:12 am
The Reasons Behind Why Online Casinos Are a Worldwide Trend
Online casinos have modernized the casino gaming landscape, offering an exceptional degree of comfort and variety that conventional casinos fall short of. Over time, a large audience worldwide have chosen the thrill of virtual gambling because of its always-open nature, engaging traits, and constantly growing collections of titles.
One of the most compelling reasons of online casinos is the incredible diversity of entertainment options at your disposal. Whether you like playing on classic one-armed bandits, playing through plot-filled visual slot games, or strategizing in strategy-based games like poker, internet-based gambling sites provide endless choices. A large number of platforms even include live casino options, letting you to engage with human game hosts and other players, all while soaking in the authentic environment of a land-based casino from the comfort of your home.
If you’re unfamiliar with the world of virtual gambling or would like to discover trusted platforms, why not become part of our growing gaming forum? It’s a space where enthusiasts discuss stories, assisting you to improve your gaming journey. Dive into the experience and start your journey now: https://www.facebook.com/profile.php?id=61568692273712 .
In addition to diversity, virtual gaming providers excel accessibility.
thumb latch trigger
December 7, 2024 @ 10:21 am
Hey there! I know this is kind of off topic but I
was wondering if you knew where I could locate a captcha plugin for my comment form?
I’m using the same blog platform as yours and I’m having trouble finding one?
Thanks a lot!
Stop by my web-site; thumb latch trigger
Derekapaky
December 7, 2024 @ 10:35 am
кухни на заказ в спб – возможность обустроить кухню под ваши индивидуальные потребности.
香港現金網
December 7, 2024 @ 10:42 am
Way cool! Some extremely valid points! I appreciate you writing this article and also the rest of the site is also very good.
ArmandoWal
December 7, 2024 @ 10:42 am
ciprofloxacin over the counter: cipro for sale – buy cipro cheap
SXJFPOG
December 7, 2024 @ 10:54 am
Huntingtons disease. Blackberry. Googledocs. Alexander ovechkin. England. Jason mamoa. Virginia. Carthage. Heart rate. Chief. Chris. La chargers. Christopher columbus. Engineer. Discern. Corey seager. Force. Delaware. Focus movie. Zodiac sign. Great white. Crescent wrench. Rap. Successful. Kansas. Gabes. Brittany howard. Apologize. Columbus day. Applicable. Mrs. doubtfire. Perjury. Olympics. Rishi sunak. Moonlighting. Promiscuous meaning. Samsara. Nyrangers. Malta. Fiji. Rainbows. Amizon. Katherine johnson. Ascent. Africa cup of nations standings. Finger monkey. Joan jett. https://site-363914.tv53.ru/
Alien. Suriname. Godfather cast. Craven. Twin peaks restaurant. Grouse. Josh shapiro. Movies with timothee chalamet. Pho. Uranium. Atheist. The waterboy. Purdue university. The anthem. Meet the fockers. Catapult. Tarpon springs. Vultures. Ridiculous. Mayflower. Jayson tatum stats. Trenton. Government shutdown. Chicago illinois. Eid al adha. Dia. Indianapolis. Frasier cast. Mahatma gandhi. Statistics. Check mark. Kim kardashian kids. Cheaper by the dozen cast. Ah. Windsor castle. Dictatorship. Santa clara university. Sydney. Adam brody. Einstein. Prague. Bolero. Masonry. Intolerable acts. Scarce. Seasonal affective disorder. Ringo starr.
Social - paibe
December 7, 2024 @ 11:19 am
Why Online Casinos Remain So Popular
Digital casinos have reshaped the betting market, providing an unmatched level of comfort and diversity that traditional establishments don’t provide. Over the past decade, countless gamblers around the world have welcomed the excitement of online gaming as a result of its ease of access, appealing qualities, and ever-expanding catalogs of games.
One of the main appeals of internet-based platforms is the astounding selection of gaming experiences provided. Whether you are a fan of spinning vintage one-armed bandits, trying out narrative-rich modern slot games, or playing smart in card and board games like Texas Hold’em, internet-based gambling sites feature infinite opportunities. Several sites even introduce real-time gaming experiences, allowing you to participate with live hosts and other players, all while taking in the engaging atmosphere of a physical gaming house without leaving your home.
If you’re exploring for the first time with the world of internet-based gaming or are looking to learn about proven options, why not engage with our lively interactive platform? It’s a place where gaming aficionados share tips, making it easier for you to enjoy more of your gambling adventure. Dive into the conversation and visit us now: https://au.pinterest.com/casibom1/ .
In addition to diversity, digital casino services thrive in seamless entry.
rin
December 7, 2024 @ 12:11 pm
Exceptional post however , I was wondering if
you could write a litte more on this topic?
I’d be very grateful if you could elaborate a little bit further.
Bless you!
vivod iz zapoya v stacionare_abkn
December 7, 2024 @ 12:16 pm
вывод из запоя в стационаре [url=superjackson.ukrbb.net/viewtopic.php?f=28&t=9748]вывод из запоя в стационаре[/url] .
vivod iz zapoya v stacionare_eqml
December 7, 2024 @ 12:18 pm
лечения наркозависимости в стационаре [url=https://www.justforum.bestforums.org/viewtopic.php?f=26&t=4771]лечения наркозависимости в стационаре[/url] .
vivod iz zapoya v stacionare_fdOa
December 7, 2024 @ 12:18 pm
быстрый вывод из запоя в стационаре [url=www.turforum.borda.ru/?1-8-0-00001917-000-0-0-1730818279/]быстрый вывод из запоя в стационаре[/url] .
Dnrtmmi
December 7, 2024 @ 12:18 pm
Как безопасно купить диплом колледжа или ПТУ в России, что важно знать
vivod iz zapoya v stacionare_fiSt
December 7, 2024 @ 12:19 pm
перед выводом из запоя в стационаре [url=http://astana.forum24.ru/?1-1-0-00000091-000-0-0-1730817787]перед выводом из запоя в стационаре[/url] .
vivod iz zapoya v stacionare_wdpt
December 7, 2024 @ 12:19 pm
вывод из запоя самара стационар [url=https://recordrpservak.ukrbb.net/viewtopic.php?f=31&t=1166/]вывод из запоя самара стационар[/url] .
Social - paibe
December 7, 2024 @ 12:25 pm
The Reasons Behind Why Online Casinos Are a Worldwide Trend
Digital casinos have modernized the betting market, offering an exceptional degree of convenience and variety that brick-and-mortar gambling houses fall short of. Over time, countless gamblers internationally have chosen the adventure of virtual gambling in light of its anytime, anywhere convenience, appealing qualities, and widening range of offerings.
One of the key draws of virtual gambling hubs is the sheer selection of entertainment options at your disposal. Whether you are a fan of playing on retro one-armed bandits, immersing yourself in narrative-rich thematic slots, or strategizing in strategy-based games like Blackjack, online platforms offer infinite choices. Many casinos furthermore feature live dealer games, letting you to communicate with actual dealers and gaming peers, all while immersing yourself in the engaging ambiance of a real casino without leaving your home.
If you’re new with the world of digital casinos or want to learn about reputable operators, why not join our vibrant online hub? It’s a place where players post insights, guiding you to improve your casino activities. Discover the connections and see it here now: https://www.facebook.com/profile.php?id=61568692273712 .
Apart from the game range, virtual gaming providers thrive in accessibility.
porn video slots
December 7, 2024 @ 12:36 pm
Your means of describing all in this paragraph is actually pleasant, every one be capable of easily be aware of it,
Thanks a lot.
OSCQXDT
December 7, 2024 @ 12:39 pm
Value. Swiss airlines. Barry manilow. Gyro. Wrath of man. Plasma. Christ. Daisy. Tulane university. Kayleigh mcenany. Ny time. Dubious. Daily wordle. Yosemite national park. Chakra. Blobfish. Joro spider. Nazareth. Kalimba. Merida. Skype. Brittany murphy. Luca guadagnino. Veracity. Jj abrams. Veterinarian. Banff national park. Dolly parton age. Coast guard. Bougainvillea. American hustle. Muir woods. Io. Yogi berra. Augusta. Spam. English. Helen. Offset. Ginger beer. Sardine. Color wheel. Capitol hill. Nerdle. Baton. Extrapyramidal symptoms. Disneyworld. Fire. Reuters. Xi jinping. Proprietary. Ice cube. Korea. William howard taft. https://site-163438.tv53.ru/
Associative property. Perpendicular. Evan longoria. Dictionary online. Charleston. Genghis khan. University of san francisco. Johnny depp. Universal studios hollywood. Diabetes insipidus. The house that jack built. Van halen. However synonym. Baptism. Home alone 2 cast. Pirates bay. Water lily. Calendar. Basal ganglia. Jack nicholson movies. Eddie redmayne. Family guy. Thanksgiving day 2023. Mm. Feud. Zodiac. United states courts of appeals news. The bronx. Unit 731. Mi state. Great barrier reef. Monsters inc cast. Toy theater. Collegeboard. Monrovia. Daniel. Sam shepard. Brimstone. Gulf. Rockets. Rhombus. Situation. Claude monet. Obsolete. Prose.
LouisGepsy
December 7, 2024 @ 12:44 pm
География поставок компании «Технокор» не ограничивается регионом: за более чем 30 лет работы мы освоили всю территорию России – от Калининграда до острова Сахалин. Компания «Технокор» является дилером известных отечественных и зарубежных производителей – МТЗ, LOVOL и других https://ooo-tehnokor.promportal.su
Lazrhhn
December 7, 2024 @ 1:06 pm
Легальная покупка диплома о среднем образовании в Москве и регионах
Cazrdqx
December 7, 2024 @ 1:16 pm
Официальная покупка диплома вуза с сокращенной программой в Москве
JamesPurry
December 7, 2024 @ 1:22 pm
Эффективное управление временем – ключ к успеху в современном мире. Освойте искусство тайм-менеджмента, чтобы успевать больше, уставать меньше и достигать амбициозных целей. От простых техник до продвинутых методик – каждый найдет свой путь к продуктивности напоминалка
Jacobgaugh
December 7, 2024 @ 1:31 pm
фабрика кухня — Производственные кухни по индивидуальным проектам.
Social - paibe
December 7, 2024 @ 1:32 pm
The Reasons Behind Why Online Casinos Are Becoming So Popular
Digital casinos have revolutionized the casino gaming market, providing an unmatched level of comfort and range that conventional gambling houses don’t provide. Over the past decade, countless gamblers globally have turned to the excitement of internet-based gaming thanks to its availability, appealing qualities, and constantly growing range of offerings.
One of the key draws of online gaming options is the vast array of titles at your disposal. Whether you prefer spinning vintage slot machines, trying out plot-filled video-based games, or exercising tactics in card and board games like Texas Hold’em, casino websites deliver countless choices. Many casinos moreover include interactive dealer games, letting you to engage with real dealers and other players, all while enjoying the authentic vibes of a brick-and-mortar establishment without leaving your home.
If you’re exploring for the first time with the world of online gaming or are looking to delve deeper into trusted platforms, why not sign up for our lively community? It’s a platform where fans exchange experiences, assisting you to enhance your casino activities. Explore the community and learn more now: https://t.me/casibomgiris_net .
Apart from the game range, internet-based gambling hubs thrive in seamless entry.
Jaimeget
December 7, 2024 @ 1:35 pm
buy generic clomid no prescription [url=http://clomid.store/#]how to buy clomid without prescription[/url] where buy generic clomid
Social - paibe
December 7, 2024 @ 2:41 pm
What Makes Online Casinos Are a Global Phenomenon
Online casinos have revolutionized the casino gaming market, delivering an exceptional degree of ease and selection that physical casinos struggle to rival. Throughout the last ten years, countless gamblers globally have welcomed the adventure of online gaming due to its availability, engaging traits, and widening game libraries.
One of the biggest attractions of internet-based platforms is the unparalleled array of games provided. Whether you are a fan of rolling traditional fruit machine slots, playing through story-driven modern slot games, or strategizing in strategy-based games like Roulette, casino websites boast countless choices. Many casinos moreover include real-time gaming experiences, enabling you to engage with human game hosts and fellow gamblers, all while experiencing the engaging feel of a physical gaming house from the comfort of your home.
If you’re new with the world of virtual gambling or hope to find out more about reputable operators, why not become part of our growing gaming forum? It’s a destination where fans post experiences, making it easier for you to get the most out of your gambling adventure. Check out the conversation and start your journey now: https://www.instagram.com/1winbr_com/ .
Beyond variety, digital casino services shine availability.
Ayrlp - Recovery
December 7, 2024 @ 2:52 pm
Thanks for sharing your thoughts on Ayrlp – Recovery.
Regards
Click Here
December 7, 2024 @ 3:05 pm
Thank you for the good writeup. It in fact was a amusement account it.
Look advanced to far added agreeable from you! By the way,
how can we communicate?
toto macau
December 7, 2024 @ 3:10 pm
Hello there, You’ve done an incredible job. I will certainly digg it and personally recommend to my friends.
I am confident they will be benefited from this web site.
Trefgfp
December 7, 2024 @ 3:34 pm
Купить диплом о среднем образовании в Москве и любом другом городе
Leonardsiz
December 7, 2024 @ 3:39 pm
кухня заказать – Легко и быстро закажите кухню, которая станет сердцем вашего дома.
Angelhob
December 7, 2024 @ 3:44 pm
https://www.pilot-partner.ru/ – Лучшие фотографии для вашего бизнеса от профессионалов.
Social - paibe
December 7, 2024 @ 3:47 pm
Reasons Why Online Casinos Are Becoming a Global Phenomenon
Internet-based gambling hubs have changed the gaming scene, delivering a level of convenience and range that conventional gambling houses struggle to rival. Over time, countless gamblers worldwide have welcomed the pleasure of online gaming due to its ease of access, engaging traits, and widening catalogs of games.
One of the key draws of virtual gambling hubs is the astounding array of games at your disposal. Whether you prefer engaging with classic reel games, exploring narrative-rich visual slot games, or playing smart in classic casino games like Blackjack, internet-based gambling sites boast infinite opportunities. Several sites moreover offer live casino options, making it possible for you to interact with live hosts and fellow gamblers, all while experiencing the immersive vibes of a land-based casino in your own space.
If you’re a beginner with the world of internet-based gaming or want to discover reliable sites, why not sign up for our active social network? It’s a hub where gamblers discuss stories, assisting you to maximize your virtual play. Discover the conversation and visit us now: https://www.instagram.com/1winbr_com/ .
Adding to the extensive catalog, digital casino services shine availability.
부산 결혼식 사회자 MC 섭외
December 7, 2024 @ 4:08 pm
I think what you posted was very logical. But, what about this?
what if you composed a catchier post title? I mean, I
don’t wish to tell you how to run your blog, but what if you added a
headline that makes people want more? I mean Filling in the gaps of
HIV is a little boring. You could look at Yahoo’s front page and watch how
they create news titles to grab people to
open the links. You might try adding a video or
a pic or two to get readers excited about what you’ve written. In my opinion, it would bring your posts a little bit more interesting.
Parking Lot Contractor near me
December 7, 2024 @ 4:11 pm
My developer is trying to convince me to move to .net from PHP.
I have always disliked the idea because of the costs.
But he’s tryiong none the less. I’ve been using WordPress on a number of
websites for about a year and am concerned about switching to another platform.
I have heard fantastic things about blogengine.net.
Is there a way I can transfer all my wordpress content into it?
Any help would be really appreciated!
Mazrrhh
December 7, 2024 @ 4:18 pm
Как приобрести аттестат о среднем образовании в Москве и других городах
Agustinodome
December 7, 2024 @ 4:37 pm
Для веб-проектов, работающих на VPS, обеспечивается высокая скорость загрузки страниц, что является ключевым фактором для улучшения пользовательского опыта и SEO-показателей сайта бесплатные хостинги для сайтов
Ralphkesee
December 7, 2024 @ 4:38 pm
Инверторы играют ключевую роль в системах солнечной энергетики, позволяя преобразовывать постоянный ток, генерируемый солнечными панелями, в переменный, который может быть использован для питания бытовых приборов, освещения и других потребностей дома, офиса или производственного объекта. Это делает их незаменимым компонентом для создания эффективных и автономных систем электроснабжения https://solar.biz.ua/invertory-ibp/gibridnyj-invertor-ibp-3000vt-24v-mppt-na-3-kvt-ismppt-bf-3000-axioma-energy.html
essencial natura masculino
December 7, 2024 @ 4:41 pm
I am sure this piece of writing has touched all the internet people, its really really good
paragraph on building up new website.
Social - paibe
December 7, 2024 @ 4:49 pm
Reasons Why Online Casinos Are Becoming an International Sensation
Online casinos have modernized the betting industry, providing an exceptional degree of ease and selection that physical venues can’t match. Over time, a growing community across the globe have turned to the adventure of online gaming in light of its ease of access, appealing qualities, and ever-expanding catalogs of games.
One of the strongest selling points of internet-based platforms is the sheer range of titles at your disposal. Whether you love interacting with vintage slots, trying out narrative-rich visual slot games, or exercising tactics in classic casino games like Texas Hold’em, virtual venues deliver endless opportunities. Several sites also feature real-time gaming experiences, making it possible for you to communicate with actual dealers and fellow gamblers, all while soaking in the immersive vibes of a real casino from anywhere you want.
If you’re a beginner with the world of digital casinos or want to discover reliable sites, why not participate in our vibrant community? It’s a place where gamblers offer experiences, making it easier for you to enhance your virtual play. Discover the experience and see it here now: https://au.pinterest.com/casibom1/ .
Besides the wide selection, online casinos excel availability.
jav hd.vn
December 7, 2024 @ 5:03 pm
Greetings from Idaho! I’m bored to tears at work
so I decided to check out your website on my iphone during
lunch break. I really like the information you provide here and can’t wait
to take a look when I get home. I’m amazed at how fast your blog loaded
on my phone .. I’m not even using WIFI, just 3G .. Anyhow,
superb blog!
carbon offset programs
December 7, 2024 @ 5:04 pm
Great post! We are linking to this great content on our website.
Keep up the great writing.
online
December 7, 2024 @ 5:06 pm
Do you mind if I quote a couple of your articles as long as I provide
credit and sources back to your website?
My website is in the very same niche as yours and my users would really
benefit from some of the information you provide here. Please let me know if
this okay with you. Regards!
ArmandoWal
December 7, 2024 @ 5:35 pm
purchase cipro: buy cipro without rx – buy cipro without rx
Social - paibe
December 7, 2024 @ 5:52 pm
Why Online Casinos Are Becoming a Worldwide Trend
Online casinos have modernized the betting world, delivering a level of user-friendliness and selection that land-based gambling houses don’t provide. Over the past decade, millions of players worldwide have welcomed the adventure of digital casino play in light of its availability, thrilling aspects, and progressively larger collections of titles.
One of the biggest attractions of internet-based platforms is the astounding selection of gaming experiences at your disposal. Whether you are a fan of rolling classic one-armed bandits, playing through theme-based thematic slots, or mastering skills in traditional table offerings like Baccarat, virtual venues feature countless opportunities. Numerous services even present live casino options, enabling you to connect with real dealers and other players, all while experiencing the immersive environment of a real casino right at home.
If you’re exploring for the first time with the world of online gaming or are looking to find out more about safe services, why not participate in our dynamic gaming forum? It’s a hub where enthusiasts discuss insights, enabling you to get the most out of your online casino experience. Discover the connections and visit us now: https://www.facebook.com/profile.php?id=61568958127704 .
Besides the wide selection, digital casino services stand out seamless entry.
Jacobgaugh
December 7, 2024 @ 6:05 pm
https://www.armor-games.ru/ — Перейдите на сайт, чтобы узнать больше о кухнях на заказ.
Derekapaky
December 7, 2024 @ 6:21 pm
кухня на заказ – идеальное решение для реализации индивидуальных дизайнерских идей.
Jimmieclict
December 7, 2024 @ 6:33 pm
http://lisinoprilus.com/# 1 lisinopril
Social - paibe
December 7, 2024 @ 6:54 pm
How Online Casinos Are Becoming a Worldwide Trend
Digital casinos have changed the casino gaming landscape, offering a level of user-friendliness and range that land-based venues struggle to rival. In recent years, millions of players worldwide have welcomed the adventure of digital casino play in light of its availability, thrilling aspects, and continuously increasing selection of games.
One of the strongest selling points of virtual gambling hubs is the astounding selection of choices provided. Whether you prefer rolling classic slots, exploring theme-based modern slot games, or exercising tactics in table games like Blackjack, casino websites provide numerous opportunities. Numerous services even present interactive dealer games, making it possible for you to connect with live hosts and co-players, all while soaking in the authentic environment of a physical gaming house from the comfort of your home.
If you’re exploring for the first time with the world of online gaming or would like to find out more about safe services, why not become part of our active interactive platform? It’s a destination where fans post tips, enabling you to enjoy more of your online casino experience. Check out the community and see it here now: https://www.facebook.com/profile.php?id=61568958127704 .
Apart from the game range, online casinos are known for availability.
4health dog food reviews
December 7, 2024 @ 6:58 pm
Greetings! I know this is kinda off topic but
I was wondering if you knew where I could locate a captcha
plugin for my comment form? I’m using the same blog platform as yours
and I’m having trouble finding one? Thanks a lot!
Xazrmrl
December 7, 2024 @ 7:01 pm
Как не попасть впросак при покупке диплома колледжа или ПТУ в России
rent a rv
December 7, 2024 @ 7:28 pm
What a data of un-ambiguity and preserveness of valuable know-how concerning unexpected emotions.
Social - paibe
December 7, 2024 @ 7:55 pm
Why Online Casinos Remain Highly Preferred Worldwide
Online casinos have changed the casino gaming landscape, offering an unmatched level of convenience and diversity that traditional casinos fall short of. Throughout the last ten years, a growing community globally have welcomed the pleasure of virtual casinos thanks to its anytime, anywhere convenience, appealing qualities, and constantly growing game libraries.
One of the strongest selling points of virtual gambling hubs is the unparalleled diversity of gaming experiences ready to play. Whether you love spinning classic slot machines, playing through story-driven thematic slots, or playing smart in table games like Blackjack, online platforms deliver numerous opportunities. A large number of platforms even introduce real-time gaming experiences, giving you the chance you to engage with live hosts and co-players, all while immersing yourself in the realistic vibes of a land-based casino from the comfort of your home.
If you’re a beginner with the world of online gaming or seek to find out more about reliable sites, why not sign up for our active community? It’s a place where fans share insights, enabling you to enhance your gambling adventure. Discover the conversation and see it here now: https://t.me/casibomgiris_net .
In addition to diversity, virtual gaming providers excel accessibility.
Leonardsiz
December 7, 2024 @ 8:01 pm
https://www.tomason-russia.ru/ – Сделайте шаг к созданию кухни вашей мечты.
link slotraja777
December 7, 2024 @ 8:05 pm
I’m very pleased to discover this site. I want to to thank you for your time due to this fantastic read!!
I definitely really liked every bit of it and i also have you book-marked to
check out new information on your web site.
WaltonSer
December 7, 2024 @ 8:50 pm
[url=][/url]
Временная регистрация в Санкт-Петербурге: Быстро и Легально!
Ищете, где оформить временную регистрацию в СПб?
Мы гарантируем быстрое и легальное оформление без очередей и лишних документов.
Ваше спокойствие – наша забота!
Минимум усилий • Максимум удобства • Полная легальность
Свяжитесь с нами прямо сейчас!
Временная регистрация в СПб
[url=][/url]
Social - paibe
December 7, 2024 @ 8:57 pm
What Makes Online Casinos Are an International Sensation
Internet-based gambling hubs have transformed the gaming market, offering an exceptional degree of comfort and diversity that physical casinos struggle to rival. Over the past decade, a vast number of enthusiasts across the globe have chosen the excitement of virtual gambling thanks to its always-open nature, captivating elements, and ever-expanding selection of games.
One of the most compelling reasons of digital gambling sites is the astounding variety of games at your disposal. Whether you are a fan of interacting with vintage reel games, diving into plot-filled video-based games, or testing your strategy in traditional table offerings like Blackjack, internet-based gambling sites feature infinite choices. Many casinos moreover include interactive dealer games, letting you to interact with real dealers and co-players, all while enjoying the authentic atmosphere of a land-based casino without leaving your home.
If you’re exploring for the first time with the world of internet-based gaming or hope to learn about proven options, why not join our dynamic gaming forum? It’s a hub where gaming aficionados exchange experiences, assisting you to get the most out of your casino activities. Join the conversation and start your journey now: https://www.facebook.com/profile.php?id=61568481962080 .
Apart from the game range, virtual gaming providers shine availability.
Ремонт телевизоров Samsung
December 7, 2024 @ 9:05 pm
Предлагаем услуги профессиональных инженеров офицальной мастерской.
Еслли вы искали ремонт телевизоров samsung в москве, можете посмотреть на сайте: ремонт телевизоров samsung в москве
Наши мастера оперативно устранят неисправности вашего устройства в сервисе или с выездом на дом!
strut bracket
December 7, 2024 @ 9:12 pm
It’s really a cool and useful piece of info. I’m satisfied that you shared this useful info
with us. Please stay us up to date like this. Thanks for sharing.
my webpage; strut bracket
Social - paibe
December 7, 2024 @ 10:01 pm
The Reasons Behind Why Online Casinos Remain a Global Phenomenon
Online casinos have modernized the betting industry, delivering an unmatched level of comfort and breadth that brick-and-mortar gambling houses fall short of. In recent years, a vast number of enthusiasts across the globe have embraced the fun of internet-based gaming as a result of its always-open nature, exciting features, and continuously increasing selection of games.
One of the main appeals of virtual gambling hubs is the sheer range of gaming experiences on offer. Whether you love interacting with vintage slots, immersing yourself in plot-filled video slots, or testing your strategy in classic casino games like Texas Hold’em, internet-based gambling sites offer numerous options. Plenty of operators furthermore include live casino options, making it possible for you to interact with real dealers and opponents, all while enjoying the authentic feel of a land-based casino from the comfort of your home.
If you’re a beginner with the world of online gaming or want to discover proven options, why not become part of our growing social network? It’s a platform where gamblers exchange experiences, making it easier for you to enjoy more of your gaming journey. Check out the discussions and see it here now: https://www.facebook.com/profile.php?id=61568958127704 .
Adding to the extensive catalog, digital casino services excel seamless entry.
Jaimeget
December 7, 2024 @ 10:29 pm
azithromycin zithromax [url=https://azithromycinus.com/#]zithromax antibiotic[/url] zithromax online pharmacy canada
Angelhob
December 7, 2024 @ 10:29 pm
фотосъемка ювелирных изделий – Детальная визуализация каждого элемента ваших изделий.
escort osmanbey
December 7, 2024 @ 10:32 pm
merhaba en kaliteli şişli escort osmanbey escort sitesi
Jacobgaugh
December 7, 2024 @ 10:34 pm
фабрика кухня — Надежная фабрика, создающая кухни на заказ по вашим пожеланиям.
Social - paibe
December 7, 2024 @ 11:05 pm
How Online Casinos Have Become So Popular
Digital casinos have transformed the gambling market, providing an unmatched level of comfort and selection that land-based venues don’t provide. In recent years, countless gamblers around the world have embraced the pleasure of online gaming thanks to its always-open nature, appealing qualities, and progressively larger selection of games.
One of the strongest selling points of online gaming options is the incredible range of gaming experiences on offer. Whether you love spinning vintage fruit machine slots, diving into story-driven visual slot games, or playing smart in strategy-based games like poker, virtual venues feature countless options. Several sites also introduce real-time gaming experiences, enabling you to communicate with real dealers and gaming peers, all while immersing yourself in the engaging ambiance of a real casino from anywhere you want.
If you’re exploring for the first time with the world of online gaming or seek to discover trusted platforms, why not become part of our dynamic interactive platform? It’s a destination where players discuss experiences, making it easier for you to enhance your online casino experience. Dive into the connections and see it here now: https://www.facebook.com/profile.php?id=61568958127704 .
In addition to diversity, virtual gaming providers thrive in ease of access.
Click Here
December 7, 2024 @ 11:16 pm
I’m no longer certain the place you are getting your info, but good
topic. I must spend a while learning more or understanding more.
Thanks for excellent info I was searching for this information for my
mission.
เว็บสล็อต
December 7, 2024 @ 11:16 pm
fantastic issues altogether, you simply won a logo new reader.
What would you suggest in regards to your post that you simply made
some days in the past? Any positive?
Click Here
December 7, 2024 @ 11:59 pm
Hello, i think that i saw you visited my weblog so i came to “return the
favor”.I am attempting to find things to improve my website!I suppose its ok to use a few of your ideas!!
Social - paibe
December 8, 2024 @ 12:07 am
How Online Casinos Are a Worldwide Trend
Virtual gambling platforms have reshaped the betting world, providing a level of convenience and selection that brick-and-mortar gambling houses struggle to rival. Over the past decade, a vast number of enthusiasts across the globe have turned to the excitement of internet-based gaming as a result of its always-open nature, exciting features, and ever-expanding collections of titles.
One of the main appeals of online gaming options is the incredible range of entertainment options on offer. Whether you like interacting with traditional slot machines, trying out story-driven modern slot games, or playing smart in traditional table offerings like poker, casino websites feature limitless possibilities. Several sites even introduce live gaming streams, allowing you to participate with live hosts and opponents, all while immersing yourself in the immersive feel of a real casino without leaving your home.
If you’re unfamiliar with the world of internet-based gaming or want to find out more about reputable operators, why not join our dynamic community? It’s a platform where enthusiasts share experiences, helping you to enhance your online casino experience. Join the connections and see it here now: https://www.facebook.com/profile.php?id=61569299482300&is_tour_dismissed .
Besides the wide selection, digital casino services are known for ease of access.
sex 12 tuổi
December 8, 2024 @ 12:21 am
Does your site have a contact page? I’m having trouble locating it but, I’d like to send you an e-mail.
I’ve got some suggestions for your blog you might be interested in hearing.
Either way, great site and I look forward to seeing it grow over
time.
8Kbet
December 8, 2024 @ 12:37 am
When some one searches for his required thing, thus he/she wants to be available that in detail, so that thing is maintained over
here.
Leonardsiz
December 8, 2024 @ 12:41 am
кухни санкт петербург – Стильные кухонные гарнитуры, выполненные профессионалами в Петербурге.
Derekapaky
December 8, 2024 @ 12:48 am
кухня заказать легко и удобно с профессиональными мастерами, которые учтут все детали.
Social - paibe
December 8, 2024 @ 1:10 am
How Online Casinos Are a Global Phenomenon
Online casinos have modernized the gaming scene, providing a unique kind of accessibility and range that brick-and-mortar casinos are unable to replicate. Over the past decade, a vast number of enthusiasts around the world have turned to the adventure of virtual gambling as a result of its accessibility, thrilling aspects, and progressively larger catalogs of games.
One of the biggest attractions of internet-based platforms is the incredible diversity of titles provided. Whether you prefer rolling vintage slots, exploring story-driven visual slot games, or playing smart in strategy-based games like Baccarat, internet-based gambling sites feature countless opportunities. Several sites additionally introduce real-time gaming experiences, enabling you to engage with real dealers and fellow gamblers, all while enjoying the realistic atmosphere of a traditional gambling venue right at home.
If you’re exploring for the first time with the world of internet-based gaming or would like to explore safe services, why not sign up for our dynamic community? It’s a space where fans post reviews, helping you to enjoy more of your casino activities. Explore the community and visit us now: https://www.facebook.com/profile.php?id=61569299482300&is_tour_dismissed .
Apart from the game range, online casinos stand out seamless entry.
DRTEYT783
December 8, 2024 @ 1:41 am
Very good information. Lucky me I discovered your website by
accident (stumbleupon). I’ve bookmarked it for later!
Social - paibe
December 8, 2024 @ 2:13 am
What Makes Online Casinos Have Become a Global Phenomenon
Internet-based gambling hubs have revolutionized the casino gaming industry, offering a unique kind of ease and range that conventional casinos are unable to replicate. Throughout the last ten years, a vast number of enthusiasts across the globe have welcomed the adventure of internet-based gaming thanks to its anytime, anywhere convenience, engaging traits, and widening selection of games.
One of the biggest attractions of virtual gambling hubs is the incredible variety of games available. Whether you are a fan of playing on retro slots, playing through plot-filled visual slot games, or mastering skills in classic casino games like Baccarat, casino websites provide endless possibilities. A large number of platforms moreover present real-time gaming experiences, giving you the chance you to connect with professional croupiers and fellow gamblers, all while enjoying the lifelike feel of a land-based casino without leaving your home.
If you’re a beginner with the world of online gaming or are looking to explore reputable operators, why not sign up for our vibrant community? It’s a place where fans discuss insights, guiding you to maximize your casino activities. Dive into the experience and see it here now: https://x.com/upX_Top .
Beyond variety, virtual gaming providers excel accessibility.
PTE in Sydney
December 8, 2024 @ 2:31 am
I am sure this article has touched all the internet
users, its really really fastidious paragraph on building up new website.
Jacobgaugh
December 8, 2024 @ 3:03 am
https://www.mtucizone.ru/ — Наши проекты, советы и вдохновение для вашей кухни.
Ремонт телевизоров Philips Москва
December 8, 2024 @ 3:11 am
Предлагаем услуги профессиональных инженеров офицальной мастерской.
Еслли вы искали ремонт телевизоров philips цены, можете посмотреть на сайте: ремонт телевизоров philips адреса
Наши мастера оперативно устранят неисправности вашего устройства в сервисе или с выездом на дом!
arnavutköy eskort
December 8, 2024 @ 3:15 am
cazibeli escort bayan arıyorsanız arnavutköy escort sitesini ziyaret etmelisiniz
Social - paibe
December 8, 2024 @ 3:16 am
The Reasons Behind Why Online Casinos Remain a Global Phenomenon
Virtual gambling platforms have transformed the betting landscape, offering an exceptional degree of user-friendliness and variety that brick-and-mortar establishments fall short of. Recently, millions of players internationally have welcomed the excitement of virtual gambling thanks to its anytime, anywhere convenience, exciting features, and ever-expanding selection of games.
One of the biggest attractions of internet-based platforms is the vast array of games at your disposal. Whether you like engaging with retro slot machines, diving into narrative-rich video slots, or testing your strategy in card and board games like Baccarat, casino websites offer countless entertainment avenues. Many casinos also offer interactive dealer games, making it possible for you to interact with human game hosts and co-players, all while soaking in the engaging ambiance of a brick-and-mortar establishment without leaving your home.
If you’re a beginner with the world of virtual casino play or hope to find out more about reliable sites, why not engage with our vibrant gaming forum? It’s a space where enthusiasts post reviews, helping you to enjoy more of your casino activities. Explore the discussions and visit us now: https://www.facebook.com/profile.php?id=61568958127704 .
Adding to the extensive catalog, digital casino services thrive in seamless entry.
Jimmieclict
December 8, 2024 @ 3:22 am
https://cytotec.top/# buy cytotec online fast delivery
8Kbet
December 8, 2024 @ 3:25 am
Having read this I believed it was very enlightening. I appreciate
you finding the time and effort to put this informative article together.
I once again find myself personally spending way too much time both reading
and commenting. But so what, it was still worth it!
Erwin
December 8, 2024 @ 3:33 am
wonderful publish, very informative. I wonder why the opposite experts of this sector do not understand
this. You must continue your writing. I’m sure, you have a huge readers’ base already!
Kevinswind
December 8, 2024 @ 3:44 am
Инверторы играют ключевую роль в системах солнечной энергетики, позволяя преобразовывать постоянный ток, генерируемый солнечными панелями, в переменный https://solar.biz.ua/invertory-ibp/gibridnyj-invertor-ibp-3000vt-24v-mppt-na-3-kvt-ismppt-bf-3000-axioma-energy.html
DFGHHKKK
December 8, 2024 @ 4:20 am
Hi there to every one, as I am truly keen of reading this weblog’s post to be updated regularly.
It contains pleasant data.
Social - paibe
December 8, 2024 @ 4:21 am
How Online Casinos Are Becoming a Worldwide Trend
Internet-based gambling hubs have changed the betting world, offering a unique kind of accessibility and selection that conventional venues can’t match. Recently, a large audience worldwide have turned to the adventure of internet-based gaming because of its accessibility, appealing qualities, and constantly growing range of offerings.
One of the most compelling reasons of online gaming options is the sheer array of choices at your disposal. Whether you like playing on traditional slots, diving into theme-based visual slot games, or strategizing in strategy-based games like Blackjack, online platforms provide infinite options. A large number of platforms furthermore present live casino options, allowing you to interact with professional croupiers and co-players, all while immersing yourself in the realistic feel of a brick-and-mortar establishment in your own space.
If you’re a beginner with the world of virtual gambling or are looking to explore reputable operators, why not sign up for our active online hub? It’s a platform where players post tips, helping you to enhance your gambling adventure. Discover the connections and start your journey now: https://www.instagram.com/1winbr_com/ .
Adding to the extensive catalog, online casinos are known for ease of access.
Breakfast seminyak cafe bali
December 8, 2024 @ 5:16 am
An interesting discussion is definitely worth comment.
There’s no doubt that that you need to publish more about this subject
matter, it may not be a taboo subject but typically people do not talk about
these issues. To the next! Many thanks!!
Social - paibe
December 8, 2024 @ 5:24 am
Why Online Casinos Are So Popular
Virtual gambling platforms have changed the gaming scene, offering a unique kind of user-friendliness and diversity that traditional casinos are unable to replicate. Throughout the last ten years, millions of players across the globe have welcomed the excitement of digital casino play as a result of its anytime, anywhere convenience, captivating elements, and ever-expanding catalogs of games.
One of the strongest selling points of virtual gambling hubs is the unparalleled selection of games on offer. Whether you like rolling traditional reel games, exploring engaging modern slot games, or mastering skills in classic casino games like poker, virtual venues feature countless possibilities. Numerous services furthermore introduce interactive dealer games, letting you to engage with human game hosts and opponents, all while experiencing the realistic environment of a brick-and-mortar establishment in your own space.
If you’re exploring for the first time with the world of virtual casino play or are looking to find out more about trusted platforms, why not become part of our lively gaming forum? It’s a platform where enthusiasts exchange insights, guiding you to improve your online casino experience. Explore the experience and check it out now: https://au.pinterest.com/casibom1/ .
In addition to diversity, virtual gaming providers shine accessibility.
Angelhob
December 8, 2024 @ 5:27 am
фотосъемка одежды на моделях – Профессиональная съемка одежды на моделях для онлайн-магазинов и брендов.
Sanford Auto Glass Shop
December 8, 2024 @ 5:38 am
Your style is really unique compared to other people I
have read stuff from. Thanks for posting when you have the opportunity,
Guess I will just book mark this page.
visit website
December 8, 2024 @ 6:04 am
I couldn’t refrain from commenting. Exceptionally well written!
Leonardsiz
December 8, 2024 @ 6:18 am
кухни на заказ — Индивидуальное изготовление кухонь для вашего интерьера.
web site
December 8, 2024 @ 6:24 am
Cheers. I value this!
Social - paibe
December 8, 2024 @ 6:28 am
The Reasons Behind Why Online Casinos Have Become Highly Preferred Worldwide
Virtual gambling platforms have reshaped the casino gaming landscape, delivering a unique kind of user-friendliness and diversity that physical gambling houses struggle to rival. Recently, a growing community globally have turned to the thrill of online gaming due to its ease of access, engaging traits, and widening selection of games.
One of the key draws of digital gambling sites is the incredible variety of titles at your disposal. Whether you love interacting with vintage slots, playing through engaging video slots, or testing your strategy in table games like Texas Hold’em, internet-based gambling sites offer infinite entertainment avenues. Many casinos also present live gaming streams, letting you to engage with real dealers and fellow gamblers, all while immersing yourself in the engaging ambiance of a land-based casino without leaving your home.
If you’re exploring for the first time with the world of online gaming or are looking to learn about proven options, why not sign up for our growing interactive platform? It’s a space where gaming aficionados exchange experiences, making it easier for you to enhance your virtual play. Explore the discussions and start your journey now: https://www.facebook.com/profile.php?id=61568742522948 .
Beyond variety, internet-based gambling hubs excel seamless entry.
Robertvox
December 8, 2024 @ 6:30 am
where can i buy cipro online: buy cipro online – buy ciprofloxacin over the counter
MetBevarate
December 8, 2024 @ 6:43 am
скачать block strike много денег [url=https://apk-smart.com/igry/strelyalki/1380-vzlomannyj-block-strike-mod-mnogo-deneg.html]https://apk-smart.com/igry/strelyalki/1380-vzlomannyj-block-strike-mod-mnogo-deneg.html[/url] скачать block strike много денег
P.S Live ID: K89Io9blWX1UfZWv3ajv
P.S.S [url=http://qlxjhvcwebpin.mex.tl/?gb=1#top]Программы и игры для Андроид телефона[/url] [url=https://homecare82.com/bbs/board.php?bo_table=online&wr_id=317439]Программы и игры дл[/url] [url=https://forum.home-visa.ru/viewtopic.php?f=69&t=1311765]Программы и игры для Андроид телефона[/url] 125_777
Mazrcoh
December 8, 2024 @ 7:04 am
Купить диплом экономиста – оптимальное решение
Jaimeget
December 8, 2024 @ 7:16 am
lisinopril 5mg tablets [url=https://lisinoprilus.com/#]where can i buy zestril[/url] where can i buy lisinopril
Social - paibe
December 8, 2024 @ 7:32 am
What Makes Online Casinos Are a Global Phenomenon
Online casinos have reshaped the casino gaming landscape, offering a unique kind of ease and selection that physical venues can’t match. Over time, a growing community internationally have adopted the fun of virtual casinos due to its anytime, anywhere convenience, appealing qualities, and continuously increasing selection of games.
One of the strongest selling points of online casinos is the incredible range of games provided. Whether you prefer playing on old-school fruit machine slots, diving into theme-based modern slot games, or testing your strategy in classic casino games like poker, virtual venues deliver limitless opportunities. Many casinos moreover present live dealer games, making it possible for you to participate with professional croupiers and other players, all while immersing yourself in the lifelike ambiance of a physical gaming house from anywhere you want.
If you’re new with the world of virtual casino play or want to delve deeper into safe services, why not join our dynamic online hub? It’s a platform where enthusiasts exchange reviews, helping you to get the most out of your casino activities. Join the discussions and see it here now: https://www.facebook.com/profile.php?id=61568481962080 .
Apart from the game range, virtual gambling platforms excel availability.
Cazrxmf
December 8, 2024 @ 7:34 am
Как правильно купить диплом колледжа и пту в России, подводные камни
Jacobgaugh
December 8, 2024 @ 7:43 am
https://www.armor-games.ru/ — Перейдите на сайт, чтобы узнать больше о кухнях на заказ.
Derekapaky
December 8, 2024 @ 7:48 am
https://www.huskytaxi.ru/ – ваш путь к стильной и функциональной кухне.
vreau sa cumpar permis de conducere original
December 8, 2024 @ 7:48 am
Hey! I know this is kinda off topic however , I’d figured I’d
ask. Would you be interested in trading links or maybe guest writing a blog
post or vice-versa? My website covers a lot of the same subjects as yours
and I think we could greatly benefit from each other.
If you are interested feel free to shoot me an email.
I look forward to hearing from you! Superb blog by the way!
Jamessauby
December 8, 2024 @ 7:54 am
Гимнастика для спины, занятия Красивая осанка, Здоровая спина, Йога, Гамаки, Стретчинг, Шпагат, Пилатес, Zumba, Восточные танцы, массаж в Студии фитнес на профсоюзной улице
GPS
December 8, 2024 @ 8:16 am
Whoa! This blog looks exactly like my old one! It’s
on a completely different topic but it has pretty much the same page layout and design. Great choice of
colors!
chemical cooling tower
December 8, 2024 @ 8:17 am
Thank you for sharing your thoughts. I truly appreciate your efforts and I will be waiting
for your further write ups thanks once again.
loker bekasi
December 8, 2024 @ 8:17 am
You really make it seem so easy with your presentation but I find this topic to be actually something that I
think I would never understand. It seems too complicated and extremely broad for
me. I am looking forward for your next post,
I’ll try to get the hang of it!
Social - paibe
December 8, 2024 @ 8:36 am
Reasons Why Online Casinos Remain a Global Phenomenon
Online casinos have revolutionized the gaming market, providing a unique kind of ease and diversity that physical establishments don’t provide. Over the past decade, a growing community across the globe have welcomed the thrill of digital casino play in light of its accessibility, appealing qualities, and ever-expanding collections of titles.
One of the strongest selling points of virtual gambling hubs is the sheer variety of games at your disposal. Whether you enjoy spinning retro fruit machine slots, diving into engaging video-based games, or mastering skills in card and board games like Baccarat, online platforms provide numerous possibilities. Numerous services additionally include live gaming streams, allowing you to communicate with real dealers and opponents, all while soaking in the engaging vibes of a physical gaming house from the comfort of your home.
If you’re just starting with the world of virtual gambling or hope to learn about trusted platforms, why not sign up for our growing interactive platform? It’s a space where enthusiasts share insights, assisting you to maximize your gambling adventure. Join the discussions and visit us now: https://x.com/upX_Top .
Besides the wide selection, digital casino services are known for accessibility.
Social - paibe
December 8, 2024 @ 9:40 am
The Reasons Behind Why Online Casinos Are Becoming a Worldwide Trend
Virtual gambling platforms have modernized the gambling industry, offering a unique kind of ease and breadth that conventional venues can’t match. Over time, countless gamblers worldwide have turned to the pleasure of virtual gambling because of its anytime, anywhere convenience, exciting features, and widening catalogs of games.
One of the key draws of internet-based platforms is the unparalleled selection of titles at your disposal. Whether you love spinning traditional fruit machine slots, trying out story-driven video-based games, or mastering skills in traditional table offerings like Texas Hold’em, casino websites boast countless entertainment avenues. A large number of platforms additionally feature live casino options, giving you the chance you to connect with real dealers and gaming peers, all while immersing yourself in the realistic vibes of a land-based casino in your own space.
If you’re exploring for the first time with the world of virtual gambling or want to find out more about proven options, why not join our active gaming forum? It’s a platform where players exchange reviews, making it easier for you to improve your online casino experience. Dive into the conversation and visit us now: https://www.instagram.com/mystake_top/ .
Apart from the game range, virtual gaming providers stand out accessibility.
solid wood flooring austin
December 8, 2024 @ 9:46 am
Installing the correct flooring can totally change a space.
Whether it is actually wood, ceramic tile, or rug, the choice of
flooring can determine the area’s character.
Företagslån utan säkerhet
December 8, 2024 @ 10:08 am
Inspiring quest there. What happened after? Good luck!
Click Here
December 8, 2024 @ 10:22 am
Hi there! Do you know if they make any plugins
to protect against hackers? I’m kinda paranoid about losing everything I’ve worked
hard on. Any suggestions?
index
December 8, 2024 @ 10:27 am
It’s hard to find educated people about this topic, but you sound like you know what you’re
talking about! Thanks
Social - paibe
December 8, 2024 @ 10:44 am
What Makes Online Casinos Are Becoming Highly Preferred Worldwide
Virtual gambling platforms have transformed the gaming industry, providing an unmatched level of user-friendliness and selection that brick-and-mortar venues fall short of. Recently, millions of players worldwide have chosen the thrill of internet-based gaming because of its always-open nature, captivating elements, and progressively larger game libraries.
One of the key draws of virtual gambling hubs is the unparalleled diversity of entertainment options provided. Whether you like engaging with traditional one-armed bandits, trying out plot-filled video slots, or testing your strategy in classic casino games like Roulette, digital casinos provide numerous options. Plenty of operators additionally present interactive dealer games, allowing you to interact with professional croupiers and opponents, all while enjoying the engaging vibes of a real casino from the comfort of your home.
If you’re new with the world of internet-based gaming or want to explore proven options, why not join our active social network? It’s a platform where players offer tips, guiding you to improve your virtual play. Join the experience and start your journey now: https://www.facebook.com/profile.php?id=61569299482300&is_tour_dismissed .
Apart from the game range, internet-based gambling hubs excel ease of access.
vivod iz zapoya v stacionare_taKn
December 8, 2024 @ 11:05 am
лечению наркозависимости в стационаре [url=http://gonochki.forum24.ru/?1-10-0-00001144-000-0-0-1730819125/]лечению наркозависимости в стационаре[/url] .
Leonardsiz
December 8, 2024 @ 11:08 am
кухни санкт петербург — Широкий выбор кухонь в Санкт-Петербурге на заказ.
register mamiqq
December 8, 2024 @ 11:15 am
Hello there! I simply would like to give you a huge thumbs up for the great info you have right here
on this post. I will be returning to your website for
more soon.
Lazrtll
December 8, 2024 @ 11:32 am
Как получить диплом о среднем образовании в Москве и других городах
vivod iz zapoya v stacionare_hzOn
December 8, 2024 @ 11:35 am
выводу из запоя в стационаре [url=http://www.bisound.com/forum/showthread.php?p=1221507]выводу из запоя в стационаре[/url] .
vivod iz zapoya v stacionare_lyKt
December 8, 2024 @ 11:41 am
лечение наркозависимости в стационаре [url=chesskomi.borda.ru/?1-6-0-00000052-000-0-0-1730818609]лечение наркозависимости в стационаре[/url] .
vivod iz zapoya v stacionare_gzOt
December 8, 2024 @ 11:46 am
вывод из запоя в стационаре самара [url=www.pelsh.forum24.ru/?1-8-0-00000132-000-0-0-1730818865]вывод из запоя в стационаре самара[/url] .
vivod iz zapoya v stacionare_fdpi
December 8, 2024 @ 11:46 am
лечению наркозависимости в стационаре [url=www.aqvakr.forum24.ru/?1-7-0-00011599-000-0-0-1730819037]лечению наркозависимости в стационаре[/url] .
Social - paibe
December 8, 2024 @ 11:51 am
How Online Casinos Are Becoming a Global Phenomenon
Digital casinos have reshaped the gaming scene, providing a unique kind of accessibility and diversity that physical venues don’t provide. Over the past decade, a growing community worldwide have turned to the adventure of virtual gambling due to its always-open nature, captivating elements, and ever-expanding selection of games.
One of the main appeals of digital gambling sites is the astounding array of choices available. Whether you like interacting with retro fruit machine slots, diving into engaging modern slot games, or exercising tactics in traditional table offerings like Roulette, virtual venues boast infinite opportunities. Many casinos moreover feature live dealer games, making it possible for you to communicate with live hosts and opponents, all while taking in the authentic ambiance of a land-based casino in your own space.
If you’re new with the world of internet-based gaming or want to delve deeper into safe services, why not sign up for our growing community? It’s a hub where players post experiences, making it easier for you to get the most out of your online casino experience. Dive into the community and see it here now: https://www.instagram.com/1bet_be/ .
Beyond variety, virtual gaming providers stand out ease of access.
토토사이트 먹튀검증
December 8, 2024 @ 12:13 pm
메이저 보증업체를 선택하면, 믿을 수 있는 환경에서
안전한 놀이터의 다양한 게임을 즐길 수 있습니다.
이러한 사이트들은 사용자에게 안전하고 즐거운 경험을 제공하여, 스포츠토토 게임에 대한 신뢰성을 높이는
데 큰 역할을 합니다. 홈페이지를 방문해주세요.
impression sur bache
December 8, 2024 @ 12:16 pm
My relatives every time say that I am killing my time here at
net, but I know I am getting experience all the time by reading thes nice articles.
auto glass facts
December 8, 2024 @ 12:19 pm
I’m now not certain the place you’re getting your information,
however great topic. I must spend a while finding out
much more or understanding more. Thanks
for fantastic info I used to be on the lookout for this information for my
mission.
Jacobgaugh
December 8, 2024 @ 12:28 pm
кухни на заказ — Закажите уникальную кухню, идеально подходящую вашему интерьеру.
ZacharyNoila
December 8, 2024 @ 12:39 pm
Просторный тренажерный зал. Fitness Беляево оборудован 150+ единицами современных тренажеров. Самые популярные из них представлены в нескольких экземплярах, чтобы вы не тратили время на ожидание в очереди пилатес беляево
Sazrisg
December 8, 2024 @ 12:49 pm
Официальная покупка диплома вуза с сокращенной программой в Москве
Social - paibe
December 8, 2024 @ 12:57 pm
How Online Casinos Are Becoming an International Sensation
Digital casinos have changed the betting market, delivering an exceptional degree of accessibility and range that land-based casinos are unable to replicate. Over time, a growing community around the world have embraced the excitement of internet-based gaming in light of its availability, engaging traits, and progressively larger collections of titles.
One of the biggest attractions of internet-based platforms is the vast diversity of gaming experiences at your disposal. Whether you love playing on classic slots, immersing yourself in narrative-rich video-based games, or exercising tactics in table games like poker, digital casinos feature endless options. Many casinos moreover introduce live casino options, enabling you to connect with human game hosts and opponents, all while immersing yourself in the lifelike environment of a real casino without leaving your home.
If you’re exploring for the first time with the world of virtual gambling or would like to explore safe services, why not become part of our vibrant gaming forum? It’s a hub where enthusiasts post tips, enabling you to get the most out of your online casino experience. Join the experience and see it here now: https://www.instagram.com/kwz.kz_official/ .
Besides the wide selection, digital casino services are known for constant connectivity.
Jacobgaugh
December 8, 2024 @ 1:31 pm
кухни на заказ цены — узнайте актуальные предложения на сайте.
Doncaster escorts agency
December 8, 2024 @ 1:33 pm
I’ve been browsing online more than 2 hours today, yet I never found any interesting article like yours.
It’s pretty worth enough for me. In my view,
if all webmasters and bloggers made good content
as you did, the web will be much more useful than ever before.
Richardvak
December 8, 2024 @ 1:56 pm
[url=][/url]
Временная регистрация в СПб: Быстро и Легально!
Ищете, где оформить временную регистрацию в Санкт-Петербурге?
Мы гарантируем быстрое и легальное оформление без очередей и лишних документов.
Ваше спокойствие – наша забота!
Минимум усилий • Максимум удобства • Полная легальность
Свяжитесь с нами прямо сейчас!
Временная регистрация
[url=][/url]
Social - paibe
December 8, 2024 @ 2:04 pm
The Reasons Behind Why Online Casinos Are Becoming Highly Preferred Worldwide
Virtual gambling platforms have transformed the casino gaming world, offering a unique kind of user-friendliness and range that physical establishments can’t match. In recent years, millions of players globally have welcomed the excitement of virtual gambling because of its ease of access, engaging traits, and ever-expanding catalogs of games.
One of the most compelling reasons of internet-based platforms is the incredible variety of games on offer. Whether you love playing on vintage slots, playing through narrative-rich video slots, or playing smart in traditional table offerings like Blackjack, virtual venues provide numerous possibilities. Plenty of operators even feature live gaming streams, allowing you to connect with human game hosts and other players, all while taking in the lifelike ambiance of a real casino from anywhere you want.
If you’re new with the world of virtual casino play or seek to learn about proven options, why not engage with our dynamic gaming forum? It’s a hub where players offer insights, helping you to enhance your casino activities. Discover the conversation and see it here now: https://www.facebook.com/profile.php?id=61568481962080 .
In addition to diversity, digital casino services excel ease of access.
Situs Glowin88
December 8, 2024 @ 2:11 pm
Since the admin of this site is working, no hesitation very quickly it will be renowned, due to its feature contents.
Angelhob
December 8, 2024 @ 2:43 pm
http://www.bagroo.ru – Откройте сайт для детальной информации о профессиональной фотосъемке товаров.
Social - paibe
December 8, 2024 @ 3:08 pm
Reasons Why Online Casinos Have Become an International Sensation
Digital casinos have revolutionized the gaming industry, providing an unmatched level of comfort and selection that land-based venues fall short of. Recently, a vast number of enthusiasts across the globe have chosen the thrill of virtual gambling in light of its ease of access, captivating elements, and continuously increasing catalogs of games.
One of the main appeals of digital gambling sites is the astounding selection of games available. Whether you are a fan of rolling old-school fruit machine slots, diving into narrative-rich thematic slots, or testing your strategy in traditional table offerings like poker, online platforms provide countless possibilities. Many casinos furthermore present live casino options, allowing you to engage with human game hosts and opponents, all while experiencing the authentic feel of a traditional gambling venue in your own space.
If you’re new with the world of online gaming or seek to explore proven options, why not become part of our dynamic community? It’s a platform where enthusiasts post tips, helping you to improve your gaming journey. Check out the experience and start your journey now: https://au.pinterest.com/casibom1/ .
Besides the wide selection, digital casino services excel availability.
Concrete Contractor Company near me
December 8, 2024 @ 3:13 pm
Heya i’m for the first time here. I found this board and I find It truly useful & it helped me out much.
I hope to give something back and aid others like you helped me.
Jaimeget
December 8, 2024 @ 4:02 pm
purchase cipro [url=https://ciprofloxacin.cheap/#]cipro pharmacy[/url] buy generic ciprofloxacin
Social - paibe
December 8, 2024 @ 4:14 pm
The Reasons Behind Why Online Casinos Are Becoming a Global Phenomenon
Internet-based gambling hubs have revolutionized the casino gaming world, offering a level of comfort and variety that brick-and-mortar gambling houses don’t provide. In recent years, a vast number of enthusiasts around the world have adopted the thrill of virtual gambling due to its always-open nature, thrilling aspects, and progressively larger collections of titles.
One of the strongest selling points of digital gambling sites is the incredible array of titles on offer. Whether you are a fan of engaging with retro reel games, immersing yourself in engaging video-based games, or playing smart in classic casino games like Blackjack, virtual venues feature countless entertainment avenues. Plenty of operators additionally feature interactive dealer games, enabling you to interact with human game hosts and gaming peers, all while immersing yourself in the realistic environment of a real casino right at home.
If you’re just starting with the world of digital casinos or would like to learn about reliable sites, why not participate in our dynamic online hub? It’s a platform where gaming aficionados discuss reviews, making it easier for you to maximize your gaming journey. Check out the experience and see it here now: https://www.instagram.com/sweet_bonanzacom/ .
In addition to diversity, digital casino services are known for ease of access.
Leonardsiz
December 8, 2024 @ 4:15 pm
http://www.tomason-russia.ru – Найдите свою идеальную кухню в нашем каталоге.
Derekapaky
December 8, 2024 @ 5:02 pm
larson-holz-spb.ru – платформа для обновления вашего кухонного интерьера.
Order MDMA pills Amsterdam
December 8, 2024 @ 5:12 pm
Post writing is also a excitement, if you know afterward you can write or else it is difficult to write.
Social - paibe
December 8, 2024 @ 5:18 pm
Reasons Why Online Casinos Are Becoming a Worldwide Trend
Virtual gambling platforms have changed the casino gaming landscape, providing an unmatched level of convenience and selection that conventional establishments fall short of. Throughout the last ten years, a large audience internationally have turned to the pleasure of internet-based gaming thanks to its ease of access, captivating elements, and constantly growing catalogs of games.
One of the most compelling reasons of online casinos is the unparalleled variety of choices on offer. Whether you like spinning traditional slot machines, immersing yourself in story-driven video-based games, or strategizing in classic casino games like Baccarat, internet-based gambling sites provide numerous choices. Plenty of operators additionally feature live casino options, giving you the chance you to engage with actual dealers and opponents, all while experiencing the immersive feel of a traditional gambling venue from anywhere you want.
If you’re new with the world of internet-based gaming or want to find out more about trusted platforms, why not engage with our growing online hub? It’s a space where gaming aficionados exchange reviews, helping you to enjoy more of your online casino experience. Discover the experience and learn more now: https://t.me/casibomgiris_net .
In addition to diversity, online casinos excel accessibility.
Josephmob
December 8, 2024 @ 5:26 pm
А вы любите стейки? [url=https://obzornov.ru/]купить стейк спб[/url]
Jacobgaugh
December 8, 2024 @ 5:27 pm
кухня на заказ недорого москва — Кухни высокого качества по доступным ценам в столице.
nohu90.productions
December 8, 2024 @ 5:42 pm
Hello, Neat post. There’s a problem with your website
in web explorer, may check this? IE nonetheless is the market chief and a good component to other folks will leave out your magnificent writing due to this problem.
Matthewpok
December 8, 2024 @ 6:17 pm
https://cytotec.top/# п»їcytotec pills online
buy cytotec in usa [url=http://cytotec.top/#]п»їcytotec pills online[/url] buy cytotec pills
Social - paibe
December 8, 2024 @ 6:21 pm
How Online Casinos Have Become a Global Phenomenon
Internet-based gambling hubs have modernized the casino gaming industry, offering a level of accessibility and breadth that physical casinos fall short of. Throughout the last ten years, a large audience around the world have welcomed the adventure of virtual gambling as a result of its accessibility, captivating elements, and continuously increasing collections of titles.
One of the key draws of internet-based platforms is the incredible array of choices available. Whether you are a fan of playing on traditional reel games, diving into theme-based thematic slots, or testing your strategy in card and board games like Roulette, casino websites offer infinite opportunities. Plenty of operators furthermore introduce live gaming streams, letting you to connect with human game hosts and fellow gamblers, all while enjoying the engaging atmosphere of a land-based casino from anywhere you want.
If you’re a beginner with the world of digital casinos or seek to delve deeper into safe services, why not sign up for our vibrant gaming forum? It’s a destination where gamblers share experiences, assisting you to enjoy more of your casino activities. Discover the conversation and visit us now: https://www.facebook.com/profile.php?id=61568958127704 .
In addition to diversity, internet-based gambling hubs shine constant connectivity.
working private instagram viewer
December 8, 2024 @ 6:21 pm
There are various tools and websites that claim to allow
users to view private Instagram profiles, but it’s important
to door these past caution. Many of these tools can be unreliable, may require personal information, or could violate Instagram’s terms of
service. Additionally, using such tools can compromise your own security or
guide to scams. The safest and most ethical exaggeration to view a working private instagram viewer profile is to send a follow demand directly to the user.
Always prioritize privacy and devotion in your online interactions.
tree removal canberra
December 8, 2024 @ 6:38 pm
Tree removal in Canberra, Australia, is a regulated activity that involves the removal of trees from private or
public land. The process is governed by local council regulations, environmental considerations, and specific guidelines designed to preserve the region’s biodiversity and urban canopy.
The key points to consider when it comes to tree removal in Canberra include
the following:
1. Regulations and Permits
In Canberra, tree removal is governed by the Environmental Protection and Biodiversity Conservation Act as well as the Tree Protection Act 2005 and
other ACT Government regulations. Generally, removal of trees on private property may require a permit
from the ACT Government’s Environment, Planning and Sustainable Development Directorate (EPSDD) if the tree is deemed significant.
Significant trees are those that have historical, environmental, or aesthetic value, or if they are part of a heritage listing.
The guidelines aim to prevent the unnecessary loss of
trees, particularly those that contribute to Canberra’s urban forest and ecological health.
For private properties, landowners must contact the local council to determine whether a permit is required.
In some cases, tree removal can be approved if it poses a risk to structures,
human safety, or public health. However, permits are typically needed for the removal of larger, protected species or trees in certain zones.
2. Reasons for Tree Removal
There are various reasons why trees might need to be removed in Canberra, such as:
Disease or Pest Infestation: Some trees may be infested by pests or affected
by diseases, making them unsafe or unsightly.
Structural Damage: Trees may pose a risk to property or infrastructure, especially if their
roots threaten foundations, sidewalks, or underground utilities.
Safety Concerns: If a tree is dead, decaying, or unstable, it could be a safety
hazard during storms or high winds.
Land Development: Trees may need to be removed for new construction, landscaping projects, or other developments.
3. Replacement Trees and Mitigation
If a significant tree is removed, it may be required by
the ACT Government to plant a replacement tree or carry out mitigation measures.
The intention is to maintain the ecological balance, support biodiversity, and preserve Canberra’s green urban spaces.
The council often requires a replacement tree that meets specific size, species, and planting guidelines to ensure
it will thrive.
4. Tree Removal Companies
Tree removal should be performed by a certified arborist or tree removal company, especially
in urban settings where skillful handling is necessary to
avoid damaging surrounding property or people. These professionals are trained to assess the tree’s
health, determine the safest method of removal, and follow local laws.
When hiring a tree removal company in Canberra, it’s important to ensure they have the
necessary insurance, licenses, and qualifications.
5. Environmental Considerations
Canberra has a strong commitment to maintaining its natural environment, which includes preserving its tree canopy
and supporting biodiversity. Urban trees help with stormwater management, air quality,
cooling effects, and providing habitat for local wildlife. Tree removal is
generally considered a last resort, with efforts made to save and protect trees
wherever possible.
6. Cost of Tree Removal
The cost of tree removal in Canberra can vary depending on several factors, including the
size of the tree, its location (proximity to structures or power lines), and the
complexity of the removal. Costs can also be influenced by whether the tree requires additional work, such
as stump grinding, debris removal, or site restoration.
Jacobgaugh
December 8, 2024 @ 6:42 pm
nmphoto.ru — ваш надежный партнер в создании кухни.
order prednisone 10mg without prescription
December 8, 2024 @ 7:14 pm
where to get generic prednisone no prescription get generic prednisone without insurance cost of cheap prednisone without insurance
prednisone 10 mg prednisonegeqi com order generic prednisone without rx how to get prednisone online
i need to buy prednisone 20mg
prednisone without a dr prescription can i order cheap prednisone price order generic prednisone without a prescription
where to get cheap prednisone pill how can i get prednisone where to get cheap prednisone price
Social - paibe
December 8, 2024 @ 7:23 pm
Reasons Why Online Casinos Have Become Highly Preferred Worldwide
Internet-based gambling hubs have changed the casino gaming scene, offering a level of ease and diversity that conventional gambling houses struggle to rival. Over time, a growing community worldwide have adopted the excitement of internet-based gaming due to its accessibility, appealing qualities, and progressively larger game libraries.
One of the main appeals of online casinos is the sheer selection of games provided. Whether you like rolling traditional slots, trying out narrative-rich visual slot games, or strategizing in traditional table offerings like Blackjack, internet-based gambling sites deliver infinite entertainment avenues. Several sites even feature live gaming streams, enabling you to communicate with human game hosts and opponents, all while immersing yourself in the authentic atmosphere of a brick-and-mortar establishment from the comfort of your home.
If you’re a beginner with the world of online gaming or want to learn about trusted platforms, why not engage with our growing community? It’s a hub where gaming aficionados offer experiences, guiding you to enhance your gambling adventure. Discover the conversation and visit us now: https://au.pinterest.com/casibom1/ .
In addition to diversity, virtual gaming providers are known for constant connectivity.
Explore
December 8, 2024 @ 7:30 pm
Awesome post.
what is a safe dose of prednisone long term
December 8, 2024 @ 7:41 pm
buying prednisone tablets where to get cheap prednisone without dr prescription generic prednisone without a prescription
order cheap prednisone pill get prednisone without rx how to get prednisone without insurance
prednisone buy online uk
5mg prednisone every day cheapest way to buy prednisone where to buy generic prednisone without a prescription
buy prednisone online what is a safe dose of prednisone long term buying prednisone pill
Lou
December 8, 2024 @ 7:49 pm
Wow, this paragraph is good, my younger sister is analyzing
such things, thus I am going to let know her.
can you get prednisone without insurance
December 8, 2024 @ 8:12 pm
buy prednisone worldwide buy cheap prednisone for sale purchase prednisone sale
how to buy prednisone onlinenwithout prescription prednisone cost goodrx where to buy cheap prednisone pill
can i buy generic prednisone without prescription
how can i get generic prednisone without a prescription where can i get generic prednisone without insurance apo prednisone overseas pharmacies
80 mg prednisone high dose prednisone side effects buy prednisone 10mg for sale
Social - paibe
December 8, 2024 @ 8:26 pm
How Online Casinos Are Becoming Highly Preferred Worldwide
Online casinos have modernized the casino gaming scene, offering an unmatched level of convenience and breadth that land-based casinos are unable to replicate. Throughout the last ten years, a vast number of enthusiasts around the world have welcomed the adventure of virtual casinos due to its availability, engaging traits, and progressively larger range of offerings.
One of the key draws of online casinos is the vast diversity of games ready to play. Whether you enjoy spinning old-school slots, immersing yourself in plot-filled thematic slots, or strategizing in traditional table offerings like poker, casino websites feature endless entertainment avenues. Numerous services even feature interactive dealer games, giving you the chance you to participate with professional croupiers and co-players, all while soaking in the engaging feel of a land-based casino right at home.
If you’re exploring for the first time with the world of virtual gambling or are looking to find out more about proven options, why not join our growing online hub? It’s a place where fans offer tips, making it easier for you to improve your online casino experience. Dive into the discussions and visit us now: https://www.linkedin.com/in/my-stake-a096b427a/ .
Adding to the extensive catalog, online casinos stand out seamless entry.
türk escort
December 8, 2024 @ 8:27 pm
ortalama olarak en üst düzey kaliteli escortların bulunduğu türk rus özbek escort sitesi olan https://seyrantepescort.com/
how to get prednisone
December 8, 2024 @ 8:41 pm
how much is too prednisone cost of cheap prednisone without rx canadian prednisone
prednisone acquisto can you buy prednisone tabs without doctors priscription can you take prednisone long term
buy prednisone online usa
generic prednisone no prescription prednisone average cost prednisone without perscription
over the counter prednisone pills get cheap prednisone without insurance can i buy cheap prednisone without a prescription
escort şişli
December 8, 2024 @ 8:58 pm
bağlanabileceğiniz kaliteli şişli escort sitesi
Smooth Jazz CDs
December 8, 2024 @ 9:06 pm
Write more, thats all I have to say. Literally,
it seems as though you relied on the video to make your point.
You definitely know what youre talking about, why waste your intelligence on just posting videos
to your weblog when you could be giving us something enlightening to read?
Social - paibe
December 8, 2024 @ 9:31 pm
The Reasons Behind Why Online Casinos Are Becoming a Worldwide Trend
Internet-based gambling hubs have changed the casino gaming market, providing a level of accessibility and range that physical establishments fall short of. Throughout the last ten years, a large audience worldwide have turned to the pleasure of internet-based gaming thanks to its always-open nature, engaging traits, and progressively larger game libraries.
One of the biggest attractions of internet-based platforms is the vast range of entertainment options ready to play. Whether you enjoy rolling old-school slots, playing through plot-filled thematic slots, or mastering skills in traditional table offerings like Roulette, digital casinos feature numerous opportunities. Several sites moreover include real-time gaming experiences, letting you to participate with real dealers and co-players, all while immersing yourself in the realistic environment of a real casino without leaving your home.
If you’re exploring for the first time with the world of digital casinos or would like to discover reliable sites, why not sign up for our active social network? It’s a space where gaming aficionados offer reviews, guiding you to get the most out of your gambling adventure. Join the experience and learn more now: https://www.instagram.com/1winbr_com/ .
In addition to diversity, online casinos shine constant connectivity.
Sazraby
December 8, 2024 @ 9:59 pm
Сколько стоит диплом высшего и среднего образования и как это происходит?
winston salem auto glass
December 8, 2024 @ 10:02 pm
Great weblog right here! Additionally your website a
lot up fast! What host are you using? Can I get your
associate hyperlink in your host? I want my website
loaded up as fast as yours lol
onexxxslots
December 8, 2024 @ 10:04 pm
Используйте 1xslots бонус код для увеличения выигрышей.
Jacobgaugh
December 8, 2024 @ 10:16 pm
http://www.armor-games.ru — Узнайте больше о наших услугах и кухнях на сайте.
Jamescaurf
December 8, 2024 @ 10:20 pm
buy Kamagra: cheap kamagra – sildenafil oral jelly 100mg kamagra
Myths About Auto Glass Replacement
December 8, 2024 @ 10:20 pm
I’ve been surfing online more than three hours lately, yet I never discovered any interesting article like yours.
It is pretty worth sufficient for me. Personally,
if all web owners and bloggers made excellent content material as you
probably did, the net might be much more useful
than ever before.
Social - paibe
December 8, 2024 @ 10:36 pm
Why Online Casinos Are Becoming a Worldwide Trend
Digital casinos have changed the gaming landscape, providing a level of convenience and range that traditional casinos don’t provide. In recent years, countless gamblers across the globe have adopted the fun of virtual gambling thanks to its accessibility, exciting features, and widening game libraries.
One of the strongest selling points of online gaming options is the sheer selection of entertainment options on offer. Whether you enjoy rolling retro fruit machine slots, trying out story-driven modern slot games, or strategizing in table games like Texas Hold’em, virtual venues offer infinite possibilities. Plenty of operators additionally introduce interactive dealer games, letting you to participate with professional croupiers and other players, all while immersing yourself in the immersive feel of a real casino in your own space.
If you’re new with the world of digital casinos or are looking to delve deeper into safe services, why not become part of our lively interactive platform? It’s a hub where players share reviews, helping you to get the most out of your online casino experience. Discover the connections and learn more now: https://au.pinterest.com/casibom1/ .
Adding to the extensive catalog, online casinos stand out ease of access.
Angelhob
December 8, 2024 @ 11:07 pm
фотограф предметная съемка – Услуги профессионала для качественной съемки ваших товаров.
Social - paibe
December 8, 2024 @ 11:44 pm
The Reasons Behind Why Online Casinos Are Becoming So Popular
Internet-based gambling hubs have modernized the betting world, providing a unique kind of comfort and variety that conventional casinos fall short of. In recent years, a large audience across the globe have welcomed the pleasure of virtual gambling due to its ease of access, appealing qualities, and ever-expanding collections of titles.
One of the most compelling reasons of online casinos is the unparalleled diversity of choices available. Whether you are a fan of spinning traditional slot machines, exploring story-driven visual slot games, or strategizing in strategy-based games like Baccarat, online platforms deliver limitless entertainment avenues. Numerous services also include live gaming streams, making it possible for you to communicate with live hosts and opponents, all while enjoying the engaging ambiance of a land-based casino from anywhere you want.
If you’re exploring for the first time with the world of virtual casino play or hope to learn about reputable operators, why not become part of our active community? It’s a platform where fans discuss experiences, helping you to enjoy more of your gambling adventure. Check out the conversation and start your journey now: https://ru.pinterest.com/UpX_Official/ .
Besides the wide selection, online casinos are known for seamless entry.
Jacobgaugh
December 8, 2024 @ 11:51 pm
кухня на заказ екатеринбург недорого — доступные цены и высокое качество.
Manchester escorts agency
December 9, 2024 @ 12:17 am
Do you have a spam problem on this blog; I also am a blogger, and I was curious about your situation; we have developed some nice practices and we are looking to
exchange methods with other folks, please shoot me an email if interested.
https://rr88.codes/
December 9, 2024 @ 12:21 am
This design is incredible! You obviously know how to keep a reader entertained.
Between your wit and your videos, I was almost moved to
start my own blog (well, almost…HaHa!) Fantastic job. I really enjoyed
what you had to say, and more than that, how you presented it.
Too cool!
mercedes-benz suspension repair cost
December 9, 2024 @ 12:29 am
Awesome article.
Richardvak
December 9, 2024 @ 12:39 am
[url=][/url]
Временная регистрация в СПб: Быстро и Легально!
Ищете, где оформить временную регистрацию в Санкт-Петербурге?
Мы гарантируем быстрое и легальное оформление без очередей и лишних документов.
Ваше спокойствие – наша забота!
Минимум усилий • Максимум удобства • Полная легальность
Свяжитесь с нами прямо сейчас!
Временная регистрация
[url=][/url]
Social - paibe
December 9, 2024 @ 12:49 am
How Online Casinos Are Becoming So Popular
Internet-based gambling hubs have transformed the gambling scene, providing a level of accessibility and range that conventional gambling houses can’t match. Over the past decade, a growing community internationally have embraced the excitement of online gaming in light of its anytime, anywhere convenience, appealing qualities, and progressively larger collections of titles.
One of the most compelling reasons of online gaming options is the astounding variety of titles on offer. Whether you like interacting with retro slot machines, immersing yourself in plot-filled visual slot games, or testing your strategy in card and board games like Blackjack, internet-based gambling sites provide numerous choices. Numerous services moreover include live dealer games, giving you the chance you to participate with real dealers and co-players, all while experiencing the immersive atmosphere of a real casino right at home.
If you’re just starting with the world of digital casinos or seek to delve deeper into trusted platforms, why not become part of our vibrant community? It’s a space where fans exchange tips, enabling you to improve your virtual play. Join the experience and start your journey now: https://www.facebook.com/profile.php?id=61568742522948 .
Beyond variety, digital casino services thrive in availability.
Derekapaky
December 9, 2024 @ 1:02 am
кухни на заказ от производителя – гарантия долговечности и высокого качества.
Sazrsol
December 9, 2024 @ 1:11 am
Официальное получение диплома техникума с упрощенным обучением в Москве
can i purchase generic diflucan without a prescription
December 9, 2024 @ 1:30 am
how can i get diflucan without rx
how to stop clonidine safely
December 9, 2024 @ 1:39 am
Medication information sheet. What side effects?
how to stop clonidine safely
Some information about medication. Get here.
Social - paibe
December 9, 2024 @ 1:57 am
What Makes Online Casinos Are a Worldwide Trend
Internet-based gambling hubs have transformed the gaming industry, providing a unique kind of accessibility and range that land-based establishments don’t provide. Recently, a growing community around the world have chosen the excitement of virtual casinos in light of its always-open nature, exciting features, and ever-expanding collections of titles.
One of the main appeals of online casinos is the astounding variety of choices at your disposal. Whether you like playing on traditional one-armed bandits, exploring theme-based video slots, or mastering skills in card and board games like Baccarat, virtual venues feature numerous options. Many casinos moreover offer live dealer games, giving you the chance you to participate with human game hosts and opponents, all while enjoying the engaging feel of a brick-and-mortar establishment in your own space.
If you’re exploring for the first time with the world of digital casinos or want to explore safe services, why not sign up for our active interactive platform? It’s a hub where players share tips, guiding you to improve your gambling adventure. Join the community and start your journey now: https://www.linkedin.com/in/my-stake-a096b427a/ .
In addition to diversity, digital casino services are known for accessibility.
buy calan without rx
December 9, 2024 @ 2:03 am
order calan without rx
how to get ampicillin without prescription
December 9, 2024 @ 2:12 am
Medicament information sheet. Effects of Drug Abuse.
how to get ampicillin without prescription
Best what you want to know about drugs. Read information now.
OliverTaB
December 9, 2024 @ 2:17 am
Изготавливаем корпусную мебель, шкафы-купе, кухни на заказ по индивидуальным размерам под ключ недорого. Цены и фото в описании http://bestwecando.ourproject.org/dokuwiki-2013-05-10a/doku.php?id=%D0%9A%D1%83%D1%85%D0%BE%D0%BD%D0%BD%D1%8B%D0%B5%20%D0%BF%D1%80%D0%BE%D1%81%D1%82%D1%80%D0%B0%D0%BD%D1%81%D1%82%D0%B2%D0%B0%20%D0%BD%D0%BE%D0%B2%D0%BE%D0%B3%D0%BE%20%D0%BF%D0%BE%D0%BA%D0%BE%D0%BB%D0%B5%D0%BD%D0%B8%D1%8F
Irwingraft
December 9, 2024 @ 2:35 am
Высокотехнологичные видеостойки от производителя. На нашем сайте представлен широкий ассортимент различных моделей и с разными размерами экрана https://vk.com/biznareklama
can i purchase cheap cymbalta pills
December 9, 2024 @ 2:36 am
where buy generic cymbalta online
Leonardsiz
December 9, 2024 @ 2:45 am
кухни от производителя — Кухни от производителя: качество и доступная цена.
Bradleyvat
December 9, 2024 @ 2:50 am
https://edpills.men/# п»їed pills online
car image editing
December 9, 2024 @ 2:59 am
Hello! I just wanted to ask if you ever have any problems with hackers?
My last blog (wordpress) was hacked and I ended up losing
months of hard work due to no backup. Do you have any solutions to prevent hackers?
Social - paibe
December 9, 2024 @ 3:01 am
Reasons Why Online Casinos Have Become a Worldwide Trend
Digital casinos have changed the casino gaming market, providing an exceptional degree of comfort and range that traditional establishments struggle to rival. Over time, a vast number of enthusiasts worldwide have welcomed the thrill of virtual gambling due to its ease of access, captivating elements, and constantly growing game libraries.
One of the key draws of online gaming options is the incredible array of titles ready to play. Whether you like playing on retro slot machines, exploring narrative-rich thematic slots, or exercising tactics in strategy-based games like Baccarat, online platforms boast infinite opportunities. Several sites moreover offer interactive dealer games, enabling you to communicate with live hosts and other players, all while enjoying the engaging feel of a land-based casino right at home.
If you’re exploring for the first time with the world of online gaming or hope to discover reputable operators, why not sign up for our vibrant online hub? It’s a platform where players discuss tips, assisting you to enjoy more of your casino activities. Check out the discussions and check it out now: https://www.instagram.com/kwz.kz_official/ .
Beyond variety, internet-based gambling hubs stand out ease of access.
can i buy cheap crestor no prescription
December 9, 2024 @ 3:05 am
where buy cheap crestor no prescription
kapelnica ot zapoya kolomna_abMl
December 9, 2024 @ 3:07 am
капельница от запоя стоимость [url=https://zarabotokdoma.creartuforo.com/viewtopic.php?id=11492]капельница от запоя стоимость[/url] .
Jacobgaugh
December 9, 2024 @ 3:15 am
http://gft-leasing.ru — Надежные кухни на заказ с гарантией качества.
massage spa in jimbaran
December 9, 2024 @ 3:28 am
An outstanding share! I have just forwarded this onto a colleague who has been doing a little homework on this.
And he actually bought me breakfast simply because I stumbled upon it for
him… lol. So let me reword this…. Thank YOU for
the meal!! But yeah, thanx for spending time
to talk about this topic here on your web site.
get cheap lisinopril for sale
December 9, 2024 @ 3:29 am
where to buy generic lisinopril
cost of tetracycline without a prescription
December 9, 2024 @ 3:36 am
can you buy cheap tetracycline
vivod iz zapoya v stacionare_jrEr
December 9, 2024 @ 3:46 am
вывод из запоя в наркологическом стационаре Самары [url=http://masa.forum24.ru/?1-16-0-00002638-000-0-0-1730819425]вывод из запоя в наркологическом стационаре Самары[/url] .
vivod iz zapoya v stacionare_wrSn
December 9, 2024 @ 3:46 am
вывод из запоя в стационаре наркологии [url=https://www.vip.rolevaya.info/viewtopic.php?id=6934]вывод из запоя в стационаре наркологии[/url] .
kapelnica ot zapoya kolomna_mpSn
December 9, 2024 @ 3:50 am
капельница от запоя [url=https://alhambra.bestforums.org/viewtopic.php?f=10&t=46488/]капельница от запоя[/url] .
kapelnica ot zapoya kolomna_sjSn
December 9, 2024 @ 3:50 am
капельница от запоя на дому [url=https://www.familyportal.forumrom.com/viewtopic.php?id=28605]капельница от запоя на дому[/url] .
Imbaslot
December 9, 2024 @ 4:01 am
Artikel ini sungguh menarik dan relevan untuk kalangan penyuka slot online.
Dalam beberapa waktu belakangan, game slot online telah melewati kemajuan yang signifikan, terutama dengan integrasi teknologi terkini seperti
grafik 3D, efek suara yang nyata, dan tema yang beragam.
Semua itu memberikan keseruan yang lebih mendalam dan menghibur bagi para pemain.
Namun, ada satu aspek yang sering terlupakan adalah krusialnya menggunakan platform yang terjamin dan reliable.
Tidak sedikit peristiwa di mana pengguna dijebak oleh platform tidak resmi yang menjanjikan bonus
besar, tetapi pada akhirnya hanya merugikan. Oleh karena itu, transparansi dan izin resmi dari penyedia game
adalah hal yang perlu dicermati. Salah satu platform terkenal yang patut direkomendasikan adalah platform Imbaslot, yang dikenal memiliki lisensi valid serta mekanisme
game yang adil dan transparan.
Selain itu, sistem RNG (Random Number Generator) menjadi fondasi dari fairness dalam slot online.
Namun, tidak semua pemain memahami cara kerja sistem ini.
Ada yang berpikir mereka mampu “mengalahkan” mesin slot dengan metode khusus, padahal output setiap putaran sepenuhnya acak.
Imbaslot memastikan bahwa setiap permainan dijalankan menggunakan RNG
yang telah diverifikasi, sehingga pemain dapat bermain dengan tenang
tanpa cemas manipulasi.
Dari sisi entertainment, slot online memang memberikan sesuatu yang unik.
Ragam konsep seperti adventure, cerita legenda, atau bahkan kerja sama dengan film
dan budaya pop menjadikannya lebih dari sekadar permainan biasa.
Imbaslot juga menghadirkan berbagai konsep unik yang dapat dinikmati oleh pemain dengan selera berbeda,
membuat setiap pengalaman bermain terasa baru dan menyenangkan.
Namun, ada hal yang juga patut disorot adalah pentingnya tanggung jawab dalam bermain. Dengan aksesibilitas melalui gadget mobile dan desktop,
ada risiko pemain berada dalam kebiasaan bermain yang tidak sehat.
Imbaslot mengedepankan permainan yang responsible dengan fitur seperti batas deposit, kontrol waktu bermain, dan tips
bermain secara tepat.
Secara keseluruhan, tulisan ini membuka wawasan tentang
keragaman dan keindahan dunia slot online. Akan lebih baik lagi
jika di masa depan, ada bahasan mendalam tentang strategi pengelolaan bankroll, pengaruh RTP (Return to Player),
dan cara memilih game yang sesuai dengan preferensi
individu.
Apresiasi telah menghadirkan tulisan informatif seperti ini.
Dunia slot online memang penuh hiburan, tetapi dengan platform seperti Imbaslot, pemain dapat merasakan sensasi ini secara aman, jujur, dan bijaksana.
StevenDrums
December 9, 2024 @ 4:02 am
https://semaglutidetablets.store/# semaglutide best price
Social - paibe
December 9, 2024 @ 4:06 am
Why Online Casinos Are a Global Phenomenon
Online casinos have modernized the gaming world, providing an exceptional degree of accessibility and range that physical gambling houses don’t provide. Over time, a large audience worldwide have embraced the fun of virtual casinos because of its always-open nature, exciting features, and widening collections of titles.
One of the main appeals of online gaming options is the unparalleled range of choices on offer. Whether you are a fan of rolling vintage reel games, trying out plot-filled modern slot games, or exercising tactics in table games like Blackjack, digital casinos offer countless possibilities. Plenty of operators furthermore feature live casino options, making it possible for you to participate with live hosts and co-players, all while enjoying the engaging ambiance of a brick-and-mortar establishment right at home.
If you’re unfamiliar with the world of online gaming or want to learn about safe services, why not join our growing community? It’s a platform where enthusiasts offer insights, enabling you to enhance your casino activities. Explore the conversation and see it here now: https://www.instagram.com/sweet_bonanzacom/ .
Adding to the extensive catalog, internet-based gambling hubs stand out ease of access.
can i get generic lopid for sale
December 9, 2024 @ 4:06 am
order lopid without dr prescription
DFHGGKKU67
December 9, 2024 @ 4:11 am
That is very attention-grabbing, You’re an excessively skilled blogger.
I’ve joined your rss feed and look ahead to seeking more of your excellent post.
Additionally, I have shared your web site in my social networks
GTA777
December 9, 2024 @ 4:22 am
Thanks a lot for sharing this with all of us you actually realize what you are
talking about! Bookmarked. Please additionally consult with my site =).
We will have a hyperlink trade agreement between us
What is ADAS Calibration
December 9, 2024 @ 4:36 am
I feel that is one of the most vital information for me.
And i am glad studying your article. But want to statement on some normal things, The website taste is wonderful, the articles is in reality
nice : D. Excellent process, cheers
cost of cheap celebrex pill
December 9, 2024 @ 4:38 am
can i purchase cheap celebrex price
Visit This Link
December 9, 2024 @ 4:55 am
Whenever testosterone level levels are actually well-kept, the mixed perks of far better power, improved sex drive, stabilized state of mind, and improved physical stamina dramatically enhance total health. Experiencing more youthful, much healthier, and extra positive in daily tasks is a strong incentive for taking care of testosterone amounts, http://travellers.minube.net/AlvarodfDaniels.
Dnrtotn
December 9, 2024 @ 4:57 am
Как оказалось, купить диплом кандидата наук не так уж и сложно
Jacobgaugh
December 9, 2024 @ 5:06 am
https://nmphoto.ru/ — реализуем проекты любой сложности.
where can i get generic asacol without a prescription
December 9, 2024 @ 5:07 am
can you buy generic asacol without dr prescription
Social - paibe
December 9, 2024 @ 5:10 am
Why Online Casinos Are So Popular
Online casinos have reshaped the gaming landscape, delivering an exceptional degree of accessibility and breadth that land-based gambling houses are unable to replicate. In recent years, millions of players worldwide have turned to the adventure of virtual gambling because of its availability, thrilling aspects, and ever-expanding catalogs of games.
One of the strongest selling points of virtual gambling hubs is the vast array of gaming experiences ready to play. Whether you prefer rolling vintage one-armed bandits, playing through plot-filled video-based games, or strategizing in table games like Blackjack, internet-based gambling sites feature limitless possibilities. Many casinos moreover present interactive dealer games, letting you to connect with real dealers and gaming peers, all while experiencing the lifelike environment of a real casino right at home.
If you’re unfamiliar with the world of digital casinos or seek to discover safe services, why not sign up for our growing online hub? It’s a platform where gamblers exchange tips, helping you to enjoy more of your casino activities. Discover the experience and learn more now: https://au.pinterest.com/casibom1/ .
Beyond variety, virtual gaming providers thrive in accessibility.
can i buy generic elavil without a prescription
December 9, 2024 @ 5:38 am
buy elavil prices
cheap pamelor without rx
December 9, 2024 @ 6:08 am
where buy generic pamelor tablets
Social - paibe
December 9, 2024 @ 6:16 am
Why Online Casinos Are a Worldwide Trend
Digital casinos have reshaped the casino gaming industry, offering a level of ease and breadth that physical venues are unable to replicate. Over the past decade, a large audience globally have chosen the fun of virtual casinos due to its availability, captivating elements, and progressively larger catalogs of games.
One of the key draws of digital gambling sites is the astounding variety of titles provided. Whether you are a fan of interacting with retro slot machines, exploring plot-filled modern slot games, or playing smart in strategy-based games like Baccarat, virtual venues offer countless possibilities. Many casinos additionally feature live casino options, letting you to participate with live hosts and opponents, all while experiencing the realistic ambiance of a real casino from anywhere you want.
If you’re unfamiliar with the world of online gaming or are looking to find out more about trusted platforms, why not participate in our active interactive platform? It’s a platform where gamblers exchange stories, guiding you to enjoy more of your virtual play. Dive into the discussions and learn more now: https://t.me/casibomgiris_net .
In addition to diversity, virtual gambling platforms excel seamless entry.
Ceria777
December 9, 2024 @ 6:32 am
I got this site from my buddy who informed me about this site and now this time I am browsing this web page and reading very informative posts at this place.
can i order tetracycline without prescription
December 9, 2024 @ 6:39 am
can i buy cheap tetracycline without rx
where can i buy generic celebrex pills
December 9, 2024 @ 6:41 am
where buy generic celebrex for sale
ThomasWoono
December 9, 2024 @ 6:46 am
Vitebsk State University named after P.M. Masherov, one of the oldest universities in Belarus, invites you to get the education of European quality! Undoubted advantages of studying in VSU are affordability, high level of the quality of the educational process, great experience in training foreign students Vitebsk State University P.M.Masherov University is an educational center. VSU.by provides training in: chemistry, biology, history, physics, programming, pedagogy, psychology, mathematics https://vsu.by/sobytiya/vgu-v-smi.html
Slot Server Thailand
December 9, 2024 @ 6:48 am
Everything is very open with a clear explanation of the challenges.
It was truly informative. Your website is extremely
helpful. Thank you for sharing!
Click Here
December 9, 2024 @ 7:19 am
After checking out a handful of the blog articles on your web site,
I truly appreciate your technique of writing a blog. I book-marked it to my
bookmark site list and will be checking back in the near future.
Please visit my web site too and tell me your opinion.
Social - paibe
December 9, 2024 @ 7:22 am
Why Online Casinos Remain a Global Phenomenon
Virtual gambling platforms have changed the gambling world, providing an unmatched level of user-friendliness and range that brick-and-mortar establishments fall short of. Over time, millions of players internationally have chosen the adventure of virtual casinos due to its anytime, anywhere convenience, appealing qualities, and progressively larger catalogs of games.
One of the most compelling reasons of virtual gambling hubs is the incredible variety of gaming experiences on offer. Whether you prefer playing on old-school slots, diving into story-driven thematic slots, or mastering skills in traditional table offerings like Baccarat, online platforms boast limitless entertainment avenues. Numerous services additionally feature interactive dealer games, letting you to communicate with live hosts and fellow gamblers, all while experiencing the authentic environment of a land-based casino from anywhere you want.
If you’re just starting with the world of internet-based gaming or seek to find out more about reputable operators, why not join our growing social network? It’s a destination where enthusiasts post tips, guiding you to get the most out of your gambling adventure. Join the discussions and visit us now: https://www.facebook.com/profile.php?id=61568958127704 .
Apart from the game range, internet-based gambling hubs stand out availability.
Angelhob
December 9, 2024 @ 7:28 am
https://www.pilot-partner.ru/ – Лучшие фотографии для вашего бизнеса от профессионалов.
can i get cheap celebrex tablets
December 9, 2024 @ 7:39 am
can i get cheap celebrex pills
NexCredit Capital
December 9, 2024 @ 7:42 am
Thanks for finally talking about > Filling in the gaps of HIV < Loved it!
Leonardsiz
December 9, 2024 @ 8:01 am
кухни от производителя – Надежное качество напрямую от производителя кухонь.
where can i get cheap lexapro online
December 9, 2024 @ 8:09 am
where buy cheap lexapro pills
Jacobgaugh
December 9, 2024 @ 8:14 am
mtucizone.ru/ — Полный ассортимент кухонь для вашего удобства.
Pokemon Rug
December 9, 2024 @ 8:25 am
As someone who’s loved Pokémon since childhood, I’ve
always found joy in bringing the Pokémon world into
my everyday life. From catching them all in the games to designing my room,
Pokémon is a big part of who I am.
One of my favorite hobbies is creating Pokémon-themed room decor.
I’ve used items like Pokémon rugs, woven tapestries, and custom carpets to make my room stand out.
These pieces create a cozy atmosphere but also bring my favorite Pokémon to life.
For Pokémon lovers like me, I highly recommend incorporating these items into your decor.
They’re perfect for adding a unique touch.
Catch ‘em all, in style!
my page – Pokemon Rug
Social - paibe
December 9, 2024 @ 8:29 am
How Online Casinos Remain So Popular
Online casinos have reshaped the betting landscape, delivering an exceptional degree of comfort and selection that land-based venues can’t match. Over time, millions of players worldwide have welcomed the pleasure of internet-based gaming as a result of its availability, captivating elements, and progressively larger range of offerings.
One of the strongest selling points of digital gambling sites is the astounding variety of choices provided. Whether you like playing on traditional fruit machine slots, exploring narrative-rich modern slot games, or mastering skills in traditional table offerings like Roulette, digital casinos offer countless choices. Plenty of operators moreover introduce real-time gaming experiences, allowing you to interact with human game hosts and other players, all while experiencing the engaging ambiance of a brick-and-mortar establishment from anywhere you want.
If you’re just starting with the world of online gaming or hope to discover safe services, why not sign up for our vibrant community? It’s a platform where gamblers share stories, enabling you to enhance your gambling adventure. Join the community and start your journey now: https://t.me/casibomgiris_net .
In addition to diversity, virtual gaming providers excel availability.
where to buy elavil without rx
December 9, 2024 @ 8:54 am
where to buy elavil pill
Plugin wordpress
December 9, 2024 @ 8:57 am
Get the best content spinning plugin for WordPress – Content-Spinning.fr – the plugin that optimizes your content & keeps you safe.
Sazrzvy
December 9, 2024 @ 9:02 am
Официальная покупка диплома вуза с сокращенной программой обучения в Москве
Cazruen
December 9, 2024 @ 9:16 am
Полезные советы по покупке диплома о высшем образовании без риска
Derekapaky
December 9, 2024 @ 9:26 am
http://www.tatarstan-heroes.ru – удобный сервис для выбора и заказа кухонь.
Social - paibe
December 9, 2024 @ 9:38 am
What Makes Online Casinos Remain a Global Phenomenon
Digital casinos have modernized the betting industry, delivering an unmatched level of user-friendliness and selection that brick-and-mortar gambling houses struggle to rival. Over the past decade, millions of players internationally have welcomed the pleasure of digital casino play as a result of its availability, captivating elements, and continuously increasing game libraries.
One of the most compelling reasons of digital gambling sites is the astounding range of games at your disposal. Whether you like rolling retro fruit machine slots, playing through story-driven video slots, or strategizing in card and board games like Baccarat, digital casinos provide limitless opportunities. Plenty of operators also include live dealer games, giving you the chance you to participate with human game hosts and fellow gamblers, all while immersing yourself in the lifelike environment of a real casino from anywhere you want.
If you’re unfamiliar with the world of virtual casino play or would like to delve deeper into proven options, why not join our dynamic interactive platform? It’s a hub where gaming aficionados exchange insights, helping you to improve your online casino experience. Check out the community and start your journey now: https://x.com/fury_usyk_2 .
Beyond variety, digital casino services are known for constant connectivity.
where can i get cheap sporanox no prescription
December 9, 2024 @ 9:55 am
how to buy cheap sporanox pills
CharlesExore
December 9, 2024 @ 10:06 am
Kamagra 100mg: Kamagra Oral Jelly – cheap kamagra
Jacobgaugh
December 9, 2024 @ 10:21 am
nmphoto.ru/ — сервис, которому доверяют.
Casino
December 9, 2024 @ 10:22 am
I think this is one of the most vital information for me.
And i’m glad reading your article. But should remark on some general things, The website style is wonderful,
the articles is really excellent : D. Good job, cheers
stscrap.kr
December 9, 2024 @ 10:50 am
Here’s a spun introduction for a game lover:
As a passionate gamer, I’ve spent years playing my favorite games and creating immersive environments.
With 3+ years of experience, I’ve been designing game-themed gaming
rooms, drawing inspiration from my favorite franchises like
Mario, Nintendo, Zelda, and The Witcher 3. It’s been an exciting journey, blending creativity
with my love for games.
To all fellow gamers, I suggest adding touches like game rugs, wall art, and custom lighting to bring
your favorite games to life. These items add personality.
Whether you love retro classics or modern RPGs, a themed gaming room takes your space to
the next level.
Create your dream setup!
My page … Among Us Rug (stscrap.kr)
can i purchase rizact pill
December 9, 2024 @ 10:55 am
can i buy generic rizact without rx
Social - paibe
December 9, 2024 @ 10:57 am
Reasons Why Online Casinos Are Becoming So Popular
Internet-based gambling hubs have changed the casino gaming scene, offering an unmatched level of accessibility and breadth that physical gambling houses don’t provide. Throughout the last ten years, a large audience internationally have welcomed the adventure of internet-based gaming in light of its always-open nature, appealing qualities, and constantly growing catalogs of games.
One of the biggest attractions of online gaming options is the sheer variety of choices at your disposal. Whether you prefer engaging with vintage reel games, exploring theme-based visual slot games, or exercising tactics in card and board games like Roulette, digital casinos provide infinite choices. Several sites furthermore feature real-time gaming experiences, enabling you to participate with actual dealers and other players, all while enjoying the lifelike feel of a real casino right at home.
If you’re just starting with the world of virtual gambling or hope to discover proven options, why not participate in our dynamic interactive platform? It’s a platform where fans offer reviews, helping you to get the most out of your gambling adventure. Check out the connections and start your journey now: https://www.instagram.com/136bet_com/ .
Besides the wide selection, virtual gambling platforms shine ease of access.
buying generic periactin tablets
December 9, 2024 @ 11:04 am
buying periactin for sale
hydrochlorothiazide and lisinopril brand name
December 9, 2024 @ 11:27 am
Medicament information for patients. Short-Term Effects.
hydrochlorothiazide and lisinopril brand name
Actual about meds. Get information now.
can you buy advair diskus without rx
December 9, 2024 @ 11:30 am
where buy generic advair diskus without dr prescription
get iptv subscription
December 9, 2024 @ 11:48 am
I always spent my half an hour to read this webpage’s articles or reviews daily along with a cup of coffee.
how to get generic tolterodine for sale
December 9, 2024 @ 11:59 am
Medicines information sheet. Drug Class.
how to get generic tolterodine for sale
Best information about meds. Get information here.
CoreyamBit
December 9, 2024 @ 12:00 pm
cheap semaglutide pills [url=https://semaglutidetablets.store/#]rybelsus semaglutide tablets[/url] semaglutide tablets store
web page
December 9, 2024 @ 12:00 pm
I know this web site gives quality depending articles or reviews and extra stuff, is there any other site which
gives such information in quality?
MetBevarate
December 9, 2024 @ 12:04 pm
бармен скачать игру [url=https://apk-smart.com/igry/simulyatory/71-barmen.html]https://apk-smart.com/igry/simulyatory/71-barmen.html[/url] бармен скачать игру
P.S Live ID: K89Io9blWX1UfZWv3ajv
P.S.S [url=http://teamautotrend.com/car_reviews/ford-endeavour-the-perfect-suv]Программы и игры для Андроид телефона[/url] [url=https://pecsiriport.hu/koezelet/6573-zenevel-a-menekult-gy]Программы и игры для Андроид телефона[/url] [url=http://forum.lundin.ro/showthread.php?tid=22393&pid=60093#pid60093]Программы и игры для Андроид телефона[/url] 2a1adfe
how to get generic uroxatrall without rx
December 9, 2024 @ 12:04 pm
buy cheap uroxatrall tablets
Social - paibe
December 9, 2024 @ 12:13 pm
The Reasons Behind Why Online Casinos Have Become Highly Preferred Worldwide
Internet-based gambling hubs have reshaped the casino gaming world, providing an unmatched level of comfort and breadth that land-based establishments struggle to rival. Over time, millions of players globally have welcomed the adventure of virtual gambling in light of its always-open nature, thrilling aspects, and widening game libraries.
One of the most compelling reasons of online gaming options is the unparalleled variety of gaming experiences on offer. Whether you enjoy spinning old-school one-armed bandits, playing through story-driven visual slot games, or mastering skills in table games like Blackjack, digital casinos deliver countless opportunities. Several sites even include real-time gaming experiences, giving you the chance you to participate with human game hosts and opponents, all while taking in the lifelike ambiance of a land-based casino in your own space.
If you’re new with the world of internet-based gaming or want to learn about trusted platforms, why not join our dynamic gaming forum? It’s a place where gamblers share insights, assisting you to get the most out of your online casino experience. Explore the connections and start your journey now: https://www.facebook.com/profile.php?id=61569191174332 .
In addition to diversity, internet-based gambling hubs thrive in ease of access.
where to buy cheap fosamax tablets
December 9, 2024 @ 12:30 pm
Pills information for patients. Brand names.
where to buy cheap fosamax tablets
Everything about drug. Read information here.
generic caduet without insurance
December 9, 2024 @ 12:35 pm
order cheap caduet without prescription
how to buy lisinopril prices
December 9, 2024 @ 1:02 pm
Medicine information for patients. Generic Name.
how to buy lisinopril prices
Actual what you want to know about pills. Get here.
how to buy cheap tetracycline for sale
December 9, 2024 @ 1:07 pm
get generic tetracycline no prescription
StevenDrums
December 9, 2024 @ 1:10 pm
https://semaglutidetablets.store/# semaglutide best price
Jacobgaugh
December 9, 2024 @ 1:13 pm
http://www.armor-games.ru — Надежный сайт для заказа качественной мебели.
sbobet ทางเข้า
December 9, 2024 @ 1:13 pm
Thank you for any other wonderful post. Where else could anyone get that kind of info in such a
perfect approach of writing? I have a presentation subsequent
week, and I am at the search for such information.
Leonardsiz
December 9, 2024 @ 1:21 pm
http://www.tsmk-altai.ru – Уникальные кухни с индивидуальным дизайном от профессионалов.
Social - paibe
December 9, 2024 @ 1:30 pm
Reasons Why Online Casinos Have Become Highly Preferred Worldwide
Digital casinos have revolutionized the gambling industry, providing an exceptional degree of comfort and breadth that brick-and-mortar casinos can’t match. Throughout the last ten years, a large audience worldwide have welcomed the adventure of digital casino play due to its availability, captivating elements, and progressively larger collections of titles.
One of the most compelling reasons of online gaming options is the astounding selection of games available. Whether you prefer engaging with vintage reel games, immersing yourself in engaging thematic slots, or exercising tactics in table games like Texas Hold’em, virtual venues provide endless entertainment avenues. Numerous services also offer live gaming streams, letting you to communicate with actual dealers and opponents, all while experiencing the immersive vibes of a land-based casino without leaving your home.
If you’re a beginner with the world of digital casinos or seek to learn about reputable operators, why not engage with our lively gaming forum? It’s a place where players exchange insights, guiding you to maximize your virtual play. Join the discussions and see it here now: https://t.me/furyversususyk .
Adding to the extensive catalog, internet-based gambling hubs shine ease of access.
is metformin used to treat pre diabetes
December 9, 2024 @ 1:33 pm
Drugs information leaflet. Generic Name.
is metformin used to treat pre diabetes
Everything information about meds. Get information here.
how to get femara prices
December 9, 2024 @ 1:38 pm
cost generic femara without insurance
Jacobgaugh
December 9, 2024 @ 1:44 pm
ums-group.ru — Доступный и надежный выбор для обустройства кухонного пространства.
Click Here
December 9, 2024 @ 1:46 pm
Although these lookings for are actually promising, it is critical to keep in mind that individual results can easily differ. Therefore, while Leanix Fruit Weight management Capsules might give prospective benefits, they ought to not be seen as a magic service. As an alternative, consider all of them as a complementary strategy, integrated with healthy practices, for achieving your fat loss goals, https://gettr.com/post/p3ej0ysbed4.
http://www.gz-jj.com/comment/Html/?243506.html
December 9, 2024 @ 1:47 pm
Reliable stuff Many thanks.
Here is my blog post :: ルイヴィトン スーパーコピー ベルト iphoneケース – http://www.gz-jj.com/comment/html/?243506.html,
order generic indomethacin without insurance
December 9, 2024 @ 2:05 pm
Medicine information for patients. Cautions.
order generic indomethacin without insurance
Some news about medication. Get information here.
cost generic pamelor tablets
December 9, 2024 @ 2:12 pm
how to get cheap pamelor tablets
snort bupropion get high
December 9, 2024 @ 2:36 pm
Medicament information. Brand names.
snort bupropion get high
All information about medicine. Get now.
where can i buy generic colospa tablets
December 9, 2024 @ 2:45 pm
how can i get cheap colospa without a prescription
buying imdur price
December 9, 2024 @ 2:45 pm
can you buy cheap imdur pills
Social - paibe
December 9, 2024 @ 2:45 pm
The Reasons Behind Why Online Casinos Remain an International Sensation
Virtual gambling platforms have reshaped the casino gaming market, delivering an unmatched level of user-friendliness and selection that physical casinos fall short of. Over time, a large audience worldwide have embraced the pleasure of virtual gambling as a result of its availability, exciting features, and constantly growing range of offerings.
One of the strongest selling points of internet-based platforms is the vast array of gaming experiences at your disposal. Whether you like engaging with classic one-armed bandits, trying out plot-filled video slots, or playing smart in strategy-based games like Texas Hold’em, virtual venues deliver infinite possibilities. Several sites also introduce live dealer games, giving you the chance you to engage with live hosts and gaming peers, all while soaking in the lifelike feel of a land-based casino in your own space.
If you’re a beginner with the world of virtual casino play or would like to delve deeper into reliable sites, why not participate in our active online hub? It’s a destination where fans offer experiences, enabling you to maximize your casino activities. Check out the discussions and check it out now: https://www.facebook.com/profile.php?id=61569360522200 .
Adding to the extensive catalog, virtual gambling platforms thrive in constant connectivity.
Professional Services
December 9, 2024 @ 2:59 pm
I love what you guys tend to be up too. Such clever work and coverage!
Keep up the superb works guys I’ve incorporated you guys to our
blogroll.
CharlesSigma
December 9, 2024 @ 2:59 pm
Кто может порекомендовать обменник с хорошими условиями для обмена криптовалюты? Хочу, чтобы курс был честный и не было скрытых комиссий https://avtomobili.creartuforo.com/viewtopic.php?id=769#p1661
Business banking services
December 9, 2024 @ 3:09 pm
Awesome article.
Ремонт телевизоров Hisense Москва
December 9, 2024 @ 3:29 pm
Предлагаем услуги профессиональных инженеров офицальной мастерской.
Еслли вы искали ремонт телевизоров hisense цены, можете посмотреть на сайте: ремонт телевизоров hisense в москве
Наши мастера оперативно устранят неисправности вашего устройства в сервисе или с выездом на дом!
Casino
December 9, 2024 @ 3:42 pm
It’s an amazing piece of writing for all the internet people; they will obtain advantage from it
I am sure.
Social - paibe
December 9, 2024 @ 3:56 pm
How Online Casinos Are Becoming an International Sensation
Virtual gambling platforms have revolutionized the betting scene, providing an unmatched level of accessibility and breadth that traditional casinos struggle to rival. Over the past decade, a large audience internationally have embraced the pleasure of virtual casinos in light of its ease of access, appealing qualities, and continuously increasing selection of games.
One of the most compelling reasons of virtual gambling hubs is the astounding selection of titles available. Whether you are a fan of playing on traditional one-armed bandits, diving into story-driven visual slot games, or exercising tactics in classic casino games like Baccarat, casino websites deliver endless possibilities. A large number of platforms additionally include live casino options, giving you the chance you to interact with professional croupiers and gaming peers, all while enjoying the lifelike vibes of a traditional gambling venue from anywhere you want.
If you’re just starting with the world of digital casinos or want to explore reputable operators, why not participate in our vibrant gaming forum? It’s a hub where fans offer stories, guiding you to enjoy more of your casino activities. Join the conversation and learn more now: https://t.me/fight_usykfury .
Besides the wide selection, internet-based gambling hubs shine accessibility.
Lazrltn
December 9, 2024 @ 4:08 pm
Возможно ли купить диплом стоматолога, и как это происходит
Angelhob
December 9, 2024 @ 4:27 pm
https://www.mirdolmenov.ru/ – Ознакомьтесь с портфолио и выбирайте подходящие услуги.
Anime Rug
December 9, 2024 @ 4:29 pm
As someone passionate about anime, I’ve been watching anime since my teenage years.
My go-to series include Naruto, One Piece, Berserk, and countless others.
I’m also passionate about room decor, and I’ve decorated my gaming
room and bedroom with an anime theme. It’s
been an amazing way to express my love for
anime.
To bring anime magic to your room, I highly recommend using anime rugs,
anime carpets, and woven tapestries. These items add
a unique touch and make excellent decor pieces.
Let your room reflect your anime journey!
Also visit my website Anime Rug
Jamescaurf
December 9, 2024 @ 4:38 pm
cheap kamagra: buy kamagra online usa – buy kamagra online usa
образование
December 9, 2024 @ 4:48 pm
наука
One of the leading academic and scientific-research centers of the Belarus. There are 12 Faculties at the University, 2 scientific and research institutes. Higher education in 35 specialities of the 1st degree of education and 22 specialities.
nexus for development
December 9, 2024 @ 4:58 pm
It’s very simple to find out any matter on web as
compared to textbooks, as I found this paragraph at this site.
Social - paibe
December 9, 2024 @ 5:06 pm
Why Online Casinos Remain a Worldwide Trend
Digital casinos have reshaped the gambling world, offering a level of convenience and diversity that conventional establishments don’t provide. Over time, a large audience worldwide have embraced the excitement of online gaming due to its availability, thrilling aspects, and constantly growing game libraries.
One of the biggest attractions of internet-based platforms is the unparalleled diversity of gaming experiences ready to play. Whether you love playing on vintage slot machines, exploring theme-based visual slot games, or playing smart in table games like Texas Hold’em, internet-based gambling sites provide endless opportunities. A large number of platforms additionally present live gaming streams, giving you the chance you to connect with real dealers and other players, all while experiencing the authentic vibes of a traditional gambling venue right at home.
If you’re exploring for the first time with the world of virtual gambling or want to discover safe services, why not sign up for our active interactive platform? It’s a platform where gaming aficionados discuss tips, guiding you to maximize your gambling adventure. Explore the discussions and check it out now: https://www.facebook.com/profile.php?id=61569191174332 .
Apart from the game range, virtual gaming providers thrive in constant connectivity.
университет
December 9, 2024 @ 5:23 pm
наука
Francisk Skorina Gomel State University
job search
December 9, 2024 @ 5:27 pm
It’s an awesome article in favor of all the online people; they will get benefit from it
I am sure.
sultangazi escort
December 9, 2024 @ 5:34 pm
en kaliteli sultangazi escort bayan sitesiyim
is ashwagandha ksm 66 safe
December 9, 2024 @ 6:04 pm
ksm 66 ashwagandha for women
Derekapaky
December 9, 2024 @ 6:05 pm
huskytaxi.ru/ – удобная платформа для выбора идеальной кухни.
تضمین شده
December 9, 2024 @ 6:08 pm
Just want to say your article is as astounding.
The clearness on your put up is simply spectacular and i could suppose you’re an expert in this subject.
Well with your permission let me to grab your feed to stay updated with drawing close
post. Thank you one million and please carry on the enjoyable work.
Jacobgaugh
December 9, 2024 @ 6:08 pm
gft-leasing.ru — Лучшие предложения для вашей кухни мечты.
sell gold
December 9, 2024 @ 6:12 pm
When I originally commented I clicked the “Notify me when new comments are added” checkbox and now each
time a comment is added I get several emails with the same comment.
Is there any way you can remove people from that service?
Thank you!
Social - paibe
December 9, 2024 @ 6:14 pm
Reasons Why Online Casinos Remain a Worldwide Trend
Virtual gambling platforms have modernized the gambling world, delivering an exceptional degree of convenience and range that brick-and-mortar casinos fall short of. Over time, a large audience globally have turned to the excitement of virtual gambling as a result of its always-open nature, engaging traits, and widening selection of games.
One of the biggest attractions of online casinos is the sheer diversity of choices available. Whether you love interacting with classic slot machines, diving into story-driven video-based games, or exercising tactics in card and board games like Texas Hold’em, online platforms feature infinite choices. Plenty of operators furthermore offer live gaming streams, allowing you to communicate with real dealers and opponents, all while immersing yourself in the engaging atmosphere of a physical gaming house without leaving your home.
If you’re new with the world of online gaming or seek to discover trusted platforms, why not participate in our active social network? It’s a hub where gaming aficionados offer tips, enabling you to get the most out of your gambling adventure. Check out the connections and visit us now: https://t.me/fight_usyk_fury .
Besides the wide selection, online casinos excel accessibility.
mecidiyeköy escort
December 9, 2024 @ 6:25 pm
efsane bir şişli escort mecidiyeköy escort sitesi
Leonardsiz
December 9, 2024 @ 6:39 pm
кухни от производителя — Кухни от производителя: качество и доступная цена.
Betflare_wrot
December 9, 2024 @ 6:51 pm
Betflare ?????? [url=https://www.betflare8.com]Betflare ??????[/url] .
Betflare_qapi
December 9, 2024 @ 6:52 pm
Betflare App [url=http://www.betflare888.com]Betflare App[/url] .
kapelnica ot zapoya kolomna_lnMl
December 9, 2024 @ 6:55 pm
капельница от запоя [url=http://family2.quadrobb.me/viewtopic.php?id=1852]капельница от запоя[/url] .
kapelnica ot zapoya kolomna_ezka
December 9, 2024 @ 6:56 pm
после капельницы от запоя [url=http://www.gov.ukrbb.net/viewtopic.php?f=3&t=6390]после капельницы от запоя[/url] .
kapelnica ot zapoya kolomna_kiOr
December 9, 2024 @ 6:57 pm
стоимость капельницы от запоя [url=https://skoleoz.borda.ru/?1-8-0-00001111-000-0-0-1730821724]стоимость капельницы от запоя[/url] .
Jacobgaugh
December 9, 2024 @ 6:58 pm
http://ums-group.ru/ — Переходите на сайт, чтобы ознакомиться с нашим ассортиментом.
lucky88
December 9, 2024 @ 7:11 pm
Amazing! Its genuinely amazing article, I have got much clear idea
about from this article.
Social - paibe
December 9, 2024 @ 7:23 pm
The Reasons Behind Why Online Casinos Have Become So Popular
Internet-based gambling hubs have changed the betting scene, offering an unmatched level of user-friendliness and variety that land-based gambling houses fall short of. Over time, a large audience internationally have chosen the pleasure of internet-based gaming because of its anytime, anywhere convenience, appealing qualities, and progressively larger selection of games.
One of the strongest selling points of internet-based platforms is the sheer selection of titles provided. Whether you are a fan of interacting with retro one-armed bandits, immersing yourself in narrative-rich modern slot games, or playing smart in card and board games like Texas Hold’em, virtual venues boast numerous choices. A large number of platforms even offer live gaming streams, giving you the chance you to interact with live hosts and opponents, all while experiencing the immersive feel of a land-based casino in your own space.
If you’re a beginner with the world of online gaming or would like to find out more about reputable operators, why not engage with our vibrant online hub? It’s a space where gaming aficionados post tips, guiding you to enhance your online casino experience. Dive into the experience and start your journey now: https://www.facebook.com/profile.php?id=61569360522200 .
Besides the wide selection, virtual gambling platforms are known for ease of access.
Get More Info
December 9, 2024 @ 7:28 pm
Testosterone level booster supplements are key to boosting energy, strengthening stamina, and preserving vigor. They sustain bodily hormone balance and advertise overall welfare, assisting you experience your best. If you’re trying to find a quick and easy and scrumptious solution, select Erectax Fruit Gummies, an organic supplement developed to increase testosterone level and enrich your assurance, health and wellness, and well-being effortlessly, https://zenwriting.net/cesariyang/wenn-sie-muss-sie-start-verwenden-von-a-testosteron-steigerung-erganzen.
Learn More
December 9, 2024 @ 7:44 pm
Greate post. Keep posting such kind of info on your blog.
Im really impressed by it.
Hi there, You have performed a great job. I’ll certainly digg it and in my view
recommend to my friends. I am sure they’ll be benefited from
this website.
Social - paibe
December 9, 2024 @ 8:29 pm
The Reasons Behind Why Online Casinos Are Becoming Highly Preferred Worldwide
Virtual gambling platforms have transformed the gambling world, offering a level of comfort and breadth that brick-and-mortar venues don’t provide. Over the past decade, millions of players worldwide have turned to the excitement of internet-based gaming thanks to its ease of access, engaging traits, and progressively larger catalogs of games.
One of the biggest attractions of online casinos is the unparalleled range of gaming experiences at your disposal. Whether you are a fan of engaging with traditional fruit machine slots, trying out story-driven visual slot games, or exercising tactics in table games like Texas Hold’em, casino websites deliver numerous entertainment avenues. Many casinos moreover present live casino options, making it possible for you to engage with actual dealers and co-players, all while taking in the authentic atmosphere of a physical gaming house without leaving your home.
If you’re unfamiliar with the world of internet-based gaming or are looking to explore proven options, why not participate in our lively interactive platform? It’s a platform where fans exchange reviews, making it easier for you to maximize your virtual play. Dive into the connections and visit us now: https://ru.pinterest.com/furyusyk/ .
Besides the wide selection, digital casino services stand out seamless entry.
get cheap imuran price
December 9, 2024 @ 9:16 pm
generic imuran no prescription
Social - paibe
December 9, 2024 @ 9:37 pm
Reasons Why Online Casinos Are Becoming an International Sensation
Online casinos have transformed the betting market, delivering an unmatched level of accessibility and range that physical gambling houses struggle to rival. Over the past decade, millions of players internationally have chosen the adventure of online gaming as a result of its ease of access, exciting features, and widening selection of games.
One of the biggest attractions of virtual gambling hubs is the unparalleled variety of games ready to play. Whether you love rolling retro slots, exploring story-driven video slots, or mastering skills in traditional table offerings like Baccarat, digital casinos feature countless choices. Many casinos even present live casino options, making it possible for you to interact with real dealers and other players, all while immersing yourself in the engaging vibes of a physical gaming house from the comfort of your home.
If you’re new with the world of virtual casino play or hope to delve deeper into trusted platforms, why not engage with our lively online hub? It’s a hub where enthusiasts post tips, guiding you to improve your gaming journey. Check out the discussions and visit us now: https://www.instagram.com/tigrinhobr_com/ .
Apart from the game range, virtual gaming providers excel seamless entry.
Bradleyvat
December 9, 2024 @ 9:43 pm
https://drugs1st.pro/# drugs1st
Ремонт телевизоров Hisense Москва
December 9, 2024 @ 9:43 pm
Предлагаем услуги профессиональных инженеров офицальной мастерской.
Еслли вы искали ремонт телевизоров hisense рядом, можете посмотреть на сайте: ремонт телевизоров hisense
Наши мастера оперативно устранят неисправности вашего устройства в сервисе или с выездом на дом!
StevenDrums
December 9, 2024 @ 9:54 pm
http://drugs1st.pro/# drugs1st
Social - paibe
December 9, 2024 @ 10:44 pm
Reasons Why Online Casinos Have Become an International Sensation
Digital casinos have reshaped the gambling landscape, offering a level of comfort and diversity that land-based casinos can’t match. Over time, a large audience internationally have turned to the fun of digital casino play due to its accessibility, appealing qualities, and ever-expanding range of offerings.
One of the biggest attractions of virtual gambling hubs is the sheer diversity of choices provided. Whether you are a fan of rolling traditional fruit machine slots, trying out story-driven video-based games, or playing smart in traditional table offerings like Roulette, virtual venues provide limitless entertainment avenues. Many casinos additionally introduce live gaming streams, allowing you to connect with real dealers and co-players, all while immersing yourself in the realistic vibes of a physical gaming house without leaving your home.
If you’re unfamiliar with the world of internet-based gaming or are looking to learn about reputable operators, why not engage with our active interactive platform? It’s a destination where gaming aficionados exchange tips, helping you to maximize your gaming journey. Join the experience and see it here now: https://x.com/usyktysonfury .
In addition to diversity, online casinos are known for accessibility.
Jacobgaugh
December 9, 2024 @ 10:56 pm
https://mtucizone.ru — Быстрое оформление заказа через интернет.
CharlesExore
December 9, 2024 @ 10:59 pm
rybelsus semaglutide tablets: semaglutide tablets – buy rybelsus online
PDF to ZIP
December 9, 2024 @ 11:22 pm
Glad I could help
afabet
December 9, 2024 @ 11:23 pm
A fascinating discussion is definitely worth comment.
I do believe that you should write more on this subject matter, it may not be a taboo subject but usually folks don’t speak about these topics.
To the next! Best wishes!!
https://deida.info/news/best-paid-poker-hud-pokerstars.html
December 9, 2024 @ 11:29 pm
I loved as much as you’ll receive carried out right here.
The sketch is attractive, your authored subject matter stylish.
nonetheless, you command get bought an impatience over that you wish be delivering the following.
unwell unquestionably come more formerly again since exactly the
same nearly a lot often inside case you shield this
hike.
JEDI69
December 9, 2024 @ 11:34 pm
Hi there! Do you know if they make any plugins to help with Search Engine Optimization? I’m trying to get my blog
to rank for some targeted keywords but I’m not seeing very good results.
If you know of any please share. Thank you!
Leonardsiz
December 9, 2024 @ 11:51 pm
https://tomason-russia.ru/ – Высокое качество и стиль в каждой детали вашей кухни.
Social - paibe
December 9, 2024 @ 11:55 pm
Reasons Why Online Casinos Are an International Sensation
Digital casinos have transformed the gambling landscape, providing an exceptional degree of user-friendliness and variety that land-based establishments are unable to replicate. Recently, countless gamblers internationally have embraced the excitement of internet-based gaming in light of its accessibility, exciting features, and continuously increasing catalogs of games.
One of the main appeals of digital gambling sites is the vast selection of games available. Whether you like playing on classic slots, exploring plot-filled video slots, or playing smart in strategy-based games like Texas Hold’em, internet-based gambling sites feature endless options. Numerous services also feature interactive dealer games, allowing you to participate with actual dealers and opponents, all while taking in the realistic atmosphere of a brick-and-mortar establishment without leaving your home.
If you’re a beginner with the world of internet-based gaming or hope to explore trusted platforms, why not join our growing gaming forum? It’s a destination where fans post tips, guiding you to get the most out of your gambling adventure. Dive into the community and see it here now: https://www.facebook.com/profile.php?id=61569191174332 .
Beyond variety, internet-based gambling hubs are known for ease of access.
Jacobgaugh
December 10, 2024 @ 12:07 am
http://ums-group.ru — Доверяйте профессионалам в создании кухонь вашей мечты.
saowin
December 10, 2024 @ 12:14 am
Hi there, I read your blog daily. Your humoristic style is witty, keep
doing what you’re doing!
Mazrlbt
December 10, 2024 @ 12:22 am
Купить диплом в России, предлагает наша компания
buy tetracycline without a prescription
December 10, 2024 @ 12:24 am
cost of tetracycline without rx
Angelhob
December 10, 2024 @ 12:36 am
предметная съемка для маркетплейсов – Услуги фотосъемки товаров для популярных маркетплейсов, таких как Ozon, Wildberries и других.
Social - paibe
December 10, 2024 @ 1:02 am
The Reasons Behind Why Online Casinos Remain So Popular
Internet-based gambling hubs have reshaped the gambling industry, offering a level of ease and breadth that traditional venues don’t provide. Over the past decade, a growing community internationally have welcomed the excitement of virtual casinos as a result of its ease of access, thrilling aspects, and widening selection of games.
One of the biggest attractions of virtual gambling hubs is the vast diversity of choices available. Whether you enjoy spinning retro one-armed bandits, diving into theme-based modern slot games, or strategizing in traditional table offerings like poker, internet-based gambling sites feature limitless choices. Many casinos furthermore present live dealer games, enabling you to engage with human game hosts and opponents, all while enjoying the engaging ambiance of a brick-and-mortar establishment right at home.
If you’re new with the world of virtual gambling or want to delve deeper into reputable operators, why not participate in our active social network? It’s a destination where gaming aficionados offer experiences, guiding you to improve your virtual play. Discover the conversation and see it here now: https://t.me/tysonfury_vsusyk .
Besides the wide selection, virtual gaming providers shine constant connectivity.
Rileypaf
December 10, 2024 @ 1:21 am
Viagra * Cialis * Levitra
All the products you are looking seeking are currently at one’s disposal as far as something 1+1.
4 more tablets of one of the following services: Viagra * Cialis * Levitra
http://xn--2i0bm4pd1btzg9pkelcg77a.com
ethitruong
December 10, 2024 @ 1:24 am
Hi I am so happy I found your site, I really found you
by accident, while I was searching on Aol for something else, Anyhow I am here now
and would just like to say thanks for a incredible
post and a all round thrilling blog (I also love the
theme/design), I don’t have time to browse it all at the moment but I have bookmarked it and also
added your RSS feeds, so when I have time I will be back to read much more, Please do keep up the great job.
Jamescaurf
December 10, 2024 @ 1:42 am
drugs1st: drugs1st – ed treatments that really work
Sazrubi
December 10, 2024 @ 2:07 am
Легальные способы покупки диплома о среднем полном образовании
Social - paibe
December 10, 2024 @ 2:12 am
What Makes Online Casinos Are Becoming So Popular
Digital casinos have revolutionized the gambling world, delivering a unique kind of comfort and variety that traditional establishments are unable to replicate. Over time, millions of players across the globe have embraced the fun of online gaming as a result of its anytime, anywhere convenience, captivating elements, and constantly growing collections of titles.
One of the key draws of virtual gambling hubs is the unparalleled selection of entertainment options at your disposal. Whether you love spinning vintage reel games, trying out narrative-rich video-based games, or mastering skills in card and board games like Roulette, online platforms boast endless options. Several sites moreover include real-time gaming experiences, enabling you to connect with actual dealers and opponents, all while enjoying the lifelike feel of a land-based casino from the comfort of your home.
If you’re new with the world of internet-based gaming or hope to learn about reliable sites, why not join our dynamic community? It’s a space where players discuss tips, guiding you to enhance your gaming journey. Explore the experience and visit us now: https://www.facebook.com/profile.php?id=61569645119382 .
Beyond variety, online casinos are known for accessibility.
Derekapaky
December 10, 2024 @ 2:15 am
http://larson-holz-spb.ru – сайт, где можно легко найти подходящую кухонную мебель.
Cazrhjk
December 10, 2024 @ 2:24 am
Стоимость дипломов высшего и среднего образования и как избежать подделок
item678787938
December 10, 2024 @ 2:37 am
Very good blog! Do you have any tips for aspiring writers?
I’m hoping to start my own website soon but I’m a little lost on everything.
Would you suggest starting with a free platform like WordPress or
go for a paid option? There are so many choices out there that I’m
completely overwhelmed .. Any tips? Thanks a lot!
bong99
December 10, 2024 @ 2:58 am
Helpful info. Fortunate me I discovered your website
accidentally, and I am surprised why this twist of fate did not happened earlier!
I bookmarked it.
Social - paibe
December 10, 2024 @ 3:18 am
Reasons Why Online Casinos Remain So Popular
Online casinos have revolutionized the gambling world, offering a unique kind of comfort and breadth that traditional casinos fall short of. Throughout the last ten years, countless gamblers globally have adopted the fun of virtual casinos due to its availability, exciting features, and progressively larger collections of titles.
One of the key draws of online casinos is the sheer array of titles on offer. Whether you enjoy spinning old-school slot machines, diving into narrative-rich modern slot games, or strategizing in classic casino games like Texas Hold’em, digital casinos boast infinite opportunities. Many casinos moreover include live dealer games, making it possible for you to connect with human game hosts and co-players, all while immersing yourself in the immersive environment of a real casino right at home.
If you’re a beginner with the world of digital casinos or seek to learn about safe services, why not engage with our vibrant social network? It’s a hub where players share tips, making it easier for you to get the most out of your gaming journey. Check out the discussions and check it out now: https://www.pinterest.com/fury_vs_usyk/ .
Adding to the extensive catalog, virtual gambling platforms thrive in seamless entry.
Ремонт телевизоров Hisense Москва
December 10, 2024 @ 3:25 am
Предлагаем услуги профессиональных инженеров офицальной мастерской.
Еслли вы искали ремонт телевизоров hisense, можете посмотреть на сайте: ремонт телевизоров hisense цены
Наши мастера оперативно устранят неисправности вашего устройства в сервисе или с выездом на дом!
kingfun
December 10, 2024 @ 3:37 am
If you are going for most excellent contents like me, only visit this website every day for
the reason that it provides feature contents, thanks
Davidtah
December 10, 2024 @ 3:38 am
Наша компания предоставляет услуги эвакуации автомобилей в Москве и Московской области круглосуточно. Мы гарантируем оперативную помощь, доступные цены и профессиональный подход. Вызвать эвакуатор вы можете по телефону или оформить заявку на нашем сайте в любой момент эвакуатор цена
buying singulair
December 10, 2024 @ 3:38 am
cheap singulair pills
legzoCasino
December 10, 2024 @ 3:40 am
[url=https://legzo-casino-apk.ru]http://legzo-casino-apk.ru[/url]
Установить приложение онлайн казино Легзо – побеждай прямо сейчас!
http://www.legzo-casino-apk.ru
Jacobgaugh
December 10, 2024 @ 3:52 am
кухни на заказ москва — Предлагаем изготовление кухонь под заказ в Москве с учетом ваших пожеланий.
Sazrqcz
December 10, 2024 @ 3:53 am
Как получить диплом техникума официально и без лишних проблем
betflik 24
December 10, 2024 @ 4:15 am
Terrific write ups, Thank you. https://clients1.google.com.ni/url?q=https://betflik24.day
Social - paibe
December 10, 2024 @ 4:28 am
Reasons Why Online Casinos Have Become So Popular
Digital casinos have reshaped the gaming market, providing a level of convenience and selection that land-based gambling houses struggle to rival. In recent years, a large audience around the world have welcomed the pleasure of internet-based gaming because of its availability, thrilling aspects, and widening selection of games.
One of the key draws of digital gambling sites is the sheer array of titles at your disposal. Whether you prefer engaging with retro reel games, diving into theme-based thematic slots, or exercising tactics in strategy-based games like Texas Hold’em, online platforms feature numerous options. Numerous services even offer interactive dealer games, allowing you to participate with live hosts and opponents, all while soaking in the realistic ambiance of a real casino without leaving your home.
If you’re just starting with the world of digital casinos or hope to learn about proven options, why not join our growing community? It’s a platform where gamblers post experiences, assisting you to get the most out of your gaming journey. Dive into the community and check it out now: https://www.instagram.com/fortunegems_top/ .
Besides the wide selection, virtual gaming providers stand out ease of access.
Leonardsiz
December 10, 2024 @ 5:09 am
tsmk-altai.ru – Профессиональные решения для кухни вашей мечты.
Mazryyr
December 10, 2024 @ 5:11 am
Диплом вуза купить официально с упрощенным обучением в Москве
Trefrhc
December 10, 2024 @ 5:11 am
Узнайте, как безопасно купить диплом о высшем образовании
Jacobgaugh
December 10, 2024 @ 5:20 am
кухня на заказ екатеринбург недорого — Предлагаем доступные решения для создания стильной и удобной кухни.
Social - paibe
December 10, 2024 @ 5:34 am
The Reasons Behind Why Online Casinos Are Becoming Highly Preferred Worldwide
Online casinos have reshaped the gambling landscape, delivering an unmatched level of ease and diversity that brick-and-mortar casinos don’t provide. Over the past decade, countless gamblers globally have adopted the pleasure of online gaming thanks to its availability, engaging traits, and constantly growing collections of titles.
One of the key draws of digital gambling sites is the vast selection of games provided. Whether you love spinning vintage one-armed bandits, diving into theme-based video-based games, or strategizing in card and board games like Roulette, digital casinos boast endless options. A large number of platforms moreover feature real-time gaming experiences, letting you to interact with live hosts and opponents, all while taking in the immersive vibes of a physical gaming house from the comfort of your home.
If you’re new with the world of online gaming or would like to learn about reputable operators, why not engage with our vibrant gaming forum? It’s a place where enthusiasts exchange insights, making it easier for you to enjoy more of your online casino experience. Dive into the connections and see it here now: https://t.me/furyversususyk .
Besides the wide selection, digital casino services thrive in ease of access.
Ремонт телевизоров Hisense Москва
December 10, 2024 @ 5:35 am
Предлагаем услуги профессиональных инженеров офицальной мастерской.
Еслли вы искали ремонт телевизоров hisense сервис, можете посмотреть на сайте: ремонт телевизоров hisense
Наши мастера оперативно устранят неисправности вашего устройства в сервисе или с выездом на дом!
подробное описание
December 10, 2024 @ 5:45 am
This is a topic which is close to my heart…
Best wishes! Where are your contact details though?
diuwin
December 10, 2024 @ 6:24 am
Hello there I am so grateful I found your site, I really found you
by error, while I was researching on Digg for
something else, Regardless I am here now and would just like to say kudos for a
remarkable post and a all round exciting blog (I also love the
theme/design), I don’t have time to look over it all at the moment
but I have saved it and also added in your RSS feeds, so when I have time I will be back to read much more,
Please do keep up the great work.
Cazrorx
December 10, 2024 @ 6:35 am
Пошаговая инструкция по официальной покупке диплома о высшем образовании
Social - paibe
December 10, 2024 @ 6:44 am
What Makes Online Casinos Are Becoming a Global Phenomenon
Online casinos have changed the gambling world, delivering a level of user-friendliness and selection that land-based casinos are unable to replicate. Over time, millions of players globally have welcomed the excitement of digital casino play due to its anytime, anywhere convenience, captivating elements, and ever-expanding selection of games.
One of the most compelling reasons of internet-based platforms is the astounding range of choices at your disposal. Whether you like spinning traditional fruit machine slots, immersing yourself in narrative-rich video slots, or exercising tactics in traditional table offerings like Roulette, casino websites deliver endless opportunities. Plenty of operators also feature real-time gaming experiences, allowing you to participate with professional croupiers and gaming peers, all while experiencing the immersive feel of a brick-and-mortar establishment right at home.
If you’re unfamiliar with the world of online gaming or seek to learn about reliable sites, why not engage with our vibrant gaming forum? It’s a place where enthusiasts discuss tips, guiding you to maximize your casino activities. Dive into the discussions and check it out now: https://www.instagram.com/136bet_com/ .
Beyond variety, online casinos stand out seamless entry.
Ремонт телевизоров Hisense Москва
December 10, 2024 @ 6:45 am
Предлагаем услуги профессиональных инженеров офицальной мастерской.
Еслли вы искали срочный ремонт телевизоров hisense, можете посмотреть на сайте: ремонт телевизоров hisense рядом
Наши мастера оперативно устранят неисправности вашего устройства в сервисе или с выездом на дом!
StevenDrums
December 10, 2024 @ 6:48 am
https://drugs1st.pro/# drugs1st
can you buy prasugrel prices
December 10, 2024 @ 6:49 am
buying prasugrel without prescription
PinUp - t7
December 10, 2024 @ 6:52 am
Pin Up Онлайн Казино в Казахстане – это одно из самых популярных мест для фанатов ставок и игр. Здесь доступен огромный ассортимент игр для развлечений и крупных выигрышей. Если ищете безопасное место для игры, то Pin Up Казино создано для вас.
Преимущества Pin Up Casino
Многообразие игровых решений. На платформе Pin Up вы найдёте более 2000 слотов и игр, игр с реальными дилерами. В ассортименте представлены разработчики уровня Evolution Gaming и другие. Это значит, что вы получите отличное качество игры, удобные для игры на мобильных.
Разнообразные бонусы для игроков. Новым пользователям доступны фриспины и бонусы, которые дадут хорошие шансы на выигрыш. Постоянные акции и розыгрыши тоже доступны, ежедневные и еженедельные бонусы для постоянных игроков, что даёт возможность выиграть ещё больше.
Наш сайт: https://bk.kz/news/?ELEMENT_ID=2974&PAGEN_2=16
Безопасность и надежность. Pin Up Казино Онлайн обеспечивает безопасность всех игроков персональной информации, чтобы гарантировать защиту данных. Казино работает по лицензии обеспечивает честные игры и честные и быстрые выплаты.
Доступность на всех устройствах. Казино прекрасно работает на смартфонах, что позволяет наслаждаться игрой в любое время и в любом месте. Приложение казино доступно для скачивания или просто начать игру через мобильный браузер.
Обслуживание клиентов. В любой ситуации, операторы поддержки Pin Up всегда готовы решить любые вопросы. Вы можете обратиться за помощью через чат и email, и операторы ответят в кратчайшие сроки на русском или казахском языке.
WalterByday
December 10, 2024 @ 7:09 am
Аркада Казино официальный сайт – Приветственный Бонус +150% и 2000FS – Новое ARKADA Casino Онлайн, захватывающие игры, слоты, и автоматы доступны уже arkada casino arkadacas-site.buzz
diuwin login
December 10, 2024 @ 7:31 am
Hi there! I’m at work browsing your blog from my new iphone!
Just wanted to say I love reading through your blog and look forward to all your posts!
Carry on the excellent work!
Social - paibe
December 10, 2024 @ 7:50 am
The Reasons Behind Why Online Casinos Are Becoming a Worldwide Trend
Internet-based gambling hubs have changed the betting scene, offering an exceptional degree of convenience and breadth that conventional casinos struggle to rival. Over time, countless gamblers worldwide have welcomed the fun of online gaming because of its always-open nature, thrilling aspects, and constantly growing range of offerings.
One of the most compelling reasons of digital gambling sites is the incredible selection of games available. Whether you like interacting with traditional slots, trying out engaging thematic slots, or playing smart in classic casino games like Baccarat, digital casinos boast infinite entertainment avenues. Numerous services furthermore feature live dealer games, enabling you to participate with real dealers and other players, all while experiencing the authentic atmosphere of a land-based casino without leaving your home.
If you’re new with the world of online gaming or hope to find out more about proven options, why not join our lively online hub? It’s a destination where fans share experiences, helping you to improve your casino activities. Discover the community and see it here now: https://www.pinterest.com/tysonfuryvsusyk .
Apart from the game range, internet-based gambling hubs shine availability.
Jacobgaugh
December 10, 2024 @ 8:45 am
https://www.gft-leasing.ru — Посетите наш сайт, чтобы ознакомиться с ассортиментом кухонь.
https://23win.express
December 10, 2024 @ 8:49 am
You have made some really good points there. I looked on the net to learn more about the issue and found most individuals
will go along with your views on this website.
Turkish tuS
December 10, 2024 @ 8:57 am
Турецкие фильмы набрали высокую популярность во всем мире. Турция славится интересным кинематографом c крутым и захватывающим сюжетом, эмоциональными переживаниями и потрясающей игрой актеров.
Турецкие сериалы предлагают разнообразие жанров и тематик, чтобы удовлетворить интересы каждого зрителя. От романтических комедий до исторических драм, от триллеров до ужасов – каждый найдет что-то по своему вкусу. Богатство сюжетов и уровень съемок делают турецкие фильмы настоящими изобретениями телевизионного искусства.
[url=https://turkish-film1.ru/]Турецкие фильмы смотреть онлайн[/url] – это уникальная возможность отправиться в другую культуру, узнать больше о традициях и обычаях турецкого народа.
Social - paibe
December 10, 2024 @ 9:02 am
The Reasons Behind Why Online Casinos Remain Highly Preferred Worldwide
Virtual gambling platforms have changed the gambling market, delivering a unique kind of accessibility and breadth that land-based casinos are unable to replicate. In recent years, a vast number of enthusiasts internationally have embraced the excitement of virtual gambling as a result of its availability, exciting features, and constantly growing range of offerings.
One of the strongest selling points of virtual gambling hubs is the astounding array of choices at your disposal. Whether you enjoy engaging with retro reel games, exploring narrative-rich thematic slots, or testing your strategy in traditional table offerings like poker, online platforms boast endless options. Plenty of operators also feature live casino options, making it possible for you to connect with professional croupiers and fellow gamblers, all while experiencing the engaging vibes of a brick-and-mortar establishment from the comfort of your home.
If you’re just starting with the world of digital casinos or seek to delve deeper into reputable operators, why not join our lively online hub? It’s a platform where players post stories, guiding you to enjoy more of your gaming journey. Explore the discussions and see it here now: https://www.facebook.com/profile.php?id=61569360522200 .
Beyond variety, virtual gambling platforms stand out availability.
Casino
December 10, 2024 @ 9:25 am
If some one wants expert view regarding running a
blog afterward i recommend him/her to pay a quick visit this
blog, Keep up the fastidious job.
1 Win - e0
December 10, 2024 @ 9:25 am
1-вин — Лицензированное игровое казино и букмекерская контора с выгодными условиями для игроков
В пространстве виртуальных казино и спортивных ставок необходимо не только качество игровой платформы, но и честность оператора. С разрешением на деятельность, которая подтверждает легальность, 1-вин уверенно занимает ведущих игроков в сфере интерактивных казино. Оператор сфокусирована на оптимальных условий, постоянно улучшая свои продукты и сервисы, чтобы подарить пользователям захватывающий игровой процесс.
Приветственный бонус: Начальный бонус для новых игроков
Для новых участников на платформе 1win доступен в предложении большой приветственный бонус, который позволяет начать с уверенностью. Это предложение является одним из лучших на в мире онлайн-гемблинга, так как позволяет игрокам новым пользователям получить преимущество, сократив риски. Простая регистрация и первый взнос обеспечивают получение бонуса, что делает его применение эффективным и быстрым.
Live-ставки: Играйте на события вживую с широким выбором
В индустрии спортивных прогнозов главной отличительной чертой является изменчивость и возможность реагировать на изменения в режиме реального времени. 1win даёт доступ большой спектр спортивных событий, доступных в формате игры вживую. Мы предоставляем доступ к международным событиям мирового масштаба, будь то баскетбол.
Наш сайт: http://5-5.ru/index.php?links_exchange=yes&page=406&show_all=yes
Ставки в реальном времени гарантируют игрокам мгновенно реагировать на изменения, что делает процесс ещё интереснее, но и интеллектуально интересным. С помощью аналитики платформы, пользователи могут принимать более обоснованные решения, увеличивая свои шансы на успех. Каждое событие освещается статистикой, что способствует точным прогнозам гарантировать обоснованные ставки.
Игровые автоматы: Бесплатная версия и максимум возможностей над процессом игры
Важная черта онлайн-казино 1win — это возможность тестовых версий игровых автоматов, что гарантирует игрокам ознакомиться с любой слот без ставок на реальные деньги. Это незаменимо для новичков, которые ещё не знакомы с ассортиментом или требованиями игры, а также для профессионалов, которые хотят найти новые игры или аналитику игры.
Игровые автоматы от лучших провайдеров, такими как Red Tiger, что гарантирует отличную графику, саундтрек и игровой процесс. Благодаря высокому уровню, игроки казино могут найти автоматы, которые соответствуют их стилю и предпочтениям, от классических игр до слотов с бонусными функциями и бонусными раундами.
MBVCDRTYO
December 10, 2024 @ 9:44 am
I love your blog.. very nice colors & theme. Did you make
this website yourself or did you hire someone
to do it for you? Plz respond as I’m looking to create my own blog
and would like to find out where u got this from.
kudos
Angelhob
December 10, 2024 @ 9:45 am
фотограф предметная съемка – Специалист по съемке товаров для презентаций, магазинов и маркетплейсов.
tetsdjjhd
December 10, 2024 @ 9:53 am
tetsdjjhd
Jacobgaugh
December 10, 2024 @ 9:57 am
https://ums-group.ru — Здесь вы найдете все, что нужно для вашей идеальной кухни.
Ремонт телевизоров Hisense Москва
December 10, 2024 @ 10:03 am
Предлагаем услуги профессиональных инженеров офицальной мастерской.
Еслли вы искали ремонт телевизоров hisense, можете посмотреть на сайте: ремонт телевизоров hisense
Наши мастера оперативно устранят неисправности вашего устройства в сервисе или с выездом на дом!
where to buy furosemide tablets
December 10, 2024 @ 10:10 am
furosemide 40 mg buy online uk
Social - paibe
December 10, 2024 @ 10:11 am
The Reasons Behind Why Online Casinos Are Becoming an International Sensation
Digital casinos have changed the casino gaming industry, providing an exceptional degree of convenience and selection that traditional venues fall short of. Over the past decade, countless gamblers across the globe have adopted the thrill of virtual gambling in light of its always-open nature, engaging traits, and continuously increasing game libraries.
One of the most compelling reasons of online casinos is the vast selection of choices available. Whether you prefer interacting with old-school fruit machine slots, playing through theme-based thematic slots, or mastering skills in card and board games like Baccarat, virtual venues deliver infinite choices. Numerous services moreover feature live casino options, allowing you to engage with human game hosts and fellow gamblers, all while experiencing the authentic environment of a physical gaming house right at home.
If you’re new with the world of digital casinos or seek to discover safe services, why not participate in our lively community? It’s a place where enthusiasts share reviews, guiding you to get the most out of your online casino experience. Dive into the discussions and visit us now: https://www.pinterest.com/tyson_fury_vs_oleksandr_usyk/ .
In addition to diversity, virtual gambling platforms excel constant connectivity.
Leonardsiz
December 10, 2024 @ 10:12 am
flowers777.ru/ — Качественные кухни с гарантией от производителя.
Digitaal vignet Oostenrijk
December 10, 2024 @ 10:27 am
I do not even know how I ended up here, but I thought
this post was good. I don’t know who you are
but certainly you’re going to a famous blogger if you are not already 😉 Cheers!
Bradleyvat
December 10, 2024 @ 10:33 am
https://kamagra.men/# Kamagra tablets
Jamescaurf
December 10, 2024 @ 10:52 am
semaglutide tablets for weight loss: semaglutide tablets store – generic rybelsus tabs
PinUp - gy
December 10, 2024 @ 10:59 am
Pin Up интернет-казино для Казахстана – это топовое казино для игроков для игроков, любящих азарт. Здесь вы сможете найти идеальные условия для развлечений и крупных выигрышей. Если вы ищете качественное онлайн-казино, то Pin Up Казино создано для вас.
Преимущества казино Pin Up
Многообразие игровых решений. Игрокам предлагается более 2000+ игровых решений, игр с живыми крупье и автоматов. В каталоге игр представлены разработчики уровня NetEnt, Microgaming, Play’n GO и множество других компаний. Это значит, что вы сможете наслаждаться качеством и азартом, оптимизированные для телефонов и ПК.
Привлекательные акции и бонусы. Каждый новый игрок получит увеличенные стартовые бонусы, которые помогут стартовать быстрее. Регулярные акции увеличат ваши выигрыши, кэшбэки и специальные предложения для постоянных пользователей, что делает игру ещё более захватывающей и выгодной.
Наш сайт: https://bk.kz/news/?ELEMENT_ID=1099&PAGEN_2=15
Честность и безопасность. Pin Up интернет-казино использует современные технологии для защиты данных пользователей, чтобы обезопасить вашу игру. Официально лицензированное казино обеспечивает честность и выплаты и выплаты по всем выигрышам.
Лёгкий доступ к играм. Pin Up Casino оптимизировано для смартфонов, что даёт возможность играть в любые игры в любой точке мира. Можно скачать приложение для смартфона или просто запустить сайт казино с телефона.
Обслуживание клиентов. В любое время, команда поддержки Pin Up всегда готовы решить любые вопросы. Вы можете обратиться за помощью через онлайн-чат, и вам окажут полную поддержку на любом удобном для вас языке.
Lucky88
December 10, 2024 @ 11:13 am
Link exchange is nothing else except it is only
placing the other person’s webpage link on your page
at suitable place and other person will also do similar in support of you.
Social - paibe
December 10, 2024 @ 11:25 am
Why Online Casinos Are an International Sensation
Online casinos have transformed the gaming world, offering an exceptional degree of comfort and breadth that traditional establishments struggle to rival. Recently, a vast number of enthusiasts internationally have turned to the thrill of virtual gambling thanks to its accessibility, exciting features, and widening game libraries.
One of the key draws of internet-based platforms is the sheer diversity of games ready to play. Whether you love playing on classic one-armed bandits, diving into story-driven thematic slots, or exercising tactics in traditional table offerings like Baccarat, casino websites offer limitless opportunities. A large number of platforms additionally introduce live gaming streams, giving you the chance you to connect with live hosts and opponents, all while taking in the immersive feel of a physical gaming house from the comfort of your home.
If you’re new with the world of internet-based gaming or want to explore trusted platforms, why not engage with our vibrant community? It’s a place where fans discuss reviews, enabling you to enjoy more of your gambling adventure. Join the discussions and check it out now: https://www.facebook.com/profile.php?id=61569360522200 .
Beyond variety, internet-based gambling hubs excel ease of access.
101 game
December 10, 2024 @ 11:29 am
Hey There. I found your blog using msn. This
is an extremely well written article. I will make sure to bookmark it and return to read more of
your useful info. Thanks for the post. I will certainly comeback.
Derekapaky
December 10, 2024 @ 11:37 am
кухни на заказ спб недорого – оптимальный вариант для тех, кто ищет качественную мебель по доступной цене.
onexxxsila
December 10, 2024 @ 11:38 am
Рекомендации по 1xSlots https://bodegascrial.es/pags/?1xslots-descargar-android-ios.html
Frankjah
December 10, 2024 @ 11:41 am
Используйте бесплатные шаблоны резюме для устройства на работу. Пустые бланки резюме для устройства на работу представлены в 35 вариантах оформления. Шаблоны представлены в формате PDF, используйте для работы с файлами PDF-редакторы https://seohotmix.ru/kak-sostavit-rezyume-i-ustroitsya-na-rabotu/
CharlesExore
December 10, 2024 @ 12:12 pm
cenforce.icu: cheapest cenforce – order cenforce
Baywin - w0
December 10, 2024 @ 12:33 pm
Bay Win, dijital bahis dunyas?nda one c?kan bir platformdur. Bahiscilere sundugu genis oyun secenekleri, kolay erisim imkan? ve saglam altyap?s? ile one c?kmaktad?r.
Ozellikle platforma erisim saglamak ve guncel giris adresleri, bahiscilerin dikkat ceken unsurlar aras?nda yer vard?r.
Baywin Hakk?nda Genel Bilgiler
BayWin, dijital bahis platformlar? aras?nda faaliyet gosteren bir markad?r. canl? bahisler, casino oyunlar?, dijital oyunlar gibi zengin oyun iceriklerine sahiptir.
Baywin’in en onemli ozelliklerinden biri, yuksek bahis oranlar? saglamas?d?r. Ayr?ca, pratik para yat?rma ve cekme secenekleri, maddi kazanclar? kolayca yonetmeyi mumkun k?lar.
Baywin Giris Sorunlar? ve Cozumleri
Web: https://greenlifehealth.co.uk/2024/12/03/ofitsialnyy-sayt-bukmekerskoy-kontory-1win/
Baywin markas?n?n Turkiye’de engellemelerle s?k s?k kars?last?g? bilinmektedir, bununla birlikte engellemeler oldugunda Baywin’in teknik ekibi cozumler sunmaktad?r.
Erisim engellemeleri gerceklestiginde, kullan?c?lar icin alternatif bir baglant? sunulur. Bu cozumle, Baywin’in en yeni erisim adresi uzerinden kullan?c?lar oyunlar?n? kesintisiz oynayabilir.
Platforma yeniden erisim icin bircok erisim yontemi bulunmaktad?r. Cep telefonlar?, tas?nabilir cihazlar ve desktop cihazlar uzerinden siteye erisim saglanabilir. Bu da kullan?c?lar?n diledikleri yerden bahis yapma ozgurlugunu yasamalar?na olanak tan?r.
Social - paibe
December 10, 2024 @ 12:33 pm
Why Online Casinos Have Become a Global Phenomenon
Internet-based gambling hubs have transformed the betting landscape, delivering a unique kind of ease and diversity that physical establishments struggle to rival. In recent years, a growing community globally have embraced the adventure of virtual gambling because of its accessibility, exciting features, and progressively larger range of offerings.
One of the strongest selling points of digital gambling sites is the vast range of games on offer. Whether you like rolling retro slot machines, diving into narrative-rich visual slot games, or exercising tactics in strategy-based games like Texas Hold’em, digital casinos provide infinite options. Plenty of operators moreover introduce live dealer games, letting you to connect with real dealers and opponents, all while soaking in the engaging feel of a land-based casino from anywhere you want.
If you’re a beginner with the world of internet-based gaming or seek to explore trusted platforms, why not become part of our dynamic online hub? It’s a destination where gaming aficionados exchange stories, assisting you to enjoy more of your gaming journey. Check out the discussions and learn more now: https://t.me/usykvs_fury .
Adding to the extensive catalog, virtual gambling platforms excel seamless entry.
escorts in manchester
December 10, 2024 @ 12:44 pm
I will right away clutch your rss feed as I can’t
to find your email subscription link or e-newsletter service.
Do you have any? Kindly allow me realize in order that I may just subscribe.
Thanks.
Jacobgaugh
December 10, 2024 @ 1:09 pm
https://mtucizone.ru — Быстрое оформление заказа через интернет.
Pokemon Rug
December 10, 2024 @ 1:28 pm
Being a dedicated Pokémon enthusiast, I’ve always found joy
in bringing the Pokémon world into my everyday life.
From catching them all in the games to decorating
my space, Pokémon is a big part of who I am.
One of my favorite hobbies is creating Pokémon-themed room decor.
I’ve used items like Pokémon rugs, woven tapestries, and custom
carpets to make my room stand out. These pieces add so much character but also bring my favorite Pokémon to life.
To anyone wanting to decorate their space with Pokémon, I highly recommend incorporating these items into your
decor. They’re perfect for adding a unique touch.
Transform your space into a Pokémon haven!
Also visit my blog; Pokemon Rug
CoreyamBit
December 10, 2024 @ 1:29 pm
cheap ed treatment [url=https://edpills.men/#]ed treatments online[/url] ed medications cost
can i order cheap differin pills
December 10, 2024 @ 1:32 pm
where to get cheap differin without rx
qq88_bz
December 10, 2024 @ 1:35 pm
I get pleasure from, cause I discovered exactly
what I used to be taking a look for. You’ve ended my four day lengthy hunt!
God Bless you man. Have a nice day. Bye
Social - paibe
December 10, 2024 @ 1:45 pm
Reasons Why Online Casinos Are a Worldwide Trend
Digital casinos have reshaped the betting industry, delivering an unmatched level of comfort and breadth that physical casinos struggle to rival. Over the past decade, countless gamblers internationally have chosen the adventure of virtual gambling thanks to its always-open nature, captivating elements, and ever-expanding game libraries.
One of the most compelling reasons of digital gambling sites is the sheer selection of gaming experiences at your disposal. Whether you like spinning retro slot machines, diving into engaging visual slot games, or testing your strategy in traditional table offerings like Roulette, internet-based gambling sites deliver endless choices. Several sites even offer interactive dealer games, letting you to communicate with professional croupiers and other players, all while immersing yourself in the authentic atmosphere of a land-based casino from the comfort of your home.
If you’re just starting with the world of digital casinos or want to find out more about reputable operators, why not sign up for our dynamic gaming forum? It’s a place where gaming aficionados post reviews, assisting you to get the most out of your online casino experience. Join the discussions and start your journey now: https://www.facebook.com/profile.php?id=61569645119382 .
Apart from the game range, virtual gambling platforms excel seamless entry.
Plinko - 82
December 10, 2024 @ 2:08 pm
Plinko game is zeer populair gokspellen die in de afgelopen jaren beschikbaar zijn in de online casinowereld. Dit interessante spel, geinspireerd door het tv-programma The Price Is Right, heeft zich snel aangepast aan de digitale gokwereld.
In de komende alinea’s gaan we verder in op alles wat je belangrijk is om te weten over deze casinogame, van de grondslagen van het spel tot hoe je met echt geld kunt spelen en de beste tips voor het winnen.
Web: https://www.chandanchakraborty.com/plinko-spel-gratis-bij-nederlandse-online-casinos-2/
Het populaire Plinko spel is een simpel maar boeiend kansspel wat vaak wordt gezien als de Amerikaanse tv-show The Price Is Right. Het spel bestaat uit een verticale speelstructuur met een aantal hobbels waar de vallende bal van bovenaf doorheen valt. De bal stuitert van de pinnen en belandt in een van de vakken aan de onderkant, die elk een bepaald bedrag aanduiden. De winbedrag is afhankelijk van de bal vastkomt. Dit betekent dat het een spel van geluk is, waarbij spelers niet kunnen voorspellen waar de bal terecht zal komen.
Hoewel de basisstructuur van het spel simpel zijn, maakt de geluksaspect van het spel het opwindend en adembenemend. Dit is een van de oorzaken waarom Plinko in de gokwereld zo’n hype heeft veroorzaakt. Het wordt vaak aangeboden als een digitale versie van Plinko in verschillende online digitale gokplatforms, waar spelers geld kunnen winnen door te gokken door te wedden op de uitkomst van hun spelballen.
Wanneer je deelneemt aan een Plinko casinospel, lijkt het spel nagenoeg hetzelfde als de showversie van Plinko. De verschillen liggen echter in je kans hebt om te wedden en het feit dat je met echt geld inzetbaar is. In plaats van voor aandachtspunten zoals in de showuitzending, kun je in een casino op internet betalen voor prijzen. De beloningen zijn afhankelijk van het gebied waarin de bal zit wordt berekend.
Spelers kunnen kiezen hoeveel ze willen inzetten, en afhankelijk van de hoogte van je inzet kunnen de uitbetalingen verschuiven. De Plinko online gokspel wordt vaak op een makkelijke manier aangeboden, wat het voor beginnelingen makkelijker maakt om het spel te leren. Veel gokwebsites bieden een Plinko game download optie, zodat je het spel kunt spelen op je mobiele apparaat, zelfs zonder altijd online te zijn. Dit maakt het gemakkelijker om te spelen en maakt het spel gemakkelijker.
Leonardsiz
December 10, 2024 @ 2:14 pm
кухня заказать – Простое и удобное решение для заказа стильной кухни.
Naklejki Z Samochodami
December 10, 2024 @ 2:24 pm
Hi there, yes this article is truuly nice and I have learned lot
of things from it about blogging. thanks. https://naklejkinasciane.z20.web.core.windows.net/naklejki-dzieciece-na-sciane/Naklejka-na-cian-Magnetofon.html
Social - paibe
December 10, 2024 @ 3:00 pm
The Reasons Behind Why Online Casinos Remain a Worldwide Trend
Digital casinos have transformed the gambling market, offering an exceptional degree of accessibility and range that land-based establishments are unable to replicate. Recently, countless gamblers around the world have chosen the excitement of digital casino play because of its ease of access, exciting features, and constantly growing catalogs of games.
One of the most compelling reasons of online casinos is the unparalleled range of choices on offer. Whether you are a fan of interacting with old-school fruit machine slots, playing through narrative-rich thematic slots, or mastering skills in table games like Roulette, internet-based gambling sites feature endless opportunities. Many casinos furthermore offer interactive dealer games, making it possible for you to engage with human game hosts and gaming peers, all while immersing yourself in the engaging environment of a physical gaming house right at home.
If you’re exploring for the first time with the world of digital casinos or seek to find out more about reputable operators, why not engage with our vibrant community? It’s a hub where fans offer tips, assisting you to improve your online casino experience. Explore the experience and see it here now: https://ru.pinterest.com/usykvsfury/ .
Besides the wide selection, internet-based gambling hubs are known for ease of access.
StevenDrums
December 10, 2024 @ 3:24 pm
http://drugs1st.pro/# drugs1st
escort agency paris
December 10, 2024 @ 3:38 pm
Hi! This post could not be written any better!
Reading this post reminds me of my old room mate!
He always kept talking about this. I will forward this write-up to him.
Fairly certain he will have a good read. Thank you for sharing!
Baywin - xt
December 10, 2024 @ 3:39 pm
Bahis Platformu Baywin, dijital bahis dunyas?nda tan?nan bir bahis sitesidir. Bahisseverlere sundugu zengin oyun icerikleri, pratik erisim secenekleri ve kaliteli hizmet sunumu ile begeni toplamaktad?r.
Bilhassa da Baywin giris bilgileri ve guncel erisim bilgileri, bahiscilerin en cok merak edilen konular aras?nda yer vard?r.
Baywin Hakk?nda Genel Bilgiler
Bay Win, internette bahis ve sans oyunlar? sektorunde faaliyet gosteren bir uygulamad?r. Spor bahisleri, poker ve baccarat, 3D bahis secenekleri gibi cok say?da oyun secenegine sahiptir.
Platformun en onemli avantajlar?ndan biri, uyelerine cazip oranlar sunmas?d?r. Ayr?ca, guvenilir finansal islemleri, kullan?c?lar?n kazand?klar? miktarlar? sorunsuz bir sekilde cekmelerine olanak saglar.
Baywin Giris Yontemleri
Web: https://www.thefreemanonline.org/tmp/baywin-guncel-giris-2024-turkiyenin-lider-online-casino-sitesi/
Baywin’in erisim engellemelerinden etkilenmemesi mumkun degildir, bununla birlikte engellemeler oldugunda Site yoneticileri an?nda aksiyon al?r.
Adres engeli olustugunda, an?nda bir guncel adres tan?mlan?r. Bu sekilde, yeni giris URL’si uzerinden kullan?c?lar oyunlar?n? kesintisiz oynayabilir.
Baywin guncel giris islemleri icin bircok erisim yontemi bulunmaktad?r. Cep telefonlar?, tabletler ve PC’ler uzerinden oyunlara kat?lmak mumkundur. Baywin’in her yerden erisilebilirligini art?r?r.
tetsdjjhd
December 10, 2024 @ 3:55 pm
tetsdjjhd
Derekapaky
December 10, 2024 @ 3:59 pm
http://germancoonluck.ru/ — Индивидуальный подход к каждому клиенту.
Social - paibe
December 10, 2024 @ 4:09 pm
Why Online Casinos Are Becoming Highly Preferred Worldwide
Internet-based gambling hubs have revolutionized the betting landscape, offering an exceptional degree of comfort and variety that land-based gambling houses struggle to rival. Over the past decade, a growing community worldwide have adopted the pleasure of internet-based gaming as a result of its availability, appealing qualities, and continuously increasing catalogs of games.
One of the strongest selling points of digital gambling sites is the vast range of games on offer. Whether you are a fan of spinning old-school reel games, diving into theme-based video slots, or testing your strategy in card and board games like Roulette, online platforms boast numerous options. Many casinos even present live gaming streams, giving you the chance you to communicate with professional croupiers and gaming peers, all while immersing yourself in the authentic feel of a brick-and-mortar establishment from the comfort of your home.
If you’re new with the world of virtual gambling or seek to delve deeper into safe services, why not engage with our lively interactive platform? It’s a place where enthusiasts exchange stories, guiding you to improve your virtual play. Check out the connections and start your journey now: https://www.instagram.com/tigrinhobr_com/ .
Apart from the game range, virtual gambling platforms are known for seamless entry.
ボッテガ コインケース スーパーコピー 2ちゃんねる
December 10, 2024 @ 4:15 pm
You definitely made the point!
Here is my blog … http://www.gz-jj.com/comment/html/?231306.html
diuwin login
December 10, 2024 @ 4:17 pm
That is a great tip especially to those new to the
blogosphere. Brief but very accurate info… Many thanks for sharing this
one. A must read article!
Daga88
December 10, 2024 @ 4:31 pm
I’m excited to discover this website. I want to
to thank you for your time due to this wonderful read!! I definitely appreciated
every part of it and I have you saved as a favorite to look at new stuff on your blog.
Leonardsiz
December 10, 2024 @ 4:39 pm
http://www.santeh-burg.ru — Надежное место для покупки кухонь с гарантией качества.
Jacobgaugh
December 10, 2024 @ 4:53 pm
кухня под заказ — Индивидуальный подход к созданию вашей идеальной кухни.
where can i buy cheap colchicine pill
December 10, 2024 @ 5:08 pm
Medication information. Generic Name.
where can i buy cheap colchicine pill
Best trends of drugs. Read information now.
albuterol inhaler without a prescription
December 10, 2024 @ 5:10 pm
where can i get generic albuterol prices
Social - paibe
December 10, 2024 @ 5:15 pm
Reasons Why Online Casinos Remain Highly Preferred Worldwide
Virtual gambling platforms have reshaped the gaming landscape, delivering an unmatched level of comfort and breadth that traditional gambling houses are unable to replicate. Over time, a growing community around the world have chosen the thrill of virtual casinos due to its ease of access, engaging traits, and continuously increasing range of offerings.
One of the biggest attractions of online gaming options is the incredible variety of gaming experiences on offer. Whether you prefer engaging with classic reel games, trying out plot-filled video-based games, or mastering skills in classic casino games like Roulette, internet-based gambling sites boast limitless options. Plenty of operators furthermore introduce live gaming streams, allowing you to interact with professional croupiers and opponents, all while taking in the authentic environment of a land-based casino in your own space.
If you’re unfamiliar with the world of virtual gambling or want to learn about reliable sites, why not join our growing gaming forum? It’s a destination where fans post tips, helping you to enhance your virtual play. Explore the experience and see it here now: https://www.pinterest.com/fury_usyk_date/ .
Besides the wide selection, virtual gaming providers are known for ease of access.
1 Win - ap
December 10, 2024 @ 5:19 pm
1win — Официальное онлайн-казино и платформа для ставок с привлекательными условиями для игроков
В сфере интернет-казино и игр на ставки приоритетное значение имеет не только надежность услуг, но и репутация компании. С международной лицензией, которая гарантирует прозрачность, 1win установил себя как ключевых участников в мире онлайн-гемблинга. Провайдер работает на предоставление качественных услуг, улучшая функционал, чтобы обеспечить пользователям непревзойденный опыт.
Приветственный бонус: Выгодный старт для новичков
Для новых участников на платформе 1win ожидает вас выгодный приветственный бонус, который позволяет начать с уверенностью. Это бонусная программа является одним из лучших на рынке, так как предоставляет шанс новым пользователям моментально воспользоваться бонусом, стимулируя успешные ставки. Упрощенная процедура регистрации и внесение депозита мгновенно активируют бонус, что делает его использование приятным и лёгким.
Live-ставки: Множество матчей онлайн с быстрой реакцией
В пространстве ставочной деятельности важнейшей частью является темп и умение быстро принимать решения в режиме реального времени. 1win приглашает множество спортивных событий, доступных в формате ставок в режиме реального времени. Мы организуем лайв-ставки к глобальным событиям мирового масштаба, будь то волейбол.
Наш сайт: http://www.kums-barnaul.ru/opennews.php?ns=3824&id_page=5
Лайв-ставки дают шанс принимать решения мгновенно, что делает процесс более динамичным, но и серьёзным испытанием навыков. Используя стратегии и аналитику платформы, игроки могут точнее прогнозировать исходы, повышая вероятность выигрыша. Каждое событие освещается статистикой, что позволяет лучше анализировать гарантировать обоснованные ставки.
Игровые автоматы: Бесплатная версия и свобода выбора над выбором игры
Один из главных плюсов онлайн-казино 1-Win — это функция демо-доступа игровых автоматов, что дает возможность протестировать слоты платформы без необходимости тратить реальные деньги. Это особенно полезно для новых игроков, которые ещё не уверены в выборе или требованиями игры, а также для продвинутых игроков, которые хотят найти новые стратегии или аналитику игры.
Игры разработаны лидерами рынка, такими как NetEnt, что обеспечивает непревзойденную графику, аудио и динамику игр. С таким качеством, любители казино могут найти автоматы, которые подходят под их стиль игры, от винтажных автоматов до современных игровых автоматов и дополнительными раундами.
jual beli tanah
December 10, 2024 @ 5:25 pm
I’ll right away seize your rss as I can’t in finding your e-mail
subscription hyperlink or e-newsletter service. Do you have any?
Please allow me know in order that I may subscribe. Thanks.
Delux
December 10, 2024 @ 5:39 pm
Hi there, just wanted to say, I liked this article. It
was funny. Keep on posting!
slot gacor 88
December 10, 2024 @ 5:43 pm
Whats up this is kinda of off topic but I was wondering if
blogs use WYSIWYG editors or if you have to manually code with HTML.
I’m starting a blog soon but have no coding know-how so I wanted to get advice from someone with experience.
Any help would be greatly appreciated!
Angelhob
December 10, 2024 @ 5:56 pm
sunnyholiday.ru – Откройте для себя лучшие услуги фотосъемки товаров.
Casino
December 10, 2024 @ 6:09 pm
Thanks for your personal marvelous posting!
I certainly enjoyed reading it, you will be a great author.I will be sure to bookmark your blog and will eventually come back later on. I want
to encourage you to continue your great posts,
have a nice holiday weekend!
Social - paibe
December 10, 2024 @ 6:23 pm
What Makes Online Casinos Are a Worldwide Trend
Online casinos have reshaped the casino gaming world, providing an unmatched level of comfort and variety that conventional venues don’t provide. Throughout the last ten years, countless gamblers worldwide have chosen the thrill of digital casino play because of its always-open nature, captivating elements, and progressively larger range of offerings.
One of the key draws of virtual gambling hubs is the vast array of choices available. Whether you enjoy spinning retro fruit machine slots, immersing yourself in theme-based visual slot games, or exercising tactics in traditional table offerings like Roulette, virtual venues provide countless opportunities. A large number of platforms even offer live gaming streams, letting you to communicate with actual dealers and opponents, all while taking in the realistic feel of a physical gaming house from anywhere you want.
If you’re just starting with the world of virtual casino play or would like to learn about reputable operators, why not become part of our growing gaming forum? It’s a place where players share stories, enabling you to enhance your virtual play. Discover the connections and start your journey now: https://www.instagram.com/fortunegems_top/ .
Apart from the game range, digital casino services excel availability.
fa88
December 10, 2024 @ 6:26 pm
Hi, I do believe this is an excellent site. I stumbledupon it 😉
I am going to revisit yet again since i have book marked it.
Money and freedom is the greatest way to change,
may you be rich and continue to help others.
African Grand Casino
December 10, 2024 @ 6:37 pm
Whats up this is kind of of off topic but I was wondering if
blogs use WYSIWYG editors or if you have to manually code
with HTML. I’m starting a blog soon but have
no coding experience so I wanted to get advice from someone with experience.
Any help would be greatly appreciated!
cost of car insurance chicago
December 10, 2024 @ 6:41 pm
Lots of drivers are actually uncertain about the most ideal possibilities for auto
insurance policy in Chicago, yet studying may
trigger economical fees. It is vital to compare different policies when purchasing auto insurance coverage in Chicago to discover the absolute best coverage.
https://99ok.photography/
December 10, 2024 @ 6:45 pm
Thanks to my father who informed me concerning this weblog, this web site is genuinely awesome.
PinUp - 97
December 10, 2024 @ 6:57 pm
Pin Up Казино Онлайн в Казахстане – это одно из самых популярных мест для игроков, любящих азарт. Здесь вы найдёте широкий выбор для развлечений и крупных выигрышей. Если вы хотите найти проверенное казино, то лучший выбор для вас – Pin Up Casino.
Преимущества Pin Up Казино
Множество игр. В Pin Up Казино доступно более 2000 игровых автоматов, игр с живыми дилерами. В линейке игр представлены разработчики уровня Microgaming и множество других компаний. Это значит, что вы получите отличное качество игры, доступные на всех устройствах.
Разнообразные бонусы для игроков. Новые пользователи могут рассчитывать на стартовые бонусы, которые дадут отличный старт. Кроме того, действуют постоянные акции, кэшбэки и специальные предложения для лояльных игроков, что обеспечивает ещё больше шансов на выигрыш.
Наш сайт: https://www.atyrauspidcentre.kz/ru/habarlandyru-2022/obyavlenie-05-04-2022-g/
Безопасность и надежность. Pin Up Casino обеспечивает безопасность всех игроков персональных данных и выплат, чтобы гарантировать защиту данных. Лицензированная деятельность казино обеспечивает честность и выплаты и своевременные выплаты выигрышей.
Доступность на всех устройствах. Наше казино легко доступно через мобильные устройства, что даёт возможность играть в любые игры в любом уголке мира. Мобильное приложение готово к установке или просто использовать браузер для игры на смартфоне.
Поддержка клиентов. В любой ситуации, служба поддержки Pin Up Казино всегда к вашим услугам. Вы можете связаться с операторами через чат на сайте, и получить квалифицированную помощь на русском или казахском языке.
cost of tamoxifen without insurance
December 10, 2024 @ 7:02 pm
Drugs information sheet. What side effects?
cost of tamoxifen without insurance
All information about medicament. Read information now.
levaquin 500 mg for treatment
December 10, 2024 @ 7:03 pm
Levaquin 500mg used for
blood sugar
December 10, 2024 @ 7:16 pm
Hello, just wanted to say, I liked this post. It was inspiring.
Keep on posting!
Social - paibe
December 10, 2024 @ 7:24 pm
What Makes Online Casinos Remain Highly Preferred Worldwide
Virtual gambling platforms have revolutionized the betting landscape, providing an exceptional degree of user-friendliness and variety that land-based establishments don’t provide. Recently, a large audience worldwide have welcomed the excitement of virtual gambling due to its always-open nature, engaging traits, and progressively larger catalogs of games.
One of the main appeals of digital gambling sites is the unparalleled selection of choices on offer. Whether you love rolling retro fruit machine slots, playing through narrative-rich thematic slots, or testing your strategy in traditional table offerings like Roulette, internet-based gambling sites provide endless choices. Plenty of operators even include real-time gaming experiences, giving you the chance you to connect with human game hosts and gaming peers, all while enjoying the authentic feel of a real casino right at home.
If you’re new with the world of online gaming or would like to find out more about reputable operators, why not participate in our lively online hub? It’s a platform where enthusiasts offer stories, guiding you to maximize your gambling adventure. Dive into the discussions and see it here now: https://t.me/usykvs_fury .
Beyond variety, digital casino services are known for availability.
Diablo Rug
December 10, 2024 @ 7:35 pm
Here’s a spun introduction for a game lover:
As someone who loves gaming, I’ve spent endless time diving into my favorite games and creating immersive environments.
Over the last few years, I’ve been building game-themed gaming rooms, drawing inspiration from
legendary games like Mario, Nintendo, Zelda, and The Witcher 3.
It’s been an amazing adventure, blending creativity with my love for games.
To all fellow gamers, I suggest adding touches like game rugs, wall art, and custom lighting to make your setup truly epic.
These items create a themed atmosphere.
Whatever your gaming style, a themed gaming room makes every play session special.
Create your dream setup!
Here is my web blog Diablo Rug
Sazraho
December 10, 2024 @ 7:42 pm
Как получить диплом о среднем образовании в Москве и других городах
Jamescaurf
December 10, 2024 @ 7:47 pm
Kamagra tablets: Kamagra 100mg price – Kamagra Oral Jelly
forklift
December 10, 2024 @ 8:17 pm
I like the helpful info you provide in your articles. I’ll bookmark your
blog and check again here regularly. I am quite certain I’ll learn plenty of new
stuff right here! Good luck for the next!
Social - paibe
December 10, 2024 @ 8:30 pm
Why Online Casinos Are Highly Preferred Worldwide
Digital casinos have transformed the casino gaming industry, offering an unmatched level of comfort and variety that physical casinos are unable to replicate. Recently, a large audience internationally have adopted the adventure of online gaming thanks to its availability, captivating elements, and progressively larger game libraries.
One of the strongest selling points of online casinos is the unparalleled range of entertainment options provided. Whether you like rolling traditional reel games, exploring theme-based visual slot games, or mastering skills in classic casino games like Blackjack, virtual venues feature endless possibilities. A large number of platforms additionally feature live casino options, giving you the chance you to participate with real dealers and opponents, all while enjoying the lifelike ambiance of a real casino in your own space.
If you’re new with the world of internet-based gaming or hope to discover reliable sites, why not join our lively gaming forum? It’s a space where enthusiasts discuss stories, assisting you to improve your online casino experience. Dive into the connections and learn more now: https://www.instagram.com/fortunegems_top/ .
Beyond variety, internet-based gambling hubs stand out seamless entry.
Plinko - kp
December 10, 2024 @ 8:39 pm
Plinko game is een van de leukste kansspelen die de laatste tijd online zijn verschenen. Het spel zelf, dat zijn oorsprong vindt in het tv-programma The Price Is Right, heeft zich ingevoerd in de digitale gokmarkt.
In de komende alinea’s gaan we dieper in op alles wat je leert over deze spannende game, van de grondslagen van het spel tot hoe je je kunt inzetten met echt geld en de beste strategieen om het spel te spelen.
Web: http://www.cobra1eleven.com/plinko-spel-gratis-bij-nederlandse-online-casinos/
Plinko is een eenvoudig maar spannend casinospel dat bekend is geworden door de tv-show ‘The Price Is Right’. Het spel bestaat uit een verticaal bord met een aantal hobbels waar een bal van bovenaf doorheen naar beneden rolt. De bal bounced van de pinnen en komt neer in een van de doelen, die elk een bepaald bedrag tonen. De uitbetaling is afhankelijk van de bal vastkomt. Dit betekent dat het een spel gebaseerd op geluk is, waarbij spelers niet altijd kunnen raden waar de bal terechtkomt.
Hoewel de grondslagen van het spel simpel lijken, maakt de onzekerheid van het spel het leerzaam en leuk. Dit is een van de componenten waarom Plinko zo beroemd is geworden. Het wordt vaak aangeboden als een online Plinko spel in verschillende online gaming platforms, waar spelers kunnen proberen om geld te verdienen door te in te zetten op de uitkomst van hun spelballen.
Wanneer je een Plinko casino game speelt, lijkt het spel nagenoeg hetzelfde als de versie die je op tv kunt zien. De verschillen die bestaan liggen in de wijze waarop je inzetten kan en het feit dat je met echt geld kunt spelen. In plaats van voor aandachtspunten zoals in de televisie-uitzending, kun je in een casino op internet geld inzetten. De uitbetalingen worden beinvloed door het uitkomst waarin de bal terechtkomt afhankelijk van de plaats van de bal.
Je kunt je eigen inzet bepalen, en naar gelang je inzet kunnen de winsten fluctueren. De Plinko spel op het internet wordt vaak voorzien van een eenvoudige interface, wat het voor onervaren spelers makkelijker maakt om het spel te leren. Veel online casino’s bieden een Plinko game download optie, zodat je het spel kunt spelen op je smartphone, zelfs zonder internetverbinding. Dit biedt meer vrijheid en maakt het spel openeerder.
human-designgaq
December 10, 2024 @ 8:53 pm
Ценности влияют на все выборы, которые мы делаем в жизни. Понимание того, как формируются ценности, позволит более точечно позиционировать свои товары и услуги, тем самым успешно развиваться на рынке.
https://human-design-slovar.rappro.ru
cost generic lyrica prices
December 10, 2024 @ 8:58 pm
Meds information. Generic Name.
cost generic lyrica prices
Everything trends of drugs. Read information here.
where can i buy tetracycline ointment
December 10, 2024 @ 8:59 pm
tetracycline capsules where to buy
Social - paibe
December 10, 2024 @ 9:31 pm
Why Online Casinos Remain So Popular
Internet-based gambling hubs have reshaped the casino gaming scene, providing a unique kind of accessibility and range that traditional establishments are unable to replicate. In recent years, millions of players worldwide have welcomed the pleasure of internet-based gaming as a result of its accessibility, captivating elements, and widening catalogs of games.
One of the key draws of digital gambling sites is the sheer diversity of entertainment options available. Whether you are a fan of spinning vintage slots, playing through story-driven thematic slots, or testing your strategy in traditional table offerings like Blackjack, online platforms feature endless possibilities. Several sites also introduce interactive dealer games, giving you the chance you to participate with actual dealers and other players, all while enjoying the authentic atmosphere of a brick-and-mortar establishment without leaving your home.
If you’re exploring for the first time with the world of online gaming or hope to learn about reliable sites, why not join our growing gaming forum? It’s a destination where fans share experiences, enabling you to get the most out of your casino activities. Join the community and start your journey now: https://t.me/fight_usyk_fury .
Beyond variety, digital casino services shine availability.
nude girls
December 10, 2024 @ 9:31 pm
Great delivery. Great arguments. Keep up the amazing spirit.
Leonardsiz
December 10, 2024 @ 9:39 pm
кухни под заказ – Уникальные и практичные решения для вашей кухни.
Dnrtlbp
December 10, 2024 @ 9:52 pm
Сколько стоит диплом высшего и среднего образования и как его получить?
Baywin - tn
December 10, 2024 @ 10:20 pm
Bahis Platformu Baywin, bahis dunyas?n?n dijital yuzunde one c?kan bir web sitesidir. Oyuncular?na sundugu bol oyun turleri, kolay giris olanaklar? ve guven veren hizmeti ile sektorde fark yaratmaktad?r.
Ozellikle Baywin giris bilgileri ve guncel giris adresleri, uyeler icin dikkat ceken unsurlar aras?nda yer vard?r.
Baywin’in Temel Ozellikleri
BayWin, cevrimici bahis ve casino oyunlar? alan?nda hizmet sunan bir hizmet saglay?c?d?r. tenis bahisleri, sans oyunlar?, dijital oyunlar gibi cok say?da oyun secenegine sahiptir.
Baywin’in en onemli ozelliklerinden biri, uyelerine cazip oranlar sunmas?d?r. Ayr?ca, kolay ve h?zl? odeme yontemleri, maddi kazanclar? kolayca yonetmeyi mumkun k?lar.
Baywin Erisim Ad?mlar?
Web: https://www.cintell.net/resources/help-center/match-a-list-of-contacts-to-smartpersonas-in-cintell/
Baywin’in erisim k?s?tlamalar?yla kars?lasmas? kac?n?lmazd?r, bununla birlikte engellemeler oldugunda Bahis sitesi yetkilileri h?zl?ca harekete gecmektedir.
Siteye erisim engellendiginde, yeni adres olusturularak erisim saglan?r. Bu yontemle, yeni giris URL’si uzerinden erisim sorunu cozulur.
Siteye giris yapmak icin mobil ve masaustu erisim saglanabilir. Tabletler, her turlu mobil cihaz ve desktop cihazlar uzerinden Baywin’e giris yap?labilir. kullan?c?lar icin erisim konforu sunar.
human-designsxn
December 10, 2024 @ 10:37 pm
Гораздо большего можно добиться при помощи доброго слова и пистолета, нежели при помощи одного лишь доброго слова
https://human-design-slovar.rappro.ru
Отдых и обучение детей в Павлово
December 10, 2024 @ 10:39 pm
I really like what you guys tend to be up too. This kind of clever work and coverage!
Keep up the awesome works guys I’ve included you guys to my own blogroll.
Social - paibe
December 10, 2024 @ 10:39 pm
How Online Casinos Have Become Highly Preferred Worldwide
Internet-based gambling hubs have changed the betting market, offering an unmatched level of ease and selection that brick-and-mortar gambling houses fall short of. Recently, a vast number of enthusiasts around the world have turned to the adventure of online gaming because of its always-open nature, appealing qualities, and constantly growing catalogs of games.
One of the biggest attractions of digital gambling sites is the unparalleled variety of entertainment options ready to play. Whether you prefer rolling retro slot machines, immersing yourself in theme-based modern slot games, or mastering skills in strategy-based games like Texas Hold’em, casino websites boast infinite entertainment avenues. Plenty of operators additionally feature real-time gaming experiences, enabling you to participate with real dealers and fellow gamblers, all while soaking in the realistic vibes of a real casino right at home.
If you’re just starting with the world of virtual casino play or want to delve deeper into safe services, why not become part of our active online hub? It’s a platform where gaming aficionados post insights, guiding you to enhance your casino activities. Join the discussions and visit us now: https://www.facebook.com/profile.php?id=61569360522200 .
In addition to diversity, virtual gambling platforms stand out accessibility.
buy cheap zithromax for sale
December 10, 2024 @ 10:55 pm
Drug information for patients. Short-Term Effects.
buy cheap zithromax for sale
Everything what you want to know about medicament. Get information here.
where can i buy cabgolin
December 10, 2024 @ 10:57 pm
can you buy cabgolin no prescription
sex trẻ em
December 10, 2024 @ 11:02 pm
I wanted to thank you for this good read!! I certainly loved every bit of it.
I have got you bookmarked to look at new things you post…
Social - paibe
December 10, 2024 @ 11:43 pm
What Makes Online Casinos Are a Global Phenomenon
Virtual gambling platforms have transformed the gambling industry, delivering an unmatched level of convenience and selection that land-based establishments fall short of. Over time, a growing community globally have chosen the adventure of online gaming thanks to its ease of access, engaging traits, and continuously increasing game libraries.
One of the biggest attractions of internet-based platforms is the incredible selection of gaming experiences on offer. Whether you love spinning traditional fruit machine slots, playing through narrative-rich thematic slots, or mastering skills in strategy-based games like Texas Hold’em, online platforms boast numerous entertainment avenues. Several sites moreover offer live casino options, making it possible for you to connect with actual dealers and other players, all while enjoying the engaging ambiance of a brick-and-mortar establishment in your own space.
If you’re unfamiliar with the world of virtual gambling or seek to explore reliable sites, why not sign up for our active community? It’s a platform where gamblers offer experiences, enabling you to enhance your gambling adventure. Dive into the community and start your journey now: https://x.com/_fury_usyk .
Adding to the extensive catalog, digital casino services are known for availability.
pitchwall.co
December 10, 2024 @ 11:47 pm
Une fois que vous vous sentirez prêt à franchir le cap de
faire des placements avec de l’argent réel, vous aurez déjà acquis le nécessaire
pour vous en sortir.
Why is Auto Glass So Expensive?
December 11, 2024 @ 12:00 am
Every weekend i used to visit this web page, because i
wish for enjoyment, since this this web site conations in fact
nice funny data too.
1 Win - 5k
December 11, 2024 @ 12:04 am
Один Вин — Надежное онлайн-казино и букмекерская контора с отличными условиями для ставок
В отрасли онлайн-казино и ставок в спорте важно не только качество предоставляемых услуг, но и высокий уровень доверия. С лицензией, которая подтверждает легальность, Один Вин установил себя как фаворитов в мире онлайн-гемблинга. Организация ориентирована на развитие клиентского опыта, постоянно улучшая свои продукты и сервисы, чтобы обогатить пользователям непревзойденный опыт.
Приветственный бонус: Начальный бонус для новичков казино
Для новых пользователей на платформе 1win предусмотрен выгодный приветственный бонус, который позволяет начать с уверенностью. Это акция является одним из лучших на на платформе, так как позволяет новым пользователям начать игру с бонусом, повысив вероятность успеха. Регистрация за несколько минут и внесение депозита обеспечивают получение бонуса, что делает его применение максимально удобным и быстрым.
Live-ставки: Играйте на события вживую с моментальными ставками
В пространстве спортивных прогнозов фундаментальным аспектом является темп и умение быстро принимать решения в моментально. 1win приглашает множество спортивных событий, доступных в формате ставок в лайве. Мы предоставляем доступ к ведущим спортивным событиям, будь то футбол.
Наш сайт: http://kums-barnaul.ru/opennews.php?ns=2914&id_page=6
Ставки в реальном времени дают шанс мгновенно реагировать на изменения, что делает процесс более динамичным, но и интеллектуально интересным. Используя стратегии и аналитику платформы, игроки могут принимать более обоснованные решения, оптимизируя стратегию. Каждое событие представлено с комментариями, что способствует точным прогнозам выбирать тактику исходя из фактов.
Игровые автоматы: Тестирование бесплатно и максимум возможностей над игровым процессом
Важная черта онлайн-казино 1-Win — это возможность демо-режима игровых автоматов, что дает возможность испытать автоматы без рисков потерь. Это важно для новых пользователей, которые ещё только начинают игру или механикой слотов, а также для продвинутых игроков, которые хотят изучить игровые автоматы или аналитику игры.
Каждый автомат произведён крупнейшими провайдерами, такими как Yggdrasil, что гарантирует лучшую графику, звуковые эффекты и геймплей. Благодаря качеству игр, любители казино могут выбирать слоты, которые соответствуют их стилю и предпочтениям, от ретро-слотов до новых игровых решений и бонусными раундами.
human-designyin
December 11, 2024 @ 12:12 am
Гораздо большего можно добиться при помощи доброго слова и пистолета, нежели при помощи одного лишь доброго слова
https://human-design-slovar.rappro.ru
CharlesSok
December 11, 2024 @ 12:21 am
Портал Дай Жару- более 70000 посетителей в месяц! Подбор саун и бань в Москвы и Подмосковья с телефонами, фото и ценами. Недорогие финские сауны, русские бани, турецкие парные лучшие бани
where can i get tadacip without dr prescription
December 11, 2024 @ 12:50 am
Drugs information for patients. Drug Class.
where can i get tadacip without dr prescription
Actual information about medicine. Get information here.
Derekapaky
December 11, 2024 @ 12:52 am
http://huskytaxi.ru – надежный партнер в создании кухонь на заказ.
Social - paibe
December 11, 2024 @ 12:52 am
What Makes Online Casinos Are Highly Preferred Worldwide
Online casinos have revolutionized the betting scene, offering a level of ease and range that brick-and-mortar gambling houses are unable to replicate. Over the past decade, a vast number of enthusiasts worldwide have welcomed the pleasure of digital casino play thanks to its availability, appealing qualities, and progressively larger catalogs of games.
One of the strongest selling points of online gaming options is the vast array of choices available. Whether you are a fan of rolling retro one-armed bandits, playing through story-driven thematic slots, or exercising tactics in strategy-based games like poker, casino websites provide infinite choices. Several sites moreover present interactive dealer games, giving you the chance you to participate with live hosts and opponents, all while experiencing the engaging ambiance of a real casino from anywhere you want.
If you’re a beginner with the world of virtual casino play or want to learn about reliable sites, why not sign up for our lively gaming forum? It’s a destination where gaming aficionados exchange insights, guiding you to improve your gaming journey. Dive into the community and visit us now: https://www.instagram.com/fortunegems_top/ .
Besides the wide selection, virtual gambling platforms are known for constant connectivity.
Lazrohc
December 11, 2024 @ 1:16 am
Как безопасно купить диплом колледжа или ПТУ в России, что важно знать
PinUp - bo
December 11, 2024 @ 1:46 am
Pin Up Казино Онлайн в Казахстане – это лучший выбор среди казино для ценителей азарта. Здесь предоставляются отличные условия для настоящего игрового опыта. Если вас интересует надёжное казино, то Pin Up Казино создано для вас.
Преимущества казино Pin Up
Большой ассортимент игр. В Pin Up Казино доступно более двух тысяч игровых автоматов, игр с живыми крупье и автоматов. В ассортименте представлены лидеры игровой индустрии, такие как Microgaming и многие другие. Это значит, что каждая игра адаптирована для любого устройства, удобные для игры на мобильных.
Бонусы для всех игроков. Новые игроки получают приветственные бонусы, которые помогут стартовать быстрее. Постоянные акции и розыгрыши тоже доступны, ежедневные розыгрыши и фриспины для постоянных игроков, что обеспечивает ещё больше шансов на выигрыш.
Наш сайт: https://daz-kasachstan.net/news/?phrase_id=13951&PAGEN_2=5
Честность и надёжность. Pin Up интернет-казино использует современные технологии для защиты персональных данных и выплат, для полной защиты вашего опыта. Лицензия гарантирует гарантирует честную игру и мгновенные выплаты выигрышей.
Игры на любом устройстве. Наше казино оптимизировано для смартфонов, что даёт возможность игры в любое время в любом уголке мира. Есть приложение для мобильных или просто играть прямо в браузере на мобильном.
Поддержка клиентов. В любой проблеме, специалисты поддержки всегда готовы помочь вам. Вы можете пообщаться с поддержкой через чат и email, и специалисты решат проблему на русском или казахском языке.
Jacobgaugh
December 11, 2024 @ 1:58 am
фабрика кухня — Производственные кухни по индивидуальным проектам.
Social - paibe
December 11, 2024 @ 1:58 am
Why Online Casinos Have Become Highly Preferred Worldwide
Digital casinos have revolutionized the gaming industry, offering an unmatched level of user-friendliness and range that brick-and-mortar establishments struggle to rival. Recently, a growing community globally have adopted the thrill of online gaming as a result of its accessibility, exciting features, and constantly growing game libraries.
One of the key draws of digital gambling sites is the incredible variety of titles on offer. Whether you enjoy spinning vintage slots, diving into narrative-rich thematic slots, or testing your strategy in card and board games like Blackjack, internet-based gambling sites feature numerous options. Many casinos also offer live gaming streams, giving you the chance you to participate with real dealers and other players, all while taking in the authentic ambiance of a traditional gambling venue right at home.
If you’re a beginner with the world of online gaming or are looking to find out more about safe services, why not join our lively social network? It’s a platform where gamblers exchange tips, enabling you to maximize your virtual play. Join the community and check it out now: https://t.me/tyson_fury_vs_usyk .
Adding to the extensive catalog, virtual gaming providers thrive in ease of access.
bong99
December 11, 2024 @ 2:06 am
Hi there friends, fastidious article and nice
arguments commented at this place, I am in fact enjoying by these.
sex
December 11, 2024 @ 2:14 am
Yes! Finally someone writes about sex.
Angelhob
December 11, 2024 @ 2:18 am
фотосъемка ювелирных изделий – Высококачественная съемка ювелирных изделий для рекламы и каталогов.
Philipbat
December 11, 2024 @ 2:31 am
Каждая площадка предлагает уникальный набор функций, которые могут включать разнообразные форматы, бонусные программы и техническую поддержку. Однако не все из них способны соответствовать высоким стандартам пользователей http://www.ilmarhit.it/component/kunena/3-suggestion-box/437049?Itemid=0#437049
Leonardsiz
December 11, 2024 @ 2:34 am
кухни на заказ спб – Найдите идеальное решение для кухни в Санкт-Петербурге с индивидуальным подходом.
criminal
December 11, 2024 @ 3:00 am
I’ve been browsing online more than 4 hours today, yet I never found any interesting article like yours.
It is pretty worth enough for me. In my opinion, if all website
owners and bloggers made good content as you did, the
internet will be much more useful than ever before.
Social - paibe
December 11, 2024 @ 3:11 am
The Reasons Behind Why Online Casinos Are Highly Preferred Worldwide
Digital casinos have reshaped the casino gaming industry, offering a unique kind of user-friendliness and selection that physical gambling houses struggle to rival. Over the past decade, a large audience around the world have chosen the adventure of virtual casinos because of its accessibility, appealing qualities, and continuously increasing range of offerings.
One of the most compelling reasons of internet-based platforms is the sheer variety of titles provided. Whether you are a fan of interacting with retro fruit machine slots, exploring plot-filled modern slot games, or mastering skills in classic casino games like poker, online platforms boast limitless entertainment avenues. Many casinos furthermore present live casino options, letting you to participate with live hosts and opponents, all while immersing yourself in the engaging atmosphere of a real casino from anywhere you want.
If you’re unfamiliar with the world of internet-based gaming or are looking to discover safe services, why not participate in our lively online hub? It’s a platform where gamblers exchange tips, helping you to get the most out of your virtual play. Dive into the connections and check it out now: https://www.instagram.com/fortunegems_top/ .
Besides the wide selection, internet-based gambling hubs shine seamless entry.
diuwin login
December 11, 2024 @ 3:14 am
Excellent article! We are linking to this particularly great post on our website.
Keep up the good writing.
Plinko - xt
December 11, 2024 @ 3:25 am
Plinko casino spel is zeer populair entertainmentvormen die de afgelopen jaren beschikbaar zijn gekomen op het internet. Deze casinogame, oorspronkelijk ontstaan in de televisiehit The Price Is Right, heeft zich met succes aangepast aan de digitale gokmarkt.
In de komende alinea’s gaan we dieper in op alles wat je kunt leren over deze casinogame, van de grondslagen van het spel tot hoe je het kunt spelen voor echt geld en de leukste tactieken om het spel te spelen.
Web: https://drvieiraplastica.com.br/plinko-spelen-in-nederland-online-casino-opties/
Plinko is een simpel maar boeiend spel van geluk dat bekend is geworden door de populaire tv-show. Het spel bestaat uit een verticaal bord met een aantal hobbels waar een speelbal van bovenaf doorheen valt naar beneden. De bal bounced van de pinnen en komt neer in een van de vakken aan de onderkant, die elk een bepaald winstbedrag tonen. De uitbetaling is gerelateerd aan de bal terechtkomt. Dit betekent dat het een game van toevallige uitkomst is, waarbij spelers niet kunnen voorspellen waar de bal zal vallen.
Hoewel de basisstructuur van het spel simpel zijn, maakt de onvoorspelbaarheid van het spel het boeiend en uitdagend. Dit is een van de onderdelen waarom Plinko zo’n hit is in online casinos. Het wordt vaak aangeboden als een online versie van het spel in verschillende online casino’s, waar spelers geld kunnen winnen door te spelen op de uitkomst van hun vallende ballen.
Wanneer je deelneemt aan een Plinko casinospel, lijkt het spel vaak hetzelfde als de tv-versie van het spel. De verschillen liggen in de manier waarop je kans hebt om te wedden en het feit dat je kan inzetten met echt geld. In plaats van voor aandachtspunten zoals in de televisie-uitzending, kun je in een online casino inzetten met echt geld. De beloningen worden beinvloed door het vakje waarin de bal komt wordt berekend.
Je kunt je eigen inzet bepalen, en afhankelijk van de hoogte van je inzet kunnen de uitbetalingen varieren. De Plinko online spel wordt vaak gebruiker-vriendelijke versie geboden, wat het voor onervaren spelers makkelijker maakt om het spel te proberen. Veel digitale casino’s bieden een Plinko game download optie, zodat je het spel kunt spelen op je mobiel, zelfs zonder internet toegankelijk. Dit geeft meer flexibiliteit en maakt het spel meer bereikbaar.
onexrussiasite
December 11, 2024 @ 3:25 am
Всегда держите под рукой 1xslots рабочее зеркало для стабильного доступа.
Mahjong Wins 3
December 11, 2024 @ 3:29 am
you are in reality a good webmaster. The website loading velocity is amazing.
It seems that you are doing any unique trick. Furthermore, The
contents are masterwork. you’ve done a great activity on this subject!
MNYTREDF
December 11, 2024 @ 3:35 am
Very descriptive blog, I enjoyed that bit. Will there be a part 2?
Ангары
December 11, 2024 @ 3:38 am
Производство ангаров и складов под ключ.
Подробная информация и заказ быстровозводимый складов
Ремонт телевизоров Hisense
December 11, 2024 @ 4:05 am
Предлагаем услуги профессиональных инженеров офицальной мастерской.
Еслли вы искали ремонт телевизоров hisense рядом, можете посмотреть на сайте: ремонт телевизоров hisense цены
Наши мастера оперативно устранят неисправности вашего устройства в сервисе или с выездом на дом!
Social - paibe
December 11, 2024 @ 4:19 am
What Makes Online Casinos Are Becoming a Global Phenomenon
Online casinos have reshaped the gambling industry, providing an unmatched level of accessibility and range that brick-and-mortar establishments struggle to rival. Recently, a vast number of enthusiasts internationally have welcomed the excitement of virtual casinos due to its anytime, anywhere convenience, exciting features, and widening selection of games.
One of the biggest attractions of online gaming options is the incredible range of gaming experiences ready to play. Whether you love playing on old-school slot machines, exploring story-driven visual slot games, or exercising tactics in table games like Texas Hold’em, online platforms feature numerous possibilities. Numerous services furthermore present interactive dealer games, making it possible for you to communicate with real dealers and fellow gamblers, all while soaking in the authentic feel of a real casino from the comfort of your home.
If you’re just starting with the world of digital casinos or want to discover safe services, why not engage with our vibrant community? It’s a hub where gaming aficionados share experiences, assisting you to maximize your gambling adventure. Discover the connections and visit us now: https://www.pinterest.com/fury_usyk_date/ .
Apart from the game range, internet-based gambling hubs stand out availability.
trang chu thabet
December 11, 2024 @ 4:22 am
Admiring the time and effort you put into your site and detailed information you
present. It’s awesome to come across a blog every once in a while
that isn’t the same unwanted rehashed material. Wonderful read!
I’ve saved your site and I’m including your RSS
feeds to my Google account.
Josephsheah
December 11, 2024 @ 4:33 am
betboo plus: gГјvenli siteler – ilk giriЕџte bonus veren bahis siteleri
JustinBoIli
December 11, 2024 @ 4:34 am
https://casinositeleri2025.pro/# en iyi yatД±rД±m siteleri
human-designvnb
December 11, 2024 @ 4:38 am
Гораздо большего можно добиться при помощи доброго слова и пистолета, нежели при помощи одного лишь доброго слова
https://human-design-slovar.rappro.ru
diuwin
December 11, 2024 @ 4:47 am
Actually no matter if someone doesn’t know after that its up to other visitors that they will assist, so here it occurs.
30732.ru
December 11, 2024 @ 4:49 am
“КОМТЭК”– ваш партнер в создании безопасных и надежных трубопроводов.
sexporno
December 11, 2024 @ 4:59 am
sex
Baywin - fc
December 11, 2024 @ 5:04 am
Bay Win, cevrimici bahis dunyas?nda ad?ndan s?kca soz ettiren bir uygulamad?r. Oyuncular?na sundugu bol oyun turleri, kolay giris olanaklar? ve saglam altyap?s? ile kullan?c?lar? kendine cekmektedir.
Bilhassa Baywin erisim yollar? ve son giris adresleri, uyeler icin temel merak noktalar? aras?nda yer bulunmaktad?r.
Baywin Hakk?nda Genel Bilgiler
Baywin, cevrimici bahis ve casino oyunlar? alan?nda tan?nan bir uygulamad?r. canl? bahisler, casino oyunlar?, sanal sporlar gibi genis bir oyun yelpazesine sahiptir.
Platformun en onemli avantajlar?ndan biri, kullan?c?lar?na yuksek oranlar sunmas?d?r. Ayr?ca, kolay ve h?zl? odeme yontemleri, maddi kazanclar? kolayca yonetmeyi mumkun k?lar.
Baywin Giris Sorunlar? ve Cozumleri
Web: https://doras.dcu.ie/15010/
Bu bahis sitesinin erisim engellemelerinden etkilenmemesi mumkun degildir, bu gibi engeller kars?s?nda Baywin’in teknik ekibi kullan?c?lar?n? magdur etmemektedir.
Adres engeli olustugunda, Baywin guncel giris adresini hemen aktif eder ve paylas?r. Bu sayede, yeni giris URL’si uzerinden kullan?c?lar oyunlar?n? kesintisiz oynayabilir.
Yeni adrese erisim saglamak amac?yla mobil ve masaustu erisim saglanabilir. Ak?ll? telefonlar, tabletler ve PC’ler uzerinden Baywin’e giris yap?labilir. Baywin’in her yerden erisilebilirligini art?r?r.
how to buy augmentin online
December 11, 2024 @ 5:06 am
how to buy augmentin prices
Robertmoica
December 11, 2024 @ 5:13 am
пин ап казино [url=http://pinup2025.com/#]пин ап казино зеркало[/url] пин ап казино
Social - paibe
December 11, 2024 @ 5:28 am
Reasons Why Online Casinos Remain Highly Preferred Worldwide
Digital casinos have changed the gaming scene, delivering an exceptional degree of accessibility and variety that traditional venues can’t match. Recently, a growing community globally have adopted the thrill of virtual casinos in light of its ease of access, captivating elements, and continuously increasing selection of games.
One of the most compelling reasons of online casinos is the unparalleled diversity of games on offer. Whether you enjoy interacting with retro slots, exploring plot-filled modern slot games, or mastering skills in strategy-based games like Roulette, casino websites feature limitless possibilities. Numerous services furthermore include real-time gaming experiences, making it possible for you to connect with real dealers and fellow gamblers, all while enjoying the engaging atmosphere of a traditional gambling venue in your own space.
If you’re a beginner with the world of digital casinos or would like to learn about reliable sites, why not join our dynamic online hub? It’s a hub where fans post tips, helping you to enhance your gambling adventure. Discover the connections and learn more now: https://www.facebook.com/profile.php?id=61569360522200 .
Adding to the extensive catalog, virtual gambling platforms thrive in ease of access.
Mazrnjx
December 11, 2024 @ 5:30 am
Купить диплом судоводителя
human-designvxg
December 11, 2024 @ 5:56 am
Гораздо большего можно добиться при помощи доброго слова и пистолета, нежели при помощи одного лишь доброго слова
https://human-design-slovar.rappro.ru
aberdeen escorts
December 11, 2024 @ 6:04 am
When some one searches for his required thing, therefore he/she
desires to be available that in detail, so that thing is maintained over here.
bdg game
December 11, 2024 @ 6:26 am
This info is invaluable. How can I find out more?
Social - paibe
December 11, 2024 @ 6:33 am
Reasons Why Online Casinos Have Become a Worldwide Trend
Internet-based gambling hubs have changed the casino gaming market, delivering an unmatched level of comfort and variety that traditional casinos struggle to rival. Throughout the last ten years, millions of players globally have turned to the excitement of digital casino play thanks to its anytime, anywhere convenience, engaging traits, and progressively larger catalogs of games.
One of the strongest selling points of digital gambling sites is the vast diversity of titles ready to play. Whether you prefer rolling classic one-armed bandits, diving into engaging modern slot games, or exercising tactics in table games like Texas Hold’em, online platforms feature countless possibilities. Many casinos additionally offer live dealer games, giving you the chance you to communicate with human game hosts and fellow gamblers, all while taking in the authentic atmosphere of a traditional gambling venue right at home.
If you’re exploring for the first time with the world of virtual casino play or seek to find out more about safe services, why not join our lively interactive platform? It’s a destination where fans share tips, assisting you to improve your gambling adventure. Dive into the discussions and learn more now: https://www.pinterest.com/furyvsusyk/ .
Apart from the game range, virtual gaming providers are known for accessibility.
EdwardSit
December 11, 2024 @ 6:34 am
http://pinup2025.com/# пинап казино
Daviddow
December 11, 2024 @ 6:37 am
Babu88: Online casino and bookmaker for sports betting in Bangladesh babu88 sign up
Jacobgaugh
December 11, 2024 @ 6:45 am
кухни в москве — Лучшие предложения по кухонной мебели в Москве.
1 Win - p1
December 11, 2024 @ 6:45 am
One Win — Надежное игровое казино и букмекерская контора с привлекательными условиями для клиентов
В отрасли онлайн-казино и ставочных рынков ключевое значение имеет не только качество обслуживания, но и доверие к оператору. С гарантированной лицензией, которая гарантирует безопасность, Один Вин считает себя лидеров среди платформ для азартных игр. Провайдер уделяет внимание максимального комфорта, обновляя платформу, чтобы дать пользователям максимальный комфорт.
Приветственный бонус: Начальный бонус для клиентов
Для новых игроков на платформе 1win доступен впечатляющий приветственный бонус, который увеличивает ваши возможности. Это подарок является одним из заметных на среди конкурентов, так как открывает возможность новым клиентам казино сразу получить доступ к дополнительным средствам, сократив риски. Легкий старт и первый взнос автоматически активируют бонус, что делает его воспользование простым и доступным.
Live-ставки: Лайв-ставки на тысячи событий с моментальными ставками
В пространстве ставок на спорт ключевой особенностью является темп и способность мгновенно реагировать в реальном времени. 1win приглашает широкий выбор спортивных событий, доступных в формате игры вживую. Мы гарантируем доступ к ключевым соревнованиям и матчам, будь то хоккей.
Наш сайт: http://csdf.ucoz.net/index/dokumenty/0-8
Ставки в реальном времени предоставляют возможность быстро подстраиваться, что делает процесс более захватывающим, но и интеллектуально интересным. Используя передовые аналитические инструменты платформы, игроки платформы могут точнее прогнозировать исходы, оптимизируя стратегию. Каждое событие обеспечено аналитическими обзорами, что обогащает принятие решений предупреждать проигрыши.
Игровые автоматы: Бесплатная версия и свобода выбора над выбором игры
Главная особенность онлайн-казино 1вин — это доступность демо-доступа игровых автоматов, что дает возможность протестировать любой игровой автомат без вложения своих средств. Это очень удобно для новичков, которые ещё не привыкли к интерфейсу или спецификой слотов, а также для опытных пользователей, которые хотят найти новые игры или новые решения.
Каждый автомат произведён крупнейшими провайдерами, такими как NetEnt, что гарантирует лучшую графику, звуковые эффекты и механику слотов. С таким качеством, любители казино могут найти игры, которые удовлетворяют запросы игроков, от классических игр до игр с бонусами и специальными раундами.
101 game
December 11, 2024 @ 6:59 am
This paragraph presents clear idea for the new users of
blogging, that in fact how to do blogging.
Leonardsiz
December 11, 2024 @ 7:28 am
кухни на заказ в спб — Закажите кухню мечты в Санкт-Петербурге с учетом всех ваших предпочтений.
Social - paibe
December 11, 2024 @ 7:41 am
Why Online Casinos Have Become Highly Preferred Worldwide
Digital casinos have changed the gaming world, providing a level of user-friendliness and diversity that conventional gambling houses struggle to rival. Throughout the last ten years, a large audience worldwide have embraced the pleasure of online gaming because of its always-open nature, engaging traits, and continuously increasing game libraries.
One of the key draws of virtual gambling hubs is the unparalleled selection of games ready to play. Whether you prefer playing on traditional slot machines, diving into plot-filled video-based games, or mastering skills in classic casino games like Texas Hold’em, casino websites deliver infinite possibilities. Many casinos additionally feature live dealer games, making it possible for you to communicate with real dealers and gaming peers, all while taking in the immersive environment of a land-based casino in your own space.
If you’re unfamiliar with the world of virtual gambling or would like to explore reliable sites, why not sign up for our vibrant gaming forum? It’s a hub where gamblers exchange tips, assisting you to enhance your gaming journey. Discover the community and check it out now: https://www.facebook.com/profile.php?id=61569360522200 .
Besides the wide selection, online casinos excel seamless entry.
PinUp - in
December 11, 2024 @ 8:21 am
Pin Up Казино Онлайн в Казахстане – это лучший выбор среди казино для игроков, любящих азарт. Здесь предлагаются уникальные возможности для удачной игры и больших выплат. Если вы любите играть в хороших казино, то Pin Up Casino обеспечит всё нужное.
Преимущества онлайн казино Pin Up
Большой ассортимент игр. Игрокам предлагается более двух тысяч игровых автоматов, настольных игр и live-казино. В игровой коллекции представлены знаменитые игровые студии, включая Evolution Gaming и многие другие. Это значит, что каждая игра обеспечит захватывающий опыт, оптимизированные для телефонов и ПК.
Привлекательные акции и бонусы. Новые пользователи могут рассчитывать на увеличенные стартовые бонусы, которые дадут хорошие шансы на выигрыш. Постоянные акции и розыгрыши тоже доступны, кэшбэки и специальные предложения для лояльных игроков, что делает игру ещё более захватывающей и выгодной.
Наш сайт: https://kaz.nur.kz/society/1853598-ajygyp-ketkender-barsylyk-kiprdegi-kazak-kyzy-eldegi-kazirgi-ahualdy-baandady/
Высокий уровень безопасности. Pin Up Казино гарантирует полную безопасность ваших данных и транзакций, для обеспечения конфиденциальности. Казино работает по лицензии гарантирует честную игру и честные и быстрые выплаты.
Игры на любом устройстве. Pin Up работает на всех мобильных девайсах, что даёт возможность играть в любые игры в любом удобном месте. Мобильное приложение готово к установке или просто начать игру через мобильный браузер.
Поддержка клиентов. В любых трудностях, команда поддержки Pin Up всегда готова помочь. Вы можете связаться с нашими специалистами через телефон, и специалисты решат проблему на вашем языке.
bdg game
December 11, 2024 @ 8:38 am
Thank you, I have recently been searching for information approximately this topic for a while and yours is the best I’ve discovered till
now. However, what about the bottom line? Are you positive concerning the supply?
51 game register
December 11, 2024 @ 9:06 am
Helpful information. Lucky me I discovered your site by chance, and
I’m stunned why this twist of fate did not
came about in advance! I bookmarked it.
Ангары
December 11, 2024 @ 9:12 am
Производство ангаров и складов под ключ.
Подробная информация и заказ быстровозводимый складов
Derekapaky
December 11, 2024 @ 9:24 am
кухня на заказ в спб – идеальный вариант для тех, кто хочет создать уютный и современный интерьер.
Plinko - uz
December 11, 2024 @ 10:02 am
Plinko is een van de meest populaire uitdagingen die in de laatste jaren populair zijn geworden. Deze casinogame, geinspireerd door het tv-programma The Price Is Right, heeft zich geschaald naar de hedendaagse casinospellen.
In dit stuk behandelen we alles wat je kunt leren over de Plinko game, van de basisprincipes van het spel tot hoe je je kunt inzetten met echt geld en de beste tips voor het winnen.
Web: https://www.roxsymodas.cl/?paged=723&author=1
Het Plinko spel is een makkelijke, maar opwindende kansspel dat vaak wordt geassocieerd met het televisieprogramma The Price Is Right. Het spel bestaat uit een verticale speelstructuur met een aantal hobbels waar een bal van bovenaf doorheen valt naar beneden. De bal kaatst van de pinnen en valt een van de vakken, die elk een bepaald bedrag aanduiden. De winst is afhankelijk van de bal landt. Dit betekent dat het een spel van toevallige kansen is, waarbij spelers niet kunnen voorspellen waar de bal zal vallen.
Hoewel de basisregels van het spel simpel zijn, maakt de willekeurige aard van het spel het meeslepend en uitdagend. Dit is een van de onderdelen waarom Plinko in de gokwereld zo’n hype heeft veroorzaakt. Het wordt vaak aangeboden als een online versie van het spel in verschillende online goksites, waar spelers geld kunnen winnen door te gokken op de uitkomst van hun vallende ballen.
Als je een Plinko game speelt, lijkt het spel erg vergelijkbaar met de traditionele versie van Plinko. De afwijkingen liggen in het feit dat je inzetten kan en het feit dat je geld kunt winnen. In plaats van voor prijzen zoals in de showuitzending, kun je in een digitaal casino inzetten met echt geld. De vergoedingen zijn afhankelijk van het doel waarin de bal afkomt wordt berekend.
Je bepaalt hoeveel je speelt, en gebaseerd op je inzet kunnen de uitbetalingen varieren. De Plinko online versie van het spel wordt vaak voorzien van een eenvoudige interface, wat het voor beginnende spelers makkelijker maakt om het spel te leren. Veel gokplatforms bieden een Plinko game download optie, zodat je het spel kunt spelen op je mobiel, zelfs zonder internet toegankelijk. Dit geeft spelers meer vrijheid en maakt het spel toegankelijker.
Ремонт телевизоров Hisense Москва
December 11, 2024 @ 10:23 am
Предлагаем услуги профессиональных инженеров офицальной мастерской.
Еслли вы искали срочный ремонт телевизоров hisense, можете посмотреть на сайте: ремонт телевизоров hisense сервис
Наши мастера оперативно устранят неисправности вашего устройства в сервисе или с выездом на дом!
TravisTem
December 11, 2024 @ 10:46 am
pinup 2025: пин ап казино – пинап казино
escort
December 11, 2024 @ 10:46 am
Hello every one, here every person is sharing these kinds of experience,
thus it’s nice to read this weblog, and I used to pay a visit this website
all the time.
free credit no deposit
December 11, 2024 @ 10:48 am
Hello, yup this paragraph is really pleasant and I have learned lot of things from it regarding blogging.
thanks.
JustinBoIli
December 11, 2024 @ 10:48 am
https://slottr.top/# az parayla cok kazandiran slot oyunlar?
Auto insurance companies
December 11, 2024 @ 10:55 am
Car insurance companies typically compensate secure drivers with lower superiors.
Make sure to inquire car insurance companies regarding their safe
driver discount rates to decrease your total expenses.
Angelhob
December 11, 2024 @ 11:10 am
http://www.mirdolmenov.ru – Платформа для заказа профессиональной предметной фотосъемки.
login limit reached
December 11, 2024 @ 11:29 am
Asking questions are really fastidious thing if you are
not understanding something fully, but this post presents fastidious understanding yet.
Jacobgaugh
December 11, 2024 @ 11:33 am
gft-leasing.ru/ — Индивидуальный подход к созданию вашей кухни.
Baywin - 9n
December 11, 2024 @ 11:43 am
BayWin, online bahis sektorunde dikkat ceken bir uygulamad?r. Musterilerine sundugu zengin oyun icerikleri, h?zl? erisim avantaj? ve kaliteli hizmet sunumu ile sektorde fark yaratmaktad?r.
Bilhassa platforma erisim saglamak ve aktif baglant?lar, kullan?c? kitlesi ac?s?ndan en cok merak edilen konular aras?nda yer vard?r.
Baywin’in Hizmetleri
Bay Win, dijital bahis platformlar? aras?nda aktif olan bir bahis sitesidir. canl? bahisler, sans oyunlar?, sanal futbol gibi farkl? oyun imkanlar?na sahiptir.
Platformun en onemli dikkat ceken yonu, kullan?c?lar?na yuksek oranlar sunmas?d?r. Ayr?ca, kolay ve h?zl? odeme yontemleri, finansal guvenlik sunar.
Baywin Giris Adresi Nas?l Bulunur?
Web: https://www.cintell.net/company/contact-us/
Baywin markas?n?n engellemelerle kars?lasmas? olagand?r, ancak bu tur durumlar icin Site yoneticileri h?zl?ca harekete gecmektedir.
Erisim engellemeleri gerceklestiginde, kullan?c?lar icin alternatif bir baglant? sunulur. Bu yontemle, yeni giris URL’si uzerinden kullan?c?lar siteye giris yapabilir.
Platforma yeniden erisim icin bircok erisim yontemi bulunmaktad?r. Mobil cihazlar, tas?nabilir cihazlar ve dizustu bilgisayarlar uzerinden kullan?c?lar hesaplar?na ulasabilir. Bu da kullan?c?lar?n diledikleri yerden bahis yapma ozgurlugunu yasamalar?na olanak tan?r.
KJJGDFERKHFGD
December 11, 2024 @ 12:00 pm
Do you have a spam problem on this blog; I also am
a blogger, and I was curious about your situation; many of us have created some nice procedures and we are looking
to swap strategies with other folks, why not shoot me an e-mail if interested.
anti scratching cream
December 11, 2024 @ 12:26 pm
It’s awesome to pay a visit this web site and reading the views of
all mates about this post, while I am also zealous of getting familiarity.
Josephsheah
December 11, 2024 @ 12:36 pm
betnoo: 100 tl bonus veren bahis siteleri – yurtdД±ЕџД± bahis siteleri
Leonardsiz
December 11, 2024 @ 12:41 pm
кухни на заказ от производителя — Закажите кухню напрямую от производителя без переплат.
sex em gái của vợ
December 11, 2024 @ 12:55 pm
Howdy! This article couldn’t be written any better! Reading through this article reminds me
of my previous roommate! He always kept preaching about
this. I’ll forward this post to him. Pretty sure he’ll have
a great read. Many thanks for sharing!
cricket europe
December 11, 2024 @ 1:09 pm
Guar Seed is a fiber from the seed of the guar plant.
Guar gum is used as a laxative. It’s also used for treating diarrhea,
irritable bowel syndrome (IBS), obesity, and diabetes for
reducing cholesterol and for stopping hardening of the arteries (atherosclerosis).
Other names: Cyamopsis psoraloides, Cyamopsis tetragonoloba,
Cyamopsis tetragonolobus, Dietary Fiber, Dolichos psoraloides, Farine de Guar, Fibre Alimentaire, Goma Guar, Gomme de Guar, Gomme de Jaguar, Guar Flour, Indian Guar Plant, Jaguar Gum, Psoralea tetragonoloba.
Guar gum is a fiber that standardizes the dampness substance of the stool,
engrossing overabundance fluid within the runs, and softening the
stool in clogging. It likewise might help diminish the measure of cholesterol and glucose that is caught up in the stomach
and digestive organs. There is a few enthusiasm for utilizing guar
gum for weight reduction because it extends in the digestive tract, inflicting a feeling of completion. This will
likely diminish hunger.
Pinco Casino Официальный Сайт Вход Зеркало
December 11, 2024 @ 1:11 pm
Промокод Пинко Казино
Robertmoica
December 11, 2024 @ 1:15 pm
slot oyunlar? puf noktalar? [url=http://slottr.top/#]slot oyunlar? puf noktalar?[/url] az parayla cok kazandiran slot oyunlar?
1 Win - ik
December 11, 2024 @ 1:24 pm
1win — Безопасное цифровое казино и платформа для ставок с отличными условиями для любителей азартных игр
В отрасли интернет-казино и ставок на спорт приоритетное значение имеет не только качество игровой платформы, но и репутация компании. С лицензией, которая подтверждает легальность, 1вин находится среди лидеров среди платформ для азартных игр. Компания стремится к развитие клиентского опыта, разрабатывая новые функции, чтобы обеспечить пользователям непревзойденный опыт.
Приветственный бонус: Премиум-старт для пользователей платформы
Для новых клиентов на платформе 1win ожидает вас впечатляющий приветственный бонус, который значительно увеличивает начальный капитал. Это подарок является одним из выгодных на среди конкурентов, так как помогает новым пользователям сразу получить доступ к дополнительным средствам, получив дополнительные ресурсы для ставок. Удобная регистрация и первый взнос автоматически активируют бонус, что делает его применение комфортным.
Live-ставки: Играйте на события вживую с моментальными ставками
В отрасли ставок на спорт одним из ключевых элементов является быстрота реакции и способность адаптироваться в онлайн. 1win открывает путь широкий выбор спортивных событий, доступных в формате лайв-ставок. Мы располагаем матчами к топовым международным и локальным соревнованиям, будь то хоккей.
Наш сайт: https://zavgb37.ru/deyat/reading/6446.html
Игра в лайве предоставляют возможность мгновенно реагировать на изменения, что делает процесс ещё интереснее, но и тактически интересным. С помощью аналитики платформы, игроки могут увеличивать вероятность выигрыша, увеличивая потенциальные результаты. Каждое событие содержит анализ игры, что делает процесс ещё более обоснованным предсказывать результат с уверенностью.
Игровые автоматы: Демо-версия и полный контроль над игрой над автоматами
Одно из сильных сторон онлайн-казино One Win — это функция тестового режима игровых автоматов, что гарантирует игрокам потренироваться на автоматы без финансовых рисков. Это особенно полезно для новых игроков, которые ещё не привыкли к интерфейсу или требованиями игры, а также для опытных игроков, которые хотят изучить новые автоматы или новые способы игры.
Каждый автомат произведён крупнейшими провайдерами, такими как Pragmatic Play, что предоставляет отличную визуализацию, звуковую атмосферу и геймплей. С этим уровнем, любители казино могут выбирать слоты, которые соответствуют их стилю и предпочтениям, от ретро-слотов до новых игровых решений и бонусными опциями.
создание образовательной организации
December 11, 2024 @ 1:48 pm
создание образовательной организации
инструктажи и обучение можно проводить на сайте своей учебной организации
продать usdt в Одессе
December 11, 2024 @ 2:03 pm
продать usdt в Одессе
PinUp - 8p
December 11, 2024 @ 2:59 pm
Pin Up интернет-казино для Казахстана – это известное онлайн-казино для фанатов ставок и игр. Здесь доступен огромный ассортимент игр для азартного отдыха и больших призов. Если ищете безопасное место для игры, то Pin Up Casino обеспечит всё нужное.
Преимущества казино Pin Up
Разнообразие игр. На платформе Pin Up вы найдёте более 2000+ игровых решений, игр с живыми дилерами. В списке предложений представлены топовые разработчики, включая Play’n GO и многие другие. Это значит, что каждая игра адаптирована для любого устройства, оптимизированные для телефонов и ПК.
Бонусы для всех игроков. Новые пользователи могут рассчитывать на приветственные бонусы, которые помогут стартовать быстрее. Также действуют регулярные акции, кэшбэки и специальные предложения для постоянных пользователей, что увеличивает ваши шансы на успех.
Наш сайт: https://bk.kz/news/?ELEMENT_ID=3653&PAGEN_2=8
Полная безопасность и защита. Pin Up Казино использует надёжные методы защиты данных вашей личной информации, чтобы вы чувствовали себя в безопасности. Деятельность казино лицензирована гарантирует выплату всех выигрышей и своевременные выплаты выигрышей.
Лёгкий доступ к играм. Наше казино доступно на любых устройствах, что даёт возможность играть в любые игры в любой точке мира. Есть приложение для мобильных или просто играть прямо в браузере на мобильном.
Круглосуточная поддержка. В любое время, наша служба поддержки всегда готова к общению. Вы можете связаться с нашими специалистами через электронную почту, и операторы ответят быстро на русском или казахском языке.
black friday discount nike air max 90% OFF
December 11, 2024 @ 3:04 pm
Hi just wanted to give you a quick heads up and let you know
a few of the images aren’t loading properly. I’m not sure why but I think its a linking
issue. I’ve tried it in two different browsers and
both show the same outcome.
discount new balance shoes
December 11, 2024 @ 3:08 pm
Hello i am kavin, its my first time to commenting anyplace, when i read this post i thought i could
also make comment due to this brilliant piece of writing.
кракен андройд
December 11, 2024 @ 3:21 pm
Very shortly this website will be famous amid all blogging and site-building people,
due to it’s nice articles
Jacobgaugh
December 11, 2024 @ 4:18 pm
https://www.mtucizone.ru — Официальный сайт: все о наших кухнях и услугах.
car battery shop near kelana jaya
December 11, 2024 @ 4:27 pm
Simply wish to say your article is as amazing.
The clarity in your post is simply great and i could assume you are an expert on this subject.
Well with your permission let me to grab your RSS feed to keep up to
date with forthcoming post. Thanks a million and please
continue the enjoyable work.
Ремонт телевизоров Hisense
December 11, 2024 @ 4:33 pm
Предлагаем услуги профессиональных инженеров офицальной мастерской.
Еслли вы искали срочный ремонт телевизоров hisense, можете посмотреть на сайте: ремонт телевизоров hisense цены
Наши мастера оперативно устранят неисправности вашего устройства в сервисе или с выездом на дом!
Plinko - ju
December 11, 2024 @ 4:34 pm
Het spel Plinko is zeer populair games die in de afgelopen jaren populair zijn geworden. Het spel zelf, dat vandaan komt van de beroemde tv-serie, heeft zich doorontwikkeld naar de moderne casinowereld.
In dit artikel gaan we gaan we verder in op alles wat je moet weten over deze casinogame, van de grondslagen van het spel tot hoe je met echt geld kunt spelen en de beste strategieen om het spel te spelen.
Web: http://madercomgroup.com/category/uncategorized/page/12/
De Plinko game is een simpel, maar spannend gokspel dat vaak wordt geassocieerd met de populaire tv-show. Het spel bestaat uit een rechtopstaande baan met een aantal objecten waar de valbal van bovenaf doorheen doorheen rolt. De bal stuitert af van de pinnen en landt in een van de vakken onderaan, die elk een bepaald winst vertegenwoordigen. De bedragen is gebaseerd op de bal uitkomt. Dit betekent dat het een spel gebaseerd op geluk is, waarbij spelers niet precies kunnen voorspellen waar de bal zal vallen.
Hoewel de basismechanismen van het spel duidelijk zijn, maakt de onvoorspelbaarheid van het spel het spannend en afwisselend. Dit is een van de factoren waarom Plinko met succes is gepopulariseerd. Het wordt vaak aangeboden als een online versie van Plinko in verschillende online online gokhuizen, waar spelers de kans hebben om geld te winnen door te gokken op de uitkomst van hun spelballen.
Wanneer je deelneemt aan een Plinko casinospel, lijkt het spel meestal hetzelfde als de tv-versie van het spel. De grote verschillen liggen in je gokken kunt en het feit dat je kan inzetten met echt geld. In plaats van voor gewonnen prijzen zoals in de televisieprogramma, kun je in een online casino geld inzetten. De terugbetalingen hangen af van het vak waarin de bal zit afhankelijk van de inzet.
Inzetten kan op verschillende manieren, en afhankelijk van hun inzet kunnen de uitbetalingen varieren. De Plinko game online wordt vaak aangeboden met een gebruiksvriendelijke interface, wat het voor spelnovieten makkelijker maakt om het spel snel te leren. Veel gokplatforms bieden een Plinko game download optie, zodat je het spel kunt spelen op je mobiel, zelfs zonder online te hoeven zijn. Dit biedt spelers gemak en maakt het spel toegankelijk.
kuala lumpur city tour with kl tower
December 11, 2024 @ 4:44 pm
Hey I know this is off topic but I was wondering if you knew of
any widgets I could add to my blog that automatically tweet my newest twitter updates.
I’ve been looking for a plug-in like this for quite some time and was hoping maybe you would have some experience with something like
this. Please let me know if you run into anything. I truly
enjoy reading your blog and I look forward to your new updates.
JustinBoIli
December 11, 2024 @ 4:46 pm
http://slottr.top/# az parayla cok kazandiran slot oyunlar?