By Roger Meissen | MU Bond Life Sciences Center
As farmers spend billions of dollars spreading nitrogen on their fields this spring, researchers at the University of Missouri are working toward less reliance on the fertilizer.
Less dependence on nitrogen could start with a simple type of grass, Setaria viridis, and its relationship with bacteria. The plant promises to lay groundwork for scientists exploring the relationship between crops and the fixing nitrogen bacteria that provide them the nitrogen amount plants need daily.
“In science sometimes you have to believe because we often work with such small microorganisms and DNA that you cannot see,” said Fernanda Amaral, coauthor and MU postdoctoral fellow at Bond Life Sciences Center. “Before this research no one had actually proved such evidence that nitrogen excreted by bacteria could be incorporated into plants like this.”

Biological Nitrogen fixation — where diazotrophic bacteria fix atmospheric nitrogen and convert it to ammonium — provides a free way for plants to alter and absorb the nutrient. Farmers have long known that legumes like soybean fix nitrogen due to the symbiosis with bacteria in the soil through development of nodules on their roots, but since grasses like corn and rice don’t form this specialized structures that relationship has been trickier to explore.
Yet in fact, this team’s experiments showed the grass Setaria viridis received 100 percent of its nitrogen needs from the bacteria Azospirillum brasilense when associated with plant root surfaces.
“I believed in these bacteria’s ability, but I was really surprised that the amount of nitrogen fixed by the bacteria was 100 percent,” Amaral said. “That’s really cool, and that nitrogen can make so much of a difference in the plant.”
Worldwide farmers used more than 100 million tons of nitrogen on fields in 2011, according to the United Nations Food and Agriculture Organization. In the same year, the U.S. alone produced and imported more than $37 billion in nitrogen.
This grass can serve as a simple model for research, standing in for grass relatives such as corn, rice and sugarcane to explore a similar relationship in those crops. This research, “Robust biological nitrogen fixation in a model grass–bacterial association,” was published in the March 2015 issue of The Plant Journal.
A nutrient, a nuclear reactor and a model plant
Proving that this grass actually uses nitrogen excreted from the bacteria took some clever experiments, a global collaboration and a nuclear reactor.
MU researchers in the lab of Gary Stacey, a Bond LSC investigator, partnered with scientists in Brazil and at Brookhaven National Laboratory in New York to find a robust plant model system.
They screened more than 30 genotypes of Setaria viridis grass, looking for a strong nitrogen fixing response when colonized with three different bacteria strains. They germinated the seeds in Petri dishes and inoculated those three days after germination with a bacterial solution. Then plants were transplanted into soil containing no nutrients. By eliminating nitrogen in the soil, the scientists were able to make sure that the bacteria was the only source of nitrogen for plant.
The team settled on Azospirillum brasilense bacteria, which has been used commercially in South America to improve crop plant growth. It colonizes the surface of the roots and showed the greatest amount of plant growth when associated with plant roots.
Proving that the bacteria truly fixed the nitrogen used by the plant, required exposing plants to radioactive isotopes at Brookhaven National Laboratory. That began with Nitrogen 13, an unstable radio isotope that showed exactly where and how quickly this nutrient was taken up from the bacteria.
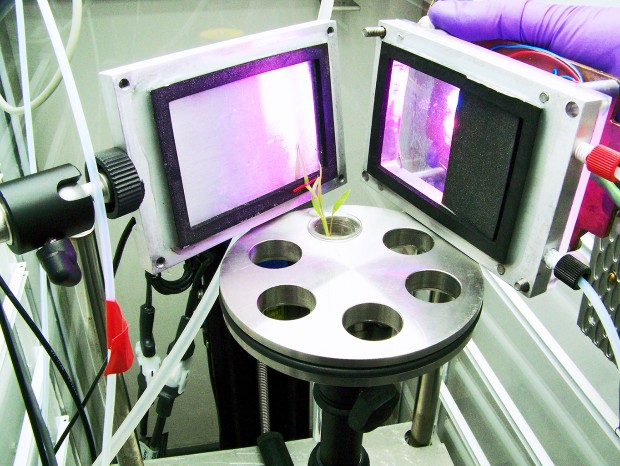
“Nitrogen 13 is really sensitive matter with a half-life of less than 10 minutes, and we first thought there wouldn’t be that much nitrogen fixed by the plant,” Amaral said. “We administered Nitrogen 13 only on the roots, quickly scanned the samples and calculated how much of the nitrogen the plants assimilated based on the decay analysis of the tracer.”
This experiment, paired with several others, showed that this model grass truly incorporated the nitrogen released by the bacteria and metabolizes it in several components.
Model (plant) citizen
But why does a type of grass that doesn’t produce food matter so much?
The answer is time and simplicity.
“Corn is really good at responding to bacterial inoculation, but it’s very big and takes a long time to produce seeds and also the genome is complex,” said Beverly Agtuca, an MU Ph.D. student who worked on the study. “Setaria viridis is a small plant that can produce a lot of seeds faster, has a pretty simple genome and can serve as a model for research.”
That makes it perfect to explore how the plant actually uses its bacterial partners, and labs around the world are already using this plant model for research.
For the Stacey lab, the next step is to pinpoint the gene in the model grass that makes this possible.
“We want to identify the genes responsible for the interaction between plant and bacteria and meanly the ones involved with the nitrogen uptake,” Fernanda said. “We hope that will allow us to improve plant growth based on the gene to further study.” We believe that our findings can stimulate others studies at this area, which seems to be a promise plant friendly way to apply for promoting a sustainable agriculture, especially to crop systems including bioenergy grass.
Amaral and Agtuca work in the lab of Gary Stacey at Bond LSC. Stacey is a Bond LSC investigator and a Curators Professor of Plant Sciences in the College of Agriculture, Food and Natural Resources at the University of Missouri. Collaborators included researchers at Brookhaven National Laboratory, State University of New York, Federal University of Paraná in Brazil and Federal University of Santa Catarina in Brazil.
Funding for this project came from the National Institute of Science and Technology- Biological Nitrogen Fixation, INCT-FBN, the Brazilian Research Council, Ciência Sem Fronteiras Program, The Department of Energy and SUNY School of Environmental Science and Forestry Honors Internship Program.