
By Samantha Kummerer | Bond LSC

By Samantha Kummerer | Bond LSC

By Samantha Kummerer | Bond LSC
Kory Floyd was stressed and having an all-around bad day, but then a coworker offered him a hug.
“That hug didn’t change anything about what had gone in my day but it changed everything about what the way that I felt,” Floyd said. “Suddenly all that stress, suddenly all that disappointment, all that worry, it didn’t go away but it seemed to lift off my shoulders, it wasn’t weighing me down.”
Floyd studies how prosocial communication, like affection, compassion, and trust, affect individuals psychologically.
He spoke specifically on communicating affection during the Life Sciences and Society Symposium on Saturday, Oct. 7.
Affection can be communicated through verbal and non-verbal communication as well as through supportive behavior. This last category is one he discovered through his research. Supportive behavior like keeping someone’s car in good condition or helping someone with a task is sometimes the most common way of expressing affection, especially between men.
The University of Arizona communications professor said he started his research by exploring an aspect of affection that confused him in his life. Growing up in a family of huggers, Floyd said he soon found out that not everyone appreciated the same type or kind of affection.
Expressing affection can be risky. Affection can be misinterpreted or not reciprocated, censured by culture, and even transmit disease.
So why bother?
For Floyd, the answer is simple, “Affection as an emotion and behavior is a fundamental human need.”
To elaborate on his belief, he pointed to how humans are born dependent on others to survive for many years. A child needs someone to feed, protect, provide for it and often make huge financial and personal sacrifices. As Floyd points out, many parents willing to do this due to love.
“I don’t think it’s necessarily a stretch to claim that an infant’s ability to get someone to love it is a matter of life or death.” He theorized. “It is a matter of that infant’s survival to get someone to feel a strong enough emotional investment that they are willing to make all the other kinds of investment that are required for that’s infant’s survival.”
As we age, affection transforms into something that feeds our need to belong.
Floyd’s studies have revealed positive associations between affection and happiness and social engagement. Other research revealed that affectionate communication is associated with mental wellness and physical health. When exposed to stressful events, highly affectionate people react with less arousal and recover faster. When compared to a less affectionate person, the more affectionate person has a better immune system, lower blood glucose levels and improved cortisol rhythm.
“It’s really difficult to thrive without some measure of affection,” Floyd concluded.
While not everyone needs the same amount of affection, it is a ubiquitous form of prosocial communication.
“None of what I described here is a prescription for you to go out and start hugging people on the sidewalk, that is not a pathway to health, that is a pathway to incarceration,” Floyd joked as he concluded his talk.
Instead, he highlighted that affection is different than other communication behaviors because it is something we reserve for only the important relationships in our lives.
“We talk to anybody, we gesture to anybody but we don’t kiss just anybody, we don’t hug just anybody,” He said. “This is a special behavior and its something that we preserve and reserve for the very special people in our lives.”
The 13th annual Life Sciences and Society Symposium, The Science of Love, started Friday, Oct. 6 and Saturday, Oct. 7. It features six experts that research various aspects of love, relationships and connection. The event will conclude on Friday, Oct. 13 with its last speaker, Jim Obergefell, who was the plaintiff in the 2015 Supreme Court case on marriage equality.
Computer scientists create applications to speed up research in the lab

By Samantha Kummerer, Bond LSC
Three years ago, Ke Gao stood uncomfortably beside rows of biomedical students and plant scientists at the Bond Life Sciences research fair. His poster wasn’t discussing the DNA of seeds or how plants transport nutrients but rather a scientific device.
“At the beginning, the visitors didn’t understand what we were presenting, but once I explained how our application can help them accelerate their research and how we can really turn their phones into a research device, they got really excited,” Gao explained.
Gao’s presentation highlighted a mobile app that transforms images of seeds into objective, quantitative data.
It started with a simple problem. Plant scientists were manually comparing hundreds and in some cases thousands, of seed photos. The process was meticulous, slow and subjective.
The solution began with a collaboration with Michele Warmund (Plant Sciences), Tommi White (MU Electron Microscopy Core) and Filiz Bunyak (Computer Science) that led to a MU Interdisciplinary Innovations Fund grant.
Gao was part of this team that developed an algorithm to turn the photos of seeds from the field into data with the touch of the button.
Gao explained the app is very similar to Instagram.
A user takes or uploads photos of seeds. Then the app calculates measurements describing shape, color and size characteristics of the seeds. This data can be emailed or stored in a database.
Some experiments need thousands of seeds analyzed; this would be a massive feat for even a group of students. With this app, hundreds of seeds can be photographed and measured from a single photo. The app analyzes each seed individually and also computes measurement averages for groups of seeds.
There are other apps that analyze seeds, but this is the first mobile application as far as the team knows. Its ability to analyze multiple seeds at once, even if they are touching is also an outstanding ability. Bunyak’s previous experience developing applications to quantify microscopy images and videos of touching and clumping cells helped them design the algorithm to make that function possible.
This isn’t just a problem for researchers in this one lab or even at the University of Missouri.
MU Computer Science professor, Filiz Bunyak, said noninvasive methods to observe and understand biology, imaging equipment and corresponding computing devices have advanced considerably in recent years, leading scientists to produce large amounts of data. The ability for researchers to analyze and quantify this large amount of complex and unstructured data, however, was still missing. Bunyak said this app began as a project to advance scientists’ capabilities to automatically analyze image-based plant phenotyping.
Further collaboration
Bunyak and her students are advancing the field of high-throughput phenotyping beyond this mobile app.
High-throughput phenotyping (HTP) refers to the process of connecting an organism’s DNA makeup to its physical characteristics; it is also a hot topic buzzing through the science community in the last five years.
Two years ago, Bond LSC scientist David Mendoza, who studies how plants collect nutrients, said he never imagined he would be doing HTP.
“The old way of doing this is growing plants on plates and, I’m not kidding, with a ruler you measure how long the roots are,” Mendoza explained of the traditional process that now seems archaic.
Now, the lab is working with computer scientists to design a robot to code the measurements for multiple roots at a single time. For a student, it would take 15 minutes, but now it’s complete in an instant.
Speed isn’t the only reward researchers are reaping.
Bunyak said computational image analysis allows researchers to come up with new ways to quantify and study data that they were not even able to do before, leading to the design of novel experimental methods.
Ruthie Angelovici is another Bond LSC researcher who uses computer scientists to aid in her research.
She said without computer imaging there would be no way for her team to do research that measures plants physical and biochemical traits. Angelovici’s lab uses Bunyak’s mobile app system but on a computer. Eight plants are photographed at once and the application keeps track of features of plants such as shape, color and area as they develop.
What is really revolutionary to Angelovici is the ability for the data of plant growth parameters to be stored and revisited without the need to re-grow. This contrasts with past experiments where researchers would scribble some notes and never be able to return.
“It’s not lost and I think that’s a big step in this field,” Angelovici said.
The collaboration is creating more than advanced tools by fostering a new way to think and approach research.
Rather than buying pre-existing software, the groups from Bond LSC utilizes the resources on campus to build their own devices.
“I would have been in front of a black box that is doing things for me and that would not have given me the tools to teach to my students,” Mendoza reflected. “Now I know what they need to learn to be competitive. Now I know what the gaps are and how they can be filled. I think that was worth it.”
Mendoza’s team publishes all the instruction to its robot online, so the technology can aid other labs in making faster discoveries at a lower price.
Angelovici compared it to buying a cake versus making a cake — at the end of the creation process, she said she would have the knowledge to do a lot of other experiments.
This new way of thinking already began to pay off this summer when her lab expanded computer software to analyze seed size.
“We only approached it because we saw how things worked together. I just pitched a project to engineering about seed collector. Again, this opened my eyes that even undergraduates can do something not so difficult for engineers, but I have no clue how to do it,” Angelovici said.
Mendoza agreed the collaboration is exciting but challenging, “You got a Ph.D. and you got a faculty position and you think you know stuff. When I started this I realized how much I don’t know, but at the same time it reminded me that it is really cool to learn something new.”
Both teams continue to work towards maximizing the functions of their individual machines, but even after the projects reach fruition the collaboration will not be over.
“On the contrary, I think we’re going to keep building more and more and better,” Mendoza said.
Nowadays, Gao no longer feels out of place at the Life Sciences fairs. Researchers from various labs come up to him and ask how they can implement his app in their own lab.
“It seems like I’m doing something that can really help people, so that’s the best part of this process,” he said.
Ruthie Angelovici is an assistant professor in the Division of Biological Sciences and is a researcher at Bond Life Sciences Center. She received her degrees in plant science from institutions in Israel — her B.S. and M.S. from Tel Aviv University, and her Ph.D. from the Weizmann Institute of Science in Rehovot. She was a postdoctoral fellow at the Weizmann Institute and at Michigan State University and has been at MU since fall of 2015.
David Mendoza is an associate professor in Plant Sciences, Life Sciences Center investigator and a member of the Interdisciplinary Plant Group. His research focuses on the mechanisms plants use to resist toxic elements or acquire nutrients. He received his Ph.D. in biochemistry from UNAM in Mexico City and continued on to do post-doc training at UC San Diego.
Filiz Bunyak is an assistant research professor in the Department of Computer Science. She received her bachelors and masters degree from Istanbul Technical University and her Ph.D. from the University of Missouri- Rolla. Her work focuses on computer imaging, image processing, and biomedical image analysis.
Ke Gao is a doctoral student in the University of Missouri’s Department of Electrical Engineering and Computer Science. He earned his bachelor’s of science from the Henan University of Science and Technology in China.
How zebrafish gained its popularity as a model organism
By Samantha Kummerer, Bond LSC
The core of many modern discoveries in developmental biology is swimming in a tank.
These are zebrafish that serve as the lab rats for Anand Chandrasekhar’s research.
Dozens of tanks containing thousands of swimming fish fill the lab in the basement of the Bond Life Sciences Center. There are baby fish, striped fish and clear fish, many genetically modified for experimental reasons, and they are studied from fertilized embryo to adolescence.
Chandrasekhar’s lab uses zebrafish to study migrating motor neurons in the brainstem that control muscle movement in the face and jaw.
Multiple types of neurons positioned precisely throughout the brain connect together to form networks. Those networks give the brain its ability to function.
“For us, it is important to study how these networks form because the brain is what makes us human,” said the Bond Life Sciences Center researcher.
During development, neurons respond to signals that enable them to move to different locations and form networks, Chandrasekhar explained. If the neurons don’t migrate properly to specific locations then the brain can’t function properly.
It is a domino effect. If the brain stem motor neurons don’t migrate correctly then the corresponding neural networks don’t form correctly and then the fish are not able to eat well.
Chandrasekhar’s lab is captivated by this migration. Different neurons move different ways and are set into motion by different signals. What are the signals to tell the neurons to stop or to go? Do these signals vary based on neuron type and species?
The lab also seeks to understand the repercussions of deficient migration using genetically modified fish.
Cell migration doesn’t just occur in the brain or with nerve cells. Cells also need to move in particular ways to form the heart and to fight infections.
A Model Swimmer
The zebrafish has long joined traditional lab rats and mice as scientists’ choice as an ideal test subject.
“When I first started out I wasn’t entirely sure I could do that (work with zebrafish), because how am I expected to handle something in water, moving around and be able to use it to really do experiments?” Chandrasekhar said. “But once you see how you handle fish, how you get them together, it just becomes one more thing that you do and then it makes you wonder how people work with mice.”
Like other model organisms, zebrafish share many genetic similarities with humans. These similarities mean researchers can investigate cancer, heart disease, muscle and tissue disorders in humans by testing and studying fish.
Mice and rats also share a large portion of DNA with humans, but the zebrafish’s unique characteristics enable experiments and observations to be cheap, efficient and fast.
One of those unique traits is a rapid development rate. This allows researchers to study important developmental stages in a single week.
“They’re incredibly efficient in terms of, I can set them up and they’ll be ready tomorrow morning,” said Ph.D. student Devynn Hummel.
The fact that embryo development in mice occurs inside their mother also makes observing early stages of development hard. Zebrafish embryos, on the other hand, grow independently, outside the mother.
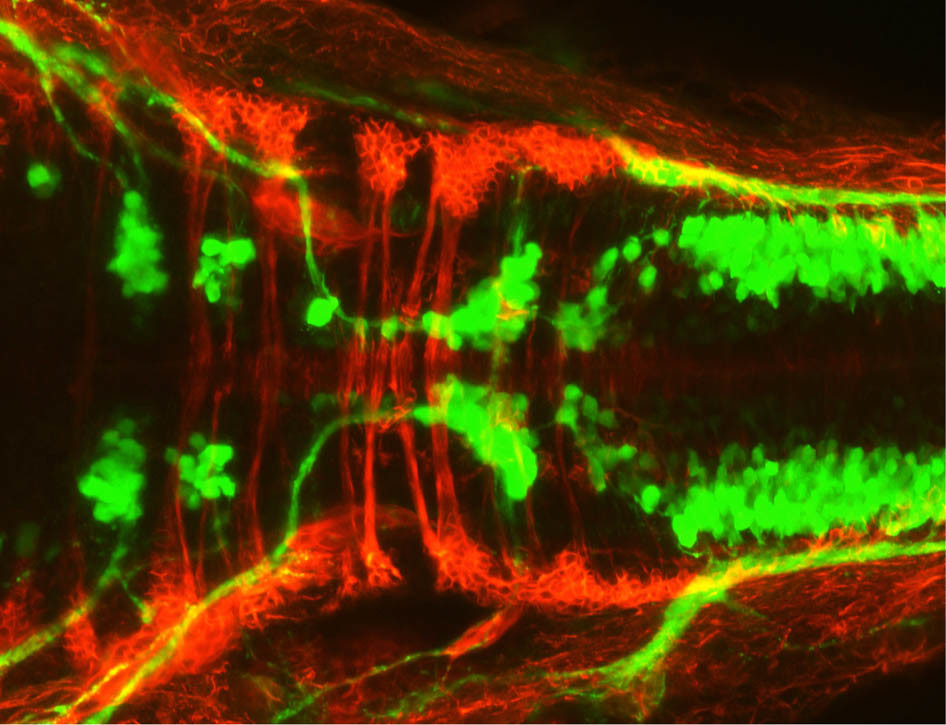
The young zebrafish’s transparency also helps researchers track cells within its body.
Hummel explained that by using fluorescently tagged molecules we are able to zero in on anything from neurons to changes in calcium concentrations, allowing fish to be used in a wide range of different studies.
“We’ve sort of engineered them to allow us to look at different tissues,” Hummel explained. “It really is remarkable the details in imaging zebrafish and what you can see. It’s truly extraordinary.”
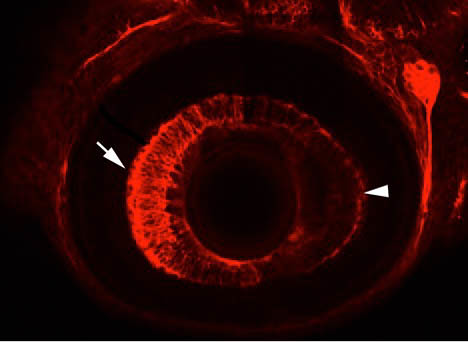
Scientists throughout the world are catching on to the advantages of the fish.
Chandrasekhar said there are significantly more zebrafish labs throughout the world than there were 20 years ago.
Despite its advantages, the zebrafish will not completely replace mice as a lab tool.
Chandrasekhar explained there are still certain experiments where mice are more advantageous. The same area of research can be explored using zebrafish, mice or even fruit flies, but the specific questions you ask change based on the tools available.
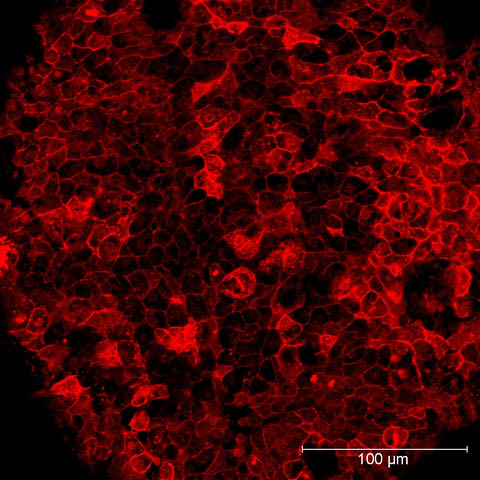
Beyond Nerves
While Chandrasekhar’s lab concentrates on neurons, understanding how the migration occurs can be applied in a different context because of shared cellular mechanisms.
“Some of these molecules are just tools that different types of cells can use in different ways to accomplish different functions,” he said. “You can study a steering wheel in a car and know that it’s allowing the wheels to turn and the same steering wheel in a truck is allowing the wheels to turn in a truck, but maybe at a different time and a different way.”
Members of Chandrasekhar’ lab already experienced a broader implication of their research when a gene they studied with a role in neuron migration provided insight on Spina bifida, a failure of the fetal spinal cord to close completely.
“We all hope the problem we study has a broader impact on human biology so there’s always a quest to dig deeper and learn something new,” Chandrasekhar said.
The lab is hoping one day soon this fishy tool will help shed even more light on the mechanisms maintaining human health. For now, the researchers and their fish will just keep swimming.
With more than 2,000 fish, the lab has no plans on slowing down.
Anand Chandrasekhar is a Biological Sciences professor at the University of Missouri. He uses mice and zebrafish to study the mechanisms involved with the development of the nervous system. His lab in Bond LSC uses cell biological and genetic methods to understand these mechanisms. He received his Ph.D. in biology from the University of Iowa.
By Samantha Kummerer, Bond LSC
What happens when a chemical engineer, a computer scientist, and an immunologist walk into a lab?
Vaccines are created faster and cheaper.
At least this trio hopes that’s the answer.
Bond Life Sciences Center computer scientist Dong Xu joined forces with immunologist Jeffery Adamovicz and chemical engineer Bret Ulery for the first time in February 2016. After 18 months and a $100,000 Bond Life Sciences startup grant, the team is closing in on a novel approach to vaccinations. They hope to start collecting initial data in the near future.
“Without this grant, it would be hard to work together,” Xu said. “Bond Life Sciences Center really played an important role for the collaboration research on campus and also for providing this seed money. They really foster these types of collaboration.”
The Problem
Efficiency has plagued the vaccination field for decades.
Traditionally, scientists create new vaccines by continuously guessing which components of the virus or bacteria to test.
“It was a plug-and-play type test system,” explained Adamovicz, who has researched vaccines for years. “We said, ‘well we could try this antigen or that antigen or that antigen and you would mix them together in different combinations and try them and then you would end up selecting one that would work.”
The process is similar to finding a needle in a haystack. It’s like scientists sort through and test every strand of hay until they discovered the needle.
As you can imagine, this method takes a long time to find something usable.
This process becomes a larger issue when viruses emerge that call for immediate measures, such as Zika or Ebola.
Creating vaccines can take a long time because sometimes a variety of different related bacteria cause illnesses. This makes that already inefficient guessing game more complicated by adding, even more, components to test.
Adamovicz explained scientists make a new vaccine each year by predicting which strain of the disease will infect the most people. This is why many people who get a flu vaccination can still get the flu — the strain they caught was not included in the vaccine.
The Solution
Xu, Ulery and Adamovicz decided to combine their strengths from their different fields to approach creating a vaccine in a new way to solve these two problems.
Adamovicz explained this type of collaboration is not routinely attempted when developing new vaccines.
Computer science and engineering enabled the team to more easily sort through thousands of potential options. Xu’s lab and his students created an algorithm to select candidates to test. Without Xu, the team would have to test tens of thousands of different antigen and peptide combinations. But with the algorithm, the team narrowed the combinations to about 10 to test.
Adamovicz said this method has the potential to create vaccines in half the normal time.
“I feel that it is a very meaningful work; that’s why I am interested in doing it because I do see the potential impact,” Xu said.

Putting a theory to work
Burkholderia is the first Guinea pig for their new approach.
The potentially deadly bacterial infection occurs in places like Thailand and northern Australia and is caused by a family of related bacteria called Burkholderia pseudomallei. But other strains of Burkholderia also cause disease in patients with Cystic Fibrosis across the globe.
“That was part of the challenge,” Adamovicz said. “It wasn’t that we could take a single bacterium and that’s all we had to worry about. We were worrying about a family of bacteria that had this variation in their genomic sequence and the proteins they produce.”
To eventually create a vaccine that could cure all strains of the bacteria, the team aimed to find a small piece of protein all Burkholderia had within their flagella, the whip-like structure of the bacteria that allows them to propel themselves.
To limit the number of segments within the protein, Adamovicz’s lab, on the biology side, determined some requirements for Xu to write an algorithm.
“It’s not to say this is like Star Trek where you just tell the computer and it does it on its own, it requires a lot of knowledgeable human input to spit out the answers,” said Adamovicz.

Adamovicz explained they wanted a vaccine that would trigger two immune responses. When harmful substances enter the body, like viruses or bacteria, the body responds in two ways. One way is through cellular immunity that responds by activating cells that already exist. The other way the body responds is by other cells making proteins called antibodies, which is called the humoral response.
The segment also had to be conservative, meaning it is less likely to mutate so there is a higher chance a vaccine would continue to be effective against the bacteria and would be effective against more members in the bacterial family. The last criteria Xu’s lab coded for ensured the protein is not similar to others found in humans.
All these limitations lowered more than three thousand possible candidates down to a testable amount and a final list of 10 candidates.
Then the research moves on to Ulery’s Lab. Here a team of chemical engineers take the identified candidates from Xu’s work and engineer targeted vaccine antigen nanoparticles termed micelles. These micelles are comprised of fat-based cores that display vaccine antigens on their surface and have been shown to induce strong immune responses.

“Most of them [subunit vaccines] are somewhat hydrophilic — water-liking peptides, portions of a protein — and we tether a fat to them and what happens is now you have something that likes water and something that doesn’t like water, and you throw it in water and they self-assemble into these small micelles,” Ulery said. “That clustering of all those peptide molecules together actually enhances the immune response to them.”

Then, the work goes back to Adamovicz to evaluate what the portions of proteins are going to make the best vaccine.
This cycle continues as all labs work to enhance and develop the design further.
“It’s a process that takes a village. It doesn’t necessarily take the whole village but you need the right people in the village to work together and that’s kind of what we’re doing,” Adamovicz explained. “We’re recognizing that there are areas in previous work that could have been better optimized.”
After months of back and forth, the team is preparing to feed infected cells some of the selected peptides, the small portions of protein. Those candidates that do well in the cell culture will be tested in mice exposed to the Burkholderia bacteria.
The team hopes this round of data collection will serve as a baseboard for future experiments. If this principle is proven in one antigen, a larger grant may enable the theory to be tested in hundreds of other antigens and applied to other diseases. Over the next four to six months, the team hopes to have a clearer vision for the future of this research.
The team credits the Bond LSC grant for bringing them together and for helping establish initial research to prove an approach like this could be impactful.
But, these types of collaborations don’t stop here.
“Previously, it was small stakeholders working on one protein their entire life, but that’s more basic research. To solve real-world problems requires many, many aspects, so the way that we are working in terms of interdisciplinary collaboration, I think that presents the future and this is already on-going in terms of an overall biology trend,” Xu explained.
Dr. Dong Xu is a professor in the University of Missouri’s Electrical Engineering and Computer Science Department and the Bond Life Sciences Center. His Digital Biology Lab develops and uses computers and software programs to help biological and medical researchers analyze large amounts of data.
Dr. Jeffrey Adamovicz is an associate professor in Veterinary Pathobiology and the director of the Laboratory of Infectious Disease Research. He works on developing vaccines and on animal models for zoonotic diseases.
Dr. Bret Ulery is an assistant professor in MU’s Chemical Engineering Department. He directs the Biomodulatory Materials Engineering Lab that creates new biomaterials for applications in immunology and regenerative medicine. He earned his doctorate degree from Iowa State University.
Gres reflects on the dissertation in HIV research
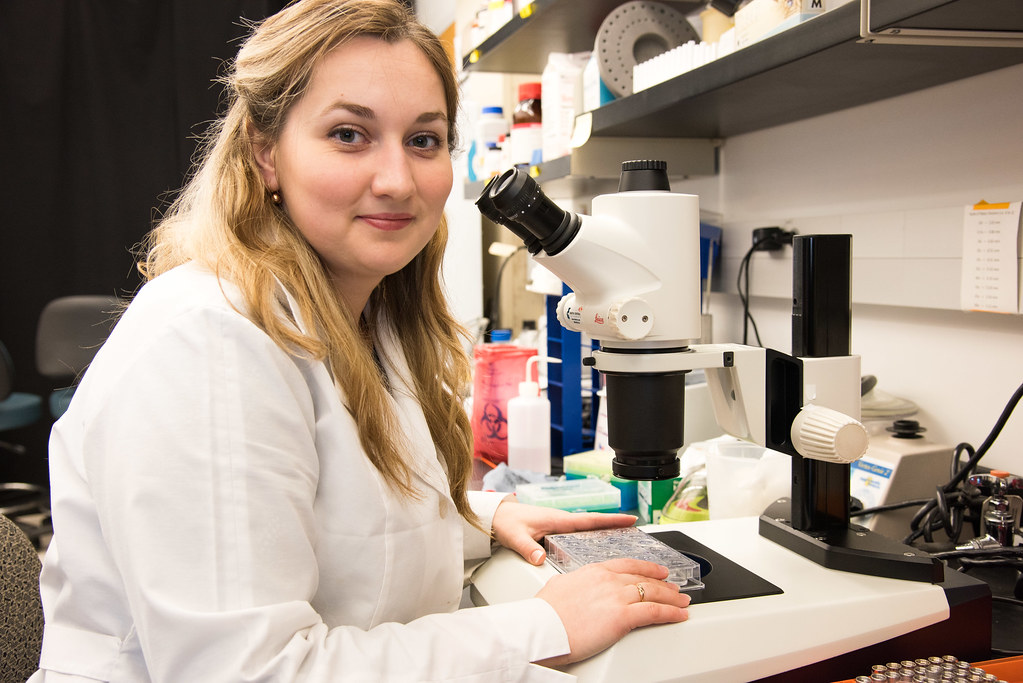
By Samantha Kummerer, Bond LSC
Ph.D. candidate Anna Gres frequently described her success at the Bond Life Sciences Center as being lucky.
“It was challenging and stressful, but I think everything worked out well for me and I was lucky in a way. I was fortunate to get to the good labs, interact with great people, attend courses, attend conferences and grow both professionally and personally,” she said.
Her mentors and colleagues use other words to describe her.
Bond LSC scientist and mentor Stefan Sarafianos called her “super spectacular” and her work “nothing short of outstanding” when introducing Gres before her dissertation defense on September 1.
When Gres came to the University of Missouri five years ago as a chemistry student, she knew she wanted to learn about crystallography, a technique that examines the precise arrangement of atoms and the geometry of molecules by looking at the image of the crystal under an X-ray beam. This interest led her to Sarafianos’ lab and into research on HIV.
“I learned a lot of new and exciting things, and I love this about research in general, you have multiple opportunities to evolve if you invest time,” Gres said of her time in Bond LSC. “I’m thrilled I decided to pursue a Ph.D. and do research. It’s definitely something I enjoy a lot.”
She studied a specific HIV protein. Her research involved using X-ray crystallography to determine the structure of the HIV-1 capsid protein. The capsid surrounds the virus’ genetic material, protecting it from a cell’s defenses while transporting the viral DNA to the cell nucleus. Her research produced one of the most complete models of the capsid protein. Previous attempts to understand the protein worked with engineered versions of it rather than the native version.
Gres’ research was the first to reveal the closest representation of the hexameric capsid lattice in intact HIV-1, a discovery that will open the door for further insights.
She explained this knowledge allows scientists to understand the viral life cycle better, study interactions of the viral protein with cellular proteins, further develop capsid-targeting antivirals and improve current ones.

Gres also studied complexes of capsid protein with host cell factors and small molecules that interact with the protein and prevent infection. Other time in the lab was devoted to exploring more than 30 mutations in the protein. Around half of the mutants still need to be completed and analyzed.
Currently, no approved drugs target this protein although it plays a critical role in the virus. Previous clinical trials failed, but recently the protein re-emerged as a promising target. Earlier this year, Gilead, a biopharmaceutical company, announced it would begin capsid inhibitor testing after initial studies revealed its potential as a treatment strategy.
When Gres finished her dissertation defense, she did so with tears in her eyes as she thanked her mentors, family, and friends for their support throughout this five-year journey.
“Work can be stressful; sometimes you need to vent because ‘the project is not working.’ It’s nice when you have people who can listen to you and calm you down and tell you it’s going to work out eventually,” she said.
Her research more than worked out and will continue to provide insights on the disease that affects millions each year.
As for Gres, she is leaving HIV research and the United States behind.
Soon she will be moving to Sweden to start a post doctorate position at the Uppsala University studying chromatin remodeling factors. Gres will bring her skills in structural biology and will transition to cryo-electron microscopy. She explained the topic would be new for her, but that’s why she chose it, so she can continue learning.
Undergrad’s passion spurred by mice muscle regeneration research

By Samantha Kummerer | Bond LSC
Uncertainty and curiosity led Rebecca Craigg to work in a lab.
As a first-generation college student with an interest in science but no idea what undergraduate research entailed, her path at the University of Missouri landed her in the Bond Life Sciences Center and the lab of D Cornelison.
“I honestly thought undergraduate research meant you just followed around someone like job shadowing,” she said laughing.
Now, after almost a year of research, the junior biology major is more than familiar with what working in a lab entails.
Craigg started working in Cornelison’s lab as an effort to figure out what kind of science she might be interested in. It was while she waited for her genetically altered mice to grow up when she found something that really sparked her interest.
Rather than spend her time cleaning lab dishes, Craigg began assisting a graduate student on a project studying the role of EphA7 in mice. EphA7 is a receptor on cells that helps mediate important events within the body. Receptors create a change in the body after receiving a signal from outside the cell. The exact hows and whys behind the EphA7 are still relatively unknown.
To unravel EphA7 the team began by breeding genetically altered mice with the gene switched off. Next, specific muscles were isolated, dissected, and preserved once the mice reached different developmental time points. Researchers thinly sliced the muscle and added immunofluorescent staining to track specific elements over a period of time. They marked EphA7 with a green fluorescent and regenerating muscle fibers with red. Imaging revealed every time green came up so did red. This led researchers to understand that EphA7 is connected to how muscles rebuild.

But this isn’t the only role the receptor plays.
Work this summer also discovered the gene is involved in the development of muscles. The team found at the end of development, the mice without EphA7 did not fully recover like they would have if they were not modified. When the team looked at marking for muscle stem cells, both the mice with the receptor and the mice without it had a depleted number of satellite cells, essentially muscle stem cells, that did not recover.
Craigg explained this discovery was puzzling.
In every other case heterozygous mice, the mice with one gene coded with EphA7, were fine. The team predicts the cells and muscles realized they were not adequate so the muscle stem cells began to become muscles. This process would leave fewer stem cells at the end of development.
Fewer muscle fibers really mean the mice are lacking normal muscle strength. Craigg said one reason for this could be EphA7’s relation to motor neuron axons. Motor neurons send signals from the nervous system to muscles to tell them to move. EphA7 is present on all motor neurons. The team hypothesized that the gene may play a role in guiding the motor axon to the muscle, thus without it, muscle size would decrease.
“We kind of know way more than we did, obviously, in that it’s involved in regeneration and all these things and that if a mouse doesn’t have it they have all these decreased numbers all over the board, but we don’t know why per say,” Craigg said.
To better understand why EphA7 functions the way it does, the team will begin to examine the relationship between motor neurons and EphA7. They will also explore if the type of muscle fiber, slow-twitch versus fast twitch, make a difference.
Craigg said each new piece of knowledge about EphA7 is a step towards better understanding what causes muscle disease in humans.
While the lab faces a lot more work ahead, it doesn’t faze Craigg. For her, the possibility of more discoveries is part of the thrill.
“We kept finding out so many new things about it. I would be counting things and I’d be like, ‘Oh my god, I can’t wait to get this data back, like, I just want to know,’” she exclaimed.
Rebecca Craigg is a junior biology major working in the lab of D Cornelison at Bond LSC.
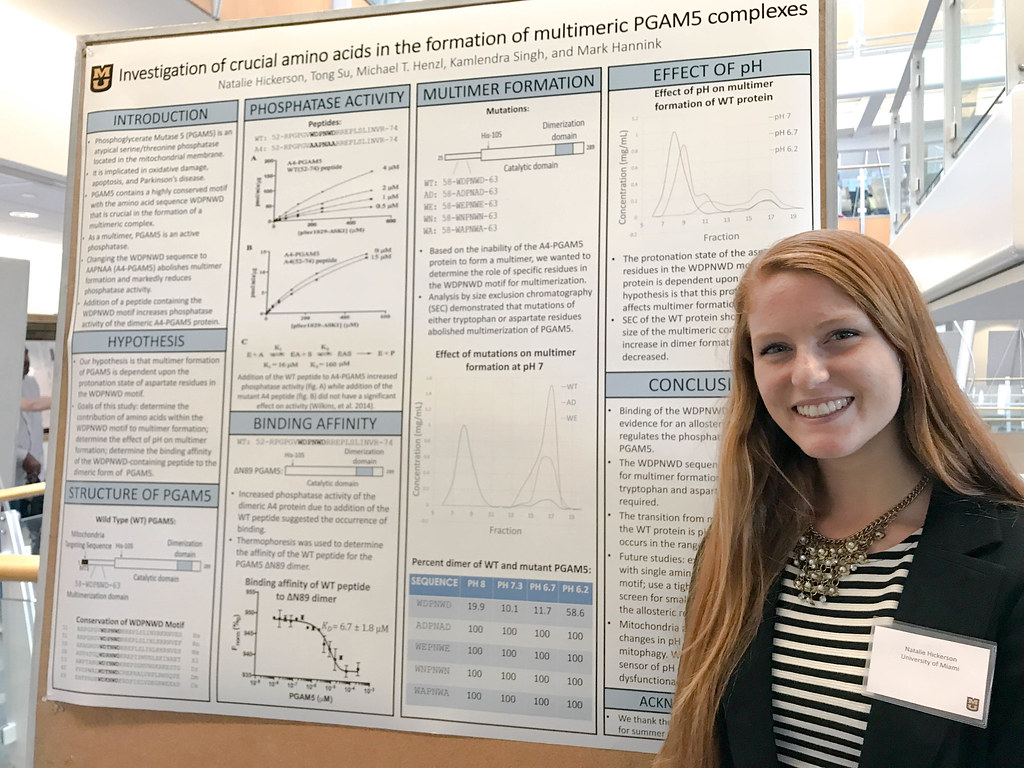
By: Samantha Kummerer| Bond LSC
It takes a lot of time and patience to be a scientist. This is something that first-time researcher Natalie Hickerson quickly discovered.
“A lot of the time things are so small. I mean you’re using such tiny volumes of DNA that you can’t see anything happening,” said the undergraduate biochemistry major.
For some, this uncertainty pair with long lab hours and multiple trial and errors would be frustrating. Hickerson, however, put in the time and asked the questions, which led her to discover the rewards of research.
“You try something a few times and it wouldn’t work and then you change one thing and suddenly it works and you get results and it was just very exciting, like ‘wow what I’m doing is real,’” she said.
Hickerson attends the University of Miami but worked as a research intern in Mark Hannink’s lab in the Bond Life Sciences Center this summer. She said she chose the University of Missouri because the program was well rounded and fit with her major.
Hannink said while Hickerson entered the lab not understanding everything, she wasn’t afraid to ask questions and that helped her catch on fast.
“What makes a scientist different is that you are an active generator of new knowledge. Instead of being a passive consumer of existing knowledge you have to become an active producer of new knowledge,” Hannink said.
The Missouri native spent two months creating new knowledge on PGAM5, an important protein involved in many mitochondrial processes and cell death.
Hickerson began by intensely studying the protein at its most basic level to determine how and why it works the way it does. Part of this process was comparing different mutations of the protein.
Previous research shows how understanding the sensitivity of PGAM5 and its changes in the cell could help in nerve-degenerate diseases like Parkinson’s and ALS. Individuals suffering from those diseases experience a loss of function or mutation in some proteins.
Part of Hickerson’s initial research aimed at figuring out what signal activates PGAM5 in a cell. A better knowledge of that process will help scientists understand how pathways function and turn off during nerve-degenerative disease.
Hannink explained if a pathway is defective, activating a different pathway may function as a type of therapy.
After months of long hours, Hickerson discovered by changing the pH levels, PGAM5 can be switched from inactive to active.
“We had suspected it to be the case but the data she provided really helped demonstrate that was true,” Hannink said.
While Hickerson is headed back to Florida to continue her pursuit of biochemistry, her findings will continue to be advanced this fall in Hannink’s lab.
“There’s a lot more to be discovered with what we were doing,” Hickerson said. “A lot of the stuff we were doing this summer was new ideas and just developing deeper knowledge on things they’ve discovered in the lab previously. So, I was looking deeper into what they already started and we did find some new things so it was neat.”
She said her experience this summer inspired her to get more involved in undergraduate research.
“It definitely gave me a much better understanding of how to work in a lab and just basic lab techniques. The overall research project gave me a lot of good foundation for that,” she said.
As Hickerson continues to learn about research, Hannink said it’s an ongoing process that he is still a part of.
“I’m doing the same thing all the time that they are learning to do,” he said. “There’s this dynamic and that active learning process also challenges me to becomes a better scientist as well.”

By Samantha Kummerer | Bond LSC
“#IAmScience because learning something new is super exciting! I love that by performing research one can contribute to the collection of knowledge.”
Johanna Morrow discovered her love for plant sciences after working in Mannie Liscum’s lab for more than a decade after college.
“I was a biology undergraduate and I didn’t really know what I wanted to do, but I knew I enjoyed research so I pursued a job with Dr. Liscum,” she said. “As time went on, I realized I was really interested in how plants are able to sense light and all the different growth and development processes that hinge on that perception.”
Her love for research brought her into the lab, but once she was there she discovered another interest.
“During the time that I worked in the lab, I found that not only did I enjoy doing bench work, but I found that I also enjoyed mentoring students and watching their passion for plant science and research flourish,” she explained.
This realization led Morrow to go back to school so she could share her love of science with more students.
Now her passion is her reality.
Morrow is working on how plants sense and respond to different types of light. She deals with mutations in the model plant Arabidopsis. She hopes by screening different variations of the plant, she can better understand what happens to a plant after it perceives light.
“Maybe we can identify new players in this mechanism or identify new ways that this could have an agronomic impact,” Morrow said.

By Samantha Kummerer | Bond LSC
“#IAmScience because I want to help unravel the mysteries of nature that will improve our futures and positively impact our planet.”
Katelynn Koskie didn’t always know she loved plants. As an undergraduate, she focused on what was above her rather than what grew below her.
“I was really interested in how galaxies interact and then I started to think, ‘you know I’ve always thought plants were really, really cool,’ and I wanted something that was a little bit more down to earth,” she said.
While she was pursuing a degree in astrophysics, she took one plant biology course and fell in love. From there she signed up for grad school and has been with plants ever since.
Koskie works with a mutated plant called hyper phototrophic hypocotyl, hph. The mutation is a variation of the lab’s model plant Arabidopsis. This variation is special. It produces more seeds, bends more under light and is stronger. It’s up to Koskie to figure out why.
That answer could have a large impact on the agriculture industry. If Koskie’s findings can be applied to crop plants like maize, farmers can grow better crops.
“Maize is more complicated than Arabidopsis, but with new techniques like CRISPR/CAS9 now it might make it a little bit easier,” she said.
She plants genetically modified seeds and then waits and observes and begins again.
It is a lot of time in the growth chamber and in the dark room, hoping the research may reveal a breakthrough.