By Sarah Kiefer
Could there be a better way to detect colon cancer than a colonoscopy? A less invasive test might depend on its association with microbes in the gut.
University of Missouri researchers looked at the overall microbiome in a rat model of human colon cancer to discern how differences in the bacteria affect adenomas, benign tumor growth that frequently is precursor to colorectal cancer. Those changes translated into a measurable difference that doctors could one day use to get a jump on this second leading cause of cancer death.
James Amos-Landgraf, an associate professor of veterinary pathobiology, worked with Lloyd W. Sumner, a professor of biochemistry and Bond Life Sciences Center researcher, on a multi-omics approach to analyze the molecules created by cell processes. By measuring large numbers of metabolites — small molecules that act as building blocks or energy sources for an organism — the team found when a unique combination of bacteria and metabolites are present, they correlate with higher susceptibility to colon cancer before any observable symptoms exist.
“In the rat model studies we could classify the severity of the cancer and tumor loads based upon earlier metabolic profiles,” Sumner said. “If you could achieve the same in people, the metabolic profiles could be used as a prognostic tool which would allow better alignment of treatments with the predictive severity. For example, you might want to be more aggressive with treatments in more severe cancer patients.”
Colorectal cancer is the second leading cause of cancer death and, currently, the only way to definitively diagnose it is to get a colonoscopy. If the adenomas are found and dealt with early, the survival rate is high.
This gut microbiome research is one of the projects Amos-Landgraf has been working on for the past 10 years, and the multi-omics approach sets it apart from the microbiome research of peers. Most experiments pick apart one or two bacteria samples and place them into an otherwise germ-free animal to see what happens, but the team wanted to analyze the entire stew of microbes present. Bacterium do not live alone, so what happens when the other bacteria recognize the new guy in the room?
“We know bacteria live in a community just like we do, and not everybody does the same job,” Amos-Landgraf said. “Just like we wouldn’t have the same lifestyle we have without someone providing our electricity, gas, or other utilities, the bacteria have requirements of their own.”
Amos-Landgraf and Sumner looked at this bigger picture, something they first partnered on in 2016. Sumner used his metabolomics expertise to profile and draw conclusions from the rat fecal samples.
“While this paper might not be foundational because other papers have noted several specific metabolites associated with cancers, it is a progressive step in the right direction towards better understanding of the metabolic relationship between the gut microbiome and forms of cancer,” Sumner said.
The analyses helped researchers find similarities and differences in small molecules present in rats with tumors in their colon versus those without.
“I think we have a scientific platform to really study these differences in more detail in a controlled system,” said Amos-Landgraf. “While we can now see a visible difference, this still leaves a lot of challenges to overcome and to answer.”
By looking in tandem at the transcriptome — the full range of messenger RNA that tells cells how to make proteins — they also saw what genes were turned on and off and correlated that with tumor presence. With tumors, they saw increases in biological processes to create bile for fat absorption, fatty acid breakdown and mucin that lubricates the gut. The team found certain species of bacteria in the gut microbiome gave way to specific metabolites and influenced genetic changes that turned off a gene that would normally suppress a tumor.
This sort of analysis is possible because of the advanced tools in MU’s Metabolomics Center.
Previously the way to study gut microbiome within the body was to look at the evidence and build a hypothesis from there in a sequential manner, which takes time and money. Now, the team can perform discovery based metabolic profiling on a larger scale with high resolution instruments that organize and categorize metabolites without having to test and re-test a hypothesis until they find a perfect fit.
“I’m an analytical chemist by training and I like instrumentation, but instruments cost money and they must solve problems,” said Sumner, who is also the director of MU’s Metabolomics Center and professor of agriculture biochemistry at the University of Missouri. “This is a project that utilizes problem solving and new knowledge, making it a measurable outcome. We just need one hundred more like it.”
Metabolic profiling is achieved through a series of a few experiments where the team uses gas and liquid chromatography to separate complex mixtures of metabolites. For example, gas chromatography uses a small, hollow tube coated on the inside that appears to be a piece of fishing wire to the naked eye. Samples are injected into the column and they move through this column to ultimately separate in a series based upon chemical interactions specific to each small molecule.
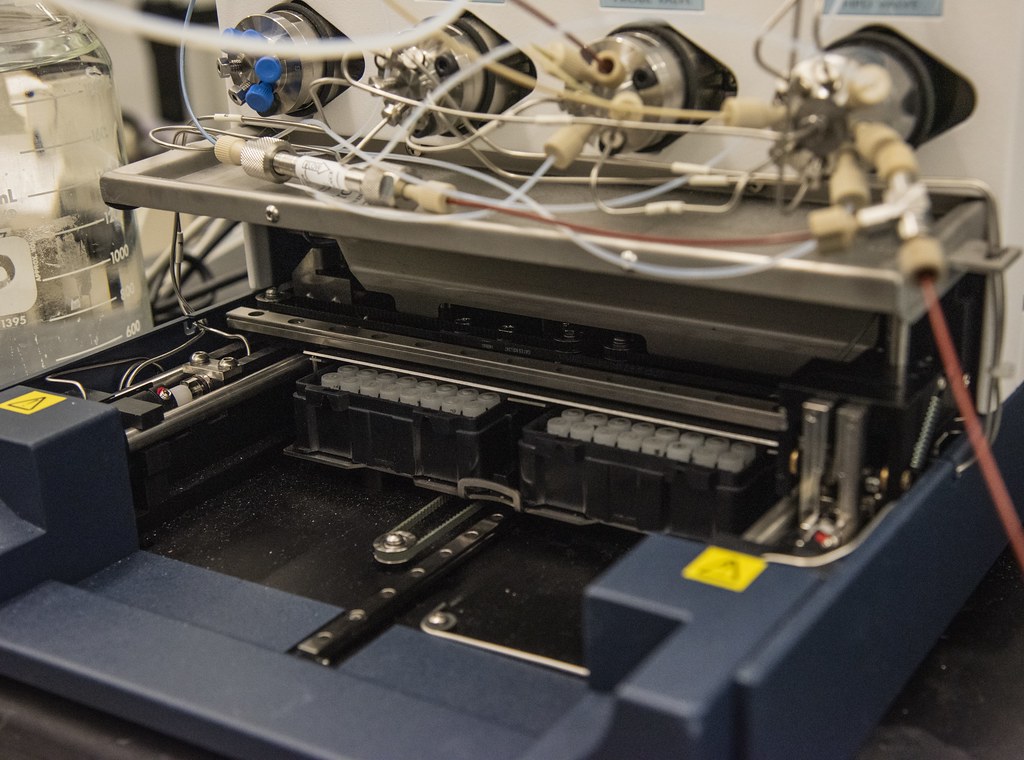
The chromatography system is coupled to a mass spectrometer that serves as a detector and weighs the molecules as they elute or are removed by washing with a solvent. The molecular weight helps to identify the metabolites. These methods provide an overview of some 1000 to 1500 metabolites that may change over time.
“A lot of my research is focused on building tools to help with the identification process of these molecules,” Sumner said. “Mass spectrometer will give us an accurate mass and molecular formula, but it doesn’t always tell us how those atoms and molecules are connected together.”
In these situations, they use nuclear magnetic resonance spectroscopy (NMR) which identifies molecules by irradiating and listening to responding radio frequency levels and matching them to a structure.
In 2022, a partnership with Stanford University led to placing a humanized microbiome into a mouse, creating a better model for studying our gut bacteria. Amos-Landgraf wants to do the same thing with their genetically modified animals to see how it affects those susceptible to development of cancer.
The next step for the team is to identify whether this metabolite correlation truly means a direct genetic relationship between these bacterial species, tumors and colon cancer.
The project could lead to more diagnostic tests in order to identify cancers early on in a patient’s history or it could be as simple as prevention through the taking of supplements or probiotics. Getting to the guts of this issue is what the project is all about, so they must take this information and dissect it to draw accurate conclusions.
“Science doesn’t happen in a vacuum and one of the reasons that people stay in science is because they want to solve a piece of the puzzle and step back to study how and why it comes together,” Amos- Landgraf said.
The paper titled, “Integrated multi-omic analyses provide insight into colon adenoma susceptibility modulation by the gut microbiota” by Susheel Bhanu Busi, Zhentian Lei, Lloyd Sumner, and James Amos-Landgraf was first published in the American Society for Microbiology journal mSystems on July 17, 2023.