Shan-Lu Liu initially thought it was a mistake when a simple experiment kept failing.
But that serendipitous accident led the Bond Life Sciences Center researcher to discover how a protein prevents mature HIV from leaving a cell.
Proceedings of the National Academy of Sciences published this research online Aug. 18.
“It’s a striking phenomena caused by this particular protein,” Liu said. “The HIV is already assembled inside the cell, ready for release, but this protein surprisingly tethers this virus from being released.”
The TIM – T-cell/transmembrane immunoglobulin and mucin – family of proteins hasn’t received much attention from HIV researchers, but recent research shows the protein family plays a critical role in viral infections. From Ebola and Dengue to Hepatitis A and HIV, these proteins aid in the entry of viruses into host cells.
But its ability to stop the virus from leaving cells remained unknown until now. Liu’s lab stumbled onto this finding in November 2011 when trying to create stable cells for a different experiment. After two months of troubleshooting the HIV lentiviral vector – where genes responsible for creating TIM-1 proteins were inserted into a cell to create a stable cell line that expresses the protein – Liu was confident the vector’s failure was not only interesting but also important.

The lab spent the next two years trying to figure out what was happening. Minghua Li, an MU Area of Pathobiology graduate student, carried out experiments that confirmed the protein’s power to inhibit HIV-1 release from cells, reducing normal viral infection. His experiments showed TIM proteins prevent normal deployment of HIV, created by an infected cell, into the body to propagate.
TIM proteins stand erect like topiary on the outside and inside surfaces of T-cells, epithelial cells and other cells. When a virus initially approaches a cell, the top of each TIM protein binds with fats – called phosphatidylserine (PS) – covering the virus surface. This allows a virus, such as Ebola virus and Dengue virus, to enter the cell, infect and replicate, building up a population inside.
But as the virus creates new copies of itself, the host cell’s machinery also incorporates TIM proteins into new viruses. That causes problems for HIV as it tries to leave the cell. Now these proteins cause the viruses to bind to each other, clumping together and attaching to the cell surface.
“We see this striking phenotype where the virus just accumulates on the cell surface,” said Liu, who is also an associate professor in the MU School of Medicine’s Department of Molecular Microbiology and Immunology. “We consider this an intrinsic property of cellular response to viral infection that holds the virus from release.”
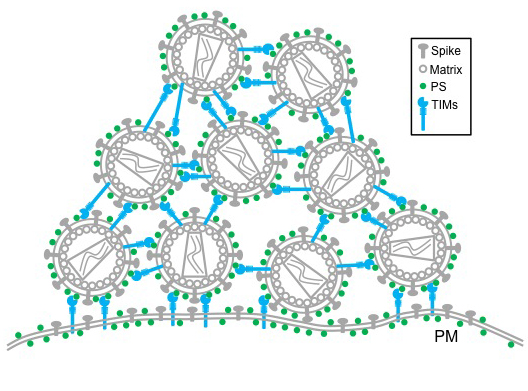
Further research is needed to determine overall benefit or detriment of this curious characteristic, but this discovery provides insight into the cell-virus interaction.
“This study shows that TIM proteins keep viral particles from being released by the infected cell and instead keep them tethered to the cell surface,” said Gordon Freeman, Ph.D., an associate professor of medicine with Harvard Medical School’s Dana-Farber Cancer Institute, who was not affiliated with the study. “This is true for several important enveloped viruses including HIV and Ebola. We may be able to use this insight to slow the production of these viruses.”
The National Institutes of Health and the University of Missouri partially supported this research. Additional collaborators include Eric Freed, PhD, senior investigator with the National Cancer Institute (NCI) HIV Drug Resistance Program; Sherimay Ablan, biologist with the NCI HIV Drug Resistance Program; Marc Johnson, PhD, Bond LSC researcher and associate professor in the MU Department of Molecular Microbiology and Immunology; Chunhui Miao and Matthew Fuller, graduate students in the MU Department of Molecular Microbiology and Immunology; Yi-Min Zheng, MD, MS, senior research specialist with the Christopher S. Bond Life Sciences Center at MU; Paul Rennert, PhD, founder and principal of SugarCone Biotech LLC in Holliston, Massachusetts; and Wendy Maury, PhD, professor of microbiology at the University of Iowa.
Read the full study on the PNAS website and browse the supplementary data for this work. See more news on this research from the MU School of Medicine.