Ever wonder why scientists love science? We asked. Happy Valentine’s Day.
Research
Harm and response

We often think of damage on a surface level.
But for plants, much of the important response to an insect bite takes place out of sight. Over minutes and hours, particular plant genes are turned on and off to fight back, translating into changes in its defenses.
In one of the broadest studies of its kind, scientists at the University of Missouri Bond Life Sciences Center recently looked at all plant genes and their response to the enemy.
“There are 28,000 genes in the plant, and we detected 2,778 genes responding, depending on the type of insect,” said Jack Schultz, Bond LSC director and study co-author. “Imagine you only look at a few of these genes, you get a very limited picture and possibly one that doesn’t represent what’s going on at all. This is by far the most comprehensive study of its type, allowing scientists to draw conclusions and get it right.”
Their results showed that the model Arabidopsis plant recognizes and responds differently to four insect species. The insects cause changes on a transcriptional level, triggering proteins that switch on and off plant genes to help defend against more attacks.
The difference in the insect
“It was no surprise that the plant responded differently to having its leaves chewed by a caterpillar or pierced by an aphid’s needle-like mouthparts,” said Heidi Appel, Bond LSC Investigator and lead author of the study. “But we were amazed that the plant responded so differently to insects that feed in the same way.”
Plants fed on by caterpillars – cabbage butterfly and beet armyworms – shared less than a quarter of their changes in gene expression. Likewise, plants fed on by the two species of aphids shared less than 10 percent of their changes in gene expression.
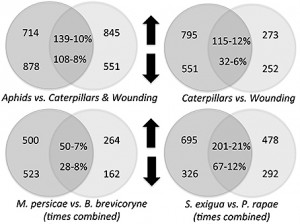
The plant responses to caterpillars were also very different than the plant response to mechanical wounding, sharing only about 10 percent of their gene expression changes. The overlap in plant gene responses between caterpillar and aphid treatments was also only 10 percent.
“The important thing is plants can tell the insects apart and respond in significantly different ways,” Schultz said. “And that’s more than most people give plants credit for.”
A sister study explored this phenomena further, led by former MU doctoral student Erin Rehrig.
It showed feeding of both caterpillars increased jasmonate and ethylene – well-known plant hormones that mediate defense responses. However, plants responded quicker and more strongly when fed on by the beet armyworm than by the cabbage butterfly caterpillar in most cases, indicating again that the plant can tell the two caterpillars apart.
The result is that the plant turns defense genes on earlier for beet armyworm.
In ecological terms, a quick defense response means the caterpillar won’t hang around very long and will move on to a different meal source.
More questions
A study this large has potential to open up a world of questions begging for answers.
“Among the genes changed when insects bite are ones that regulate processes like root growth, water use and other ecologically significant process that plants carefully monitor and control,” Schultz said. “Questions about the cost to the plant if the insect continues to eat would be an interesting follow-up study for doctoral students to explore these deeper genetic interactions.”
Frontiers in Plant Science published the primary study in its November 2014 issue. The sister study can be read here.
Big discoveries come in little (capsid) packages
It’s an understatement to say viruses are small.
But an average virus dwarfs the diminutive variety known as parvoviruses, which are among the most minuscule pathogens known to science.
Tucked inside a protective protein shell, or capsid, parvoviruses contain a single DNA strand of about 5,000 nucleotides. If parvo’s genetic material is like an hour-long stroll around your neighborhood, a bigger virus like herpes is equivalent to walking from St. Louis to Columbia, Missouri.

“I joke that we can do the whole parvovirus genome project in an afternoon, because it’s just taking it downstairs and having it sequenced,” said David Pintel, a Bond Life Sciences Center virologist and Dr. R. Phillip and Diane Acuff endowed professor in medical research at the University of Missouri. “It’s the size of one gene in the mammalian chromosome.”
But that little stretch of DNA still has plenty of tricks up its sleeve.
Pintel has spent nearly 35 years studying parvo and is one of the world’s foremost experts on the virus, but he’s still plumbing the tiny pathogen’s depths.
His lab focuses on unraveling how parvo interferes with a host cell’s lifecycle and understanding the virus’ quirky RNA processing strategies.
“Even though the virus is small, it’s not simple,” David Pintel said. “Otherwise we’d be out of business.”
Over the last two decades, parvo has become an important tool for gene therapy, an experimental technique that fights a disease by inactivating or replacing the genes that cause it. Researchers enlist a kind of parvovirus known as adeno-associated virus as a gene therapy vector, the vehicle that delivers a new gene to a cell’s nucleus. Pintel helped suss out the virus’ basic biology, an important step for developing effective gene therapy.
A varied virus
The name ‘parvo’ comes from the Latin word for ‘small.’ But the virus’ size makes it a resourceful, versatile enemy and a valuable model for understanding viruses and how they interact with hosts.
Parvoviruses fall into five main groups. They infect a broad swath of animal species from mammals such as humans and mice to invertebrates such as insects, crabs and shrimp.
Canine parvovirus, or CPV, is perhaps the best-known type.
It targets the rapidly dividing cells in a dog’s gastrointestinal tract and causes lethargy, vomiting, extreme diarrhea and sometimes death. In humans, Fifth disease, caused by parvovirus B19, is the most common. This relatively innocuous virus usually infects children and causes cold-like symptoms followed by a “slapped cheek” rash. There is no vaccine for Fifth disease, but infections typically resolve without intervention.
Reading the transcript
Pintel surveyed the whole parvo family to understand its idiosyncrasies.
To study bocavirus – a kind of parvo recently linked to a human disease – Pintel looked closely at the dog version, minute virus of canines (MVC). MCV serves as a good model for the human disease-causing virus. While examining MVC, he noticed an unexpected signal in the center of the viral genome. The signal terminates RNA encoding proteins for the virus’ shell, a vital part of the pathogen.
Finding such a misplaced signal in the middle of a stretch of RNA is like coming across a paragraph break in the middle of a sentence.
Pintel knew the virus bypassed this stop sign somehow, because the blueprint for the viral capsid lies further down the genome.
To overcome this stop sign, this particular parvovirus makes a protein found in no other virus. The protein performs double-duty for the virus: It suppresses the internal termination signal and splices together two introns, or segments of RNA that do not directly code information but whose removal is necessary for protein production. Splicing the introns together ensures that the gene responsible for producing the viral capsid is interpreted correctly.
Viruses and hosts: a game of cat and mouse
The conflict between a virus and a host is a constantly escalating battle of assault and deception.
Viruses need a host cell’s infrastructure to replicate, but have to fool or outmaneuver its defenses.
Pintel discovered one example of this trickery in mice, where parvo triggers a cellular onslaught known as the DNA damage response, or DDR. This type of parvo co-opts that defense. Normally DDR pauses the cell cycle to keep damaged DNA from being passed on to the next generation of cells, but parvo exploits that delay, buying time for the virus to multiply.
“For many, many hours the cells are just held there by the virus while the virus continues to replicate,” Pintel said. “And then that cell never survives; the virus kills the cell. It’s that holding of the cell cycle — which is part of the DNA damage response — that the virus hijacks to hold the cell cycle. It’s really cool.”
Parvo’s small size makes it especially beholden to their hosts. But that can make them particularly revelatory for researchers.
“It’s a twofold thing,” Pintel said, “Because it’s a virus that’s dependent on the cell, when you learn how the virus is doing these things, you learn how the cell does those basic processes. If we’re looking at a viral-cell interaction, yes, we’re looking at it from a viral point of view, but on the other hand we’re trying to understand the basic cellular process.”
Uncovering such nuanced interactions is a painstaking, laborious process that often goes unheralded by mass media. But those fundamental discoveries provide the building blocks upon which other researchers depend, said Femi Fasina, a postdoc in Pintel’s lab.
“When you understand basic biology, people can walk on those advancements. Although we don’t see the impact immediately, such things lead to breakthroughs that will revolutionize a lot of things.”
A small question
Despite his deepening understanding of how parvo works, there remains one debate about the virus that Pintel deems beyond the scope of his research: Are the tiny slivers of DNA that comprise parvoviruses even alive?
“I think that’s a crazy question,” he said. “It’s semantics. The virus is a genome. It goes into a cell, it doesn’t do anything until it’s inside of a cell and then it does stuff.” So whether you write parvoviruses into the book of life depends entirely on how you define the word ‘alive.’
“I put that in the realm of philosophy,” Pintel said, “not the realm of science.”
2015 Graduate Life Sciences Joint Recruitment Weekend highlights collaborative nature of research at Bond Life Sciences Center
Parkinson’s “trash pick-up problem”

It’s as if your recycling man quit his job and never came back.
Bags pile up to unexpected heights as waste continues to be generated and brought out to the curb. Day after day, the waste builds up as no one comes to pick them up.
For individuals with Parkinson’s disease, an accumulation of waste causes specific brain cells to die. The result is the onset of the disease.
But, instead of aluminum cans, plastics and paper, the waste that builds up in the brain cells of individuals with Parkinson’s is damaged mitochondria. Mitochondria are the cellular components that generate energy needed to keep cells alive.
When mitochondria is damaged and is no longer capable of making energy, it must be sent to the recycling center of a cell (called the lysosome).
Mark Hannink, a scientist at the Bond Life Sciences Center and professor of biochemistry at the University of Missouri, is peeling away the layers of this onion, one at a time.
His new research on a novel protein called PGAM5 (phosphoglycerate mutase family member 5) is pointing the way to finding a drug that can treat the disease.
Mitochondria suffer “wear and tear,” just like old cars
Mitochondria are endlessly helpful to a cell.
These powerhouses produce energy for a cell, control the cell division cycle and help regulate synapses. However, the mitochondrial proteins that produce energy eventually become damaged and no longer function properly.
“That’s part of the normal life cycle of mitochondria,” Hannink said. “Just like when the motor in an old car gives out due to wear and tear, that motor needs to be taken out and sent to the scrap dealer to be recycled and a new motor needs to be put in the car to keep it running.”
When mitochondria wear out they need to be sent to “recycling.”
“If that recycling pathway doesn’t work, the defective mitochondria will build up and will disrupt cell physiology, ultimately causing that cell to die” Hannink said.
Parkinson’s Disease is the clearest example of this recycling failure.
In early onset Parkinson’s, mutated proteins “forget” to take damaged mitochondria to the recycling center, resulting in build-up of toxic waste and, eventually, early onset of the disease.
“If the recycling mechanism isn’t functioning properly, those neurons die,” Hannink said.

Peptides behave like drug molecules
Hannink recently published research on the PGAM5 pathway in the Journal of Biological Chemistry along with MU graduate students Jordan M. Wilkins and Cyrus McConnell and fellow Biochemistry faculty member, Peter Tipton.
While its basic nature hides it from the view from the general public, this research takes a large step in the science of Parkinson’s disease.
Beyond defining the regulation of a pathway largely unstudied, their work discovered that a peptide regulates the pathway. Importantly, this peptide is able to alter the activity of the PGAM5 protein and stimulate an alternative recycling pathway for mitochondria.
Peptides are a clear signpost on a path toward drug development.
“Any time you can identify a biological process that is regulated by a peptide, that peptide becomes a lead candidate in the search for small, drug-like molecules that will act the same way,” Hannink said.
For Parkinson’s Disease, the goal is to find ways to repair the mitochondria recycling process.
“We propose that, by regulating PGAM5, it may be possible to restore mitochondrial quality control to dopaminergic neurons of patients with Parkinson’s and lessen the severity of the disease,” Hannink said.
What’s next?
While Hannink’s findings are exciting, there are also nuances to consider.
His research focuses on familial Parkinson’s disease, and it remains unknown whether sporadic Parkinson’s is also due to a defective mitochondrial recycling pathway. Sporadic Parkinson’s accounts for the vast majority of cases and typically affects older people.
It’s not clear if the PGAM5 pathway is also defective in those cases, Hannink said.
The next step of his research is to identify a small molecule that can regulate the PGAM5 protein in cells, just as the peptide did in his test-tube experiments.
Hannink thinks that development of a drug based on the PGAM5 pathway could be useful in restoring mitochondrial recycling in certain cells – like neurons affected in Parkinson’s – while blocking this recycling pathway in other cells, — like cancer cells.
The idea also needs to be tested using mice as a model system. The goal of those experiments will be to determine if the PGAM5 protein can stimulate alternative recycling pathways that can clean up and recycle damaged mitochondria pathway in neurons of mice.
“Mutant seeds” blossom in the pollen research field
The thought of pollen dispersed throughout the air might trigger horrific memories of allergies, but the drifting dander is absolutely essential to all life.
Science has long linked this element of reproduction with environmental conditions, but the reasons why and how pollen functions were less understood. Now lingering questions about the nuanced control of plants are being answered.
“Pollen is a very important part of the reproductive process and if we understand how pollen develops and how environmental stresses impinge on this process, we might be able to prevent crop loss due to high temperature or drought stress etc.,” said Shuqun Zhang, a Bond Life Sciences Center investigator.
Zhang has developed a new line of seeds that helped him and his lab identify an influential signaling pathway that triggers a chain reaction associated with normal pollen formation and function.
This research could lead to improvement to a plant’s response to disastrous environmental variables like drought to optimize pollen production and increase the production of food crops.
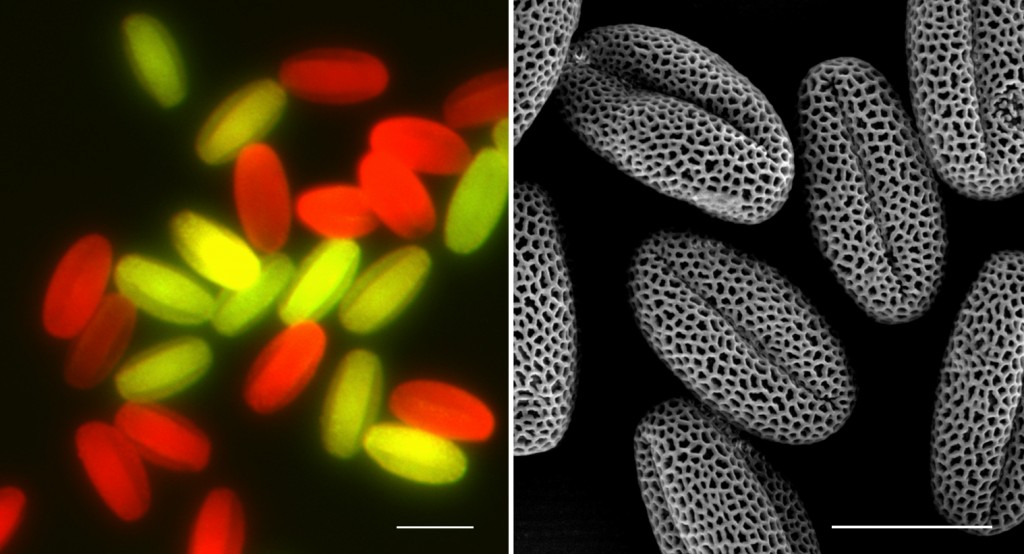
Seeds of success
Mutant seeds are the key to this work.
Instead of glowing green in the soil like you might see in a science fiction movie, they are providing important insight on plant reproduction and stress tolerance.
Zhang developed these plants from a mutant strain of Arabidopsis, a model plant used in scientific research. Certain genes were “switched off”to pinpoint where important pollen functions were signaled.
Using this mutant plant and seed system, Zhang found that WRKY34and WRKY2, two proteins that turn on/off genes, are regulated by MPK3and MPK6 “signaling” enzymes. These enzymes basically transform proteins from a non-functional state to a functional state, turning on specific duties or functions. Zhang, a professor of biochemistry at MU, began tinkering with the MPK3 and MKP6 pathways more than twenty years ago during his post-doc at Rutgers University.
Zhang’s research shows the newly identified MPK3/MPK6-WRKY34/WRKY2 pathway is a key switch in the hierarchy of the signaling system in pollen formation.
The research showed that the plant’s defense/stress response and reproductive process are linked, and the influential proteins MPK3 and MPK6 were part of the bigger WRKY34/WRKY2control pathway, which is activated in early pollen production.
The system is so useful that researchers across the country won’t stop asking for the seeds, Zhang said.
“We have a lot of requests for seeds,” Zhang said. “This is a very nice system to study pollen formation and function.”
The cascade of control
The functions of MPK3/MPK6 in plants can be compared to a “mother board” switch. The pathway — MPK3 and MPK6 —are part of a hierarchy of response, turning functions on or off. In other words, it’s a switch that controls a lot of different things. Controlling WRKY34/WRKY2 is one of the many roles played by MPK3 and MPK6.

“Whatever is plugged into it is what comes on,” Zhang said. “We are actually very, very interested in the evolutionarily context, how this came to be.”
This signaling process is just one of many in plants. MPK3 and MPK6 are two out the 20 MPKs, or MAPKs (abbreviated from Mitogen-Activated Protein Kinases) in Arabidopsis. They control plant defense, stress tolerance, growth, and development including pollen formation and functions.
“We determined that this MAPK-WRKY signaling module functions at the early stage of pollen development,” Zhang said.
The “loss of function of this pathway reduces pollen viability, and the surviving pollen has poor germination and reduced pollen tube growth, all of which reduce the transmission rate of the mutant pollen,” according to the research.
Zhang and his lab worked with the MU Division of Biochemistry and Interdisciplinary Plant Group on the research, which published in PLoS Genetics in June of this year.
A world without pollen production and defense
Without pollen, plants would not reproduce — there aren’t any Single Bars in the plant world (that we know of) — and if plant generations don’t propagate, there would be no air or food for human life to sustain.
“The factors such as heat and drought stresses cause problems to the plant’s normal developmental process and that’s how pollen fails to develop,” Zhang said. “If we understand the process, and know how environmental factors impact negatively the process, we can then make plants that can handle environmental stress better.”
Zhang and his lab continue to research the complexities of these pathways. Next on the quest is to answer how MPK3/MPK6 are involved in pollen functions such as guiding the pollen tube growth towards ovule to complete the sexual reproduction process in plants.
“It is possible that MPK3 and MPK6 are activated quickly in response to the guidance signals,” he said. “There’s still a long way to go because very few players in this process have been identified, we try to understand the biological process how they work together.” This research is in collaboration with Dr. Bruce McClure, also professor of Division of Biochemistry.
Read more:
1. PLoS Genetics (May 2014): Phosphorylation of a WRKY Transcription Factor by MAPKs is Required for Pollen Development and Function in Arabidopsis — Funded by a Hughes Research Fellowship and grants from the National Science Foundation.
2. Plant Physiology (June 2014): Two Mitogen-Activated Protein Kinases, MPK3 and MPK6, are required for Funicular Guidance of Pollen Tubes in Arabidopsis — Funded by a National Science Foundation grant and a NSF Young Investigator Award.
The only thing you need to read about Ebola today: An expert Q&A
News headlines seem to feverishly spread as if they were a pandemic of the brain.
Ebola hemorrhagic fever has been the most talked about disease of the year, appearing in thousands of headlines across the world since May. Through the noise of misinformation and sensationalism, fundamental information about the pandemic becomes harder to distinguish.
In an interview with Decoding Science on Tuesday, Shan-Lu Liu, MD, PhD, a Bond Life Sciences Center investigator who studies Ebola, weighed in on the latest news.
Liu, also an associate professor in the MU School of Medicine’s Department of Molecular Microbiology and Immunology, and his lab are particularly interested in the early behaviors of the virus in transmission and how it can navigate around the host immune response.

Q: Talk about the transmission. Ebola doesn’t spread through air, but how easily can it be transmitted through fluids?
A: It’s hard to say. It’s really not like: touch an infected person and you got it. I don’t see that could happen so easily. As an RNA virus, it’s not that stable outside of the body, unlike hepatitis B virus (HBV) where you need to boil the virus for 10 minutes and it becomes not infectious. Because Ebola is not that stable, that should not be the reason why it’s so efficient to transmit.
I think the transmission is one of the biggest things it’s, you know, I don’t think we have a complete understanding. We do know that it spreads by contact through body fluids and many people don’t realize that the handling of the deceased — that’s very dangerous. Touching broken skin or mucous membranes like the nose and mouth is dangerous.
Q: Talk about the incubation period and how that relates to symptoms and spreading of the virus.
A: The incubation time is 2-21 days. At first, the person will have flu-like symptoms, so you know, that’s why it’s hard to notice in the early stages. Some doctors or nurses say ‘just give him antibiotics send him home.’ But in stage two, you get the hemorrhage and it gets serious. The mortality rate is high, from 50 to 90 percent.
I think the fatality is definitely related to the late stages of the disease, especially with the hemorrhaging fever. The early stages are almost unnoticeable but that’s the time transmission might spread easier through contact with an infected person’s fluid. Before symptoms, the virus doesn’t spread.
Q: The Centers for Disease Control and Prevention said the virus could infect 1.4 million people in West Africa. Is this a realistic expectation?
A: You know, it could happen. It’s a prediction again, right? But I think the agency and the scientific community need to look at this prediction very carefully. What could be done in terms to prevent this from happening? It’s alarming.
Q: Last week, an article seemed to contradict with the CDC estimate. The headline: “Some good news about Ebola: It won’t spread nearly as fast as other epidemics.“ What do you make of that?
I don’t know, it’s hard for me to make a comment. Nobody knows. Things can always change. We didn’t expect to see a diagnosis in the United States — like this you know, this patient from Liberia was able to travel on a plane from virus country. Who can expect that? Anything can happen. There seem to have been some mishaps because he came from that area, right? Communication is more important now but it’s hard to predict because anything could happen.
Q: How has the Ebola virus behaved in previous outbreaks?
A: The first outbreak was in 1976 in Sudan and Congo — (Democratic Republic of Congo, known as Zaire at the time). It was from contaminated needles in a hospital and originally came from fruit bats — they are one of those animals that could transmit Ebola from animals to humans. The fruit bats transmitted the virus to primates, primates transmit to humans. It’s hard to notice in the early stages.
Editor’s note: The 1976 outbreak was the first occurrence of Ebola in humans. The outbreak affected one village, infecting 318 people that resulted in 280 deaths.
Q: Much of the media has reported a vaccine for ebola was delayed. How could this happen?
A: Drugs and vaccines are a little different. The Ebola vaccine was delayed, that’s for sure. That’s because, the vaccine on trial has to go through tedious steps to get approval and so thats why when this outbreak occurs the NIH (National Institutes of Health) decides to go ahead quickly. One of the things for ebola vaccine is um, the pharmaceutical companies and the industries are not interested in developing vaccines. Do you know why? It is not a big market. Only a hundred — or a thousand or more — people will be infected by ebola, unlike other vaccines like the HPV vaccination where 200 million people need it. The companies are not interested in developing it, because there’s no money in it.
A company needs to spend a lot of money to develop a vaccine, but they don’t see the market — the market can’t do it. But somebody needs to do it. Imagine if, if the virus spread like this, you know, unpredictable, it could be worse. In terms of therapy, the drugs and antibodies, we know they are really effective. And they are specific, so they can reach the market effectively.
Q: Will a drug be enough to prevent wide spreading of Ebola?
I think the companies and governments are speeding up to make those available. To see this prediction (the CDC 1.4 million estimate), they have to be prepared. People have put increasing attention on antibodies because a vaccine is not in the near future. So what’s the approach? A “therapeutic vaccine.” The so-called therapeutic vaccine is an antibody so you engineer, you use you know, molecular engineering technique to generate those antibodies and they can neutralize and block viral infection. It’s more realistic for Ebola and even for HIV. The HIV vaccine has failed so many times. So that’s why I think one of the new approaches is to use a new broad neutralizing antibody.
Q: Does Ebola stay in the body, like chicken pox?
A: Ebola do not cause latent infection. HIV can become latent and become chronic. So influenza virus, ebola viral infection and others normally do not lead to latency. I think for Ebola — for this type of infection — once you block the patient and clear the virus it should be good.
Q: Has the media done a good job in educating the public?
I think in terms of news coverage they are pretty careful. I looked at the news conference by the CDC director and by those doctors in Dallas, and when they make statements they are careful not to exaggerate and also give very cautious measurements. The news media need to be aware of the danger of the virus. In the meantime, you have to be aware of the possibility of being affected.
Again, I think it is a very important problem. It’s important to let the public know the situation. If you see people who have recently traveled from those West African countries, you have to be cautious — air travel is so common. But I think the media have generally done a good job.
Q: Has the government done a good job keeping the pandemic under control?
I don’t know what they do. The air travel is a problem. Intensified screening process, that should definitely be done. It’s very bad for people from the outbreak area, and I just hope that this community won’t be affected.
To control, they should be careful. A person with any sign of the disease — they need to be quickly monitored and treated.
Q: What’s the most important take-away message for the public?
A: I think it’s an important problem and we need to solve it urgently. I hope this outbreak will teach us a lesson in terms of how important emerging infectious viruses are as it comes and goes is to public health. Based on literature and reports, if people do not have obvious symptoms, they do not produce an infectious virus. The incubation time has a big range but again, we are still trying to understand the process better. Infection is a complex process. We need to better understand the viral transmission so I think for now, we need to be very cautious.
Liu and his lab do not work with the contagious Ebola virus on University of Missouri campus. All of the studies involve use of a recombinant or pseudotyped Ebola virus which is not infectious.
The search for oxidative stress treatment continues
University of Missouri research characterizes a novel compound
By Paige Blankenbuehler | Bond LSC
Your body has an invisible enemy.
One that it creates all on it’s own called oxidative stress, long thought of as an underlying cause of some of humanity’s most insidious diseases – cancer, Alzheimer’s, Parkinson’s Disease, cardiovascular disease and diabetes.
Every day, our bodies are exposed to harmful free radicals known as reactive oxygen species as a result of our environment.
But, when something goes wrong with this energy extraction process, cells become inundated with reactive oxygen compounds that cause oxidative stress. The search for drugs to treat the problem have been ongoing, and with a complicated problem like oxidative stress, it’s all about finding the right combination.
Recent research by Bond Life Sciences Center investigator and Biochemistry Department Professor, Mark Hannink, provides a new approach for addressing the problem of oxidative stress and a starting point on developing a drug in pill form.
Mark Hannink and Kimberly Jasmer, a Ph.D. student in his lab, recently helped characterize a new molecule (called HPP-4382) that provides a novel way to treat oxidative stress. Their research was done in a partnership with High Point Pharmaceuticals, LLC, of North Carolina, where this new molecule was developed.
Oxidative stress can cause damage to the building blocks of a cell, resulting in excessive cell proliferation in the case of cancer or cell death in the case of neurodegenerative diseases like Parkinson’s.
Often, the majority of stressors are actually created inside our own cells as a byproduct of how our cells extract energy from the food that we eat and the air that we breathe.

Understanding oxidative stress
Most simply, oxidative stress is an imbalance that happens when the body uses oxygen to produce energy.
Superoxide, a “promiscuous and nonspecific” compound produced as a byproduct of this process, is a highly reactive molecule that can damage DNA and other cellular components, Hannink said.
The superoxide molecule is a “free radical.” That means it’s especially promiscuous and reacts with many different types of cellular molecules, leaving destruction in its wake, he said.
That damage can lead to a long list of problems, including cancer or neurodegenerative diseases like Parkinson’s disease.
In response to oxidative stress, the cell produces protective “anti-oxidant” proteins, which help remove the harmful reactive oxygen species and minimize damage.
But a heavy anti-oxidant response could be dangerous, too. The bottom line: it’s about maintaining a fine balance between “oxidants” and “anti-oxidants”.
Search for right combination continues
A drug that corrects the imbalance of oxidative stress could one day have wide applicability.
Jasmer developed a test to measure how specific compounds altered gene expression. The genetic response to oxidative stress has both an “ON” switch and an “OFF” switch.
Using this test, Jasmer determined how each compound affected specific genetic switches and, in turn, how the response to oxidative stress is regulated.
This test helped identify which molecules might be promising candidates for treating oxidative stress, leading them to one in particular that seemed to have the desired properties: HPP-4382.
But creating effective drugs is a long process of trial and error. Once molecules have been identified that show efficacy in lab-based assays, scientists try different combinations to increase their potency and drug-like properties, and High Point is currently testing other molecules that behave like HPP-4382.
The compound serves as a good starting point for researchers who are interested in understanding how oxidative stress affects cellular processes, such as cell proliferation or cell death.“Now we have a better understanding of what this compound is doing,” Hannink said. “This compound can be used to test different ideas of how the balance between oxidants and anti-oxidants is achieved in healthy cells and how perturbation of this balance can lead to different diseases.”
This research was published in PLoS One in July and was funded by High Point Pharmaceuticals, LLC.
Holding on: Bond LSC scientist discovers protein prevents release of HIV and other viruses from infected cells
Shan-Lu Liu initially thought it was a mistake when a simple experiment kept failing.
But that serendipitous accident led the Bond Life Sciences Center researcher to discover how a protein prevents mature HIV from leaving a cell.
Proceedings of the National Academy of Sciences published this research online Aug. 18.
“It’s a striking phenomena caused by this particular protein,” Liu said. “The HIV is already assembled inside the cell, ready for release, but this protein surprisingly tethers this virus from being released.”
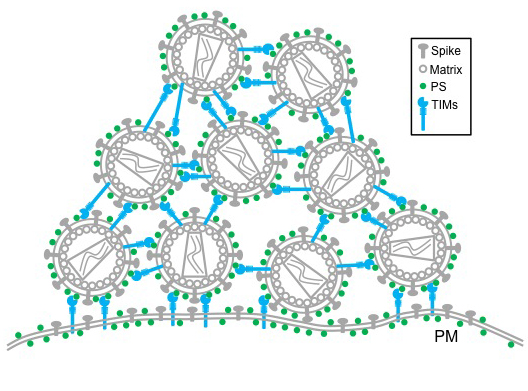
The TIM – T-cell/transmembrane immunoglobulin and mucin – family of proteins hasn’t received much attention from HIV researchers, but recent research shows the protein family plays a critical role in viral infections. From Ebola and Dengue to Hepatitis A and HIV, these proteins aid in the entry of viruses into host cells.
But its ability to stop the virus from leaving cells remained unknown until now. Liu’s lab stumbled onto this finding in November 2011 when trying to create stable cells for a different experiment. After two months of troubleshooting the HIV lentiviral vector – where genes responsible for creating TIM-1 proteins were inserted into a cell to create a stable cell line that expresses the protein – Liu was confident the vector’s failure was not only interesting but also important.

The lab spent the next two years trying to figure out what was happening. Minghua Li, an MU Area of Pathobiology graduate student, carried out experiments that confirmed the protein’s power to inhibit HIV-1 release from cells, reducing normal viral infection. His experiments showed TIM proteins prevent normal deployment of HIV, created by an infected cell, into the body to propagate.
TIM proteins stand erect like topiary on the outside and inside surfaces of T-cells, epithelial cells and other cells. When a virus initially approaches a cell, the top of each TIM protein binds with fats – called phosphatidylserine (PS) – covering the virus surface. This allows a virus, such as Ebola virus and Dengue virus, to enter the cell, infect and replicate, building up a population inside.
But as the virus creates new copies of itself, the host cell’s machinery also incorporates TIM proteins into new viruses. That causes problems for HIV as it tries to leave the cell. Now these proteins cause the viruses to bind to each other, clumping together and attaching to the cell surface.
“We see this striking phenotype where the virus just accumulates on the cell surface,” said Liu, who is also an associate professor in the MU School of Medicine’s Department of Molecular Microbiology and Immunology. “We consider this an intrinsic property of cellular response to viral infection that holds the virus from release.”
Further research is needed to determine overall benefit or detriment of this curious characteristic, but this discovery provides insight into the cell-virus interaction.
“This study shows that TIM proteins keep viral particles from being released by the infected cell and instead keep them tethered to the cell surface,” said Gordon Freeman, Ph.D., an associate professor of medicine with Harvard Medical School’s Dana-Farber Cancer Institute, who was not affiliated with the study. “This is true for several important enveloped viruses including HIV and Ebola. We may be able to use this insight to slow the production of these viruses.”
The National Institutes of Health and the University of Missouri partially supported this research. Additional collaborators include Eric Freed, PhD, senior investigator with the National Cancer Institute (NCI) HIV Drug Resistance Program; Sherimay Ablan, biologist with the NCI HIV Drug Resistance Program; Marc Johnson, PhD, Bond LSC researcher and associate professor in the MU Department of Molecular Microbiology and Immunology; Chunhui Miao and Matthew Fuller, graduate students in the MU Department of Molecular Microbiology and Immunology; Yi-Min Zheng, MD, MS, senior research specialist with the Christopher S. Bond Life Sciences Center at MU; Paul Rennert, PhD, founder and principal of SugarCone Biotech LLC in Holliston, Massachusetts; and Wendy Maury, PhD, professor of microbiology at the University of Iowa.
Read the full study on the PNAS website and browse the supplementary data for this work. See more news on this research from the MU School of Medicine.
Critical transport: Bond LSC team finds boron vital for plant stem cells, corn reproduction
Carbon’s next-door neighbor on the periodic table typically receives little attention, but when it comes to corn reproduction boron fills an important role.
According to University of Missouri scientists, tiny amounts of boron play a key part in the development of ears and tassels on every cornstalk. The July 2014 edition of the journal Plant Cell published this research.
“Boron deficiency was already known to cause plants to stop growing, but we showed a lack of boron actually causes a problem in the meristems, the stem cells of the plant,” said Paula McSteen, a Bond Life Sciences Center researcher. “That was completely unknown before, and for plant scientists that’s an important discovery.”

Meristems are a big deal to a plant. These pools of stem cells are the growing points for each plant, and every organ comes from them. They are how plants can survive for 500 or 5,000 years, continuously making new organs in the form of leaves, flowers, and seeds throughout its life.
“When you mow your grass, it keeps growing because of the meristems,” said Amanda Durbak, first author on the paper and MU biological sciences post doc. “In corn, there are actually hundreds of meristems at the tips and all sides of ears and tassels.”
But without enough boron, these growing points disintegrate, and, in corn, that means vegetation is stunted, tassels fail to develop properly and kernels don’t set on an ear. This leads to reduced yield. Missouri and the eastern half of the U.S. are typically plagued by boron-deficient soil, an essential micronutrient for crops like corn and soybeans, indicating that farmers need to supplement with boron to maximize yield.
The tassel-less mutant
The team’s discovery started with a stunted, little corn plant that just couldn’t grow tassels, only created a tiny ear and died within a few weeks. These maligned reproductive organs piqued McSteen’s interest and her team of collaborators set out to figure out which gene was affected in this mutant plant. Graduate student Kim Phillips mapped the mutation to a specific gene in the corn genome involved in transporting molecules across the plant membrane.
But, what was this defect preventing the plant from receiving? Two experiments helped find the answer.
They started by looking at similar genes in other plants and animals. Simon Malcomber of California State University-Long Beach compared the gene – named the tassel-less gene after its mutant appearance – to similar genes in other plants and animals. He found that many were known to make a protein that transports boron and a few other elements.
From field to frog
To clinch this hypothesis, McSteen looked to Bond LSC scientist Walter Gassmann and the African clawed frog. Gassmann harvests eggs from these frogs and uses them to “test” the function of genes from both plants and animals.
“What we do is we inject the frog egg with RNA made in a test tube from the corn’s DNA,” Gassmann said. “The egg is a single, living cell that will actually use the message provided by this RNA to make the boron transporting protein and put it in the egg’s membrane.”
Frog eggs don’t naturally have the ability to transport boron, so an uninjected egg in a solution of boron can’t move the element into the cell.
“Corn RNA provides the egg with instructions to make a boron transporter protein, so the boron solution should move from outside to inside the egg,” Durbak said. “The egg should swell, showing this protein moves boron, and, in fact, these eggs swell so much they explode.”
A tank and a bucket guaranteed boron was the culprit. Durbak went back to the cornfield, watering some mutant tassel-less corn with boron fertilizer and other mutant plants with only water.
Only the ones given boron recovered and grew like normal corn plants, showing that the mutant corn has difficulties obtaining enough boron under natural, low boron conditions without this boron transporter. The boron content of the plants were later tested at the MU Research Reactor and the MU Extension Soil and Plant Testing Laboratory, affirming their observations.
A closer look
But, what does boron deficiency look like on a cellular level?
To see, the team collaborated with biochemist Malcolm O’Neill at University of Georgia. He looked at the cell walls in the plant and discovered that the pectin was affected. Pectin stabilizes the plant cell wall, and many home canners know pectin for its help in making jelly and jam solidify. Pectin is strengthened when boron cross-links two carbohydrates together, giving rigidity to the plant cell wall.
“The effect is that it locks in the cellulose, so without it plant cells won’t have nearly the stability,” McSteen said. “What we think is going on is that plant meristems basically disintegrate because they don’t have the support of pectin.”
While McSteen’s team identified the gene that controls the protein for boron transport into a cell, a research team from Rutgers University identified a gene that controls the protein that transports boron out of a cell. See more about both studies in Plant Cell’s “In Brief” section.
The next step in this research is to look more closely at what happens in these boron-deficient cells early on as they develop to understand the mechanism of boron action in stem cells.
A grant from the National Science Foundation supported this research.
In addition to their Bond LSC appointments, Paula McSteen is an associate professor of biological sciences in the MU College of Arts and Sciences and Walter Gassmann is a professor of plant sciences in the MU College of Agriculture, Food and Natural Resources.




