By Paige Blankenbuehler | MU Bond Life Sciences Center
There’s a criminal on the loose, striking every day. Millions fall victim, but there’s still no way to stop it. And, in all likelihood, you have been hurt by it.
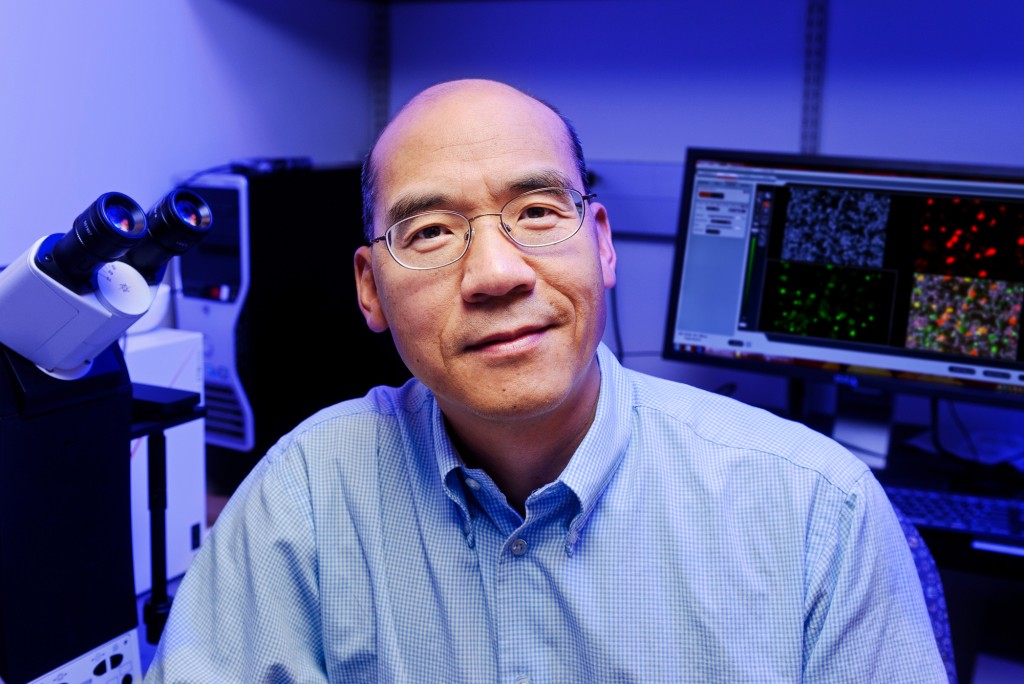
If inflammation is an unsolved criminal case of the last three decades, then Gary Weisman has been the detective. He’s certain there’s an accomplice — perhaps many — that may be triggering the discomfort.
The Bond Life Sciences Center investigator is slowly revealing what makes inflammation tick and what makes it strike. Each epiphany brings another question. He’s certain there’s a way to prevent negative effects of unsolved inflammation.

Weisman has dedicated his career to understanding the micro-processes behind inflammation. He’s become so specialized that his techniques can be as hard to crack as the case itself.
“I would not ask anyone to explain what I do,” Weisman says. Nonetheless, he’s been able to divide the process of inflammation into two categories: components that repair the body and components that lead to its destruction. This will help find inflammation’s many accomplices to figure out why humans work, and what their bodies do when they don’t work so well.
“I am interested in the meaning of life,” Weisman says. “Life has become simpler for me because the scientific method carries everywhere. I’ve become aware of how simple we are as a machine.”
Criminal or just misunderstood?
Most criminals adopt patterns, but inflammation stands as a signpost for mysterious, underlying problems.
Its effects are usually localized: an arm, a joint, the brain or a gland. You feel a temperature spike then the skin reddens in a part of your body. Later still, the skin tightens and pain comes at a snail’s pace.
Not even cells are safe. Inflammation even strikes on the molecular level.
But really, inflammation can be a good thing. It’s part of the immune system’s bag of tricks to signal the body to bring in reinforcements to fight off the invasion. Normally, inflammation corrects a physical problem, but if it is not successful in repairing a problem, inflammation can become chronic and accelerate tissue destruction.
Just like in an episode of CSI, Weisman puts the pieces of the inflammation puzzle together in his office by applying the expertise of Laurie Erb, Jean Camden and Lucas Woods — all donned in white lab coats, eyes pressed to the microscope examining evidence and building molecular evidence in the case.
The MU associate professor of biochemistry and his team have become a sort of grant-wielding wizards to sustain his pursuit of inflammation triggers. National Institutes of Health grant awards have sustained his lab for decades. The funding has come from varied sources such as the MU Food for the 21st Century Program, the Bond LSC, the Bright Focus Foundation, the American Heart Association and the Cystic Fibrosis Foundation. In recent years, research funding for Alzheimer’s disease and Sjogren’s syndrome (a disease of the salivary gland that causes dryness) have contributed, too.
But the funding source doesn’t matter because inflammation is the tie that binds.
Advancements, like recent mapping of the human genome, have moved his work forward to understand inflammation’s complexity. Each experiment he completes fills in another blank slate in the “human owner’s manual.”
“As humans, we’re so intent on the fact that we’re superior to all, but really we’re not,” Weisman says. “With the Human Genome project, we’ve come to understand that all living things have similar designs … we are on the verge of finding revolutionary solutions to preventing or reversing human diseases.”
A receptor all our own
One specific player in the body’s immune system has kept Weisman’s attention for most of his career. The P2Y2 protein is a nucleotide receptor, and his lab team members affectionately refer to it as “our receptor.”
Nucleotide receptors are regulatory molecules in red blood cells. What they regulate is nuanced, mostly undetermined and of great interest to scientists. Answering that question has become Weisman’s wheelhouse.
The body manufactures 15 different types of nucleotide receptors, all similar in construction, but each are believed to have subtly different functional roles. It’s as if Weisman and his lab is on the case of a highly organized crime ring.
“Our receptor is mainly present when inflammation occurs, and we’re trying to figure out its role in a variety of diseases,” Weisman says.
The P2Y2 receptor has been observed in Alzheimer’s patients, along with a plaque build-up in the brain, and the receptor was suspected of playing a role in the disease’s progression.
Weisman and his colleagues found that the deletion of the P2Y2 receptor in a mouse model of Alzheimer’s disease accelerates progression of plaque build-up, neurological symptoms and death. This suggests that the receptor has anti-inflammatory effects rather than being “guilty by association” with the tissue-destructive aspects of inflammation.
“It’s like I have this 30,000-piece jigsaw puzzle in front of me that I have to put together,” Weisman says. “What’s the difference between you and me? As a machine, surprisingly very little.”
This simplicity drives Weisman to continue solving the mysteries of inflammation and search for its underlying chemical processes. By understanding the body’s chemical reactions, he believes treatments can be developed to focus the immune system on repairing damaged tissues.
Through studying his receptor, Weisman is breaking up inflammation’s crime ring.