First-ever analysis compares nearly gapless genome across cat species and with humans to shine light on evolution
By Roger Meissen | Bond LSC
The tiger doesn’t know it, but a difference deep in its genome sets it apart from other cats.
This big cat preserved a distinct sense of smell thanks to a few chromosomes it uniquely retained over millennia of evolution that other feline species did not.
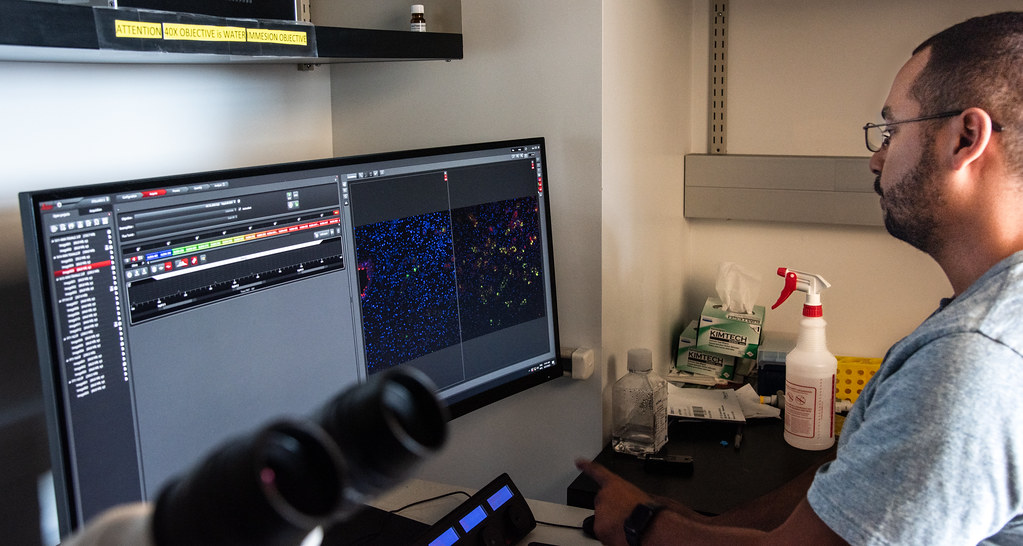
A study published in the journal Nature Genetics details this finding where scientists from the University of Missouri partnered with Texas A&M University and others to compare, for the first time, nearly gapless cat genomes across multiple species and with humans. This genome map correctly strings together nearly all chromosomes of these felines — from one end to the other without missing segments — to give an animal genetic reference that rivals the human genome.

“This study is a comprehensive look at the genome sequence structures that could be driving cat speciation, the evolutionary process by which populations became distinct species,” said Wes Warren, a Bond Life Sciences Center researcher and MU professor of genomics. “We found distinct differences between how great apes, including humans, and cat sequences evolved in a similar period of time. There were many interesting differences to consider, such as why certain chromosome regions have similar
but novel sequence landscapes even across species of cats.”
In tigers, scientists previously identified genes associated with the chemosensory system, perhaps lending them heightened senses of smell key to their survival.
“We see cats have olfactory receptors for sensing smell that are greater than most mammals but, among cats, the tiger stands out with the largest repertoire,” Warren said. “We know tigers have a solitary lifestyle, and that acute sense of smell helps males detect females for mating and lets them avoid the territory of other males to enhance their chances of survival.”
When thinking about other unique sensory features to explore, researchers also looked to the fishing cat, a rare nocturnal feline native to marshy areas in Southeast Asia. They searched for molecular signatures that would explain its sleek body, partially webbed feet and prowess for hunting fish, characteristics tailored to its life around water.

“There are specific olfactory receptors that detect waterborne odorants as well as a much larger number of receptors tailored for terrestrial smell detection; we asked if fishing cats would have more functional receptors for waterborne odorants than other cats, and that’s exactly what we saw,” Warren said. “In fishing cats, all gene copies were complete while other cats had broken copies, suggesting natural selection was at work because of their aquatic hunting behavior.”
While these illustrations are notable, scientists also learned from their extensive analysis of sequence structural variation.
What stood out overall were fewer differences in cat chromosomes — coding for traits like hair length and color, bone structure and size, fertility and senses — compared to great apes despite both families diverging into distinct species over a period of 13 million years.
Using the latest assembly techniques, the scientists pieced together the genomes of multiple cat hybrid species — the Bengal cat, the safari cat and the liger — and then compared the five lineages. New techniques severely limited genomic dark matter, which is genetic information researchers previously were unable to characterize and assign meaning to, thus enabling them to shine new light on this novel sequence structure.
Some of those pieces came from Leslie Lyons, the Gilbreath-McLorn Endowed Professor of Comparative Medicine in veterinary medicine and surgery at the MU College of Veterinary Medicine. She sequenced two of the cat hybrids nearly 25 years ago.
“I was at the National Cancer Institute as a postdoc when I sequenced the Geoffroy’s cat and Asian leopard hybrids, and I had no idea they would ever be reused for a project like this, but fortunately they were,” Lyons said. “Now, we have a cat genome that is comparable to the human genome, which is one of the best in the scientific world, so the cat has really leapt forward in terms of genomic resources.”
Structural differences in the X chromosome, in particular a region that displays features of a supergene, stood out to the team. This presumed supergene in cats, a complex interaction where a chromosomal region of neighboring genes are inherited to fix a trait with fitness consequences, could be the process of evolution that led to hybrid sterility if certain feline species cross. It is evolving faster than most of the cat genome. Much like the mule — a cross of a male donkey and a female horse — cannot produce offspring, the liger bears no offspring when lions and tigers mate. The team hypothesizes this is due to a failure in meiosis, the process that produces egg and sperm cells.
“We found multiple inversions on X chromosomes of interest in cats that include the complex DXZ4 region,” Warren said. “In these cats, we identified entire copies of this complex, repetitive region that harbors the only X-linked speciation gene identified in mammals.”
While these advances in genetics push our understanding of evolution in exciting new ways, Lyons has much more pragmatic uses for this work.
“I’m an end user,” Lyons said. “These good genetic maps allow us to find mutations that cause inherited diseases in cats, so this genome allows us to bring precision medicine to feline veterinary health care.”
Bill Murphy, paper co-lead and professor of veterinary integrative biosciences at Texas A&M, points to the future value of this work.
“Our findings will open doors for people studying feline diseases, behavior and conservation,” he said. “They’ll be working with a more complete understanding of the genetic differences that make each type of cat unique.”
Nature Genetics published “Single-haplotype comparative genomics provides insights into lineage-specific structural variation during cat evolution” Nov. 2, 2023. The study was conceptualized by Bill Murphy — VMBS professor of veterinary integrative biosciences at Texas A&M and Wes Warren — principal investigator in the Bond Life Sciences Center and professor of genomics in the College of Agriculture, Food and Natural Resources at the University of Missouri. Additional collaborations involved researchers from the University of Washington, University College Dublin, the Institute for Systems Biology in Seattle, Louisiana State University and the Guy Harvey Oceanographic Center.
This work was funded through a grant from the Morris Animal Foundation, which works to improve and protect the health of animals through scientific innovation, education and inspiration.