By Jennifer Lu |Bond LSC

When developmental plant geneticist Paula McSteen thinks about the specimens she studies, one word comes to mind: potential.
She thought it as she stood in the midst of the first corn field she ever planted as a post-doctoral fellow in corn genetics.
She thinks it as she counts kernels from corn crosses that will be sent to Hawaii, a hotspot for corn geneticists looking to add a second harvest to their research year.
And she sees it in the students she mentors as a professor of biological sciences at MU and a researcher at the Bond Life Sciences Center.
Embracing the corn
McSteen entered the field of corn genetics 21 years ago, as a post-doctoral fellow in Berkeley.
“What’s really amazing is that when you plant a field of corn, the field is just bare,” McSteen says.
“A few weeks later, you come back and your plants are this high,” she says, gesturing with her hands. “And a few weeks later, they’re this high. And a few weeks later, they’re massive and it’s all just coming from nothing. The instructions are there in the seeds, but otherwise the plant is taking nutrients and water from the soil and using the air and sun to generate sugar for growing. It’s amazing. You come back and you’ve got this whole field of corn.”
During her first corn season, McSteen remembers being surrounded as far as the eye can see by corn as tall as people. “I feel like that’s one reason why people get into corn. You’re not staring down into a microscope, you’re embracing it. It’s right there in front of you.”
The feeling hasn’t gone away.
“When I see my plants,” McSteen says, “I’m excited about what’s going on with them. What could be happening here? What’s the meaning of these results?”
McSteen, who is Irish, grew up far removed from the sunny cornfields she works in. As a child in Dublin, she wasn’t particular outdoorsy. When her family went camping by the sea over the summer holidays, McSteen spent most of her time reading books. Her favorite subject in school was science. By the time she sat for her high school leaving exam, her classes were mainly in geography, science or math.
“I’ve always been fascinated with genetics,” McSteen says. Ever since she learned about Punnett squares in high school, its puzzle-like quality has appealed to her. “I just loved that you could figure out what the prediction could be from a certain cross.”
She applied and was accepted to her top choice university, Trinity College Dublin, where she studied genetics. She went on to pursue a PhD in Norwich, England studying how snapdragons make flowers.
When it came time for her to do her post-doc, she had the choice to work on Arabidopsis, a popular plant model, or maize, a crop with many opportunities for research funding. She chose the latter. The decision changed the course of her career, from her research focus to her country of residence.
Corn brought her to California, Pennsylvania and then the University of Missouri, which has a long history of corn genetics.

Everything we do starts with mutants
McSteen studies a part of corn plants called the meristem, which is filled with stem cells that become the reproductive organs of the plant: the tassel and the ears.
The tassel, where pollen is produced, is found at the top of the cornstalk, while the ears, which are the female reproductive organs, jut out from the sides. When and how they form depend on a growth hormone called auxin.
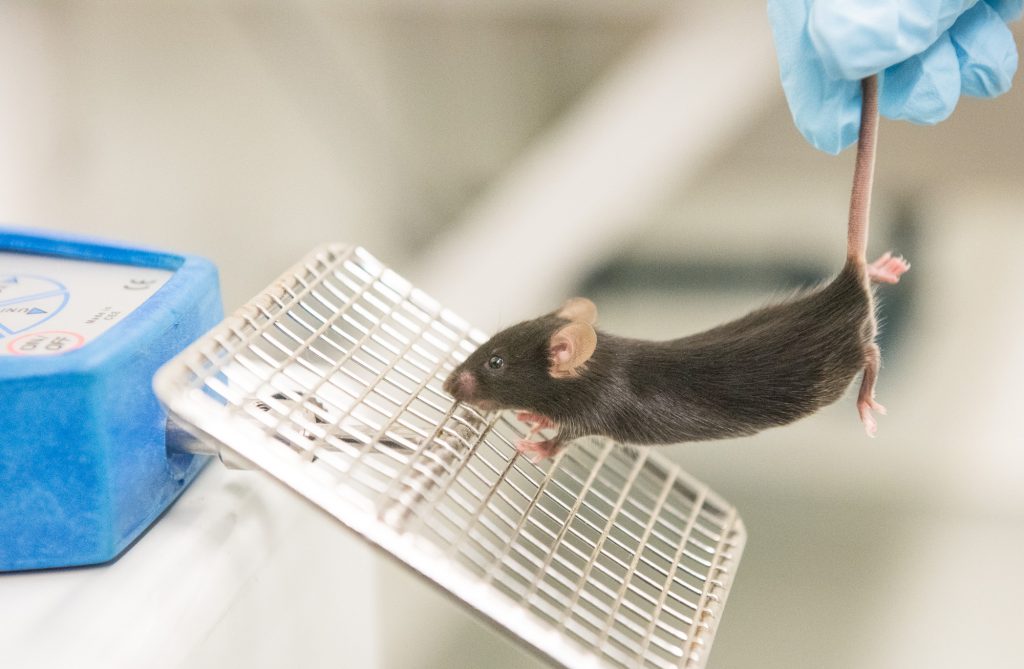
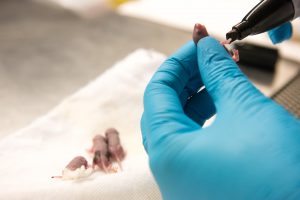
To understand auxin regulation, McSteen begins every summer with a field full of mutants. Each kernel contains a mutation, but it’s impossible to tell at first what is causing the mutation.
“To me, every single mutant is just potential. You can’t wait to find out what is mutated. You never know what you’re going to end up with.”
McSteen is interested in mutations that affect the tassels or ears. These plants produce ears with fewer kernels or tassels with fewer branches. Or they fail to make ears or tassels altogether. The defects are outward signs of problems in meristem development, and hint at disruptions to genes that are involved in how auxin is made, transported or perceived by the plant.
Once a mutant is identified, McSteen works backwards to find out which gene is causing the mutation, and where it is located on the chromosome. To date, her group has identified multiple genes related to auxin-mediated development, as well as two genes that affect the uptake or synthesis of essential nutrients.
A third project revolves around a strain of corn that produces half as many kernels as regular corn, causing it to look like grains such as rice or wheat. McSteen thinks that if they can understand what’s causing the shortfall in kernel development, it may be possible to engineer grains like rice and wheat to “double kernel” the way that corn does.
Ultimately, studying these genes help corn researchers to better understand plant development and improve yield.

You always love the organism
To keep all their experiments going, the McSteen lab plants three acres of corn every summer. Each acre contains 750 rows; each row holds 30 plants.
With the aid of a hand held planter, they drop 67,500 kernels into the soil. Then they do what McSteen calls a lopsided “planter’s shuffle” to stamp the soil down so that it covers every kernel.
“You can do a whole acre of corn in a few hours,” she said. “It’s hard work. You’re sore afterwards.”
During Missouri summers, temperatures can reach over 100 degrees Fahrenheit. It feels even hotter when it rains. The ground is either hard as a rock before it rains or so muddy after it rains that researchers have to take care not to wrench their ankles in the thick muck.
McSteen has worked on corn for so long that she’s developed severe allergies to corn pollen. It’s not uncommon among corn researchers, but her allergies prevent her from taking part in the pollination step that takes place at the height of summer.
“You’re out there in 100-degree heat getting the job done,” she says. “It’s a real bonding experience for the lab.”
To pollinate the corn, they slip paper bags over the ears and tassel of their plants. Covering the ears prevents accidental fertilization of the ears from stray pollen blown about by the wind. The bag over the tassel allows researchers to collect the yellow powder that will be used for controlled pollination.
“The next day, you bang the tassel and the pollen falls out into the bag,” McSteen says. “Then you gather it all up and you pour it on the ear.” It’s possible to pollinate about 100 plants in an hour, but you have to start early and work quickly, McSteen says. Otherwise, all the pollen is dead by noon.
McSteen’s allergies prevent her from shelling corn as well, but she’s on deck for planting and harvest time and all the other stages in between.
“If you’re a corn geneticist, you’re out there working with the plants. You always love the organism.”

A Collaborative Community
“To be a corn geneticist, you have to be very organized and plan ahead,” McSteen says. Because it takes a long time to grow several generations of corn, she’s only beginning to see the results of experiments she started years ago.
As a way to increase productivity, corn researchers send their seeds to warm places such as Mexico, Chile or Hawaii that can accommodate a winter harvest.
In the lab, McSteen chooses kernels from carefully selected mutants to ship to an island in Hawaii. There, a company will plant and harvest the corn for her, but she usually sends two of her researchers down to take care of the pollinations themselves.
McSteen counts out thirteen yellow kernels that are shiny and mold-free. “Potential,” she says, as she slips the seeds in an envelope with the cross information labeled on the front.
A three week trip to Hawaii in the winter isn’t as exotic as it sounds, says Eden Johnson, a third year graduate student in McSteen’s lab. On the island, the beaches are rocky and full of riptides. “It is literally cornfields, one diner and a stop sign. The whole island exists for corn.”
“When you’re down there in Hawaii, you hang out with the other researchers,” McSteen says. “If their field is peaking and your field is not, then you’ll go help.”
In her experience, the corn community tends to be collaborative rather than competitive. She suspects it’s because everyone recognizes that corn takes a long time to grow.
“If you find out you’re working on similar things, you’ll work together, divide the work and do it together,” McSteen says. Researchers don’t race each other to be the first one to publish. “They won’t do that because they have respect for how long it takes to grow the corn.”

Growing careers alongside corn
McSteen is as serious about mentorship as she is about corn. “It’s part of the job of being a professor”.
According to Katy Guthrie, a second year graduate student in the lab, McSteen takes a Goldilocks approach to managing her students.
Neither too hands on or too hands off, “it’s exactly what I need,” Guthrie says. “She’s kind of like my academic parent for the next five years.”
Back at Penn State, McSteen supervised a PhD student who was talented writer. “I noticed this and gave her opportunities to write.” McSteen introduced her to a science writer and encouraged her to spend a summer writing for a science publication. “Now she works for the National Academy of Sciences. I’m really proud of her, and I feel like she’ll have a big impact communicating science to the public.”
A post-doc wanted to do go into teaching, so McSteen invited her to co-teach her class. Her post-doc went on to become a teaching professor at Mizzou. Another student turned her research experience in mapping mutants in corn into a successful career at a corn company.
“I want to enable the people in my lab to reach their full potential,” McSteen says. “I always try to figure out what they want to do in the future and try and facilitate that.”